Background: HDAC1-containing NuRD complex is required for GATA-1-mediated repression and activation.
Results: GATA-1 associated with acetylated HDAC1-containing NuRD complex, which has no deacetylase activity, for gene activation.
Conclusion: Acetylated HDAC1 converts NuRD complex from a repressor to an activator during GATA-1-directed erythroid differentiation program.
Significance: HDAC1 acetylation may function as a master regulator for the activity of HDAC1 containing complexes.
Keywords: Chromatin Remodeling, Coregulator Transcription, Differentiation, Histone Deacetylase, Transcription Regulation
Abstract
Histone deacetylases (HDACs) play important roles in regulating cell proliferation and differentiation. The HDAC1-containing NuRD complex is generally considered as a corepressor complex and is required for GATA-1-mediated repression. However, recent studies also show that the NuRD complex is involved in GATA-1-mediated gene activation. We tested whether the GATA-1-associated NuRD complex loses its deacetylase activity and commits the GATA-1 complex to become an activator during erythropoiesis. We found that GATA-1-associated deacetylase activity gradually decreased upon induction of erythroid differentiation. GATA-1-associated HDAC1 is increasingly acetylated after differentiation. It has been demonstrated earlier that acetylated HDAC1 has no deacetylase activity. Indeed, overexpression of an HDAC1 mutant, which mimics acetylated HDAC1, promotes GATA-1-mediated transcription and erythroid differentiation. Furthermore, during erythroid differentiation, acetylated HDAC1 recruitment is increased at GATA-1-activated genes, whereas it is significantly decreased at GATA-1-repressed genes. Interestingly, deacetylase activity is not required for Mi2 remodeling activity, suggesting that remodeling activity may be required for both activation and repression. Thus, our data suggest that NuRD can function as a coactivator or repressor and that acetylated HDAC1 converts the NuRD complex from a repressor to an activator during GATA-1-directed erythroid differentiation.
Introduction
Histone deacetylases (HDACs)4 play important roles in diverse nuclear and cellular processes, such as developmental programming, gene expression, cell cycle progression, and cell migration. Mammalian HDACs are classified into four classes (I, II, III, and IV) based on sequence homology to the yeast histone deacetylases Rpd3 (reduced potassium dependence), Hda1 (histone deacetylase1), and Sir2 (silent information regulator 2), respectively. Although the precise cellular functions of the different HDAC enzymes are still poorly understood, evidence suggests that different members of the HDAC family have distinct functions (1–3). Importantly, it is becoming increasingly clear that class I HDAC enzymes are clinically relevant to cancer therapy (4–8). HDAC inhibitors are potential drugs for the treatment of cancer through the ability to restore normal patterns of gene expression, which may result in cell cycle arrest, differentiation, or apoptosis of tumor cells. In fact, two histone deacetylase inhibitors, SAHA and depsipeptide, have been recently approved by the United States Food and Drug Administration for treatment of cutaneous T-cell lymphoma and are in clinic trials in treating leukemia (9). Therefore, it is extremely important to understand how HDAC1 and other class I HDACs regulate the process of hematopoiesis.
HDAC1 is found in at least three distinct multiprotein complexes, including the Sin3, the CoREST, and the NuRD complexes (1). The NuRD complex has seven components, including the ATPase/helicase Mi-2, HDAC1, and HDAC2 (10, 11). The MeCP1 complex is a NuRD-related complex; it contains the additional component MBD2, a 5-cytosine-phosphoguanine methyl-binding protein (12–15). NuRD/MeCP1 is recruited to target genes through its interaction with DNA binding transcription factors. Although it is generally considered that NuRD/MeCP1 is a co-repressor complex (16–18), it can also activate genes in other cases (17, 19), suggesting that the NuRD complex can either repress or activate gene transcription. However, the mechanism of how the NuRD complex activates gene transcription remains unknown.
