Background: How DNA 5-cytosine methyltransferases (DCMTases) select their substrate nucleobase for extrusion from DNA duplex is poorly understood.
Results: The crystal structure of a pre-extrusion M.HaeIII DCMTase-substrate DNA complex is reported here.
Conclusion: M.HaeIII selects its substrate cytosine for extrusion by selectively interfering with its stacking and hydrogen bonding interactions within the DNA duplex.
Significance: This is the first structural elucidation of the target cytosine selection by a DCMTase.
Keywords: DNA-binding Protein, DNA Methylation, DNA Methyltransferase, Enzyme Structure, Nucleic Acid Structure, Protein-Nucleic Acid Interaction, X-ray Crystallography, 5-Methylcytosine, DNA Nucleobase Extrusion, Protein-DNA Interaction
Abstract
Epigenetic methylation of cytosine residues in DNA is an essential element of genome maintenance and function in organisms ranging from bacteria to humans. DNA 5-cytosine methyltransferase enzymes (DCMTases) catalyze cytosine methylation via reaction intermediates in which the DNA is drastically remodeled, with the target cytosine residue extruded from the DNA helix and plunged into the active site pocket of the enzyme. We have determined a crystal structure of M.HaeIII DCMTase in complex with its DNA substrate at a previously unobserved state, prior to extrusion of the target cytosine and frameshifting of the DNA recognition sequence. The structure reveals that M.HaeIII selects the target cytosine and destabilizes its base-pairing through a precise, focused, and coordinated assault on the duplex DNA, which isolates the target cytosine from its nearest neighbors and thereby facilitates its extrusion from DNA.
Introduction
In bacteria, cytosine methylation confers resistance of the host genome to endonucleases that destroy invading foreign DNA, whereas in eukaryotes, this covalent modification underpins the system for epigenetic regulation of gene transcription (for review, see Ref. 1). The methyl groups are introduced via DNA 5-cytosine methyltransferase enzymes (DCMTases),2 which catalyze methyl transfer from the donor co-factor S-adenosylmethionine to C5 of the target cytosine (for review, see Ref. 2). High resolution structural studies have revealed that DCMTases extrude the substrate cytosine residue from the DNA helix and introduce it into an active site pocket of the enzyme (3–5). Whereas the catalytic mechanism of DCMTases has been elucidated in some detail (6–10), the molecular mechanism by which these enzymes select a particular target cytosine and initiate its extrusion is largely unknown. Despite the large body of available structural data on various states of DCMTase/DNA recognition, none of these addresses the question of how the enzyme selects the target cytosine and promotes its extrusion from DNA helix. The targeting mechanism is particularly intriguing in the case of M.HaeIII, a bacterial DCMTase that not only extrudes the substrate cytosine, but also induces frameshifted base pairing and the formation of a large gap in the duplex DNA recognition site (4). Here we present the crystal structure of M.HaeIII in complex with a DNA substrate at a previously unobserved state, prior to extrusion of the target cytosine.
M.HaeIII recognizes the sequence 5′-GGCC in DNA and transfers a methyl group to the internal C residue; each strand is methylated sequentially, with the hemimethylated sequence being a preferred substrate. Substitution of the target cytosine by 5-fluorocytosine enables the trapping of a covalent enzyme-substrate complex (11, 12) in which the conserved catalytic nucleophile of the enzyme, Cys-71 in the case of M.HaeIII, is covalently adducted to C6 of the target cytosine, and a methyl group has been transferred from S-adenosylmethionine to C5. Immediately apparent in the structures of the first two such trapped covalent complexes, M.HhaI (3) and M.HaeIII (4), was the extrusion of the entire substrate cytosine nucleotide from the DNA helix. One aspect of the M.HhaI and M.HaeIII structures was strikingly different, however. Whereas the DNA duplex was nearly undisturbed in the M.HhaI structure save for the extruded target cytosine, the DNA conformation in the M.HaeIII structure was far from canonical, with the 3′-cytosine engaged in frameshifted base pairing (Fig. 1F, green) and a gaping hole in the duplex almost as wide as an entire base pair. In neither of these original structures was it clear how the DCMTase transitioned from the sequence-specific recognition mode to extrusion mode and catalysis of methyl transfer; the nature of this transition is particularly mysterious in the case of M.HaeIII, because of the drastic remodeling of the DNA recognition sequence. Despite the availability of dozens of structures of DCMTases bound to DNA, not a single structure has been determined at the state of sequence-specific recognition but preceding extrusion of the target cytosine. Therefore, the molecular origins of target recognition, selection of the target cytosine and management of its extrusion from DNA have gone unelucidated. Structures of a portion of mammalian DNMT1 bound to an intact target duplex have recently been reported (13), but interestingly, these studies captured the DCMTase bound to DNA in an autoinhibitory state completely distinct from the protein-DNA complex that precedes target extrusion. The aim of the present study was to capture and molecularly characterize the elusive pre-extrusion state.
