Abstract
B-RAF is mutated to a constitutively active form in 8% of human cancers including 50% of melanomas. In clinical trials, the RAF inhibitor, PLX4032 (vemurafenib), caused partial or complete responses in 48–81% of mutant B-RAF harboring melanoma patients. However, the average duration of response was 6–7 months before tumor regrowth, indicating the acquisition of resistance to PLX4032. To understand the mechanisms of resistance, we developed mutant B-RAF melanoma cells that displayed resistance to RAF inhibition through continuous culture with PLX4720 (the tool compound for PLX4032). Resistance was associated with a partial reactivation of extracellular signal-regulated kinase 1/2 (ERK1/2) signaling, recovery of G1/S cell-cycle events, and suppression of the pro-apoptotic B-cell leukemia/lymphoma 2 (Bcl-2) homology domain 3 (BH3)-only proteins, Bcl-2-interacting mediator of cell death-extra large (Bim-EL) and Bcl-2 modifying factor (Bmf). Preventing ERK1/2 reactivation with MEK (mitogen-activated protein/extracellular signal-regulated kinase kinase) inhibitors blocked G1-S cell-cycle progression but failed to induce apoptosis or upregulate Bim-EL and Bmf. Treatment with the histone deacetylase (HDAC) inhibitor, suberoylanilide hydroxamic acid, led to de-repression of Bim-EL and enhanced cell death in the presence of PLX4720 or AZD6244 in resistant cells. These data indicate that acquired resistance to PLX4032/4720 likely involves ERK1/2 pathway reactivation as well as ERK1/2-independent silencing of BH3-only proteins. Furthermore, combined treatment of HDAC inhibitors and MEK inhibitors may contribute to overcoming PLX4032 resistance.
Keywords: Bim-EL, Bmf, resistance, SAHA, melanoma
Oncogene addiction refers to the dependence of tumor cells on the continued expression of an oncogene for the maintenance of malignant properties.1 It has gained increasing recognition in recent years and provides the rationale for targeted therapeutic strategies. Notable advances are the responses of chronic myelogenous leukemia patients with BCR-ABL (breakpoint cluster region-v-abl abelson murine leukemia viral oncogene homolog 1) translocations to imatinib, breast cancer patients with amplified human epidermal receptor 2 (HER2) to trastuzumab, and mutant epidermal growth factor receptor (EGFR)-harboring non-small cell lung carcinomas patients to erlotinib and gefitinib treatments.2, 3, 4 A more recent example is the strategy to target mutations in the serine/threonine kinase B-RAF that occur in ∼50% of melanomas, 30% of thyroid carcinomas, and 14% of colorectal tumors.5 A valine to glutamic acid substitution at codon 600 (V600E) accounts for over 90% of the mutations in B-RAF and activates B-RAF kinase activity toward the MEK-extracellular signal-regulated kinase 1/2 (ERK1/2) cascade. B-RAFV600E and MEK (mitogen-activated protein/extracellular signal-regulated kinase kinase) activity are required for melanoma cell proliferation, invasion, and resistance to apoptosis in vitro,6, 7, 8, 9, 10, 11 and tumor xenograft growth in immunocompromised mice.8, 12 Furthermore, conditional melanocyte-specific expression of B-RAFV600E in mice co-operates with loss of phosphatase and tensin homolog (PTEN) or p16INK4a to induce melanoma.13, 14 Based on these preclinical data, inhibitors of mutant B-RAF have been investigated in the clinical setting.
In early studies, the RAF/receptor tyrosine kinase (RTK) inhibitor, sorafenib, failed to elicit clinical responses in melanoma and these trials were discontinued.15 More recently, the RAF inhibitor PLX4032/vemurafenib has elicited strong clinical responses in mutant B-RAF melanoma patients. In phase 1–3 trials with PLX4032, 48–81% of mutant B-RAF harboring patients demonstrated partial or complete responses in the short term.16, 17, 18 While PLX4032 gives strong palliative action, its long-term efficacy as a single agent is counteracted by the development of acquired resistance. PLX4032-treated patients gained on average 6–7 months of clincial benefit and most subsequently had tumor regrowth.19 Similar acquired resistance has been experienced with imatinib and gefitinib and has been associated with reactivation of the drug target and/or its pathway.20, 21 In many such cases, secondary mutations within the drug target that modify drug binding or permit target activation in the presence of drug have been associated with acquired resistance. By contrast, no secondary mutations have been identified, to date, in B-RAF inhibitor resistant tumors.22
A critical issue moving forward is to understand the mechanisms of resistance to PLX4032 in order to better design future combinatorial trials in melanoma. Initial findings have suggested that mutation of N-RAS, expression of B-RAF splice variants, or upregulation of platelet-derived growth factor receptor beta (PDGFRβ), insulin-like growth factor 1 receptor (IGF1R) or Cot1 is associated with acquired resistance to PLX4032 in subsets of melanoma patients.22, 23, 24, 25 Clearly other mechanisms exist22 and cell-based approaches can be used to identify alternative mechanisms of resistance for testing in the limited matched pretreatment, during treatment, and post treatment samples. Such approaches led to the identification of MET amplification in response to gefitinib26 and IGF1R and Cot1 upregulation to compensate for RAF inhibition.23, 24 Here, we undertook an in-vitro approach to identify resistance mechanisms to PLX4032/vemuafenib, using the tool compound PLX4720. We demonstrate that multiple mechanisms are involved in resistance to PLX4720, including ERK1/2 pathway reactivation and silencing of B-cell leukemia/lymphoma 2 (Bcl-2) homology domain 3 (BH3)-only protein expression.
Results
Prolonged culture of mutant B-RAF melanoma cells with PLX4720 leads to the development of resistance
The RAF inhibitor, PLX4032, elicits remarkable clinical effects in patients harboring mutant B-RAF16, 27; however, its long-term clinical efficacy is being hampered by the development of acquired resistance. To model this acquired resistance, we cultured two mutant B-RAF melanoma cell lines, WM793 and M238, in the continued presence of 5 μM PLX4720. WM793 was derived from a vertical growth phase primary tumor28 and M238 was from a skin metastasis.29 PLX4720 is the tool analog of PLX4032 and elicits effects that are indistinguishable from PLX4032.30, 31, 32 Initial treatment of mutant B-RAF melanoma cells with PLX4720 gave a cytostatic effect accompanied by cell death. However, long-term culture with PLX4720 led to the selection of cells that were capable of growth in the presence of up to 10 μM PLX4720 (Figure 1a), although their growth rates were reduced when compared with the no drug growth condition (Figure 1b). Notably, these cells, termed as WM793-Res and M238-Res, respectively, displayed larger cell size and elongated morphology (Figure 1c).
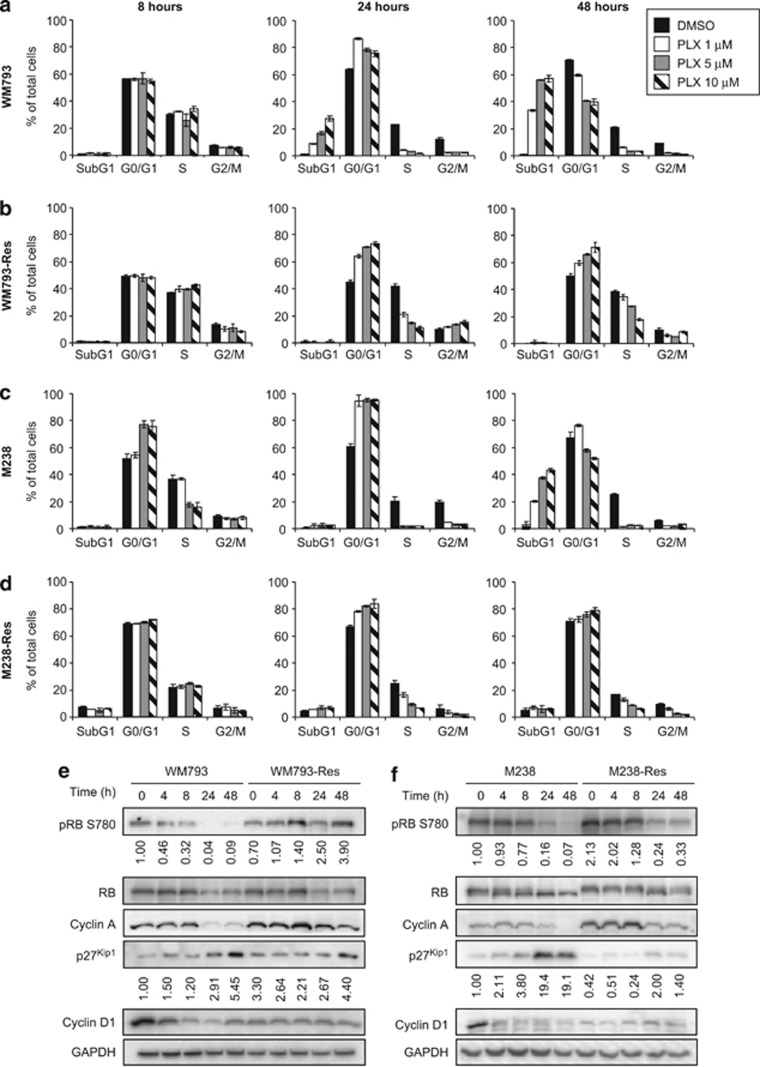
Figure 1.