The activity of HDACs may be regulated through post-translational modifications. HDAC1 can be post-translationally modified, and these modifications can modulate HDAC1 activity and protein levels (20, 21). Recently, it was reported that HDAC1 can be acetylated by histone acetyltransferase p300. Acetylated HDAC1 loses its deacetylase activity (22, 23). Interestingly, acetylated HDAC1 also inhibits the deacetylase activity of HDAC2, hence down-regulating the overall deacetylase activity of HDAC1/2-containing complexes, including the NuRD complex (24). Thus, these results uncover a novel mechanism underlying the regulation of HDAC1-containing protein complexes.
Hematopoietic lineage-specific transcription factor GATA-1 is the founding member of the GATA factor family. GATA-1 has been shown to be essential for normal hematopoiesis, especially for erythropoiesis and megakaryopoiesis (25–27). GATA-1 is a zinc finger transcription factor, and its binding motif is present in the regulatory regions of many erythroid-specific genes (28–30). The β-globin gene was the first GATA-1 target gene identified (31). GATA-1 was found to bind to the β-globin gene locus within the β-globin locus control region and the globin promoter regions (31, 32). Recent genome-wide studies revealed that GATA-1 interacts with more than a thousand genes in erythroid cells (28, 29). GATA-1 can both positively and negatively regulate these genes (29, 33). Thus, it is important to understand how GATA-1 regulates gene transcription during erythroid differentiation.
GATA-1 regulates gene transcription through associations with a variety of transcription factors and cofactors (34, 35). Recently, it was shown that GATA-1 is associated with the HDAC1-containing NuRD or MeCP1 corepressor complexes through its cofactor FOG-1 (friend of GATA-1) (36–39). It was suggested that these complexes play an important role in GATA-1-mediated repression of target genes such as GATA-2, c-Myc, and c-Kit, which are all required for the proliferation of hematopoietic progenitors (36, 37, 40–42). However, during GATA-1-mediated transcriptional activation of β-globin gene, the NuRD/MeCP1 complex is still recruited to the GATA-1 sites of the β-globin locus (40, 43, 44). Hence, the question is how the HDAC1-containing corepressor complex regulates GATA-1-mediated gene activation in erythropoiesis. In this study, we investigate the role of HDAC1 in erythroid differentiation as well as in GATA-1-mediated gene regulation. We found that HDAC1 activity is decreased during erythroid differentiation. GATA-1-associated deacetylase activity is also down-regulated accordingly by p300-mediated acetylation inactivation of HDAC1, which allows GATA-1 to activate transcription during differentiation. Our data suggest that down-regulation of GATA-1-associated deacetylase activity is required for erythroid differentiation and that acetylation of HDAC1 contributes to the conversion of the NuRD complex from repressor to activator.
EXPERIMENTAL PROCEDURES
DNA Constructs
The double GATA binding site reporter construct (pGL3 GATA-Luc) was generated by inserting two double GATA binding sites from the GATA-1 promoter into pGL3 promoter luciferase reporter vector (Promega). The GATA-1 expression vector was generated by subcloning mouse GATA-1 into the pcDNA 3.1 vector. The pcDNA HA FOG-1 was a kind gift from Gerd Blobel from the University of Pennsylvania. The expression vectors for HDAC1, HDAC1 6Q, and HDAC1 6R were described previously (22). The integrity of all constructs was confirmed by DNA sequencing.
Cell Lines and Reporter Assays
NIH 3T3 cells, the murine erythroleukemia (MEL) cell line, and erythroid precursor cell line G1E and its derivative cell line G1E-ER4 were cultured as described previously (22, 45–47). Luciferase reporter vectors (pGL3 GATA-Luc) and expression vectors were transfected into NIH 3T3 or MEL cells using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). Luciferase reporter assays were carried out using Dual-Glo luciferase reporter assay system (Promega).
