eIF3 is a large multimeric protein that forms the scaffold for other initiation factors during the complex process of eukaryotic translation initiation. Here the authors present an in vitro reconstitution of Saccharomyces cerevisiae eIF3 from its five recombinantly expressed and purified subunits. Detailed biochemical characterization confirms its suitability for future biochemical and structural analysis.
Keywords: translation, initiation, reconstitution, protein interaction
Abstract
Translation initiation in eukaryotes is a multistep process requiring the orchestrated interaction of several eukaryotic initiation factors (eIFs). The largest of these factors, eIF3, forms the scaffold for other initiation factors, promoting their binding to the 40S ribosomal subunit. Biochemical and structural studies on eIF3 need highly pure eIF3. However, natively purified eIF3 comprise complexes containing other proteins such as eIF5. Therefore we have established in vitro reconstitution protocols for Saccharomyces cerevisiae eIF3 using its five recombinantly expressed and purified subunits. This reconstituted eIF3 complex (eIF3rec) exhibits the same size and activity as the natively purified eIF3 (eIF3nat). The homogeneity and stoichiometry of eIF3rec and eIF3nat were confirmed by analytical size exclusion chromatography, mass spectrometry, and multi-angle light scattering, demonstrating the presence of one copy of each subunit in the eIF3 complex. The reconstituted and native eIF3 complexes were compared by single-particle electron microscopy showing a high degree of structural conservation. The interaction network between eIF3 proteins was studied by means of limited proteolysis, analytical size exclusion chromatography, in vitro binding assays, and isothermal titration calorimetry, unveiling distinct protein domains and subcomplexes that are critical for the integrity of the protein network in yeast eIF3. Taken together, the data presented here provide a novel procedure to obtain highly pure yeast eIF3, suitable for biochemical and structural analysis, in addition to a detailed picture of the network of protein interactions within this complex.
INTRODUCTION
Initiation of translation is the most complex step in protein synthesis. In eukaryotes, this process requires the orchestrated function of up to 12 different initiation factors, eukaryotic initiation factors (eIFs). eIF2 in complex with GTP recognizes initiator methionyl tRNA to form a ternary complex (TC) and positions it in the P-site of the small ribosomal subunit (Sonenberg and Hinnebusch 2009; Jackson et al. 2010). This process is enhanced by the action of several other initiation factors. In yeast, eIF3, eIF2, eIF5, and eIF1 are shown to bind to each other to form a multifactor complex (MFC). MFC formation was also observed in plants (Dennis et al. 2009) and in humans (Sokabe et al. 2012). This preassembled complex then binds to the 40S ribosome to form the 43S preinitiation complex (43S PIC) (Asano et al. 2000). Thus formed, 43S PIC binds to the mRNA to form 48S PIC and scans the mRNA leader region for the start codon. eIF5-stimulated GTP hydrolysis by eIF2, and subsequent release of inorganic phosphate upon start codon recognition is a key step that arrests the 48S PIC at the start codon (Algire et al. 2005). Subsequently, the 48S PIC harboring a correctly decoded start codon in the P site joins the 60S ribosomal subunit with the help of eIF5B, which leads to the dissociation of most of eIFs (see Hinnebusch 2011, and references therein).
eIF3 is the largest of the initiation factors with five core subunits in budding yeast (Tif32/eIF3a, Prt1/eIF3b, Nip1/eIF3c, Tif35/eIF3g, and Tif34/eIF3i) and 13 in humans (Asano et al. 1998; Phan et al. 1998; Damoc et al. 2007). Despite its complexity, human eIF3 shares a common core with yeast eIF3 composed of eIF3a, -b, -c, -g, and -i (Hinnebusch 2006). Mass spectrometry analysis of the human eIF3 identified three stable subcomplexes, one of which resembles that of the yeast eIF3 complex (Zhou et al. 2008). In addition to the five core subunits, yeast eIF3 complex has a loosely associated subunit, namely, Hcr1 (eIF3j). This protein promotes the binding of eIF3 to the ribosome and is involved in ribosomal biogenesis as well as proper selection of the AUG start site (Valasek et al. 2001a,b; Elantak et al. 2010).
eIF3 forms critical interactions with other initiation factors in the MFC and with the ribosome. The two larger subunits of eIF3, Tif32 and Nip1, bind to the β subunit of eIF2 and to eIF5, respectively, and they both also interact with eIF1 (Valasek et al. 2002, 2003). The extreme C terminus of Nip1 directly interacts with blades 1–3 of the small ribosomal protein Rack1/Asc1 (Kouba et al. 2012). Direct interactions between the N-terminal part of Tif32 and the ribosomal protein Rps0A and the C-terminal region of Tif32 with helices 16–18 of the 18S rRNA promote the binding of eIF3 to the 40S ribosomal subunit. These interactions suggest that eIF3 associates with that part of the 40S subunit, which is fully solvent-exposed in the 80S complex (Valasek et al. 2003).
Cryo-electron microscopy (EM) of the human eIF3 in combination with molecular modeling has revealed a five-lobed particle, which together with the eIF4G, occupies the solvent-exposed surface of the 40S ribosome (Siridechadilok et al. 2005). This proposed model of 40S binding mode of eIF3 is in accordance with that available for yeast (Valasek et al. 2003). A recent study on the reconstitution of human eIF3 has revealed that an octameric subcomplex of eIF3 comprising subunits e, f, h, k, l, and m and truncated fragments of subunits a and c exhibits a striking structural resemblance to the previously determined cryo-EM structure of native human eIF3 (Sun et al. 2011).
Apart from the above-mentioned cryo-EM reconstruction of human eIF3, little is known about its three-dimensional structure. The crystal structure of eIF3k, one of the nonconserved subunits of eIF3, was the first high-resolution structure of an eIF3 subunit. Based on this structure, protein- and RNA-binding activities were attributed to eIF3k (Wei et al. 2004). The solution structure of the RNA recognition motif (RRM) of human eIF3b in complex with a short peptide of eIF3j suggested a noncanonical RRM structure (Elantak et al. 2010); however, its yeast ortholog has all the structural features of a canonical RRM (Khoshnevis et al. 2010). The recent crystal structure analysis of yeast eIF3i/Tif34 in complex with a short peptide of eIF3b/Prt1 revealed conserved interactions necessary for proper PIC assembly (Herrmannova et al. 2011).
Translation initiation studies involving eIF3 have so far suffered from the lack of an experimental system for efficient reconstitution of the multicomponent factor. Such a reconstitution system would, on the one hand, provide new strategies for a detailed kinetic and thermodynamic dissection of translation initiation in eukaryotes and, on the other hand, facilitate structural studies of eIF3 and its interaction partners. Recently, Sun et al. (2011) described the reconstitution of human eIF3 using recombinantly expressed and purified subcomplexes. Using this system, the importance of an octameric subcomplex of eIF3 for binding to the 40S ribosomal subunit and to the hepatitis C viral (HCV) internal ribosome entry site (IRES) was demonstrated (Sun et al. 2011).
Here we report the in vitro reconstitution of the yeast eIF3 assembled from its recombinantly expressed subunits. Multi-angle light scattering (MALS) in combination with mass spectrometric analysis of the reconstituted and native eIF3 complexes showed that both complexes have a mass of ∼360 kDa, indicating the presence of one copy of each subunit in them. The recombinant eIF3 showed structural similarity with the natively purified complex as shown by size exclusion chromatography (SEC), and single-particle EM. The reconstituted complex is functional as shown by its binding to the 40S ribosomal subunit in the absence of other initiation factors. The recombinantly assembled eIF3 could also restore the translation of the luciferase mRNA in yeast extracts deficient of the native eIF3. In addition, in vitro reconstitution allowed us to study the subcomplexes of eIF3 and to investigate the thermodynamics of their interactions. The results of pull-down assays together with analytical SEC and isothermal titration calorimetry defined a stable binary complex between Tif35 and Tif34. We could show that this binary complex interacts with the rest of eIF3 mainly through the interaction between Tif34 and Prt1. The ternary complex of Prt1-Tif34-Tif35 was shown to interact with Nip1 in addition to its known binding to Tif32. Limited proteolysis of different subcomplexes of eIF3, followed by analytical SEC and mass spectrometry, helped us to map the borders of interactions between different subunits of eIF3 with high precision. Comparative analysis of the size and mass of the entire eIF3 complex with those of its various subcomplexes sheds light on the architecture of eIF3 complex. Altogether, data presented here provide a solid basis to gain further insights into the structure of eIF3 and its role in translation initiation.