WM793 and M238 cells acquire resistance to PLX4720 after long-term drug treatment. (a) Parental and resistant cells were grown in the presence of the indicated doses of PLX4720 for 3 days. Relative cell growth was measured by MTT assay. (b) Parental and resistant cells were grown in media±5 μM PLX4720 for 9 days. Drugs were replenished every 48 h. Relative cell growth was measured by MTT assay every 3 days. X axis, time points; Y axis, mean relative cell growth from triplet experiments. Error bars represent standard deviations. Left: WM793 and WM793-Res cells; Right: M238 and M238-Res cells. (c) TRITC-phalloidin staining of parental and resistant cells. (d) Parental and resistant WM793 cells were cultured in the absence of PLX4720 for 24 h. Cells were then challenged with 10 μM PLX4720 for indicated time before lysed for western blot analysis on RAF-MEK-ERK1/2 pathway proteins, as indicated. An arrow indicates the A-RAF-specific band. Quantitation of phospho-ERK bands normalized against ERK2 is shown below the blot. (e) Same as (d), except that M238 and M238-Res cells were used. (f) WM793 and WM793-Res cells were treated with 5 μM PLX4720 for indicated times. Activated Ras was pulled down from whole cell lysates of treated cells or naive Sbcl2 cells and detected by western blot with anti-Ras antibody
Impaired ERK1/2 inactivation in PLX4720-resistant cells
To understand the mechanism of acquired resistance, we initially examined the inhibitory effect of increasing doses of PLX4720 on the RAF-MEK-ERK1/2 signaling pathway in resistant cells compared with their parental counterparts. PLX4720 treatment of parental cells led to potent and durable inhibition of phospho-ERK1/2 and phospho-MEK levels (Figures 1d and e; Supplementary Figure 1). ERK1/2 pathway inhibition was accompanied by increased mobility of both B-RAF and C-RAF, consistent with a block in feedback phosphorylation.33, 34 Interestingly, extended PLX4720 treatment (24–48 h) induced a weak rebound of phospho-ERK1/2 in WM793 cells, an observation consistent with others' findings.35 This phospho-ERK1/2 rebound is not due to a quick turnover of the drug because drugs were replenished after 24 h. Resistant cells remained responsive to PLX4720 in terms of downregulating phospho-ERK1/2 and phospho-MEK levels (Figures 1d and e). However, their inhibition by PLX4720 was less complete even at high dose (10 μM) and the phospho-ERK1/2 rebound was more pronounced in resistant cells compared with parental cells. Therefore, these resistant cells displayed an impaired inactivation of ERK1/2 in response to PLX4720. No apparent increase in the expression of any of the RAF isoenzymes (Figures 1d and e) or Cot1 (data not shown) was detected in resistant cells, arguing against a compensatory upregulation of these mitogen-activated protein kinase kinase kinases (MAP3Ks) in response to PLX4720.
N-RAS and MEK mutations have been identified in resistant tumor samples from a subset of melanoma patients treated with PLX4032 or AZD6244, respectively, and in cell culture, these mutations rendered ERK1/2 signaling non-responsive to RAF or MEK inhibitors.22, 36, 37 Therefore, we sequenced exons that frequently harbor mutations in N-RAS, K-RAS, H-RAS, MEK1 and MEK2, from resistant cells but no mutations were identified in any of these genes (Supplementary Figure 2). We then compared Ras GTPase activity between parental WM793 and WM793-Res cells using the mutant N-Ras Q61K cell line, Sbcl2, as a positive control. Whereas parental WM793 cells only showed low Ras activity, elevated Ras activity was observed in resistant cells (Figure 1f). This moderate increase in Ras activity (in comparison with Sbcl2) correlates well with a partial reactivation of ERK signaling in resistant cells. In addition, Ras from resistant cells displayed similar migration characteristics on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as that in parental cells, consistent with its wild-type status as shown by DNA sequencing. The increased Ras activity could potentially result from enhanced signaling from upstream RTKs. Levels of tyrosine phosphorylated proteins were elevated in the resistant cells (Supplementary Figure 3). Two RTKs, IGF1R and PDGFRβ, have been previously implicated in acquired resistance to RAF inhibitors.22, 23 Interestingly, we observed increased PDGFRβ and decreased IGF1R expression in both resistant cell lines and their parental counterparts (Figure 1d). To test whether PDGFRβ has a role in ERK1/2 reactivation, we targeted PDGFRβ signaling by imatinib treatment or PDGFRβ small interfering ribonucleic acid (siRNA). Both approaches decreased PDGFRβ phosphorylation but showed no effect on the residual phospho-ERK1/2 level in resistant cells (Supplementary Figures 3A and B), suggesting that ERK1/2 reactivation is most likely PDGFRβ independent. The major 125 KD tyrosine phosphorylated band that is elevated in WM793-Res cells likely represents focal adhesion kinase (FAK) since a similar pattern is observed with a phospho-FAK antibody (Supplementary Figure 3A).
Resistant cells display recovery of G1-S cell-cycle events
To understand how resistant cells overcome PLX4720-induced growth arrest, we analyzed the cell-cycle profiles of parental and resistant cells treated with PLX4720 at three different doses (1, 5, and 10 μM) and times (8, 24, and 48 h) (Figure 2). Short-term (8 h) treatment of parental cells with PLX4720 had little effect on the cell-cycle profile in WM793 cells but a moderate inhibitory effect in M238 cells (Figures 2a and c, left panels). Prolonged treatment with PLX4720 (24–48 h), however, led to strong reduction of S-phase cells and the accumulation of subG1 cells at all three doses in both WM793 and M238 cells (Figures 2a and c, middle and right panels). Therefore, PLX4720 treatment elicits a G1/S blockage accompanied by cell death. Given the similar cytostatic effects of various PLX4720 doses, we then analyzed the expression of G1/S proteins at 5 μM PLX4720, a dose at which the resistant cells were evolved. The cytostatic effects of PLX4720 were associated with changes in G1/S regulators including reduced expression of cyclin D1 and cyclin A, increased expression of p27Kip1, and decreased phosphorylation of retinoblastoma (RB) protein at S780 (Figures 2e and f). Phosphorylation of S780 inactivates RB and is indicative of cycle progression.38 By contrast, PLX4720 treatment of resistant cells only partially reduced cell numbers in S phase even after 48 h (Figures 2b and d), consistent with the partial retention of phospho-ERK1/2 levels observed in these cells (Figures 1d and e). Additionally, RB phosphorylation and cyclin A levels were partially recovered and the induction of p27Kip1 was mitigated in PLX4720-treated resistant cells (Figures 2e and f). Furthermore, cyclin D1 remained at a low level in resistant cells and was unresponsive to PLX4720 treatment.
Figure 2.