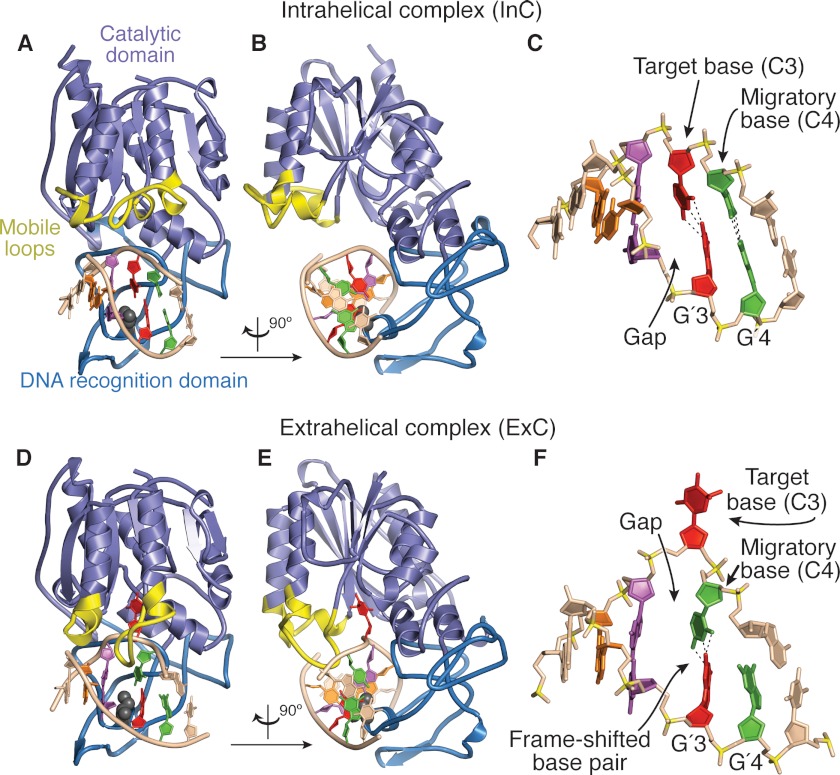
FIGURE 1.
Comparison of the structures of the pre-extrusion complex InC and the trapped catalytic complex ExC. A, overall structure of InC (this work) with various structural elements of the protein highlighted as follows: lilac, catalytic domain; sky blue, sequence-specific DNA recognition domain; yellow, mobile elements that undergo major positional shifts in InC versus ExC. The DNA is shown in schematic representation, with the four bp of the 5′-GGCC site individually color-coded (orange-to-green, 5′-to-3′ on the target strand at the top). The base pairs flanking the M.HaeIII recognition site are colored the same as the rest of the DNA. B, same as A, but rotated by 90° about the vertical axis. C, framework representation of the DNA in the InC structure, with key features denoted; color-coding as in A and B, but with backbone phosphorus atoms in mustard. D and E, two views of ExC (4), formatted as for InC in A and B. F, framework representation of the DNA in ExC, formatted as for InC in C.
EXPERIMENTAL PROCEDURES
Overexpression and Purification of M.HaeIII(C71S)
The C71S mutation was introduced into the wild-type M.HaeIII expression plasmid (11) by QuikChange protocol (14) using 5′-GTGTGATGGAATTATTGGGGGACCGCCTAGTCAATCTTGGAGTGAGGGG-3′ and its reverse complement DNA primers. M.HaeIII(C71S) was overexpressed as described previously for the wild-type M.HaeIII (11), except the cells were grown at 20 °C for ≈20 h after induction. The cells were pelleted by centrifugation for 20–30 min at 3000 × g at 4 °C and resuspended in 25 ml of cold (4 °C) lysis buffer (20 mm NaH2PO4/Na2HPO4, pH 7.25, 300 mm NaCl, 0.5 mm EDTA), frozen in liquid nitrogen, and stored at −80 °C. All further purification steps were carried out at 4 °C. Frozen cell suspension was thawed, and β-mercaptoethanol (βME) and phenylmethylsulfonyl fluoride were added to 10 mm and 1 mm final concentrations, respectively. Lysis was done by sonication. The lysate was cleared by centrifugation at 13,000 × g for 25 min, filtered through a 0.45-μm filter, and loaded onto a heparin column (Hi-Trap Heparin HP, 5 ml; GE Healthcare) pre-equilibrated with 350 mm NaCl in MH buffer (20 mm NaH2PO4/Na2HPO4, pH 7.25, 0.5 mm EDTA, 10 mm βME). The column was then washed with 25 ml of 350 mm NaCl in MH buffer and eluted with a 60-ml linear gradient from 350 to 700 mm NaCl in MH buffer. The fractions containing high concentrations of M.HaeIII were pooled and exchanged to HAP-A buffer (10 mm KH2PO4/K2HPO4, pH 7.25, 100 mm KCl, 10 mm βME) using size exclusion chromatography (HiPrep 26/10 Desalting; GE Healthcare). The fractions were combined and loaded onto a ≈1-ml hydroxyapatite column (Macro-Prep Ceramic Hydroxyapatite, type I, 40 μm; Bio-Rad), pre-equilibrated with HAP-A buffer. The column was then washed with 6 ml of HAP-A buffer and eluted with a 35-ml gradient from 100% HAP-A to 63% HAP-A plus 37% HAP-B buffer (500 mm KH2PO4/K2HPO4, pH 7.25, 100 mm KCl, 10 mm βME). The fractions containing the purest M.HaeIII were pooled, concentrated by ultrafiltration (Amicon Ultra-15, 10000 NMWL; Millipore), and further purified by gel filtration (HiLoad 16/60 Superdex 75; GE Healthcare) in 10 mm Tris-HCl, pH 7.25, 100 mm NaCl, 2% glycerol, 1 mm dithiothreitol (Crystallization Buffer).