RESULTS
eIF3 complex and its subcomplexes can be reconstituted from recombinant subunits
In order to obtain sufficient amounts of highly pure yeast eIF3 for structural and biochemical studies, the in vitro reconstitution of this complex using its recombinantly expressed subunits had to be established. For this purpose, each cDNA of all yeast eIF3 subunits was cloned into E. coli expression plasmids also encoding either an N- or C-terminal His-tag or an N-terminal GST-tag. The protein expression for each fusion construct was tested in at least three different E. coli strains, and the best combinations of strain and vector were chosen based on the highest expression level of soluble protein. The list of DNA constructs and expression strains that were selected and used for protein purification is provided in Supplemental Table S2. All of the eIF3 subunits could be purified individually to high homogeneity (Fig. 1A). The yield for the large subunits Nip1 and Tif32 was ∼2–3 mg protein per liter of culture, whereas 20–200 mg protein per liter of culture could be purified in the case of the other subunits (Prt1, Tif34, and Tif35). Apart from Tif34, all proteins had to be necessarily purified at 4°C. The preparation of Prt1 yielded a stable degradation product during the purification procedure. This fragment had preserved the C-terminal His-tag as it could still bind to the HisTrap column. Edman sequencing revealed the N terminus of this fragment to be at residue 181. This stable fragment, Prt1181C, was also cloned, purified, and used for further interaction studies with Tif34 and Tif35.
FIGURE 1.
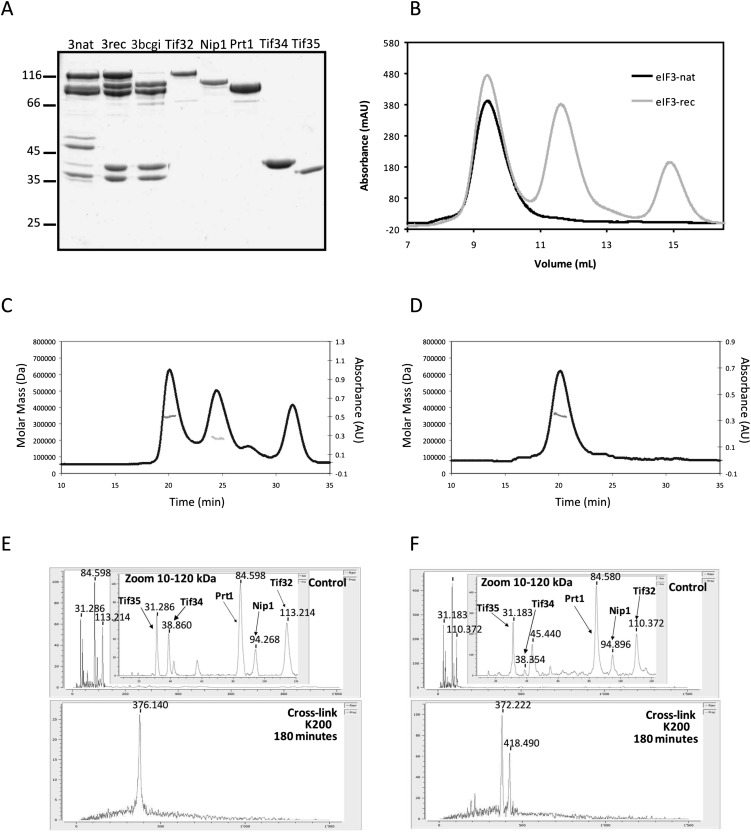
Reconstitution of yeast eIF3. (A) Comparison of reconstituted and native eIF3. Native eIF3 (3nat) copurifies with eIF5. Reconstituted eIF3 (3rec) comprises exclusively its five subunits. 3bcgi is a side product of the reconstitution of full eIF3 when Tif32 is added in lesser amounts compared with other subunits to promote the complex formation. All single recombinant subunits are shown on the gel to demonstrate the high degree of purity. (B) Size comparison between recombinant and native eIF3. Recombinant or native eIF3 were analyzed by size-exclusion chromatography (SEC). Recombinant eIF3 was prepared freshly by mixing five subunits in a nonstoichiometric ratio, therefore in addition to the peak for the full complex (between 9 and 10 mL), a second peak for Nip1-Prt1-Tif34-Tif35 complex (11–12 mL, corresponding to the third lane on A) and a third peak at ∼15 mL (corresponding to Tif34) were obtained. (C) Overlay of the SEC and MALS experiments for recombinant eIF3. The leftmost peak corresponds to recombinant eIF3. Molar mass distribution of the peak as calculated by MALS is indicated by a short line inside this peak and corresponds to a molecular weight of 340–354 kDa. The second peak corresponds to Nip1-Prt1-Tif34-Tif35 complex and corresponds to a molecular weight of 208–225 kDa. (D) Overlay of the SEC and MALS experiments for native eIF3. Molar mass distribution of the peak as calculated by MALS is indicated by a short line and corresponds to a molecular weight of 340–369 kDa. (E,F) Mass-spectrometry results for the eIF3rec (E) or eIF3nat (F). The upper panel is a zoom in the 10- to 120-kDa range of the spectrum for the control experiment without the cross-linker. The lower panel represents the whole spectrum for the cross-linked samples.
Yeast eIF3 could be reconstituted from its individual subunits by mixing Tif32 with 1.5-fold to twofold molar excess of the other subunits. Incubation time as short as 15 min was sufficient for efficient complex assembly. To compare the size of the reconstituted complex with that of the natively purified eIF3 complex, both complexes were analyzed by SEC (Fig. 1B). The elution peak corresponding to the recombinant complex showed exactly the same retention volume as that of the native complex, indicating a similar size for the two complexes.
Size of eIF3 complex and its stoichiometry
The SEC retention volumes of eIF3rec and eIF3nat (Fig. 1B) correspond to an apparent molecular weight of ∼603 kDa. The calculated mass of eIF3 is ∼360 kDa assuming one copy of each subunit in the complex. In order to get a more accurate determination of the eIF3 mass, the peaks of gel filtration column were analyzed by MALS, a technique for determining the absolute molecular mass of particles in solution. Results obtained from MALS were best fitted to a complex containing one copy of each of the subunits (Fig. 1C,D).
In addition, a complementary approach was applied to obtain the molecular weight and stoichiometry of eIF3 using high-mass MALDI mass spectrometry combined with cross-linking experiments (Nazabal et al. 2006; Bich et al. 2010). In this analysis, eIF3rec was detected as a single species with a total mass of 376,140 kDa (Fig. 1E). To account for the extra mass caused by the attached cross-linkers, the control experiment without cross-linker was performed leading to a molecular weight of eIF3rec of 359,271 kDa, which corresponds to one copy of each of the five eIF3 subunits. The analysis of eIF3nat, however, revealed the existence of two species: one composed of one copy of each eIF3 subunit and a total mass of 372,222 kDa and the other one with the total mass of 418,490 kDa, containing an extra subunit with the mass of 45.44 kDa. After subtraction of the extra-mass caused by the cross-linkers, 361,561 kDa and 406,521 kDa, respectively, were obtained for the two species (Fig. 1F). The size of the additional protein fits well with that of eIF5, which is known to copurify with eIF3 (Asano et al. 1998; Phan et al. 1998).
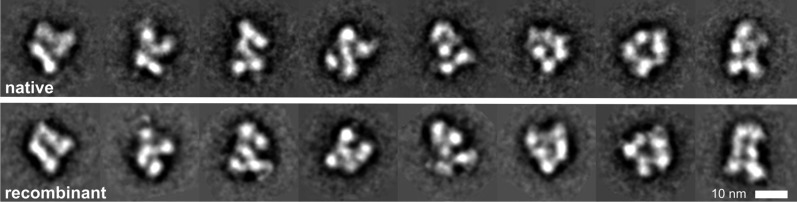
To assess the structural conservation between natively purified and recombinantly assembled eIF3 complexes, EM analysis was done for both purifications. eIF3 appears to be a compact particle with some views showing an elongated shape. The dimensions of the complex vary from 90–160 Å (width) and 140-170 Å (height). Even with the innate heterogeneity of the classification and alignment of two independent data sets in EM, we were able to identify similar views for both particles, a selection of which is shown in Figure 2. The structural features of both, eIF3nat and eIF3rec complexes from different views seem to be conserved.
FIGURE 2.
Single particle EM analysis of native and recombinant eIF3 complexes. Selected 2D class averages of negatively stained samples of native eIF3 complex (upper row) and recombinant eIF3 complex (lower row) are shown.