Resistant cells bypass G1/S growth arrest induced by PLX4720. (a) WM793 cells were treated with different doses of PLX4720 for indicated time. Cells were then harvested and stained with propidium iodide for cell-cycle analysis. (b–d) Same as (a) except WM793-Res, M238, and M238-Res cells were used. (e) WM793 and WM793-Res cells were treated as described in Figure 1d and analyzed for the expression of G1/S regulating proteins as indicated. Quantitation of phospho-RB bands (normalized against total RB) and p27Kip1 bands (normalized against GAPDH) is shown below the corresponding blots. (f) Same as (e) except M238 and M238-Res cells were used
ERK1/2 reactivation contributes to the recovery of G1-S cell-cycle events in resistant cells
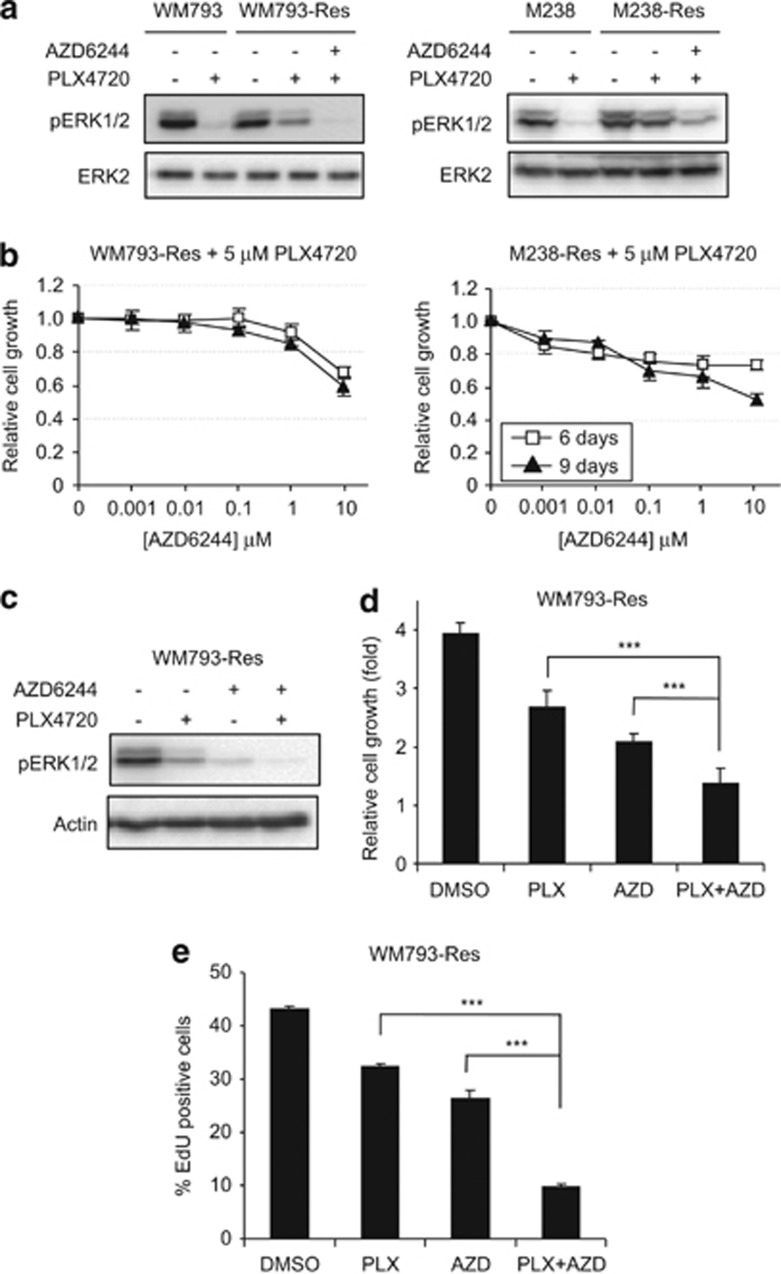
Since mutant B-RAF signaling via ERK1/2 regulates proliferation and resistant cells retained a level of phospho-ERK1/2 in the presence of PLX4720, we further analyzed the role of ERK1/2 reactivation in the growth of resistant cells. ERK1/2 phosphorylation was also partially maintained in WM793-Res cells treated with another RAF inhibitor, GDC-087939 (Supplementary Figure 4A), indicating cross-resistance to other RAF inhibitors. However, combined treatment of PLX4720 and an MEK inhibitor, AZD6244, reduced phospho-ERK1/2 staining in both WM793-Res and M238-Res cells to levels comparable to parental cells treated with PLX4720 alone (Figure 3a). In addition, co-treatment of PLX4720 and AZD6244 of resistant cells was associated with inhibition of resistant cell growth in a dose- and time-dependent manner (Figure 3b) and reduced S-phase entry, as measured by 5-ethynyl-2′-deoxyuridine (EdU) incorporation, when compared with treatment with PLX4720 alone (Figure 3d). Treatment of MEK inhibitor (AZD6244) alone also reduced the phospho-ERK1/2 level and S-phase entry when compared with dimethyl sulfoxide (DMSO) treatment, suggesting that the growth of resistant cells is still MEK dependent. However, the pathway inhibition by MEK inhibitor alone is not as complete as by the RAF/MEK inhibitor combination (Figure 3c). Accordingly, the combination treatment more effectively inhibits cell growth and S-phase entry than MEK inhibitor alone (Figures 3d and e). These data showed that the reactivated ERK1/2 signaling in PLX4720-treated resistant cells is required for the recovery of G1/S cell-cycle events.
Figure 3.
Phospho-ERK1/2 rebound contributes to the escape of PLX4720-induced cell-cycle arrest in resistant cells. (a) Parental and resistant cells were replated and cultured in the absence of PLX4720 for 24 h. Cells were then treated with DMSO or 5 μM PLX4720 or a combination of 5 μM PLX4720 and 5 μM AZD6244 for another 24 h before being lysed for western blot analysis on indicated proteins. Left: WM793 and WM793-Res; Right: M238 and M238-Res. (b) Parental and resistant WM793 cells were grown in medium containing 5 μM PLX4720 alone or a combination of 5 μM PLX4720 and indicated doses of AZD6244 for 9 days. Drugs were replenished every 48 h. Relative cells growth was measured by MTT assay on day 6 and day 9. X axis, AZD6244 doses; Y axis, mean relative cell number from triplet results. Error bars represent standard deviation. Left: WM793-Res cells; Right: M238-Res cells. (c) WM793-Res cells were treated with DMSO, 5 μM PLX4720, 5 μM AZD6244 or both for 24 h and lysed for western blot analysis. (d) Cells were grown in the presence of DMSO or indicated inhibitors (5 μM each) for 72 h. Relative cell growth (normalized against starting point) was measured by MTT assay. (e) Same as (c) except that after 24 h, cells were grown in the presence of 10 μM EdU for another 16 h followed by EdU incorporation assay. Mean percentages of EdU staining-positive cells from triplet experiments are shown. Error bars represent standard deviation. ***P<0.001, based on two-tail Student's t-test assuming unequal variance