DNA Synthesis and Purification
The self-complementary DNA oligonucleotide 5′-TpGpGpCpCpA-3′ was synthesized with an automated oligonucleotide synthesizer (MerMade 12; BioAutomation Corporation) using standard reagents, purified by urea-PAGE, desalted with C18+ reverse-phase chromatography (C18+ Sep-Pak cartridge; Waters), and annealed in Crystallization Buffer without dithiothreitol by cooling from ≈70 °C to 4 °C.
Crystallography
Protein-DNA complexes were prepared in 1:2 protein to double-stranded DNA molar ratio at 160 μm protein concentration in Crystallization Buffer with 10 mm ATP as a crystallization additive. The complex was crystallized at 4 °C by the hanging drop vapor diffusion method against the well solution consisting of 100 mm ammonium sulfate, 26–27% (v/v) pentaerythritol ethoxylate (15/4 EO/OH) (further referred to as PE, for brevity), 50 mm Bis-Tris, pH 6.5, in water. The drops were 1:1 mixture (1 + 1 to 2 + 2 μl) of the protein-DNA and well solutions. Crystallization was highly sensitive to the state of PE as the crystals grew only when aged PE was used. Specifically, we used PE from Hampton Research, HR2–745, lot 274514. For cryoprotection the crystals were quickly dipped in 39.4% (v/v) PE, 10% glycerol, 125 mm ammonium sulfate, 50 mm Bis-Tris, pH 6.5, and flash frozen in liquid nitrogen. X-ray diffraction data were collected on beamline 24-ID-E at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL). The data used for the initial model refinement were reduced and scaled with HKL2000 (15). The data used in the final stages of model refinement were reduced and scaled using XDS (16). The structure was solved by molecular replacement with PHASER (17) using a M.HaeIII model determined from a similar crystal form, which in turn was solved by molecular replacement using a M.HaeIII model from the published structure (4). The model was completed by iterative cycles of refinement in CNS (18), PHENIX (19), and manual rebuilding in Coot (20) and O (21). Noncrystallographic symmetry restrains were applied to parts of the model where justified by electron density. Data collection and refinement statistics are provided in supplemental Table S1. Ramachandran plot analysis was performed using PROCHECK (22). The coordinates have been deposited under Protein Data Bank accession code 3UBT. The DNA conformational parameters were calculated, and the base-stacking diagrams (23) in Fig. 4, A–C, were generated using 3DNA (24). Root mean square deviations between various models were calculated in Coot. The structural figures and supplemental Movie S1 were rendered using PyMOL (Schrödinger). Molecular morphing simulation was performed as described (25, 26).
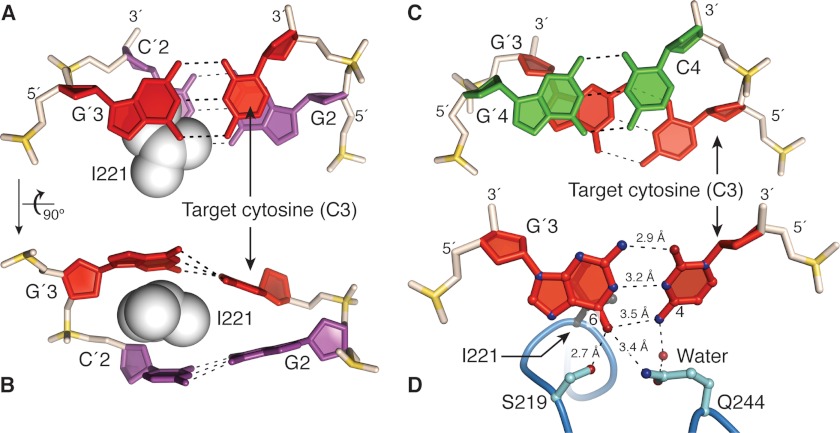
FIGURE 4.