Recombinant eIF3 binds to the 40S ribosomal subunit
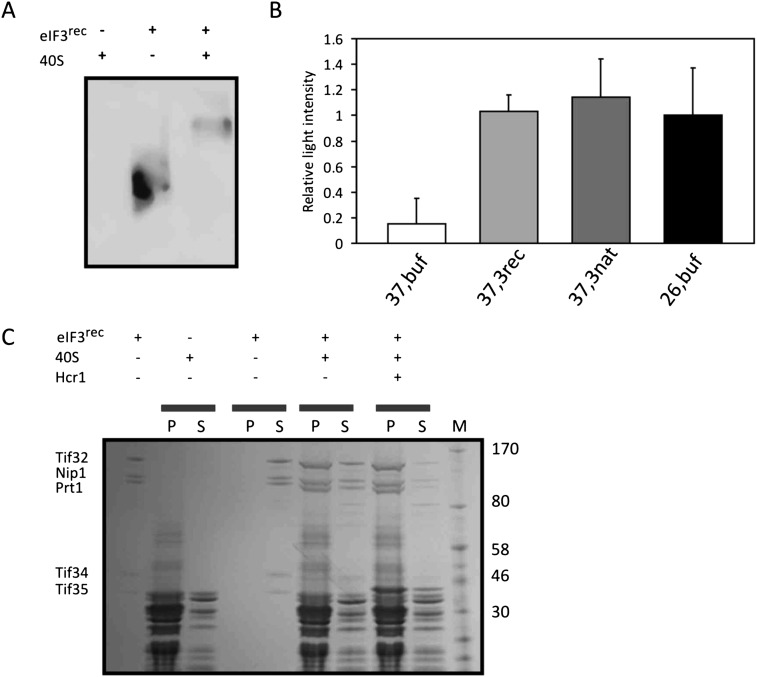
eIF3 forms a scaffold for the binding of other translation initiation factors and promotes their recruitment to the ribosome. Hence, the ability to bind to the small ribosomal subunit has been considered as an activity assay for eIF3 (Acker et al. 2007). Hcr1, a substoichiometric subunit of eIF3, is known to promote the binding of eIF3 to the 40S subunit (Nielsen et al. 2006). In order to test whether the reconstituted recombinant eIF3 exhibits ribosomal binding properties, purified yeast ribosomal 40S subunit was mixed with Hcr1 and eIF3rec in a 4:2:1 molar ratio, respectively. This molar ratio ensured that eIF3rec is saturated with Hcr1 and the ribosome. The complex was analyzed on a 5% native polyacrylamide gel followed by a Western blot and immunostaining against the His-tag of Tif32. The shift of the observed signal toward higher molecular weights upon addition of the 40S subunit and Hcr1 indicated the formation of a complex between eIF3rec and 40S subunit (Fig. 3A). To confirm this observation, we performed a cosedimentation experiment with eIF3rec, 40S, and Hcr1. The centrifugation condition was chosen in a way that 40S subunits and not eIF3rec alone would pellet. Upon mixing of 40S with eIF3rec, a large fraction of eIF3rec was found in the pellet, indicating its binding and therefore cosedimentation with the 40S (Fig. 3C). In the presence of Hcr1, an even larger portion of eIF3 was found in the ribosomal pellet, supporting previous reports on the role of Hcr1 promoting the recruitment of eIF3 to the ribosome (Nielsen et al. 2006).
FIGURE 3.
Activity tests for recombinant eIF3. (A) Analysis of the binding of eIF3 to the 40S ribosomal subunit by native PAGE and Western blot. A band shift toward higher molecular weights upon mixing 40S subunit and eIF3rec indicates the binding of eIF3 and 40S ribosome. (B) Luciferase assay performed with the cytoplasmic extracts of temperature-sensitive mutant cells treated under permissive (26°C) or nonpermissive (37°C) temperatures. To the cytoplasmic extracts, either recombinant (3rec) or native (3nat) eIF3 was added. In control experiments, only buffer was added. (C) SDS-PAGE analysis of cosedimentation experiment between eIF3rec and 40S. Mixtures were subjected to ultracentrifugation, and the total content of pellets (P) and supernatants (S) were loaded onto the gel. Under the conditions of this experiment, the majority of 40S subunits are found in the pellet, while all eIF3rec complexes remain in the supernatant. Upon mixing eIF3rec (0.1 μM) with 40S (1 μM), the factor was mainly found in the pellet, indicating binding to the 40S subunit. Addition of Hcr1 (1 μM) enhanced the binding of eIF3rec to the 40S. The leftmost and rightmost lanes show eIF3, which was used in this study and the marker, respectively.
Recombinant eIF3 initiates reporter mRNA translation
Yeast cells harboring a temperature-sensitive mutation within the PRT1 gene (known as prt1-1) have been used previously to assess the activity of eIF3 (Phan et al. 1998; Jivotovskaya et al. 2006). The cytoplasmic extracts of H1676 cells bearing this mutated PRT1 gene grown at permissive temperature were treated at 37°C to inactivate the endogenous eIF3. In vitro translation of luciferase mRNA was measured after a 1-h incubation with the cytoplasmic extracts to which either eIF3rec or eIF3nat had been added. In both cases, translation levels were restored to that of the extract that was not heat-deactivated and hence had preserved its endogenous eIF3 activity. Luciferase activities in all these cases were significantly higher than that of the heated extract to which only buffer was added as the control (Fig. 3B). Taken together, these experiments clearly demonstrate that the recombinant eIF3 complex is functional in promoting mRNA translation.
Mapping the interaction network of eIF3 complex
Tif32, Nip1, and Prt1 formed a ternary complex that turned out to be stable at salt concentrations up to 500 mM KCl. The same was observed for Tif34-Tif35 binary complex. In addition to these two subcomplexes, Prt1-Tif34-Tif35, Tif32-Nip1, and Nip1-Prt1-Tif34-Tif35 could be also assembled in vitro as stable complexes (Supplemental Fig. S1).
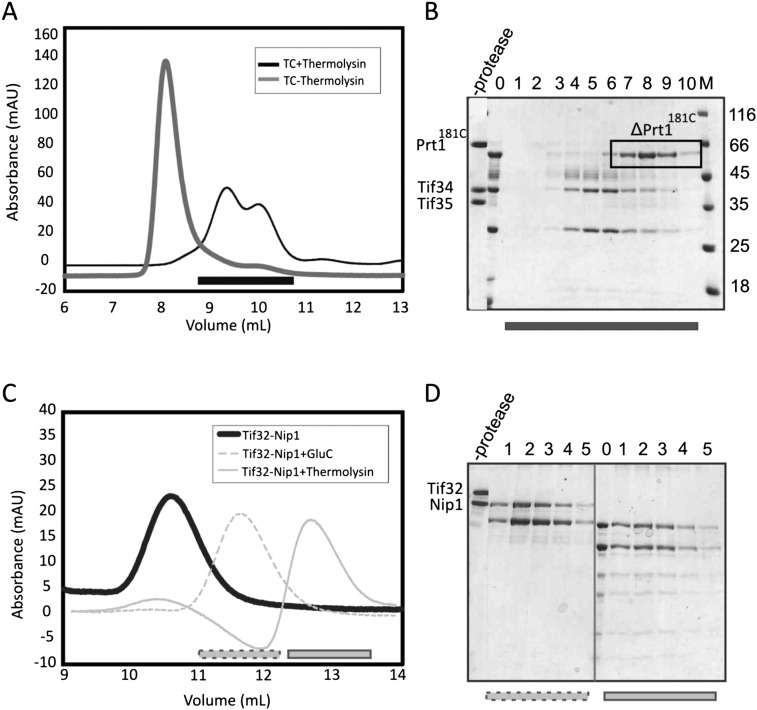
To map the protein interaction network within eIF3rec, its two major subcomplexes, Prt1181C-Tif34-Tif35 and Tif32-Nip1, respectively, were subjected to limited proteolysis using several proteases. The resulting proteolytic products were analyzed by SDS-PAGE, and the integrity of truncated subcomplexes was confirmed by analytical SEC (Fig. 4). The borders of interactions within each stable subcomplex were determined by mass spectrometry. Based on these results, we designed new fragments in order to identify the interacting domains or regions of the eIF3 subunits.
FIGURE 4.
Limited proteolysis of the subcomplexes of recombinant eIF3. (A) Analytical SEC profile of the Prt1181C/Tif34/Tif35 complex digested with thermolysin after 24 h. Resulted fragments do not form the complex as manifested by the presence of two peaks shifted to the right (dark gray) compared to the noncleaved complex (light gray). (B) SDS-PAGE analysis of the SEC profile in panel A suggests the dissociation of the complex into a truncation of Prt1181C and Tif34 in complex with a truncation of Tif35. Numbers correspond to different fractions of the peak that are marked by a black bar on the chromatogram. 0 is the sample prior to gel filtration. (C) Analytical SEC of Tif32/Nip1 complex treated either with GluC for 2 h (dashed light gray) or with thermolysin for 1 d (solid light gray). Comparison with the noncleaved complex (solid dark gray) proposes the existence of only one complex in each case, indicating the preservation of the interactions between resulted fragments. (D) SDS-PAGE analysis of gel filtration runs in panel C shows the proteolytic fragments to be bound together. Left and right panels are different fractions of the peaks after GluC (dashed gray bar on the chromatogram) and thermolysin (solid gray bar on the chromatogram) treatments, respectively. In each case, 0 indicates the sample prior to the gel filtration. In all cases, M stands for molecular weight marker, and values are in kiloDaltons.