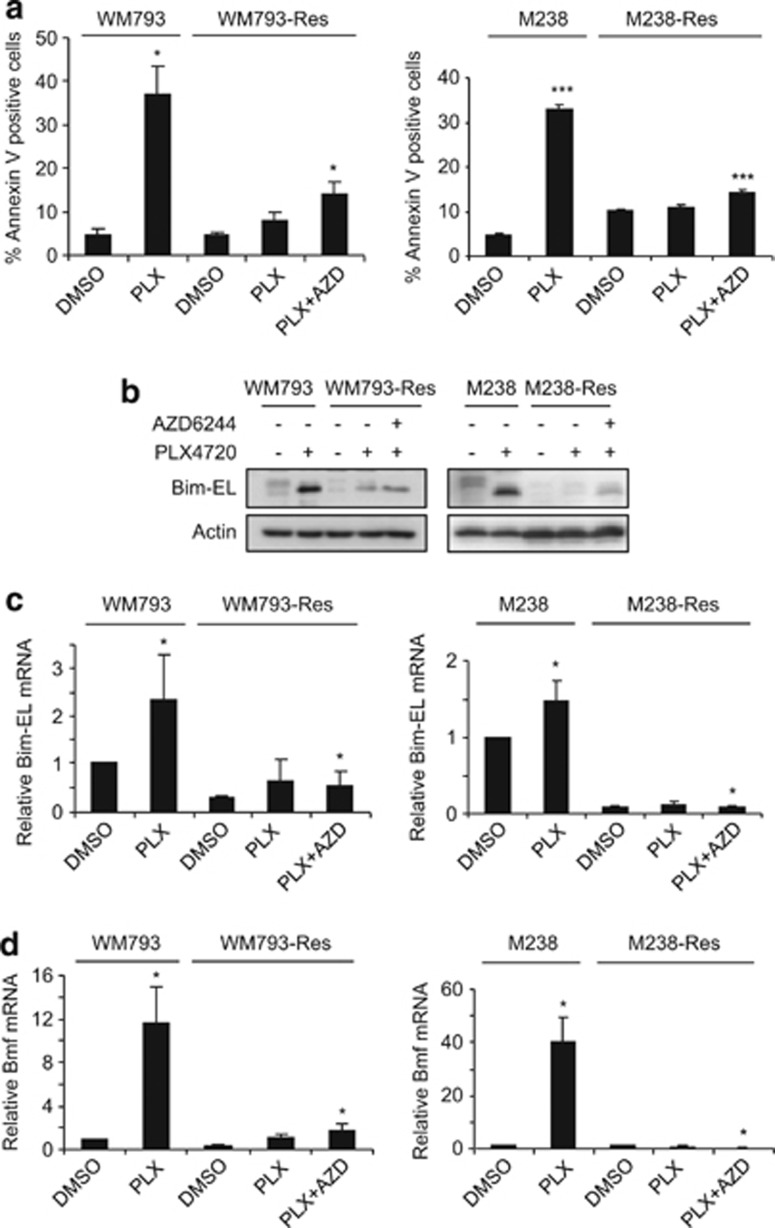
Resistant cells exhibit altered expression of Bcl-2 family proteins
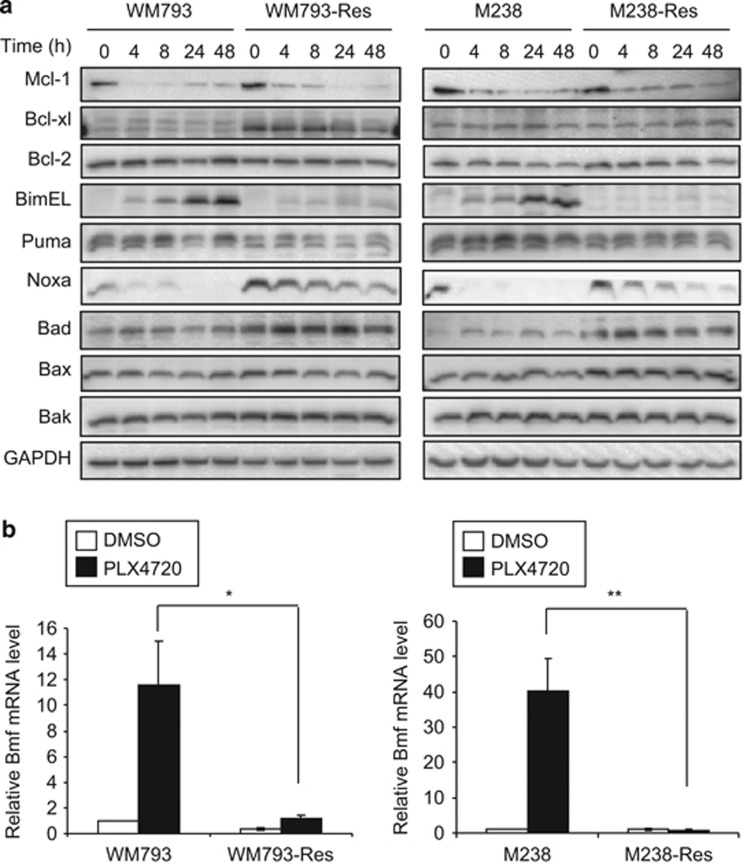
Prolonged RAF inhibition with PLX4720 also promotes apoptosis (Shao and Aplin;40 Figure 2). Notably, while PLX4720 led to a time- and dose-dependent level of cell death in parental cells, it caused only marginal cell death in resistant cells. To understand this resistance to PLX4720-induced cytotoxicity, we compared the levels of Bcl-2 family proteins in between parental and resistant cells. We used 5 μM PLX4720, a dose at which a significant level of cell death was observed (Figure 2) and at which our resistant cells were evolved. The anti-apoptotic proteins, myeloid cell leukemia sequence 1 (Mcl-1) and Bcl-2, showed similar expression profiles in response to PLX4720 treatment between parental cells and their resistant counterparts (Figure 4a). The expression of B-cell lymphoma-extra large (Bcl-xl) was increased in WM793-Res cells, but not in M238-Res cells, and depletion of Bcl-xl failed to sensitize resistant cells to PLX4720 (Supplementary Figure 5). Together, these data suggest that Bcl-xl is not a major determinant of resistance to apoptosis. We also observed changes in pro-apoptotic BH3-only proteins. Bcl-xL/Bcl-2-associated death promoter (Bad) and Noxa levels were elevated in resistant cells, which are unlikely to be responsible for cell death resistance. Notably, two BH3-only proteins, Bcl-2-interacting mediator of cell death-extra large (Bim-EL) and Bcl-2 modifying factor (Bmf) (analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) due to lack of reliable antibody against endogenous Bmf) were upregulated in parental cells but not in resistant cells following PLX4720 treatment (Figures 4a and b). The upregulation of Bim-EL and Bmf by PLX4720 treatment was mirrored in multiple B-RAFV600E melanoma cell lines (Supplementary Figure 6) and is required for PLX4720-induced apoptosis since knockdown of these two proteins conferred resistance to cell death (Supplementary Figure 7 and Shao and Aplin40).
Figure 4.
Altered expression of Bcl-2 family proteins in resistant cells. (a) Parental and resistant cells were treated with 5 μM PLX4720 for various time and lysed for western blot analysis on indicated Bcl-2 family proteins. Left: WM793 and WM793-Res cells; Right: M238 and M238-Res cells. (b) Parental and resistant cells were treated with DMSO or 5 μM PLX4720 for 24 h. Total RNA was isolated from cells for qRT-PCR analysis on Bmf. X axis, cell types. Y axis, mean relative Bmf mRNA level (normalized against DMSO-treated parental cells) from three experiments using actin mRNA level as an internal control. Error bars represent standard deviations. *P<0.05, **P<0.01, based on two-tail Student's t-test assuming unequal variance
Expression of Bim-EL and/or Bmf renders resistant cells susceptible to apoptosis
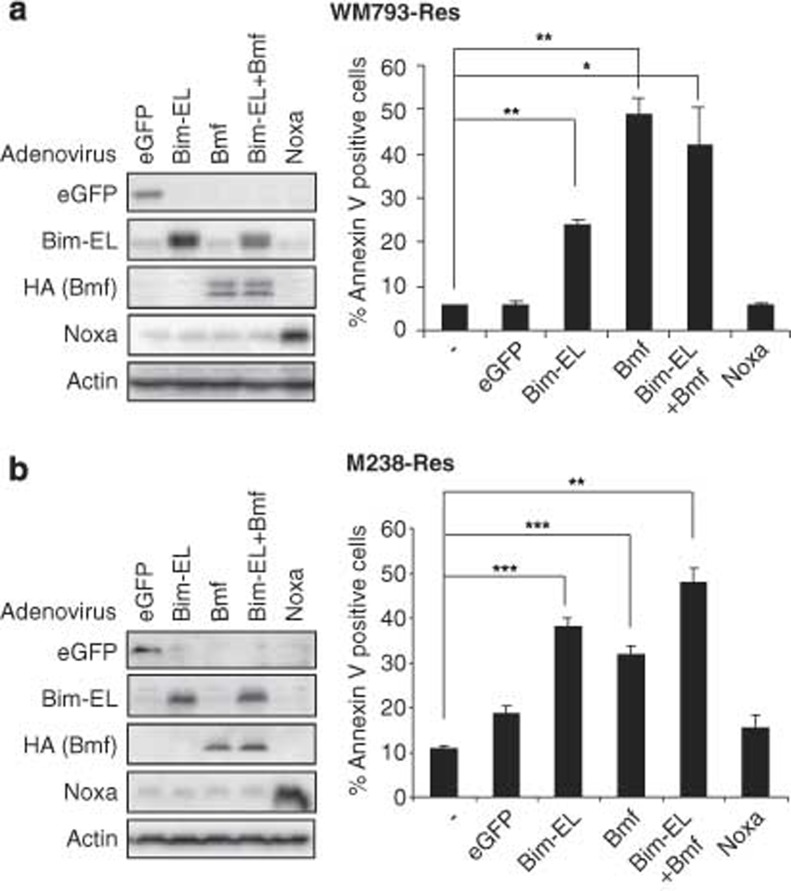
To examine whether the loss of Bim-EL and Bmf expression in resistant cells underlies the resistance to PLX4720-induced apoptosis, we tested the extent to which Bim-EL and/or Bmf expression was sufficient to induce apoptosis in WM793- and M238-resistant cell lines. We infected resistant cells with adenoviruses to express Bim-EL, Bmf, Noxa (an alternative BH3-only protein) or enhanced green fluorescence protein (eGFP), as a control (Figures 5a and b, left panels). Ectopic expression of Bim-EL or Bmf significantly increased cell death in both WM793-Res and M238-Res cells, while expression of eGFP or Noxa showed little effect (Figures 5a and b, right panels). The expression level of exogenous Bim-EL was comparable to PLX4720-induced endogenous Bim-EL level in parental cells (Supplementary Figure 8). Therefore, the cell death observed is not simply due overexpression of BH3-only proteins. These results indicate that Bim-EL and Bmf are sufficient for apoptosis in RAF inhibitor-resistant cells.