M.HaeIII-induced destabilization of the target cytosine, C3. A and B, two different close-up views of the target base pair C3/G′3 and its unstacked neighbor G2/C′2. The side chain of the intercalating residue Ile-221 is shown in space-filling representation. Note that the target cytosine C3 is partially destacked from G2, whereas the complementary nucleobases G′3 and C′2 are completely destacked through Ile-221 intercalation. Note also the distorted base pairing of the target base pair. C, close-up view of stacking between base pairs 3 and 4. Note that the two migratory cytosines, C3 and C4 are completely destacked by under-rotation of the DNA helix at that base pair step. D, direct contacts in InC to the target base pair. The interaction between G′3 and Ser-219 in InC is maintained in ExC. Gln-244 is well positioned to compete for the hydrogen bonding of the target cytosine C3 with the complementary guanine G′3. The water-mediated interaction of Gln-244 with C3 in InC, which has poor geometry, is replaced in ExC by a direct and geometrically favorable contact to the frameshifted nucleobase, C4 (data not shown). The orientation in D is slightly different from that on A for clarity.
RESULTS
Overall Structure of M.HaeIII-DNA Complex with Intrahelical Target Cytosine
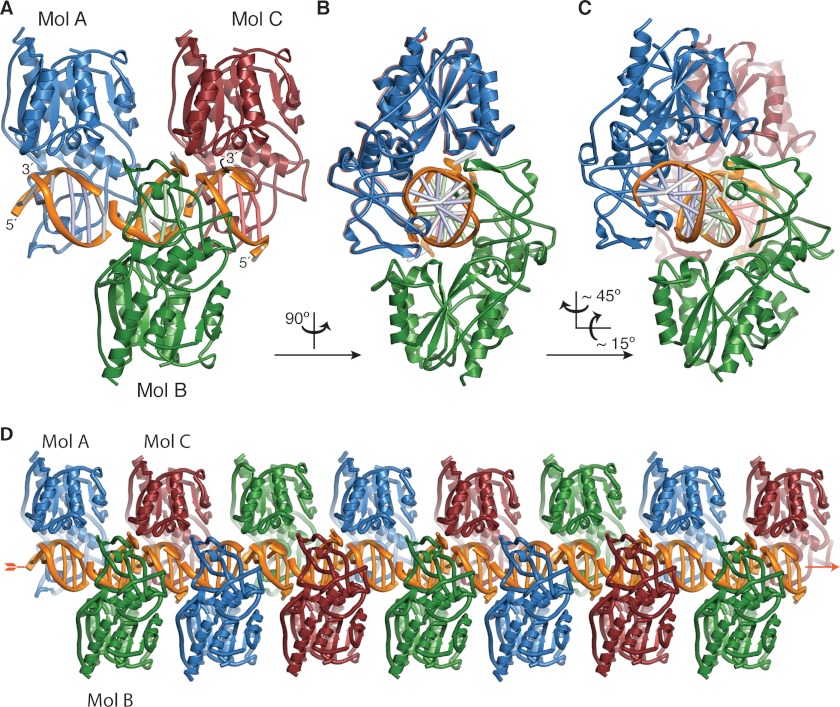
Mutation of the catalytic nucleophile of M.HaeIII, Cys-71, to Ser is known to afford a variant (C71S M.HaeIII) that lacks methyltransferase activity but retains sequence-specific DNA recognition that is independent of co-factor binding (7). To characterize the DNA recognition mode of C71S M.HaeIII, we crystallized the protein in complex with a palindromic 6-mer DNA duplex (5′-TGGCCA-3′, where 5′-GGCC-3′ is the M.HaeIII recognition sequence) and solved the structure to 2.5 Å resolution (see “Experimental Procedures”). The asymmetric unit contains three protein-DNA complexes, designated as Mol A, B, and C in Fig. 2. The DNA duplexes of Mol A–C are co-axially stacked in the asymmetric unit, and the duplexes of adjacent asymmetric units also stack, thus forming a pseudocontinuous duplex through the crystal (Fig. 2D and supplemental Fig. S1). The overall structures of the three complexes are very similar (supplemental Fig. S2, A–C), with Watson-Crick base pairing maintained throughout the M.HaeIII recognition site, a fully intrahelical target cytosine, and an absence of sequence frameshifting. The three sequence-specific M.HaeIII DNA recognition sites are nearly identical (all-atom root mean square deviation ≈ 0.6Å, supplemental Fig. S2, D–G), as are the three protein monomers in the asymmetric unit (Cα root mean square deviation ≤ 0.3 Å, supplemental Fig. S3). The only significant structural differences among Mols A–C are localized to the base pairs outside the M.HaeIII recognition site, both of which are intact in Mol A, but one of which is disrupted in Mols B and C (A5 and T′5, supplemental Fig. S2, E and G). Here we present in detail the analysis of Mol A only; the details of protein-DNA interactions are essentially identical in all three complexes. Below we refer to the structure of the present M.HaeIII-DNA complex, with its intrahelical target cytosine, as InC, whereas we denote the previously determined structure of a catalytically trapped species having an extrahelical target cytosine as ExC (4).