Tif34 bridges the interaction between Tif35 and the C-terminal region of Prt1
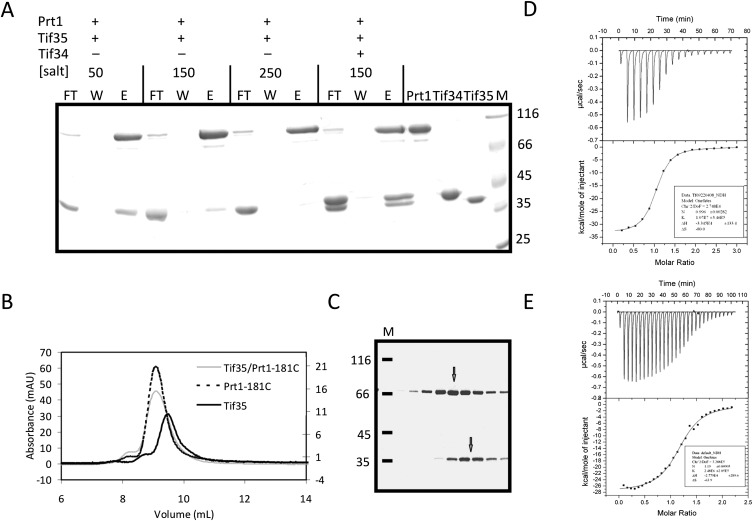
Proteolytic digestion of Prt1181C-Tif34-Tif35 ternary complex with thermolysin generated three distinct bands after 24 h of incubation, with the uppermost one corresponding to a fragment of Prt1181C lacking its C-terminal part (ΔPrt1181C). The middle and the lower bands consisted of almost full-length Tif34 and a truncated form of Tif35 (ΔTif35), respectively. Interestingly, on the gel filtration chromatography, the protease-treated sample eluted as two distinct peaks containing either Tif34-ΔTif35 or ΔPrt1181C (Fig. 4A,B). These findings therefore support previous observations that Prt1 interacts with Tif34 via its C-terminal region (Asano et al. 1998). Tif35 is reported to interact with Prt1 (Valasek et al. 2001b). However, this interaction turned out to be strictly dependent on the salt concentration. Whereas at low-salt concentrations (50 mM NaCl) Tif35 and full-length Prt1 interact with each other, increasing the salt concentration diminished their binding. At the physiological salt concentration (150 mM NaCl), the binding was weak and at 250 mM NaCl, the interaction was barely observed (Fig. 5A). The mixture of Tif35 and Prt1181C at 200 mM NaCl on an analytical SEC column had the same retention volume compared to the individual proteins (Fig. 5B), indicating that the complex either does not form or dissociates during gel filtration. SDS-PAGE analysis of the peak fractions revealed a mixture of two adjacent peaks, corresponding to each of the two proteins (Fig. 5C). This is consistent with the broadening of the observed peak for the mixture of Tif35 and Prt1181C relative to the peaks of individual proteins. Tif34, however, could bind strongly to both Tif35 and Prt1181C proteins. At 150 mM NaCl, addition of Tif34 increased the pull-down efficiency of Tif35, resulting in formation of a stable equimolar TC with full-length Prt1 (Fig. 5A). This is consistent with recent observations indicating that binding of Tif34 to Prt1 markedly stabilizes Tif35 association with the complex (Herrmannova et al. 2011). Thermodynamic analysis of the interaction of Tif34 with Tif35 and Prt1181C at physiological salt concentration revealed a higher affinity of Tif34 for Tif35 (Kd ∼ 100 nM) than for Prt1181C (Kd ∼ 400 nM) (Fig. 5D,E).
FIGURE 5.
Prt1 and Tif35 interact weakly in vitro. (A) Pull-down interaction studies between Tif35, full-length Prt1, and Tif34. Lanes FT, W, and E refer to the flow-through of the Ni-NTA beads, their last wash step, and the elution of the protein. (B) Overlay of the analytical SEC profiles of Prt1181C (dashed black), Tif35 (solid black), and Prt1181C-Tif35 mixture (solid gray) shows a broadening of the peak in the case of the mixture without any considerable shift the retention volume compared with individual components. (C) SDS-PAGE analysis of the peak of the mixture of Prt1181C-Tif35 shows that it is in fact composed of two adjacent peaks (marked by two arrows) as would be judged by the broadening of the peak. (D) The equimolar interaction of Tif35 and Tif34 is driven by the release of heat, which compensates for the decrease in entropy. (E) Interaction of Tif34 with Prt1181C is also equimolar and enthalpy driven. In both cases, the upper panels show raw data of heat effect (in μcal/sec) of injections of Tif34 into 1.5 mL of Tif35 (D) or Prt1181C (E). The lower panels show the fitted binding isotherms. The data points were obtained by integration of heat signals plotted against the molar ratio of Tif34 to either of the interaction partners in the reaction cell. The solid lines represent calculated curves using the best fit parameters obtained by nonlinear least squares fitting.
Tif32NTD anchors Nip1CTD
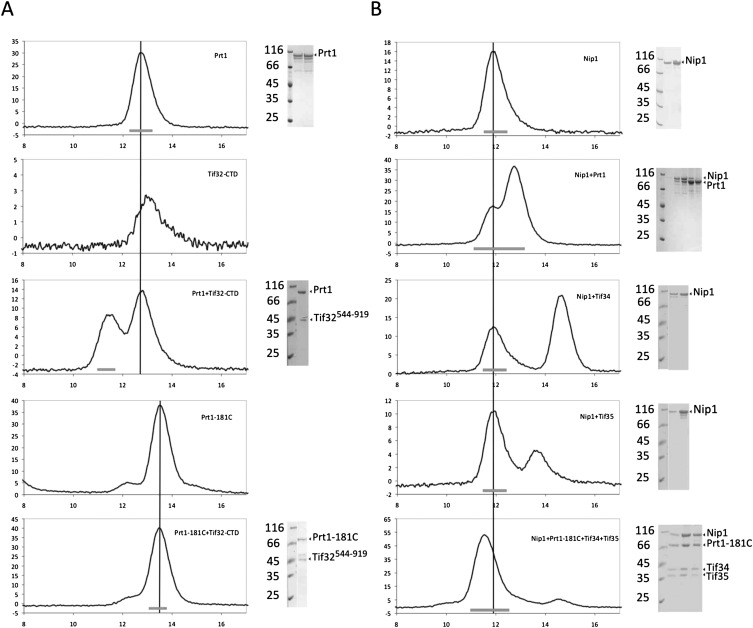
Proteolytic digestion of the Tif32-Nip1 complex with GluC generated several stable fragments, which were eluted as a complex in a single peak on the analytical SEC column (Fig. 4C,D). Mass spectrometric analysis of these fragments revealed that the N-terminal domain of Tif32 and the C-terminal domain of Nip1 are preserved. These results suggest that the N-terminal region of Tif32 serves as the interacting platform for the binding of the C-terminal part of Nip1.
For further analysis of the interactions of Tif32-Nip1 with other subunits of eIF3, Tif32NTD(1-494), Tif32CTD (544-919), and the subcomplex of Tif32NTD-Nip1CTD (240-812) were cloned, expressed, and purified. While no direct interaction was observed between the Tif32NTD-Nip1CTD complex and Prt1-Tif34-Tif35 (data not shown), Tif32CTD bound strongly to the full-length Prt1, but not to the Prt1181C (Fig. 6A).
FIGURE 6.
Mapping the interaction network of eIF3 by analytical SEC. (A) Full-length Prt1 interacts with the C-terminal region of Tif32. This interaction is not visible when the full-length Prt1 is replaced with a fragment starting from residue 181 (Prt1181C). (B) Nip1 interacts with Prt1-Tif34-Tif35 complex but not with each protein individually. Vertical lines are drawn to facilitate the comparison of the retention volumes of Nip1, Prt1, or Prt1181C in different experiments. In each case, the thick bar at the bottom of the graphs represents the fractions of the peak, which is resolved by SDS-PAGE.
Nip1 binds to the complex of Prt1-Tif34-Tif35
A yet unknown subcomplex composed of Nip1, Prt1, Tif34, and Tif35 was obtained as side product of the eIF3 reconstitution (Fig. 1B). This subcomplex contained all the subunits of eIF3 except for Tif32. To find out the interactions responsible for the formation of this subcomplex, binding of Nip1 to the individual subunits was studied using analytical SEC. Remarkably, no direct interaction between Nip1 and the other three individual proteins was observed (Fig. 6B). The complex, however, could be efficiently formed when all four proteins were present. Replacing Prt1 with Prt1181C did not abolish the subcomplex formation, excluding a possible role for the RRM of Prt1 in providing a binding site for Nip1. The retention volume of this complex on the SEC corresponds to a size of ∼250 kDa. Interestingly, this complex was able to bind efficiently to the Tif32NTD forming a ∼300-kDa complex according to SEC.