Human CD34+ Cell Culture
Frozen human cord blood CD34+ cells were purchased from STEMCELL Technologies (Vancouver, Canada). Cells were then cultured and differentiated according to the manufacturer's protocol. Briefly, cells were cultured in StemSpan SFEM medium (STEMCELL Technologies) with four recombinant human cytokines, including 100 ng/ml stem cell factor, 100 ng/ml Flt3-ligand, 20 ng/ml IL-3, and 20 ng/ml IL-6 (PeproTech, Rocky Hill, NJ). After the expansion for 7 days with 5% CO2 at 37 °C, cells were stained with anti-human PE-CD34+ antibody (BD Biosciences), and CD34+ cells were collected by flow cytometry sorting and stored in liquid nitrogen. Aliquots of CD34+ cells were thawed and cultured with 75 ng/ml each of stem cell factor, Flt-3 ligand, and thrombopoietin for 1 day and then for another 3 days, whereas cytokines in the culture medium were replaced with 10 ng/ml stem cell factor and 10 ng/ml IL3. 2 units/ml human recombinant erythropoietin (Cell Science, Canton, MA) was then added (designated as day 0). Cells were harvested at intervals as indicated.
siRNA Knockdown
HDAC1 or HDAC2 siRNA sequences were cloned into pSuper retroviral vectors (Oligoengine), and the recombinant viruses were collected and used to infect MEL cells. Retrovirus preparations and infections were performed as described (45). Single cell clones with integrated HDAC1 or HDAC2 shRNA constructs were selected to produce stable cell lines.
Immunoprecipitation, Western Analysis, and ChIP
Standard protocols were employed for immunoprecipitation and Western blot analysis (22). Chromatin immunoprecipitation (ChIP) was carried out as described (22, 48). Primers for β-globin and GATA-2 were described previously (49). Primers for the GATA-1 gene were as follows: GATA-1 HS1 forward, 5′-CCCGCTGATTCCCTTATCTATG-3′, reverse, 5′-GTGCAAGGCCCAGAAGTC 3′; GATA-1 erythroid specific promoter (IE) forward, 5′-CACCAACAGCCACAGTCG-3′, reverse, 5′-TTAGTGCTTCGGCTCGTG-3′.
Antibodies
Anti-GATA-1 (N6), anti-FOG-1 (M20), and anti-Mi2 (H242) antibodies were obtained from Santa Cruz Biotechnology. Anti-FLAG and anti-β-actin antibodies were from Sigma. HDAC1 antibodies against nonacetylated or acetylated HDAC1 were generated from rabbits injected with acetylated or nonacetylated HDAC1 C-terminal peptide. Peptide sequences were: GEGGRKNSSNF or GEGGRK(Ac)NSSNF.
RT-PCR
Total RNA was prepared from 1 × 106 cells by using the SV total RNA isolation kit (Promega). A total of 1 μg of RNA was reverse-transcribed by using the SuperScript II reverse transcriptase as suggested by the manufacturer (Invitrogen). The primers used in RT-PCR were as follows: β-globin forward, 5′-CACCTTTGCCAGCCTCAGTG-3′, reverse, 5′-GGTTTAGTGGTACTTGTGAGCC-3′; β-actin forward, 5′-GTGGGCCGCTCTAGGCACCA-3′, reverse, 5′-TGGCCTTAGGGTGCCAGGGGG-3′.
Histone Deacetylase Activity Assay
3H-Labeled acetylated histones were isolated from MEL cells as described previously (50). Deacetylation assays were carried out as described (22). Briefly, deacetylation assays were carried out by mixing nuclear extracts with 4 μg of 3H-labeled acetylated histones (10,000 cpm) in 50 μl of assay buffer (20 mm Tris HCl, pH 8.0, 150 mm NaCl, 0.5 mm EDTA, 5% glycerol) for 10 min at 30 °C followed by the addition of 50 μl of stop buffer (1.44 m HCl, 0.24 m HOAc). [3H]Acetate was extracted with 0.6 ml of ethyl acetate. After centrifugation, the upper organic phase was quantified by liquid scintillation counting.