FIGURE 2.
Association of InC protomers in the asymmetric unit and in the crystal lattice. A, the asymmetric unit of the InC crystals contains three M.HaeIII(C71S)-DNA complexes having very closely related three-dimensional structures, designated as Mol A–C. The DNA duplexes of Mol A–C are co-axially stacked in the asymmetric unit, and these trimers stack co-axially with their neighbors in the crystal lattice D. B and C, same as A, but rotated as indicated. In Mol A, all 6 bp are intact, whereas in Mols B and C, the rightmost base pair in each duplex is disrupted. In each Mol, the base pairs of the M.HaeIII recognition site are colored the same as the protomer to which they are bound, and the flanking nucleotides are colored gray. The adenine nucleobase of the disrupted flanking base pair in Mol C (top strand, 3′-end) was not modeled due to its poor electron density (see also supplemental Fig. S2).
Comparison of the Structures of M.HaeIII-DNA Complexes Having an Intrahelical versus Extrahelical Target Cytosine
Though the DNA conformation in InC has drastic differences from that of ExC, the overall structures of the two complexes are quite similar (Fig. 1). At the protein level, the most obvious difference in InC compared with ExC is a retraction away from DNA of loops N-terminal to helices C and D of the catalytic domain (yellow in Fig. 1). The portion of the catalytic domain bearing these loops is also slightly retracted from the DNA in InC versus ExC (supplemental Movie S1). Together, these structural alterations make the conformation of InC considerably more open at the interface between DNA and the catalytic domain than in ExC, and it is noteworthy that this opening results in partial disassembly of the active site in InC. These structural findings are thus consistent with the results of biochemical and biophysical studies on the closely related DCMTase M.HhaI, which showed that DNA binding precedes extrusion of the target nucleobase and initiation of catalysis (8, 10, 27–29). Upon formation of the closed, catalytically active state, the mobile loops of the catalytic domain acquire multiple contacts to both the duplex DNA backbone (Ser-79, Arg-87, Arg-81, Lys-112, and Gln-117) and the extrahelical target cytosine (Gly-68 and Ser-75); these contacts are likely to stabilize the extrahelical, frameshifted conformation of the complex (compare Fig. 3, C and D).
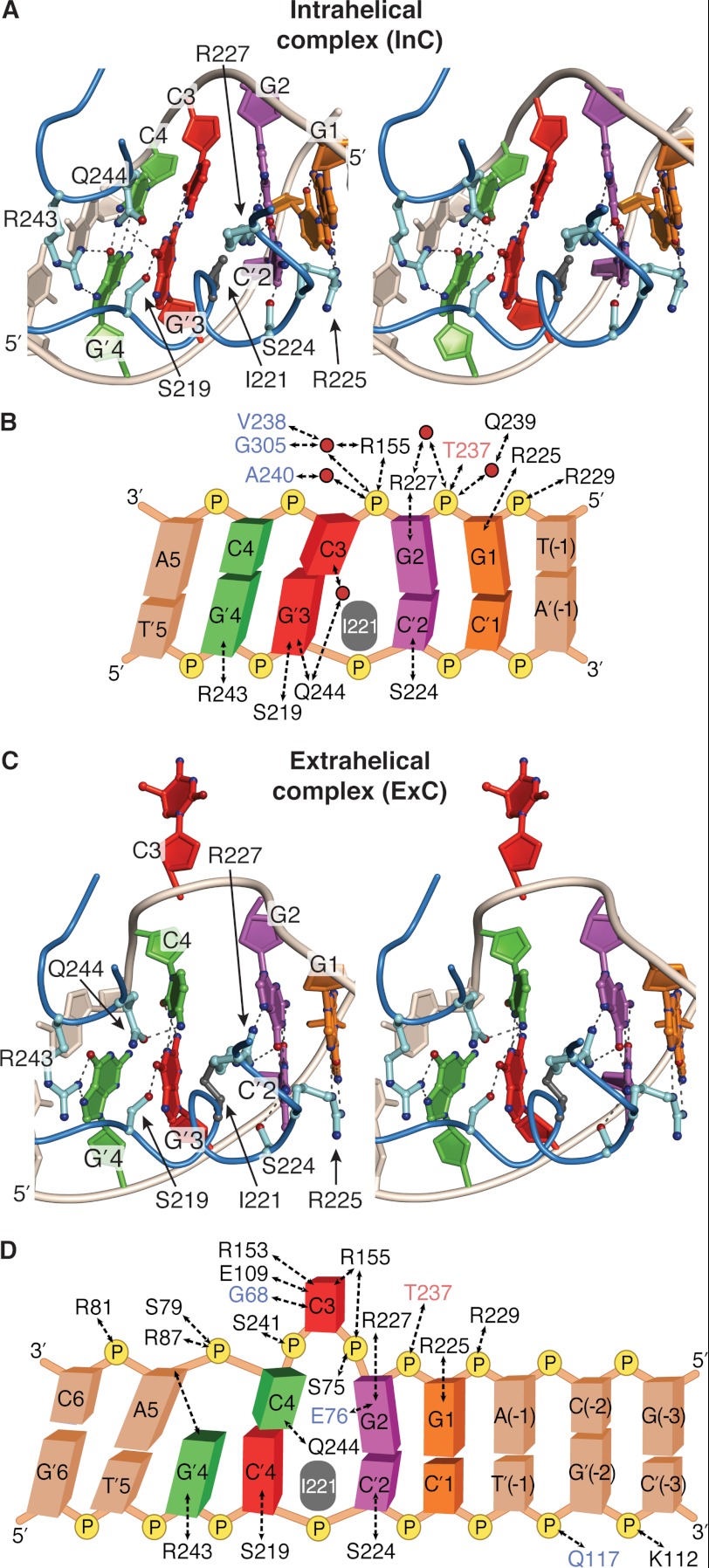
FIGURE 3.