DISCUSSION
Up to now, the majority of in vitro studies on translation initiation in Saccharomyces cerevisiae have been performed using eIF3 and proteins purified from yeast (Acker et al. 2009; Mitchell et al. 2010). This approach has advantages such as the presence of post-translational modifications that might be important for the proper function of the complex in vivo. However, the fact that substantial amounts of eIF5 copurify with the native eIF3 makes it difficult to reliably use these preparations for biochemical and functional experiments dealing with the GTPase activity of eIF2 (Acker et al. 2007; Farley et al. 2011). Moreover, since all of the subunits of eIF3 are essential for the viability of yeast cells, it is difficult to generate strains lacking subunits or domains of subunits to study their functions. In vitro reconstitution of eIF3 using recombinant subunits not only overcomes the problem of contamination by other copurifying initiation factors but also provides invaluable tools to study the structure and function of this complex. In this regard, step-wise reconstitution of the eIF3 complex allows to compare different subcomplexes structurally, for example, using single-particle cryo-EM, and to define the position of different subunits in the complex. Furthermore, in vitro reconstitution of different subcomplexes and their stable truncations increases the chance to determine the three-dimensional structure of eIF3 at high resolution by X-ray crystallography. In addition, the in vitro reconstitution allows the fluorescence-labeling of eIF3 subunits, which offers new strategies for dissecting the kinetics and thermodynamics of events during translation initiation.
We established the in vitro reconstitution of yeast eIF3 using recombinant expression of its individual subunits in E. coli. In comparison with the eIF3 preparations available so far, which were done by purification of the complex from yeast cells (Phan et al. 1998; Algire et al. 2002; Ghaemmaghami et al. 2003), from recombinant expression of the whole complex in insect cells (Fraser et al. 2004; Masutani et al. 2007) or from coexpression of different subcomplexes of mammalian eIF3 in E. coli (Sun et al. 2011), our approach provides a more robust and feasible strategy, which yields large amounts of proteins in a relatively short time. In addition, the reconstitution of yeast eIF3 from the purified individual full-length subunits can be of outstanding importance for future studies on the kinetics of translation initiation as well as for structural studies.
Comparison of the reconstituted and the native eIF3 complexes by mass spectrometry combined with cross-linking revealed that purified eIF3rec exists in one form with the approximate mass of 359 kDa. Since in mass spectrometry experiments without cross-linkers all five subunits of eIF3 were found, we conclude that one copy of each subunit must be present in the complex. For preparations of eIF3nat, however, two species were observed; a complex with one copy of each subunit (approximate mass of 361 kDa) and a complex with an additional protein of 45 kDa, most probably eIF5. These results provide further proofs for the stoichiometric ratio of the eIF3 subunits.
As proteins expressed in bacterial systems lack most of the eukaryotic-specific post-translational modifications, the question raised whether the recombinant eIF3 exhibits structural and functional features of a native complex purified from yeast. Size comparison between recombinant and native eIF3 using SEC and MALS suggested a very similar size for both complexes. Analysis of the native and recombinant complexes by single-particle EM demonstrated a high degree of structural identity between the two complexes. The reconstituted recombinant eIF3 complex is active as it binds to the 40S ribosomal subunit and also can restore in vitro translation of a reporter mRNA in cell extracts deficient of active endogenous eIF3. Both recombinant and native eIF3 complexes lead to the translation of the luciferase mRNA to the levels observed with active endogenous eIF3. Altogether, these results show that the recombinantly expressed and in vitro reconstituted eIF3 is structurally and functionally similar to the natively purified eIF3 and can therefore replace the native particle in structural and functional studies.
Using individual subunits of eIF3, the binding sites on each component of eIF3, which are important for the assembly of the complex, were identified. Subjecting Prt1181C-Tif34-Tif35 to the limited proteolysis abolished the binding of Prt1 to the other two proteins by removing its C-terminal region. This confirms previous observations on the importance of the C-terminal region of Prt1 for binding to Tif34 and Tif35 (Asano et al. 1998; Valasek et al. 2001b, Herrmannova et al. 2011). Further analysis of the binary interactions between these three proteins showed that the interaction between Tif35 and Prt1 is weak and sensitive to the salt concentration. Addition of Tif34, however, resulted in the formation of an apparently equimolar complex. These data strongly suggest that binding of Tif34 to Prt1 and Tif35 significantly strengthen the Prt1-Tif35 interaction, as reported previously (Herrmannova et al. 2011). Dissection of the thermodynamics of the binding of Tif34 to Prt1 and Tif35 revealed the formation of a stronger complex between Tif34 and Tif35, suggesting that eIF3rec may dissociate into two stable subcomplexes of Tif32-Nip1-Prt1 and Tif34-Tif35. This finding is consistent with the mass spectrometric studies on human eIF3 showing that the counterparts of Tif34 and Tif35 in human are located at the periphery of the complex, excluded from its core, and tend to leave the complex more easily (Damoc et al. 2007; Zhou et al. 2008).
Tif32, Nip1, and Prt1 form the core of eIF3 as this stable subcomplex of eIF3 resists high concentrations of salt. This is consistent with the observation that a subcomplex of eIF3 composed of Tif32 and Nip1 together with eIF5 is sufficient for the binding to the ribosome (Valasek et al. 2003). Limited proteolysis of Tif32-Nip1 yielded a stable complex comprising the N-terminal half of Tif32 and the C-terminal two-third of Nip1 (Tif32NTD-Nip1CTD), which is consistent with previous observations that delineated the Nip1 interaction site on Tif32 to be located between residues 200–600 (Valasek et al. 2002). Interestingly, both of these proteolytic fragments are known to carry PCI domains (Hofmann and Bucher 1998) that can be important for the integrity of the complex or binding to other subunits within eIF3 (Scheel and Hofmann 2005).
We identified a new subcomplex of eIF3 composed of Nip1, Prt1, Tif34, and Tif35, but a binary complex of Nip1 with any of the other three proteins could not be detected. In agreement, the formation of a stable Prt1–Nip1 binary complex was also not observed in vivo by overexpressing these two subunits alone (Hinnebusch et al. 2004), while, in contrast, a binding site of Nip1 between residues 551–641 of Prt1 was reported (Valasek et al. 2002). Hence it seems to be likely that Prt1-Tif34-Tif35 cooperatively form a composite binding site for Nip1. Alternatively, Nip1 may mainly interact with Prt1 in the context of the eIF3 complex, and the affinity of this interaction is dramatically increased by the presence of Tif34 and Tif35. Therefore, the possibility of conformational changes in the complex upon the presence of all four subunits cannot be ruled out. The fact that no interaction was detected between the binary Tif32NTD-Nip1CTD and ternary Prt1-Tif34-Tif35 complexes may suggest a possible role for part of the 240 N-terminal residues of Nip1 for this interaction. However, all attempts to purify Nip1 truncations encompassing residues prior to 240 did not result in stable protein, so no direct evidence was obtained to fully support this possibility. In addition, the C-terminal half of Tif32 was found to interact with full-length Prt1 and not with a fragment of Prt1 missing the first 180 residues, consistent with previous in vivo observations (Valasek et al. 2002).
The apparent molecular weights of both eIF3nat and eIF3rec complexes obtained by analytical SEC (∼600 kDa) are not consistent with the expected mass for the complex with one copy of each subunit, that is, a monomeric complex of ∼360 kDa mass. However, determination of the absolute masses by means of MALS and high-mass MALDI using cross-linked complexes revealed that both eIF3nat and eIF3rec complexes represent a monomeric complex with a molecular mass of ∼360 kDa. The discrepancy between the apparent size and the absolute mass can be attributed to the shape of the complex, since SEC separates molecules based on their radii of gyration rather than their masses. This suggests that the eIF3 complex does not have a perfectly globular shape, as evident also in 2D class averages of the negatively stained EM analysis. To gain further insights into the architecture of eIF3, we subjected Nip1-Prt1-Tif34-Tif35 and Tif321-494-Nip1-Prt1-Tif34-Tif35 to SEC. Interestingly, in both cases good correlations between the apparent and calculated masses were obtained. It is tempting to speculate that the C-terminal extension of Tif32, which, based on the secondary structure predictions, is composed of long α-helices, is responsible for the size ambiguity observed on the gel filtration. It is likely that Tif32NTD, Nip1, Prt1, Tif34, and Tif35 form a globular structure, and the C-terminal domain of Tif32 is protruding out of this compact complex.
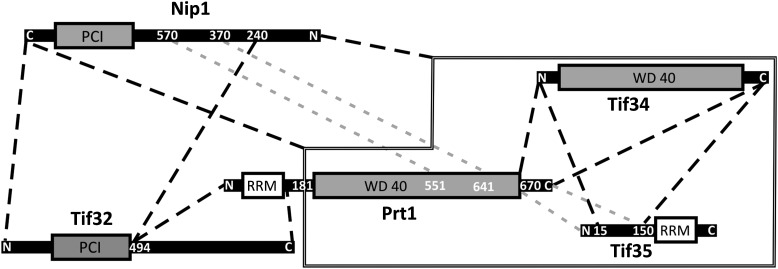
Based on the results obtained from limited proteolysis, analytical SEC, pull-down assays, mass spectrometry and MALS, we propose a model for the architecture of eIF3 (Fig. 7). In this model, we divide Prt1 into two regions; the N-terminal RNA-binding domain (from 70–170) and the C-terminal domain (Prt1181C), which is predicted to be composed of at least one WD40 motif followed by two α-helices at the C terminus. These C-terminal helices of Prt1181C interact with Tif34 and together ensure stable binding of Tif35 to both of them. In concert with Tif34 and Tif35, Prt1181C provides a binding site for Nip1. This binding is lost upon deletion of the first 240 residues of Nip1, but a fragment of Nip1 following residue 240 stably interacts with the first 494 residues of Tif32. In this model, the C-terminal half of Tif32 interacts with regions missing in the Prt1181C fragment and protrudes out of the compact core of eIF3.