Nucleosome Remodeling Assay
Nucleosome sliding assays were performed as described with minor modifications (51). Briefly, core histones were isolated from HeLa cells according to a standard protocol. A 194-bp fragment with a strong nucleosome positioning sequence was generated by PCR amplification of the plasmid pGEM-3Z-601. For reconstitution of mononucleosomes, 3.6 μg of the DNA fragment was incubated in a 0.8:1 ratio (w/w) with HeLa core histones. The reaction was dialyzed sequentially for 1 h at 4 °C against 1.25, 1.0, and 0.75 m NaCl and then against TE (10 mm Tris-HCl (pH 8.0), 0.5 mm EDTA). Nucleosome sliding assays were performed at 37 °C for 30 min in a standard volume of 10 μl containing 10 mm Tris-HCl (pH 7.6), 50 mm NaCl, 3 mm MgCl2, 1 mm β-mercaptoethanol, 0.1 μg/μl BSA, and 1 mm ATP. About 100–200 ng of DNA equivalent of assembled mononucleosomes was incubated with either purified NURF or purified Mi2 complexes. Mononucleosomes were resolved on a native 5% PAGE and stained with ethidium bromide.
RESULTS
HDAC1 Deacetylase Activity Decreased during Differentiation in MEL Cells
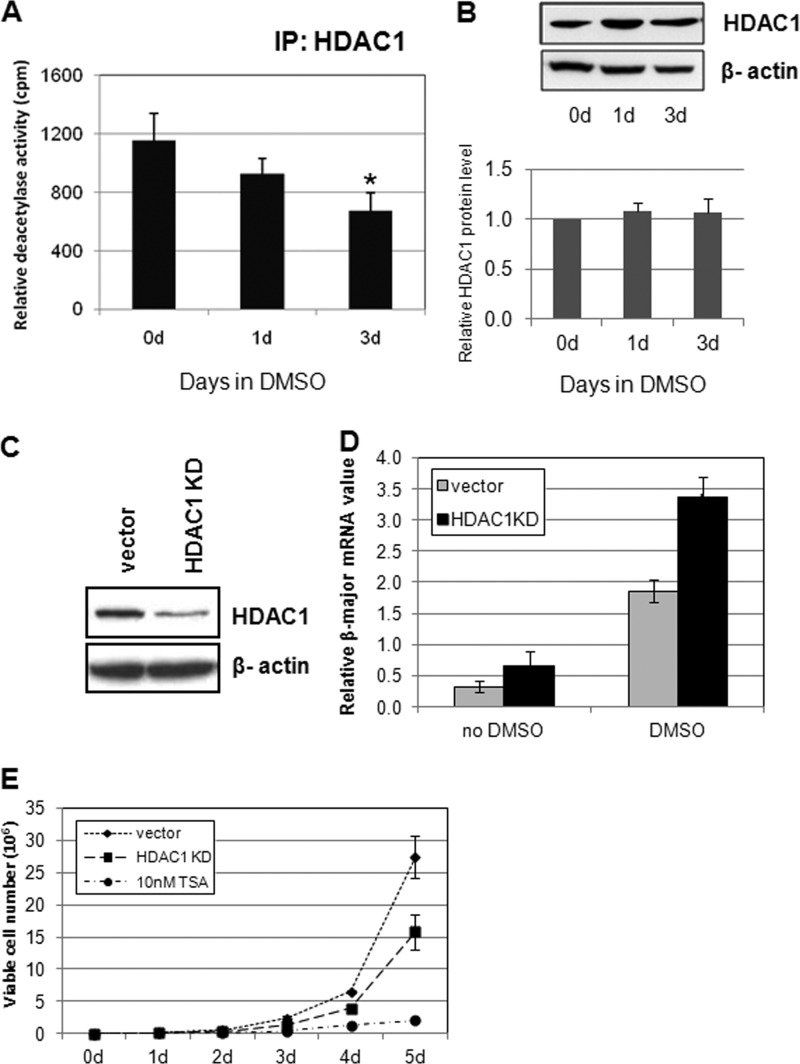
Histone deacetylase inhibitors, such as sodium butyrate or valproic acid, can promote differentiation of erythroid as well as other hematopoietic lineages (52). Therefore, it is conceivable that endogenous deacetylase activity is down-regulated during differentiation. Because HDAC1 associates with many key transcription factors important for hematopoiesis, we speculated that the HDAC1-associated deacetylase activity may be down-regulated during differentiation. The MEL cells are blocked at the late CFU-E or proerythroblast stage in the adult erythroid lineage and can be efficiently induced to undergo terminal erythroid differentiation through exposure to DMSO (53–55). First, we confirmed that differentiation of MEL cells was induced with 1.5% DMSO treatment (supplemental Fig. S1). Nuclear extracts from DMSO-treated or untreated cells were immunoprecipitated with HDAC1 antibodies, and the resulting products were subjected to deacetylation assays. Indeed, HDAC1-associated deacetylase activity was reduced during erythroid differentiation (Fig. 1A). However, the protein levels of HDAC1 remained unchanged during DMSO-induced differentiation (45) (Fig. 1B). This result shows that during erythroid differentiation, HDAC1-associated deacetylase activity is negatively modulated without changing the HDAC1 protein levels.