Figure 5.
Bim-EL or Bmf expression renders resistant cells susceptible to apoptosis. (a) Left: WM793-Res cells were infected with adenovirus harboring indicated cDNAs in the presence of 5 μM PLX4720 for 8 h and lysed for western blot analysis. Right: WM793-Res cells were infected with adenovirus containing indicated cDNAs in the presence of 5 μM PLX4720 for 24 h. Cells were harvested and stained with annexin V-APC followed by flow cytometry analysis. Mean percentage of annexin V-positive cells from three individual experiments was shown for each condition. Error bars represent standard deviation. (b) Same as (a) except M238-Res cells were used and annexin-V staining was performed after 32 h of adenovirus infection. *P<0.05, **P<0.01, ***P<0.001, based on two-tail Student's t-test assuming unequal variance
ERK1/2 reactivation does not account for the loss of BH3-only protein upregulation and apoptosis resistance in resistant cells
ERK1/2 signaling regulates both Bim-EL and Bmf40, 41; therefore, we examined whether the partial ERK1/2 reactivation observed in resistant cells represses these BH3-only proteins and confers apoptosis resistance. Parental cells were susceptible to relatively high doses of PLX4720 (5 μM), which was associated with upregulation of Bim-EL and Bmf at the protein and RNA level (Figures 6a–d). Bim-EL also displays faster migration due to loss of ERK1/2 phosphorylation, which regulates its turnover.42 Resistant cells displayed much lower annexin-V staining at the same concentration of PLX4720 (Figure 6a) and the levels of Bim-EL and Bmf were greatly reduced in resistant cells versus their parental cells (Figures 6b–d). Most importantly, elimination of phospho-ERK1/2 by combined treatment with PLX4720 and AZD6244 (both at 5 μM) only slightly enhanced apoptosis in resistant cells (Figure 6a) and failed to substantially upregulate Bim-EL and Bmf (Figures 6b–d). These data indicated that repression of Bim-EL and Bmf in resistant cells involves a mechanism that is largely independent of MEK-ERK1/2 signaling.
Figure 6.
Phospho-ERK1/2 rebound is not essential for apoptosis bypass in resistant cells. (a) Parental and resistant cells treated with DMSO or 5 μM PLX4720 or a combination of 5 μM PLX4720 and 5 μM AZD6244 for 48 h. Cells were then harvested and stained with annexin V. Quantitation of annexin V staining-positive cells from three experiments is shown. X axis, treatment conditions; Y axis, mean percentage of annexin V-positive cells. Error bars represent standard deviation. Left: WM793 and WM793-Res cells; Right: M238 and M238-Res cells. (b) Same as (a) except that after 24 h of inhibitor treatment, cells were lysed for western blot analysis on Bim-EL protein using actin as loading control. (c) Same as (b) except total RNA was isolated from cells for qRT-PCR analysis on Bim-EL. Left: WM793 and WM793-Res; Right: M238 and M238-Res. X axis, treatment conditions. Y axis, mean relative Bim-EL (normalized against DMSO-treated parental cells) from three experiments using actin mRNA level as an internal control. Error bars represent standard deviations. (d) Same as (c) except Bmf mRNA levels were analyzed. *P<0.05, ***P<0.001, based on two-tail Student's t-test assuming unequal variance
Bim-EL and Bmf gene silencing is not due to promoter DNA methylation
Promoter methylation is one common mechanism for gene silencing. Both Bim and Bmf promoter regions contain CpG islands. In Burkitt lymphoma, epigenetic silencing of Bim by promoter hypermethylation contributes to chemoresistance.43 To determine whether Bim and Bmf genes are repressed by a similar mechanism in resistant cells, we performed bisulfite DNA sequencing analysis on the promoter regions of Bim and Bmf genes in both parental and WM793-Res cells. CpG methylation was readily detected in the Bim promoter and positions of methylated CpG dinucleotides were identical in both parental and resistant cells (Supplementary Figure 9A). In addition, treatment of WM793-Res cells with 5-azacytidine failed to upregulate Bim-EL (Supplementary Figure 9B). By contrast, no CpG methylation was detected in a Bmf promoter region spanning 32 CpG dinucleotides in both parental and resistant cells (Supplementary Figure 9C). Overall, these data suggest that Bim and Bmf gene silencing is unlikely to occur through promoter DNA methylation.
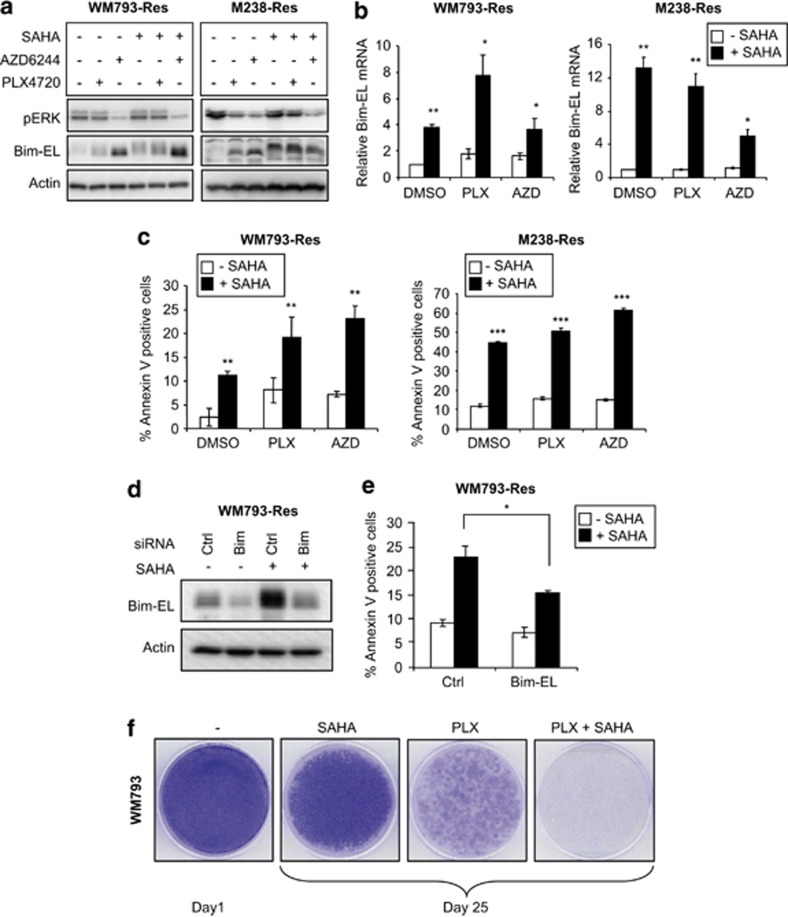
The histone deacetylase inhibitor, suberoylanilide hydroxamic acid, de-represses Bim-EL expression, sensitizes resistant cells to PLX4720/AZD6244 treatment, and inhibits the emergence of PLX4720-resistant cells
Epigenetic chromatin modification is another frequent mechanism of gene repression. Zhang et al.44, 45 have shown that HDAC inhibitors enhance cell death in a panel of adenocarcinoma and squamous cell carcinoma cell lines by increasing the expression of Bim-EL and Bmf proteins. Therefore, we asked whether histone deacetylase (HDAC) inhibition would de-repress Bim-EL and/or Bmf in resistant cells. Interestingly, treatment of the HDAC inhibitor, suberoylanilide hydroxamic acid (SAHA), increased Bim-EL mRNA and protein levels (Figures 7a and b; Supplementary Figure 10A), although Bmf mRNA levels remained unchanged (data not shown). In addition, SAHA treatment sensitized the resistant cells to PLX4720 or AZD6244 (Figure 7c). This sensitization occurred in a Bim-EL-dependent manner because depletion of Bim-EL partially rescued cell death in WM793-Res cells (Figures 7d and e). Importantly, co-treatment of SAHA and PLX4720 strongly inhibited the emergence of PLX4720-resistant colonies in a long-term survival assay (Figure 7f; Supplementary Figure 10B).