FIGURE 7.
Map of interactions between eIF3 subunits. The stable subcomplexes that could be separated by SEC are depicted by dashed lines. The interaction of Nip1 with Prt1-Tif34-Tif35 subcomplex is represented by a solid line around Prt1-Tif34-Tif35. Binding of Nip1370-570 to Prt1CTD was observed by Valasek et al. (2002), while the binding of Tif35CTD to Prt1CTD was concluded from indirect observations. Therefore these two interactions are represented by a different color (gray dashed lines).
Taken together, by using a new strategy to in vitro reconstitute active yeast eIF3, we gained insights into its architecture. These data combined with further structural studies could shed light onto the structural and mechanistic properties of eIF3 and hence eukaryotic translation initiation.
MATERIALS AND METHODS
Cloning
Coding sequences of Tif32, Nip1, Prt1, Prt1181-C, Tif34, Tif35, Tif321-494, Tif32544-919, Nip1240-C, and Hcr1 were generated by PCR amplification using yeast genomic DNA as the template and appropriate pairs of primers (Supplemental Table S1). The resulting fragments were cloned into pET15b, pET22b, pET28a (Novagen), or pGEX-6P-1 vectors (GE Healthcare) with the vector and affinity tag combination shown in Supplemental Table S2.
Protein expression and purification
For protein expression, each of the plasmids was transformed into at least three different Escherichia coli expression strains. The best expressing host in each case was selected based on the expression level and the solubility of the protein and is listed in Supplemental Table S2. T7-promoter driven protein expression was induced using either IPTG in 2×YT medium or auto-induction (Studier 2005).
Tif32 was expressed as an N-terminally His-tagged protein. Rosetta2 (DE3) cells (Novagen) expressing pET28b-TIF32 plasmid were lysed in buffer A (400 mM KCl, 50 mM HEPES/KOH at pH 7.5, 10% [v/v] glycerol, 2 mM β-mercaptoethanol, and 0.05% [v/v] Tween20) supplied with 20 mM imidazole, RNase A (Roche Applied Science) and a mixture of protease inhibitors including aprotinin, leupeptin, and pepstatin (ALP) using a microfluidizer (Microfluidics). The protein was purified using a HisTrap column (GE Healthcare). After loading the sample, the column was washed with two column volumes of buffer A, and the protein was eluted by increasing the concentration of imidazole to 500 mM in a linear gradient over 10 column volumes. Tif32 was further purified on a Superdex 200 size exclusion column (GE Healthcare) equilibrated in K-200 buffer (200 mM KCl, 20 mM HEPES/KOH at pH 7.5, 10% [v/v] glycerol, and 2 mM DTT).
Tif34 was cloned into a modified pET15b vector coding an N-terminal His10 tag followed by a TEV-cleavage site. BL21 (DE3) cells (Invitrogen) expressing this construct were lysed in 500 mM NaCl, 50 mM Tris (pH 7.5), 5% (v/v) glycerol, 20 mM imidazole, and 2 mM β-mercaptoethanol supplemented with RNase and ALP. The protein was purified using a HisTrap column in a similar way as Tif32. The N-terminal His tag of Tif34 was removed using TEV protease while dialyzing overnight against 300 mM NaCl, 30 mM Tris (pH 7.5), 5% (v/v) glycerol, and 2 mM β-mercaptoethanol. The cleaved tag and protease were subsequently removed on the HisTrap column using the same buffer as the dialysis buffer but with 200 mM NaCl. The protein was further polished on a Superdex 75 column (GE Healthcare) equilibrated in 150 mM NaCl, 10 mM HEPES/NaOH (pH 7.5), and 2 mM DTT.
Nip1 was expressed as a GST-fusion protein in a pGEX-6P-1 vector using Bl21(DE3) cells. Cells were lysed in buffer A supplemented with RNase and ALP. The protein was purified on a GSTrap column (GE Healthcare) equilibrated with the lysis buffer and eluted in lysis buffer plus 30 mM reduced glutathione. The GST tag was removed with PreScission protease (GE Healthcare) while dialyzing against dialysis200 buffer (200 mM KCl, 30 mM HEPES/KOH at pH 7.5, 10% [v/v] glycerol, and 2 mM β-mercaptoethanol). The cleaved GST moiety and the protease were removed using a source30Q column (GE Healthcare). Protein was loaded on the column equilibrated with the dialysis200 buffer and eluted with a linear gradient of KCl to the final concentration of 1 M over 20 column volumes. Nip1 was further purified on a Superdex 200 equilibrated with K-200 buffer.
Tif35 was also expressed as a GST-fusion protein in Rosetta2 (DE3) cells. Cells were lysed in 300 mM NaCl, 50 mM HEPES/NaOH (pH 7.5), 5% (v/v) glycerol, and 2 mM β-mercaptoethanol supplemented with RNase A and ALP. The fusion protein was purified on a GSTrap column equilibrated with the lysis buffer and eluted by addition of 30 mM reduced glutathione. The buffer was exchanged for 200 mM NaCl, 30 mM HEPES/NaOH (pH 7.5), 5% (v/v) glycerol, and 2 mM β-mercaptoethanol using a desalting column (GE Healthcare). The GST tag was cleaved overnight using PreScission protease. The GST protein and PreScission protease were removed on the GSTrap column. Tif35 was further polished on a Superdex 75 column equilibrated in 150 mM NaCl, 10 mM HEPES/NaOH (pH 7.5), 5% (v/v) glycerol, and 2 mM DTT.
Prt1 was expressed as a C-terminally His-tagged protein. Rosetta2 (DE3) cells expressing pET22b-Prt1 were lysed in 200 mM KCl, 50 mM HEPES/KOH (pH 7.5), 10% (v/v) glycerol, 20 mM imidazole, 0.05% (v/v) Tween 20, and 2 mM β-mercaptoethanol supplemented with RNase A and ALP. The protein was purified on a HisTrap column equilibrated with the lysis buffer and eluted by increasing the concentration of imidazole in a linear gradient over 10 column volumes to 400 mM. The eluted protein was mixed with an equal volume of 10 mM HEPES/KOH, 10% (v/v) glycerol, 2 mM β-mercaptoethanol, and 10 mM MgCl2 and incubated with 2 mM ATP overnight. The protein was further purified on a source30Q ion-exchange column equilibrated with 100 mM KCl, 30 mM HEPES/KOH (pH 7.5), 10% (v/v) glycerol, and 2 mM β-mercaptoethanol and eluted by increasing the salt to the final concentration of 1 M. The eluted protein was further purified on a Superdex 200 column equilibrated with K-200 buffer.
Tif32544-919 was expressed as a GST-tagged protein. Cells were lysed in 300 mM NaCl, 50 mM HEPES/NaOH (pH 7.5), 10% (v/v) glycerol, 0.05% (v/v) Tween 20, and 2 mM β-mercaptoethanol supplemented with RNaseA and ALP. The fusion protein was purified on a GSTrap column equilibrated with the lysis buffer and eluted by addition of 30 mM reduced glutathione. The GST tag was cleaved off overnight using PreScission protease. Protein was further purified on a Superdex 200 column equilibrated in 100 mM NaCl, 10 mM HEPES/NaOH (pH 7.5), 5% glycerol, and 2 mM DTT.
To purify Tif321-494-Nip1240-C complex, each of the fragments were expressed in Rosetta2 (DE3) cells harboring either pET15b-Tif321-494 or pGEX-6P-1-Nip1240-C. Cells were mixed together prior to the lysis. Cell lysis was carried out in 300 mM NaCl, 50 mM HEPES/NaOH (pH 7.5), 10% (v/v) glycerol, 0.05% (v/v) Tween 20, 1 mM ATP, 2 mM MgCl2, and 2 mM β-mercaptoethanol supplemented with RNase A and ALP. The complex was purified on a GSTrap column equilibrated with the lysis buffer and eluted by addition of 30 mM reduced glutathione. The complex was further purified on a HisTrap column equilibrated with 200 mM NaCl, 30 mM HEPES/NaOH, 5% glycerol, 20 mM imidazole, and 2 mM β-mercaptoethanol and eluted with increasing the imidazole concentration to 400 mM. The GST tag was removed overnight using PreScission protease. Complex was further purified on a Superdex 200 column equilibrated in 150 mM NaCl, 10 mM HEPES/NaOH (pH7.5), and 2 mM DTT.
