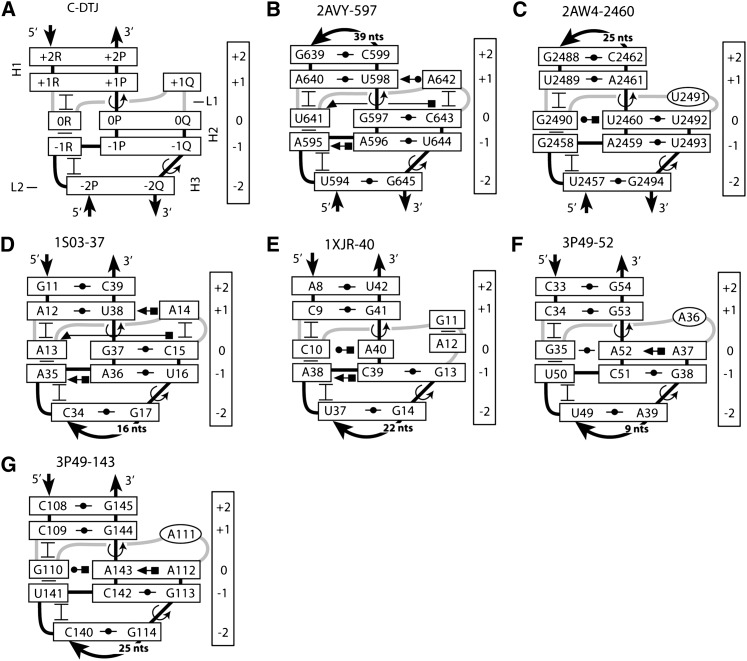
FIGURE 11.
Structure of C-DTJ motifs. (A) The general scheme of the C-DTJ motif. Like in B-DTJs, TJ1 and TJ2 are formed between base pairs [0P;0Q] and [+1R;+1P] and between base pairs [−1Q;−1P] and [−2Q;−2P], respectively. (B–G) The motifs 2AVY-597 and 2AW4-2460 are taken from the E. coli 16S and 23S rRNA, respectively (Schuwirth et al. 2005). Motif 1S03-37 is present in the mRNA of the spc operon of E. coli (Merianos et al. 2004). Motif 1XJR-40 exists in the RNA element of the stem–loop II motif (s2m) of the genome of the SARS virus (Robertson et al. 2005). Motifs 3P49-52 and 3P49-143 are found in the structure of the glycine riboswitch (Butler et al. 2011). In the presented structures, base pairs [+2P;+2R], [+1P;+1R], and [−1P;−1Q] are WC, while [−2P;−2Q] can be either UG or CG. Nucleotide 0R makes a triple with base pair [0P;0Q]. Nucleotide +1Q, if present, forms a cis-WC-SE base pair with nucleotide +1P. In structures B, D, and E, nucleotide −1R forms a trans-HG-WC base pair with −1P.