DNA sequence-specific contacts in InC and ExC. A and C, stereoview of the protein-DNA interface in InC (A) and ExC (C), focusing on sequence-specific contacts between the major groove surface of the recognition site in DNA and amino acid side chains of M.HaeIII. The base pairs are individually color-coded as in Fig. 1, although the 5′-to-3′ orientation of the duplex is opposite in Figs. 1 and 3. Amino acid side chains of residues that make sequence-specific hydrogen bonding interactions are shown, with dashed lines denoting hydrogen bonds. B and D, schematic representation of contacts in the protein-DNA interface of InC (B) and ExC (D). The residues named in black, blue, and red make the indicated hydrogen bonds solely via their side chain, main chain, or both kinds of atoms. Only those interfacial water molecules that are present in Mols A–C and that are made via a solitary water molecule are shown. The intercalating side chain of Ile-221 is shown in gray in all four panels.
Substrate DNA Recognition by M.HaeIII
A central mystery of M.HaeIII function regards how the protein could specifically recognize such drastically different DNA conformations as the B form duplex it first encounters and the drastically remodeled conformation observed in the trapped catalytic complex ExC. The M.HaeIII recognition site is rendered particularly stable by virtue of its all-G/C content, and yet in ExC, M.HaeIII manages to bring about multiple powerfully destabilizing alterations (Fig. 3D): thrusting of the Ile-221 side chain into the helical stack and complete destacking at base pairs G2/C′2 and C4/G′3, extrusion of the target cytosine C3, and frameshifting of the sequence at G′3/C4, with concomitant abandonment of pairing for G′4. The structure of InC provides clear insight into the factors that govern the formation of this highly distorted state and that target the substrate cytosine for extrusion.
Details of the protein-DNA interface in the InC structure are fully consistent with the notion that InC represents a sequence-specific recognition complex formed between M.HaeIII and a fully base-paired DNA duplex bearing distortions that poise the complex for transition into the mode of target extrusion and catalysis of methyl transfer. Remarkably, InC possesses all of the sequence-specific contacts seen in ExC, with the sole exception of those made to the target C (compare Fig. 3, A and C; see supplemental Fig. S4). The only noteworthy differences in sequence-specific contacts between InC and ExC consist of subtle adjustments in hydrogen bonding configuration between two conserved sets of interacting partners, namely Arg-243 and G′4; and the Gln-244 and C4. The sequence-specific contacts thus function to anchor the complex on DNA, providing a structural platform that isolates and focuses the leverage of the protein-DNA interface on the two migratory nucleobases, C3 and C4, both of which are free of direct contacts to M.HaeIII in InC but are extensively contacted in ExC.