Tif321-494 was expressed as an N-terminally His-tagged protein. Rosetta2 (DE3) cells expressing pET15b-Tif321-494 were lysed in 300 mM KCl, 50 mM HEPES/KOH (pH 7.5), 5% (v/v) glycerol, 20 mM imidazole, 0.05% (v/v) Tween 20, and 2 mM β-mercaptoethanol supplemented with RNase A and ALP. The protein was purified on a HisTrap column equilibrated with the lysis buffer and eluted by increasing the concentration of imidazole in a linear gradient over 10 column volumes to 400 mM. The His-tag was cleaved off overnight using TEV protease while dialyzing against 200 mM NaCl, 30 mM HEPES/NaOH, 5% glycerol, and 2 mM β-mercaptoethanol. The noncleaved proteins as well as TEV protease were removed by another HISTrap column equilibrated with the dialysis buffer. Protein was further purified on a Superdex 200 column equilibrated in 150 mM NaCl, 10 mM HEPES/NaOH (pH7.5), and 2 mM DTT.
Prt1181C was expressed as an N-terminally His-tagged protein and purified in the same way as described for Tif34. Hcr1 was purified according to the method previously described (Khoshnevis et al. 2010).
Native eIF3 and yeast ribosome purification
Native yeast eIF3 (eIF3nat) and 40S ribosomal subunits were prepared as previously described (Acker et al. 2007) using LPY201 and YAS2488 cells, respectively (kindly provided by J.R. Lorsch), with the following modifications to the purification procedure of eIF3. The harvested yeast cells were lysed in 350 mM KCl, 30 mM HEPES/KOH (pH 7.5), 10% (v/v) glycerol, 20 mM imidazole, and 2 mM β-mercaptoethanol and frozen by dripping in liquid nitrogen. Cell disruption was achieved in liquid nitrogen using a ZM 200 grinder (Retsch). The frozen lysate powder was stored at −80°C. For purification of eIF3nat, RNase A, and EDTA-free protease inhibitor cocktail (Roche) were added to the thawed lysate. Clarified lysate was loaded on a HisTrap column equilibrated with the lysis buffer, and the complex was eluted by increasing the concentration of imidazole to 500 mM. The eluted protein was mixed with equal volume of 10 mM HEPES/KOH (pH 7.5), 10% (v/v) glycerol, and 2 mM β-mercaptoethanol and loaded on a source30Q ion-exchange column equilibrated in 150 mM KCl, 20 mM HEPES/KOH (pH 7.5), 10% (v/v) glycerol, 0.5% EDTA, and 2 mM β-mercaptoethanol. The complex was eluted by increasing the KCl concentration to 1 M in a linear gradient over 20 column volumes. The complex was further purified on a Superdex 200 column equilibrated with 100 mM KCl, 20 mM HEPES/KOH (pH 7.5), 10% (v/v) glycerol, and 2 mM DTT.
In vitro reconstitutions
To assemble the Tif34-Tif35-Prt1181C complex, individual purified proteins were mixed with a molar ratio of 1:1.5:2, respectively. eIF3rec (Tif32-Prt1-Nip1-Tif35-Tif34) was reconstituted by mixing the purified proteins with a molar ratio of 1:2:2:2:3, respectively. All in vitro complex reconstitutions were carried out in K-200 buffer at 4°C, and the reconstitution mixture was incubated for 15 min before proceeding to the next step.
Mass spectrometric analysis of the complexes
All the samples measurements were performed using an Ultraflex III MALDI ToF ToF mass spectrometer (Bruker) equipped with CovalX's HM2 interaction module. Twenty microliters of each protein sample (eIF3nat and eIF3rec) was pipetted to prepare eight dilutions of the samples with final volume 10 μL. One microliter of each dilution obtained was mixed with 1 μL of a matrix composed of a recrystallize sinapinic acid matrix (10 mg/mL) in acetonitrile/water (1:1, v/v), TFA 0.1% (K200 MALDI Kit). After mixing, 1 μL of each sample was spotted on the MALDI plate (SCOUT 384). After crystallization at room temperature, the plate was introduced in the MALDI mass spectrometer and analyzed immediately. The analysis was repeated in triplicate. Each mixture prepared for the control experiment (9 μL left) was submitted to crosslinking using CovalX's K200 MALDI MS analysis kit. Nine microliters of the mixtures was mixed with 1 μL of K200 Stabilizer reagent (2 mg/mL) and incubated at room temperature. After the incubation time (180 min), samples were prepared for MALDI analysis as for the control experiments. Samples were analyzed by high-mass MALDI analysis immediately after crystallization. The MALDI ToF MS analysis was performed using CovalX's HM2 TUVO interaction module with a standard nitrogen laser and focusing on different mass ranges from 0–2000 kDa. Data were analyzed by the Complex Tracker 2.0 software (www.covalx.com).
Multi-angle light scattering
Two hundred microliters of the reconstituted or native eIF3 at a concentration of 1 mg/mL was chromatographed on an analytical Superdex 200 (10/300) gel filtration column using an Äkta purifier coupled to a miniDAWN MALS detector (Wyatt Technology). Data analysis was carried out with Astra software.
Electron microscopy
For EM, native and recombinant eIF3 complexes were applied on a GraFix gradient (Kastner et al. 2008) ranging from 10%–40% glycerol and 0%–0.15% glutaraldehyde and standard buffer conditions. Gradient fractions were negatively stained with 2% uranylactetate. Low dose images were recorded on a 4k × 4k CCD camera in a Philips CM200 FEG using an acceleration voltage of 160 kV at 121,000-fold magnification. Twofold pixel binning resulted in a final pixel size of 2.45 Å/pixel. We extracted 47,192 (native complex) and 53,339 (recombinant complex) single-particle images from 20 micrographs each using Boxer (Ludtke et al. 1999). Single particles from both data sets were analyzed independently by reference-free alignment and classification (Dube et al. 1993) using the IMAGIC-5 software (van Heel et al. 1996). After three rounds of alignment and classification, class averages of the recombinant complex were aligned to class averages of the native complexes in order to compare the aligned averages.
Ribosome binding studies
To investigate the binding of eIF3rec to the ribosome, the complex was first reconstituted, mixed with Hcr1 and the 40S ribosomal subunit, and applied on a 5% native acrylamide/poly-acrylamide gel according to the method previously described (Acker et al. 2007). The gel was blotted on the nitrocellulose membrane and immunostained against the His-tag on Tif32 with horse-radish peroxidase conjugated secondary antibody.
Cosedimentation studies
eIF3rec (0.1 μM) was mixed with 1 μM 40S in recon buffer (Acker et al. 2007) plus 2 mM DTT and incubated for 30 min at 30°C. In a parallel reaction, 1 μM Hcr1 was also added to the reaction mixture prior to the incubation. Controls containing individual reactants, eIF3rec or 40S, were also prepared. The resulting reactions were subjected to ultracentrifugation for 80 min at 400,000g on a TLA-100 rotor (Beckman Coulter). In order to reduce the pelleting of unbound eIF3, 30 μL of 20% glycerol in recon buffer was added to each tube prior to ultracentrifugation, resulting in the formation of a glycerol cushion. Next, pellets in each tube were resuspended in recon buffer while supernatants were precipitated with five volumes of acetone. Proteins in the pellets and supernatants were analyzed on 4%–12% SDS gel (Serva).
In vitro translation
Luciferase mRNA was in vitro transcribed from LUC control plasmid (Promega). Transcription template was prepared by PCR amplification using T7 promoter and a 30-mer oligo-dT. The mRNA was transcribed using T7 RNA polymerase, purified by phenol-chloroform extraction and precipitated by ethanol.
Capping of the mRNA was done using ScriptCap m7G capping system (Epicentre Biotechnologies) according to the manufacturer's instruction.
In vitro translation was performed with H1676 yeast cell lysate containing the temperature-sensitive mutation prt1-1 (Phan et al. 1998) (kind gift of Alan G. Hinnebusch). Cells were grown to optical density (OD) ∼ 1 at 25°C, resuspended in 150 mM KOAc, 20 mM HEPES/KOH (pH 7.5), 2 mM MgOAc, and 2 mM β-mercaptoethanol and dripped into liquid nitrogen. Cell rupture was performed in liquid nitrogen using a ZM 200 grinder. For each experiment, 40 μL of the lysate was thawed and clarified by centrifugation at 16,000g for 40 min and, depending on the experiment, was incubated for 6 min either at 26°C or 37°C. Reaction buffer was prepared by mixing 10 μL of 6× translation buffer (132 mM HEPES/KOH at pH 7.5, 720 mM KOAc, 12 mM MgOAc, 4 mM ATP, 0.6 mM GTP, 1 mM L-Met, 10 mM DTT) with 2 μL of 200 mM phosphoenolpyrovate (Applichem), 0.5 μL pyruvate kinase (Roche), and 1 μL of 5 mM amino acids mixture. The reaction buffer was incubated for 15 min at 37°C followed by a 5-min incubation at 26°C. Translation was started by addition of 0.5 μL Ribolock RNase inhibitor (Fermentas), 1 μL mRNA (at 2 μg/μL concentration) or water, 2 μL eIF3 (at the concentration of 10 μM) or K-200 buffer, and 40 μL lysate to the reaction buffer. After a 1-h incubation at 26°C, the reactions were transferred to the LUMITRAC200 plates (Greiner Bio One). The luminescence of each reaction was measured for 10 sec on a microtiter plate reader (FLUOstar Omega, BMG Labtech) after addition of 100 μL luminescence buffer (25 mM glycylglycine, 15 mM K2HPO4, 4 mM EGTA, 15 mM MgSO4, 4 mM ATP, 1.25 mM DTT, 0.1 mM CoA, 80 mM luciferin).