Figure 7.
Effect of the HDAC inhibitor, SAHA, on PLX4720-resistant cells. (a) Resistant cells were replated and cultured in the absence of inhibitors for 24 h. Cells were then treated with indicated inhibitors (5 μM each) for further 24 h and lysed for western blot analysis with phospho-ERK1/2, Bim-EL, and actin antibodies. Left: WM793-Res cells; Right: M238-Res cells. (b) Resistant cells were treated similarly as (a) except after 24 h of inhibitor treatment, total RNA was isolated from cells for qRT-PCR analysis on Bim-EL in WM793-Res (Left) or M238-Res (right) cells, respectively. X axis, treatment conditions; Y axis, mean relative Bim-EL mRNA level (normalized against cells treated with DMSO only) from three experiments using actin mRNA level as an internal control. Error bars represent standard deviation. (c) Same as (b) except that cells were treated with inhibitors for 48 h and harvested for annexin-V staining and flow cytometry analysis. Mean percentage of annexin V-positive cells from three experiments is shown for each treatment condition. Error bars represent standard deviation. Left: WM793-Res cells; Right: M238-Res cells. (d) WM793-Res cells were transfected with non-specific or Bim-EL siRNA for 72 h in the presence of 5 μM PLX4720 and then treated with DMSO or 5 μM SAHA for another 24 h before lysed for western blot analysis. (e) Same as (d) except that cells were treated with DMSO/SAHA for 48 h and stained with annexin V-APC for flow cytometry analysis. Mean percentage of annexin V-positive cells from three individual experiments is shown for each condition. Error bars represent standard deviation. (f) Parental WM793 cells were grown to confluence on 6 cm dishes and treated with 5 μM SAHA or 5 μM PLX4720 or both for 25 days. Medium and drugs were replenished every 2 days. At the end point, cells were stained with crystal violet. One confluent dish was stained on day 1 as a start point reference. *P<0.05, **P<0.01, ***P<0.001, based on two-tail Student's t-test assuming unequal variance
Discussion
The RAF inhibitor, PLX4032/vemurafenib, has achieved striking response rates in clinical trials in mutant B-RAFV600 patients, indicating both cytostatic and cytotoxic effects of this inhibitor. Despite this promise, the long-term clinical efficacy of PLX4032 is hampered by the emergence of resistance. Recent studies have demonstrated that ERK1/2 reactivation is a common mechanism for PLX4032 resistance in B-RAFV600E melanoma cells.22, 24, 25 We developed in-vitro cell line models of acquired resistance to PLX4720, the tool compound of PLX4032. Using these models, we show that resistance to the cytostatic action of PLX4720 is dependent on ERK1/2 signaling reactivation, while resistance to cytotoxic action is ERK1/2 signaling independent and involves silencing of two BH3-only proteins, Bim-EL and Bmf. These findings underscore the importance of ERK1/2 pathway reactivation and provide further support for concomitant treatment of RAF and MEK inhibitors to inhibit ERK1/2 reactivation and overcome the resistance to cell-cycle arrest. Notably, Bollag et al. have suggested that regression of melanoma tumors requires over 80% inhibition of ERK1/2 activation.16, 27 Our findings also support the further investigation of BH3-mimetic compounds or other therapeutic strategies to restore Bim-EL and Bmf expression in order to overcome cytotoxic resistance to PLX4032.
Our data show that ERK1/2 reactivation is required for resistant cells to overcome growth arrest induced by RAF inhibitor treatment. The lack of upregulation of RAF isoforms and mutations in RAS and MEK1/2 argue against these mechanisms being responsible for the ERK1/2 reactivation signal in this model.36, 46 Another possibility is that resistant cells dynamically switch utilization of RAF isoforms to escape pathway inhibition by PLX4720.23 This is especially relevant in the context of the elevated Ras activity observed in resistant cells (Figure 1f). Consistent with this notion, treatment with sorafenib, a pan-RAF and tyrosine receptor kinase inhibitor, eliminated ERK1/2 reactivation in resistant cells (Supplementary Figures 3A and 4B); whereas individual knockdown of RAF isoforms did not dramatically decrease ERK1/2 reactivation (Supplementary Figure 4C). Alternatively, expression of the negative feedback effectors of the ERK1/2 pathway may be lost so that Ras is hyperactivated and promotes resistance to PLX4720.47 Consistent with the latter possibility, Sprouty2, a negative regulator of Ras/ERK1/2 pathway,48 is downregulated (Supplementary Figure 4D) and Ras activity is moderately increased in resistant cells (Figure 1f). The enhanced Ras activity might also result from an upregulation of upstream RTKs such as PDGFRβ (Figures 1d and e). However, depletion of PDGFRβ or treatment of imatinib in resistant cells had little effect on phospho-ERK1/2 levels. Therefore, the basis for reactivation of the ERK1/2 axis in these cells requires more experimental validation. Interestingly, the expression of IGF1R, an RTK that has been implicated in acquired resistance to other RAF inhibitors,23 was downregulated in both resistant lines. It is possible that the role of IGF1R for resistance development may be replaced by other RTKs and is therefore no longer required.
In addition to cell-cycle arrest bypass, another characteristic of resistant cells is resistance to apoptosis. ERK1/2 signaling modulates apoptotic events through regulation of Bcl-2 family proteins in melanoma cells.49 We have shown that apoptosis resistance is due to the silencing of two BH3-only proteins, Bim-EL and Bmf, via a mechanism which is largely independent of ERK1/2 reactivation. Our results from bisulfite sequencing analysis of the Bim-EL and Bmf promoters and 5-azacytidine treatment argue against promoter methylation being sufficient to explain the silencing of Bim-EL and Bmf and, hence, resistance to PLX4720. Downregulation of transcription activators may also account for the silencing of Bim-EL and Bmf genes. The forkhead transcription factors, Forkhead box O1 (FoxO1) and forkhead box O3a (FoxO3a), are known to control the expression of Bim50 and their upregulation in response to RAF and MEK inhibitors was impaired in WM793-Res cells (Supplementary Figure 11A). However, depletion of FoxO1 or FoxO3a in parental cells failed to repress the induction of Bim-EL by PLX4720 treatment in melanoma cells (Supplementary Figure 11B), arguing against a role for these forkhead proteins in regulating Bim-EL in this system.