Selection of the Target Cytosine and Frameshifting
Although no base pairs are disrupted in InC, the duplex does exhibit significant deviations from canonical B form structure that appear specifically tailored to facilitate the extrusion of C3 and migration of C4. Specifically, the InC structure possesses a large duplex gap at the site of Ile-221 insertion, between base-pairs 2 and 3 (Fig. 3A, see also Figs. 1C and 4, A and B); in ExC, this gap resides between base pair 2 and the frameshifted C4/G′3 pair (Figs. 3C and 1F). The gap is approximately the same width in the two complexes on the nontarget strand, where Ile-221 physically blocks gap closure, but is narrower on the target strand in InC than ExC. Although x-ray structures do not per se provide energetic information, a number of features in the InC structure indicate that the target G′3/C3 base pair, and in particular the extrusion target C3, is substantially destabilized relative to canonical B form DNA. First, C3 is virtually devoid of stacking interaction with either of its nearest neighbors, as it has been pried away from G2 by the insertion of Ile-221 (Fig. 4, A and B) and completely destacked from C4 by induced helical underwinding (Fig. 4C). Second, the geometry of the target base pair deviates significantly from the norm, being separated by an unusually large distance (stretch of 0.1 Å) excessively propeller-twisted (22°), and severely buckled (−21°) (Fig. 4, B and D). Third, the hydrogen bonding partner of C3, namely G′3, is engaged in a hydrogen bonding interaction between its O6 atom and the side chain amide of Gln-244 that, because of its interaction geometry, places it in direct competition with Watson-Crick hydrogen bonding to C3 (Fig. 4D). A similar competing interaction with the target base pair of the DNA glycosylase MutM has been shown to facilitate base pair rupture and target extrusion (30). Mutation of the residue corresponding to Gln-244 (to Ala) in the M.HaeIII homolog M.NgoPII has been shown to reduce its methyl transfer activity substantially (31).
DISCUSSION
The structure of the pre-extrusion complex InC clearly suggests that M.HaeIII locates its specific recognition sequence, selects its target cytosine among the four cytosines within that sequence, and promotes extrusion of the target cytosine from DNA through active engagement in extensive protein-DNA interactions. First, at the pre-extrusion state represented by InC, M.HaeIII directly contacts the major groove surface of every nonmigratory nucleobase in the sequence except C′1, stabilizing their intrahelical conformation. M.HaeIII thrusts Ile-221 into the helical stack, distorting the target base pair and weakening the stacking of the target cytosine C3 with G2. M.HaeIII establishes a network of backbone contacts that induce underwinding of the DNA duplex at the site of the target base pair, destacking C3 from its other neighbor, C4. The protein positions Gln-244 in direct competition with Watson-Crick hydrogen bonding of the target base pair. The protein is devoid of direct interactions with the migratory nucleobases C3 and C4, which if present could stabilize their intrahelical conformation; these nucleobases are both contacted post-migration. Likewise, M.HaeIII makes no contacts to the two backbone phosphates that undergo large positional shifts, C3pC4 and C4pA5, although these are both contacted post-extrusion (Fig. 3, C and D). The phosphate on the 5′-side of the target cytosine, G2pC3, also undergoes a significant positional shift upon extrusion, but the hydrogen bonding interaction is improved in terms of distance, geometry and number of interacting atoms upon extrusion. The active mechanism of target engagement observed here for M.HaeIII represents a completely different mode from that recently reported for the human DCMTase DNMT1, where passive discrimination against an unmethylated recognition sequence was observed (13). In this structure, the elements of protein structure responsible for specific recognition of the target sequence and selection of the target cytosine do not interact directly with the DNA duplex because they are occluded by an autoinhibitory domain in the protein. The present structure leaves open the question of whether frameshifting of the M.HaeIII recognition site occurs in synchrony with base extrusion or follows it. We note in this regard that simulated interpolation of the InC to ExC transition reveals a remarkably facile pathway for simultaneous migration of both cytosines through an intermediate in which the frameshifting base is transiently base paired to both G′4 to G′3 during the hand-off from one to the other (supplemental Movie S1).
Supplementary Material
Acknowledgments
We thank Brian Bowman, Michael Eck, David Jeruzalmi, Danaya Pakotiprapha, Yan Qi, Daqi Tu, and Wilhelm Weihofen for help with crystallography; Prarthna Desai for help with protein purification and crystallization; and the entire staff at 23-ID and 24-ID beamlines of the Advanced Photon Source for help with data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant GM044853 (to G. L. V.). This work is based upon research conducted at the Advanced Photon Source on the NE-CAT and GM/CA-CAT beamlines. NE-CAT is supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. GM/CA-CAT was supported, in whole or in part, by National Institutes of Health Grants Y1-CO-1020 from the NCI and Y1-GM-1104 from the NIGMS. Use of the Advanced Photon Source was supported by the United States Department of Energy (U.S. DOE) under Contract DE-AC02-06CH11357 and by the U.S. DOE, Basic Energy Sciences, Office of Science, under Contract W-31-109-ENG-38.