Limited proteolysis
Thermolysin, elastase, GluC, aminopeptidase, carboxypeptidase, trypsin, chymotrypsin, and proteinase K (Roche) were mixed with the eIF3 subcomplexes in a ratio of 1:500 (w/w). Samples were taken at different time points and analyzed on SDS-PAGE. Major cleavage products were further analyzed by analytical SEC, and the borders of stable fragments were determined by mass spectrometry.
Interaction studies using analytical SEC
Interaction studies were performed on an analytical Superdex 75 (10/300) or Superdex 200 (10/300) column (GE Healthcare). In each case, a total amount of 50 μg protein in the maximum volume of 500 μL was loaded on the column.
Pull-down assay
Five hundred picomoles of Prt1 was mixed with 1 nmol of Tif35 in a final volume of 100 μL in 20 mM HEPES/NaOH (pH 7.4), 20 mM imidazole, 5% (v/v) glycerol, 2 mM β-mercaptoethanol, and 50, 150, or 250 mM NaCl. Reactions were incubated for 1 h with Ni-NTA beads equilibrated in the corresponding buffer. Beads were collected and washed four times, and the bound moieties were eluted with 400 mM imidazole. To test the binding of Tif34, 1 nmol of this protein was added to the other two proteins.
Isothermal titration calorimetry
Tif34, Tif35, and Prt1181C were extensively dialyzed against ITC buffer (150 mM NaCl, 10 mM HEPES/NaOH at pH 7.5, 5% [v/v] glycerol) and concentrated to 110 μM, 5.2 μM, and 10 μM, respectively. The experiments were performed on a VP-ITC calorimeter (Microcal) at 20°C. Twenty microliter aliquots of Tif34 were injected into the ITC cell containing either Tif35 or Prt1181C every 30 sec, during which the titration peak returned to the base line. To calculate the thermodynamics parameters, the titration data were analyzed using the ORIGIN software (OriginLab).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank J.R. Lorsch (Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine) and A.G. Hinnebusch (Laboratory of Gene Regulation and Development, Eunice K. Shriver NICHD, National Institutes of Health) for providing us with the yeast strains for the purification of ribosome and native eIF3, as well as prt1-1 cells for the activity assays. We thank M.V. Rodnina for her support and advice during purification of ribosomes. We thank M. Ratz for assistance, as well as H. Ghalei and A. Dickmanns, for critically reading the manuscript. S.K. was supported by a Georg-Christoph-Lichtenberg stipend granted by the State of Lower Saxony.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032532.112.
REFERENCES
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR 2007. Reconstitution of yeast translation initiation. Methods Enzymol 430: 111–145 [DOI] [PubMed] [Google Scholar]
- Acker MG, Shin BS, Nanda JS, Saini AK, Dever TE, Lorsch JR 2009. Kinetic analysis of late steps of eukaryotic translation initiation. J Mol Biol 385: 491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire MA, Maag D, Savio P, Acker MG, Tarun SZ Jr, Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, et al. 2002. Development and characterization of a reconstituted yeast translation initiation system. RNA 8: 382–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire MA, Maag D, Lorsch JR 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20: 251–262 [DOI] [PubMed] [Google Scholar]
- Asano K, Phan L, Anderson J, Hinnebusch AG 1998. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem 273: 18573–18585 [DOI] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG 2000. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev 14: 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bich C, Maedler S, Chiesa K, DeGiacomo F, Bogliotti N, Zenobi R 2010. Reactivity and applications of new amine reactive cross-linkers for mass spectrometric detection of protein-protein complexes. Anal Chem 82: 172–179 [DOI] [PubMed] [Google Scholar]
- Damoc E, Fraser CS, Zhou M, Videler H, Mayeur GL, Hershey JW, Doudna JA, Robinson CV, Leary JA 2007. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol Cell Proteomics 6: 1135–1146 [DOI] [PubMed] [Google Scholar]
- Dennis MD, Person MD, Browning KS 2009. Phosphorylation of plant translation initiation factors by CK2 enhances the in vitro interaction of multifactor complex components. J Biol Chem 284: 20615–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P, Tavares P, Lurz R, van Heel M 1993. The portal protein of bacteriophage SPP1: A DNA pump with 13-fold symmetry. EMBO J 12: 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elantak L, Wagner S, Herrmannova A, Karaskova M, Rutkai E, Lukavsky PJ, Valasek L 2010. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J Mol Biol 396: 1097–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley AR, Powell DW, Weaver CM, Jennings JL, Link AJ 2011. Assessing the components of the eIF3 complex and their phosphorylation status. J Proteome Res 10: 1481–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Lee JY, Mayeur GL, Bushell M, Doudna JA, Hershey JWB 2004. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40S ribosomal subunits in vitro. J Biol Chem 279: 8946–8956 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Herrmannova A, Daujotyte D, Yang JC, Cuchalova L, Gorrec F, Wagner S, Danyi I, Lukavsky PJ, Shivaya Valasek L 2011. Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre-initiation complex assembly. Nucleic Acids Res 40: 2294–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 2006. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem Sci 31: 553–562 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75: 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Dever TE, Sonenberg N 2004. Mechanism and regulation of protein synthesis initiation in eukaryotes. In Protein synthesis and ribosome structure (ed. KH Nierhaus, DN Wilson), pp. 241–322. Wiley-VCH, Weinheim, Germany [Google Scholar]
- Hofmann K, Bucher P 1998. The PCI domain: A common theme in three multiprotein complexes. Trends Biochem Sci 23: 204–205 [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jivotovskaya AV, Valasek L, Hinnebusch AG, Nielsen KH 2006. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol Cell Biol 26: 1355–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, et al. 2008. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat Methods 5: 53–55 [DOI] [PubMed] [Google Scholar]
- Khoshnevis S, Neumann P, Ficner R 2010. Crystal structure of the RNA Recognition motif of yeast translation initiation factor eIF3bw reveals differences to human eIF3b. PLoS ONE 5: e12748 doi: 10.1371/journal.pone.0012784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouba T, Rutkai E, Karásková M, Valásek LS 2012. The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes. Nucleic Acids Res 40: 2683–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W 1999. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Masutani M, Sonenberg N, Yokoyama S, Imataka H 2007. Reconstitution reveals the functional core of mammalian eIF3. EMBO J 26: 3373–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Walker SE, Algire MA, Park EH, Hinnebusch AG, Lorsch JR 2010. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol Cell 39: 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazabal A, Wenzel RJ, Zenobi R 2006. Immunoassays with direct mass spectrometric detection. Anal Chem 78: 3562–3570 [DOI] [PubMed] [Google Scholar]
- Nielsen KH, Valasek L, Sykes C, Jivotovskaya A, Hinnebusch AG 2006. Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast. Mol Cell Biol 26: 2984–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Zhang XL, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG 1998. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol 18: 4935–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel H, Hofmann K 2005. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics 6: 71 doi: 10.1186/1471-2105-6-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310: 1513–1515 [DOI] [PubMed] [Google Scholar]
- Sokabe M, Fraser CS, Hershey JW 2012. The human translation initiation multi-factor complex promotes methionyl-tRNAi binding to the 40S ribosomal subunit. Nucleic Acids Res 40: 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Sun C, Todorovic A, Querol-Audi J, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis CS, Hartland A, Gradia S, et al. 2011. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc Natl Acad Sci 108: 20473–20478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Hasek J, Nielsen KH, Hinnebusch AG 2001a. Dual function of eIF3j/Hcr1p in processing 20S pre-rRNA and translation initiation. J Biol Chem 276: 43351–43360 [DOI] [PubMed] [Google Scholar]
- Valasek L, Phan L, Schoenfeld LW, Valaskova V, Hinnebusch AG 2001b. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J 20: 891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Nielsen KH, Hinnebusch AG 2002. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J 21: 5886–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG 2003. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M 1996. A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- Wei Z, Zhang P, Zhou Z, Cheng Z, Wan M, Gong W 2004. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J Biol Chem 279: 34983–34990 [DOI] [PubMed] [Google Scholar]
- Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, et al. 2008. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci 105: 18139–18144 [DOI] [PMC free article] [PubMed] [Google Scholar]