Chromatin modifications such as histone acetylation/deacetylation also have important roles in gene regulation. Deacetylated histones usually associate with condensed chromatin structures and recruitment of transcriptional repression machinery. Cancer cells display altered epigenomes51 and the role of HDACs in the development of cancers such as myeloid leukemia has been well characterized.52, 53 Recent studies from Settleman et al. showed that a reversible drug-tolerant state in cancer cells is also associated with chromatin modification and treatment of HDAC inhibitors relinquishes drug tolerance.54 Notably, treatment of resistant cells with SAHA restored PLX4720 induction of Bim-EL but not of Bmf. Furthermore, SAHA enhanced apoptosis in resistant cells cultured with PLX4720 or AZD6244 and reduced the number of PLX4720-resistant colonies in a long-term drug treatment experiment. These results suggest that epigenetic chromatin remodeling may be implicated in Bim-EL repression and, therefore, contribute to PLX4720 resistance. The mechanism of Bim de-repression by SAHA in our resistant cells is still unknown. Recent studies have shown that SAHA may activate Bim transcription by increasing acetylation of histone H3 or H4 in the promoter region.55, 56 In line with this notion, the histone H4 acetylation within a Bim promoter region proximal to the start codon decreased in the resistant cells when compared with its parental counterpart and was restored upon SAHA treatment (Supplementary Figure 11C). However, whether these changes of histone H4 acetylation in resistant cells are directly involved in Bim activation by SAHA requires further study.
It is becoming apparent that resistance of melanomas to RAF inhibitors involves multiple mechanisms and that the interplay between these mechanisms warrants extensive investigation. Our work presented herein shows the presence of bifurcated resistance mechanisms to the RAF inhibitor, PLX4720. Resistance to the cytostatic effect of the inhibitor stems from reactivation of ERK1/2 signaling, whereas resistance to the cytotoxic action of the drug is due to repression of BH3-only proteins. Furthermore, a combinational treatment of MEK and HDAC inhibitors may provide clinical benefits for PLX4032-resistant patients.
Materials and Methods
Inhibitors
PLX4720 was kindly provided by Dr Gideon Bollag (Plexxikon Inc., Berkeley, CA, USA) and stock solutions were dissolved in DMSO. AZD6244, sorafenib, GDC-0879, and SAHA were purchased from Selleck Chemicals LLC (Houston, TX, USA). 5-Azacytidine was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell culture
WM793 and WM793-Res cells were cultured in MCDB 153 containing 20% Leibovitz L-15 medium, 2% FBS, 5 μg/ml insulin and penicillin/streptomycin. M238 and M238-Res cells were grown in RPMI medium with 10% FBS, 20 mM ℒ-glutamine and penicillin/streptomycin.
MTT cell growth assay
Melanoma cells were continuously grown in the aforementioned medium in the presence/absence of inhibitors for the indicated period of time. Medium and drugs were replenished every 2 days. Relative cell numbers were measured every 3 days by incubating cells with 0.5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) followed by optical density (OD) reading. OD readings were normalized against that of no-treatment group. Mean readings from three independent experiments were plotted for each time point.
Immunofluorescence
Parental and resistant cells were plated on glass coverslips. Cells were briefly washed with PBS, fixed with 3.7% paraformaldehyde for 20 min, and permeabilized with 0.2% Triton X-100 for 5 min. Coverslips were then incubated with tetramethylrhodamine B isothiocyanate (TRITC)-conjugated phalloidin (Sigma-Aldrich) diluted 1 : 2000 in 1% BSA/PBS for 1 h to visualize F-actin. Coverslips were mounted and viewed on a Nikon Eclipse Ti microscope (Melville, NY, USA) equipped with a Cool SNAP ES2 camera (Melville, NY, USA). Images were acquired using NIS Elements AR software (Nikon, Melville, NY, USA).
Western blotting
Western blotting was performed as previously described.40 Immunoreactivity was detected using peroxidase-conjugated secondary antibodies and chemiluminescence substrate (Pierce, Rockford, IL, USA). Chemiluminescence was detected using a Versadoc Imaging system (Bio-Rad, Hercules, CA, USA).
Antibodies
The following antibodies were purchased from Cell Signaling Technology (Beverley, MA, USA): Akt (#9227), phospho-Akt (Ser473, #9271), Bad (#9292), Bak (#3792), Bax (#2772), Bcl-xL (#2762), phospho-ERK1/2 (Thr202/Tyr204, #4377), MEK1 (#9124), phospho-MEK1 (#9121), PUMA (#4976), Rb (#3909), phospho-RB (Ser780, #9307) and GAPDH (#2118). A-RAF (sc-407), B-RAF (sc-5284), C-RAF/RAF-1 (sc-133), cyclin A (sc-751) and ERK1/2 (sc-094) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Bcl-2 (clone 4D7), cyclin D1 (#556470), Mcl-1 (#559027), and p27Kip1 (clone 57) were obtained from BD Transduction (San Jose, CA, USA). Anti-actin (#A2066) was purchased from Sigma-Aldrich. Anti-Bim (#AAP-330) was from Stressgen (San Diego, CA, USA). Noxa antibody (OP180) was obtained from Calbiochem/EMD4 Biosciences (Gibbstown, NJ, USA).
Quantitative RT-PCR
Total RNA was extracted from melanoma cells and reverse transcribed, as previously described.40 The primers used were Bmf (forward, 5′-gaggtacagattgcccgaaag-3′ reverse, 5′-ttcaaagcaaggttgtgca-3′) and actin (forward, 5′-tggaccaccaactgcttag-3′ reverse, 5′-gatgcagggatgatgttc-3′). Reactions were performed with SYBR Green using the MyiQ real-time PCR detection system (Bio-Rad). Relative mRNA levels were calculated using the comparative Ct (ΔCt) method.57 Quantitation of mRNA levels represents data from three independent experiments.
Annexin-V staining
Cells were trypsinized, washed in PBS and resuspended in 100 μl binding buffer.40 Next, cells were stained with 5 μl annexin V-APC (BD Biosciences) for 15 min before the addition of 400 μl binding buffer. Staining was measured by flow cytometry on a FACSCalibur (BD Biosciences) in the KCC Flow cytometry shared resource facility. Data were analyzed using Flowjo software (Three Star, Inc., Ashland, OR, USA).
EdU incorporation assay
WM793-Res cells were treated with DMSO, 5 μM PLX4720, or 5 μM PLX4720 plus 5 μM AZD6244 for 24 h before the addition of 10 μM EdU for another 16 h. Cells were then processed using the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay kit (Invitrogen, Eugene, OR, USA) for flow cytometry analysis.
Crystal violet staining
Cells grown on 6 cm dishes were washed in PBS twice and stained with crystal violet solution (1% crystal violet, 10% buffered formalin) for 15 min. After decanting the staining solution, cells were thoroughly washed in distilled water three times and air dried before taking pictures.
Ras pull-down assay
Cells were treated with 5 μM PLX4720 for indicated period of time before lysis. Active Ras-GTP was pulled down from whole cell lysates by using the Active Ras pull-down and detection kit (Thermo Scientific, Rockford, IL, USA) following manufacturer's guidance and detected by western blot.
Acknowledgments
We thank Melanie Mayberry for the sequencing of WM793-Res cells, Dr Gideon Bollag (Plexxikon, Berkeley, CA) for providing PLX4720, Dr Meenhard Herlyn (Wistar Institute, Philadelphia, PA) for WM793 cell line and Dr Antoni Ribas (UCLA, CA) for M238 cell line. This work was supported by NIH grants R01-GM067893 and R01-CA125103 (AE Aplin) and an Outrun the Sun fellowship (Y Shao). The Kimmel Cancer Center is supported by National Cancer Institute Support Grant 1P30CA56036.