This article contains supplemental Table S1, Figs. S1–S4, and Movie S1.
The atomic coordinates and structure factors (code 3UBT) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- DCMTase
- DNA 5-cytosine methyltransferase
- Bis-Tris
- bis(2-hydroxyethyl)aminotris(hydroxymethyl)methane
- βME
- β-mercaptoethanol
- ExC
- extrahelical target cytosine
- InC
- intrahelical target cytosine
- PE
- pentaerythritol ethoxylate.
REFERENCES
- 1. Goll M. G., Bestor T. H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 [DOI] [PubMed] [Google Scholar]
- 2. Verdine G. L., Norman D. P. (2003) Covalent trapping of protein-DNA complexes. Annu. Rev. Biochem. 72, 337–366 [DOI] [PubMed] [Google Scholar]
- 3. Klimasauskas S., Kumar S., Roberts R. J., Cheng X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell 76, 357–369 [DOI] [PubMed] [Google Scholar]
- 4. Reinisch K. M., Chen L., Verdine G. L., Lipscomb W. N. (1995) The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell 82, 143–153 [DOI] [PubMed] [Google Scholar]
- 5. Song J., Teplova M., Ishibe-Murakami S., Patel D. J. (2012) Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 335, 709–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zangi R., Arrieta A., Cossío F. P. (2010) Mechanism of DNA methylation: the double role of DNA as a substrate and as a cofactor. J. Mol. Biol. 400, 632–644 [DOI] [PubMed] [Google Scholar]
- 7. Chen L., MacMillan A. M., Verdine G. L. (1993) Mutational separation of DNA binding from catalysis in a DNA cytosine methyltransferase. J. Am. Chem. Soc. 115, 5318–5319 [Google Scholar]
- 8. Gerasimaite R., Merkiene E., Klimaŝauskas S. (2011) Direct observation of cytosine flipping and covalent catalysis in a DNA methyltransferase. Nucleic Acids Res. 39, 3771–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X., Bruice T. C. (2006) The mechanism of M.HhaI DNA C5 cytosine methyltransferase enzyme: a quantum mechanics/molecular mechanics approach. Proc. Natl. Acad. Sci. U.S.A. 103, 6148–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu J. C., Santi D. V. (1987) Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 262, 4778–4786 [PubMed] [Google Scholar]
- 11. Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. (1991) Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry 30, 11018–11025 [DOI] [PubMed] [Google Scholar]
- 12. Osterman D. G., DePillis G. D., Wu J. C., Matsuda A., Santi D. V. (1988) 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry 27, 5204–5210 [DOI] [PubMed] [Google Scholar]
- 13. Song J., Rechkoblit O., Bestor T. H., Patel D. J. (2011) Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331, 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Papworth C., Bauer J., Braman J., Wright D. (1996) Site-directed mutagenesis in one day with >80% efficiency. Strategies 9, 4 [Google Scholar]
- 15. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 16. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 19. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 20. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A Found. Crystallogr. 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 22. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 23. Lu X. J., Olson W. K. (2003) 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu X. J., Olson W. K. (2008) 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 3, 1213–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Echols N., Milburn D., Gerstein M. (2003) MolMovDB: analysis and visualization of conformational change and structural flexibility. Nucleic Acids Res. 31, 478–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krebs W. G., Gerstein M. (2000) The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 28, 1665–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klimasauskas S., Szyperski T., Serva S., Wüthrich K. (1998) Dynamic modes of the flipped-out cytosine during HhaI methyltransferase-DNA interactions in solution. EMBO J. 17, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Estabrook R. A., Nguyen T. T., Fera N., Reich N. O. (2009) Coupling sequence-specific recognition to DNA modification. J. Biol. Chem. 284, 22690–22696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matje D. M., Coughlin D. F., Connolly B. A., Dahlquist F. W., Reich N. O. (2011) Determinants of precatalytic conformational transitions in the DNA cytosine methyltransferase M.HhaI. Biochemistry 50, 1465–1473 [DOI] [PubMed] [Google Scholar]
- 30. Qi Y., Spong M. C., Nam K., Banerjee A., Jiralerspong S., Karplus M., Verdine G. L. (2009) Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature 462, 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Radlinska M., Kondrzycka-Dada A., Piekarowicz A., Bujnicki J. M. (2005) Identification of amino acids important for target recognition by the DNA:m5C methyltransferase M.NgoPII by alanine-scanning mutagenesis of residues at the protein-DNA interface. Proteins 58, 263–270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.