Glossary
- ERK1/2
extracellular signal-regulated kinase 1/2
- BH3
Bcl-2 homology domain 3
- Bim-EL
Bcl-2-interacting mediator of cell death-extra large
- Bmf
Bcl-2 modifying factor
- HDAC
Histone deacetylase
- SAHA
suberoylanilide hydroxamic acid
- MEK
mitogen-activated protein/extracellular signal-regulated kinase kinase
- HER2
human epidermal receptor 2
- BCR-ABL
Breakpoint cluster region-v-abl abelson murine leukemia viral oncogene homolog 1
- EGFR
epidermal growth factor receptor
- PTEN
phosphatase and tensin homolog
- PDGFRβ
platelet-derived growth factor receptor beta
- IGF1R
insulin-like growth factor 1 receptor
- MAP3K
mitogen-activated protein kinase kinase kinase
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- siRNA
small interfering ribonucleic acid
- FAK
focal adhesion kinase
- Rb
retinoblastoma 1
- EdU
5-ethynyl-2′-deoxyuridine
- DMSO
dimethyl sulfoxide
- Bcl-2
B-cell leukemia/lymphoma 2
- Bcl-xl
B-cell lymphoma-extra large
- Mcl-1
myeloid cell leukemia sequence 1
- Bad
Bcl-xL/Bcl-2-associated death promoter
- qRT-PCR
quantitative real-time polymerase chain reaction
- eGFP
enhanced green fluorescence protein
- RTK
receptor tyrosine kinase
- FoxO1
forkhead box O1
- FoxO3a
forkhead box O3a
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by C Borner
Supplementary Material
References
- Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- Baselga J, Tripathy D, Mendelsohn J, Baughman S, Benz C, Dantis L, et al. Phase II study of weekly intravenous recombinant humanized anti-p185HER2 monoclonal antibody in patients with HER2/neu-overexpressing metastatic breast cancer. J Clin Oncol. 1996;14:737–744. doi: 10.1200/JCO.1996.14.3.737. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers' and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Guilhot Fo,, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003;63:5198–5202. [PubMed] [Google Scholar]
- Collisson EA, De A, Suzuki H, Gambhir SS, Kolodney MS. Treatment of metastatic melanoma with an orally available inhibitor of the Ras-Raf-MAPK cascade. Cancer Res. 2003;63:5669–5673. [PubMed] [Google Scholar]
- Sumimoto H, Miyagishi M, Miyoshi H, Yamagata S, Shimizu A, Taira K, et al. Inhibition of growth and invasive ability of melanoma by inactivation of mutated BRAF with lentivirus-mediated RNA interference. Oncogene. 2004;23:6031–6039. doi: 10.1038/sj.onc.1207812. [DOI] [PubMed] [Google Scholar]
- Karasarides M, Chiloeches A, Hayward R, Niculescu-Duvaz D, Scanlon I, Friedlos F, et al. B-RAF is a therapeutic target in melanoma. Oncogene. 2004;23:6292–6298. doi: 10.1038/sj.onc.1207785. [DOI] [PubMed] [Google Scholar]
- Bhatt KV, Spofford LS, Aram G, McMullen M, Pumiglia K, Aplin AE. Adhesion control of cyclin D1 and p27Kip1 levels is deregulated in melanoma cells through BRAF-MEK-ERK signaling. Oncogene. 2005;12:3459–3471. doi: 10.1038/sj.onc.1208544. [DOI] [PubMed] [Google Scholar]
- Boisvert-Adamo K, Aplin AE. B-RAF and PI-3 kinase signaling protect melanoma cells from anoikis. Oncogene. 2006;25:4848–4856. doi: 10.1038/sj.onc.1209493. [DOI] [PubMed] [Google Scholar]
- Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant V599E B-Raf regulates growth and vascular development of malignant melanoma tumors. Cancer Res. 2005;65:2412–2421. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Eisen T, Ahmad T, Flaherty KT, Gore M, Kaye S, Marais R, et al. Sorafenib in advanced melanoma: a Phase II randomised discontinuation trial analysis. Br J Cancer. 2006;95:581–586. doi: 10.1038/sj.bjc.6603291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with Vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley KSM, Sondak VK. Melanoma – an unlikely poster child for personalized cancer therapy. N Engl J Med. 2010;363:876–878. doi: 10.1056/NEJMe1005370. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlyn M. Human melanoma: development and progression. Cancer Metastasis Rev. 1990;9:101–112. doi: 10.1007/BF00046337. [DOI] [PubMed] [Google Scholar]
- Sondergaard JN, Nazarian R, Wang Q, Guo D, Hsueh T, Mok S, et al. Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific Raf inhibitor PLX4032. J Transl Med. 2010;8:39. doi: 10.1186/1479-5876-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. V600EBRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci USA. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummer T, Naegele H, Reth M, Misawa Y. Identification of novel ERK-mediated feedback phosphorylation sites at the C-terminus of B-Raf. Oncogene. 2003;22:8823–8834. doi: 10.1038/sj.onc.1207185. [DOI] [PubMed] [Google Scholar]
- Ritt DA, Monson DM, Specht SI, Morrison DK. Impact of feedback phosphorylation and Raf heterodimerization on normal and mutant B-Raf signaling. Mol Cell Biol. 2010;30:806–819. doi: 10.1128/MCB.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraiso KHT, Fedorenko IV, Cantini LP, Munko AC, Hall M, Sondak VK, et al. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. Br J Cancer. 2010;102:1724–1730. doi: 10.1038/sj.bjc.6605714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci USA. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Yu Q, Sicinska E, Das M, Bronson RT, Sicinski P. Deletion of the p27Kip1 gene restores normal development in cyclin D1-deficient mice. Proc Natl Acad Sci USA. 2001;98:194–199. doi: 10.1073/pnas.011522998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich KP, Herter S, Tien J, Wong L, Berry L, Chan J, et al. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer Res. 2009;69:3042–3051. doi: 10.1158/0008-5472.CAN-08-3563. [DOI] [PubMed] [Google Scholar]
- Shao Y, Aplin A. Akt3-mediated resistance to apoptosis in B-RAF targeted melanoma cells. Cancer Res. 2010;70:6670–6681. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBrocklin MW, Verhaegen M, Soengas MS, Holmen SL. Mitogen-activated protein kinase inhibition induces translocation of Bmf to promote apoptosis in melanoma. Cancer Res. 2009;69:1985–1994. doi: 10.1158/0008-5472.CAN-08-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ.Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim J Biol Chem 200327818811–18816.Order. [DOI] [PubMed] [Google Scholar]
- Richter-Larrea JA, Robles EF, Fresquet V, Beltran E, Rullan AJ, Agirre X, et al. Reversion of epigenetically mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood. 2010;116:2531–2542. doi: 10.1182/blood-2010-02-268003. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Adachi M, Zhao X, Kawamura R, Imai K. Histone deacetylase inhibitors FK228, N-(2-aminophenyl)-4-[N-(pyridin-3-yl-methoxycarbonyl)amino- methyl]benzamide and m-carboxycinnamic acid bis-hydroxamide augment radiation-induced cell death in gastrointestinal adenocarcinoma cells. Int J Cancer. 2004;110:301–308. doi: 10.1002/ijc.20117. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Adachi M, Kawamura R, Imai K. Bmf is a possible mediator in histone deacetylase inhibitors FK228 and CBHA-induced apoptosis. Cell Death Differ. 2006;13:129–140. doi: 10.1038/sj.cdd.4401686. [DOI] [PubMed] [Google Scholar]
- Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa H, Torii S, Yasunaga T, Sprouty1 NishidaE. and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- Boisvert-Adamo K, Aplin AE. Mutant B-RAF mediates resistance to anoikis via Bad and Bim. Oncogene. 2008;27:3301–3312. doi: 10.1038/sj.onc.1211003. [DOI] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Wang J, Hoshino T, Redner RL, Kajigaya S, ETO LiuJM. fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Sternsdorf T, Tini M, Evans RM. Transcriptional regulation in acute promyelocytic leukemia. Oncogene. 2001;20:7204–7215. doi: 10.1038/sj.onc.1204853. [DOI] [PubMed] [Google Scholar]
- Sharma S, Lee D, Li B, Quinlan M, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xargay-Torrent S, Lopez-Guerra M, Saborit-Villarroya I, Rosich L, Campo E, Roue G, et al. Vorinostat-induced apoptosis in mantle cell lymphoma is mediated by acetylation of proapoptotic BH3-only gene promoters. Clin Cancer Res. 2011;17:3956–3968. doi: 10.1158/1078-0432.CCR-10-3412. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Mandai M, Oura T, Matsumura N, Hamanishi J, Baba T, et al. Identification of an ovarian clear cell carcinoma gene signature that reflects inherent disease biology and the carcinogenic processes. Oncogene. 2010;29:1741–1752. doi: 10.1038/onc.2009.470. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.