FIGURE 1.
HDAC1 deacetylase activity is down-regulated during erythroid differentiation in MEL cells. A, HDAC1-associated histone deacetylase activity is reduced upon DMSO-induced differentiation of MEL cells. MEL cells were induced with 1.5% DMSO for the indicated days. Nuclear extracts from MEL cells were immunoprecipitated (IP) with HDAC1 antibodies. The deacetylase activities of the immunoprecipitates were determined by deacetylation of 3H-labeled Ac-histone. *, significant difference when compared with day 0 (Student's t test, p < 0.05). B, nuclear extracts from MEL cells were subjected to SDS-PAGE and Western blot analysis with antibodies as indicated. A representative Western blot result is shown. HDAC1 protein level quantification was determined from three independent experiments. C, knockdown of HDAC1 in MEL cells. The siRNA duplexes for mouse HDAC1 were cloned into pSuper retroviral vectors, and the resulting viruses were used to infect MEL cells. Single colonies were selected for the most effective reduction of HDAC1. D, HDAC1 knockdown (KD) affects differentiation. MEL cells with vector, HDAC1, or HDAC2 knockdown constructs were treated with 1.5% DMSO for the indicated time periods. Total RNA was extracted, and real time RT-PCR was performed with β-major globin primers and normalized to β-actin. *, significant difference when compared with vector control (Student's t test, p < 0.01). E, HDAC1 knockdown affects MEL cell proliferation. 1 × 105 control, HDAC1, or HDAC2 knockdown MEL cells were inoculated into medium. The number of viable cells was counted daily for 5 days. The same amount of vector control cells was also treated with 10 nm TSA and tested for proliferation. Data shown are the means ± S.E. of three independent experiments.
HDAC1 Promotes Erythroid Cell Proliferation and Inhibits Differentiation
If down-regulation of HDAC1-associated deacetylase activity is important for erythroid cell differentiation, then reduction of HDAC1 levels in erythroid cells may promote differentiation. To test this hypothesis, we knocked down HDAC1 expression using shRNA-mediated gene silencing in MEL cells. The level of knockdown was verified by Western blotting analysis. The HDAC1 knockdown cells exhibited more than 50% reduction of HDAC1 protein level (Fig. 1C). These cells were then treated with DMSO, and total RNA was collected for measuring the expression of β-globin, a marker for erythroid differentiation, by real time RT-PCR. Reducing the levels of HDAC1 resulted in a significant elevation of β-globin expression in comparison with the vector control cell line (Fig. 1D), indicating that down-regulation of HDAC1 promotes erythroid differentiation in MEL cells. To test whether HDAC1 knockdown also affects the cell proliferation rate, the same number of cells was inoculated with the same volume of growth medium and sampled in 24-h intervals for 5 days. HDAC1 knockdown cells were significantly impaired in growth (Fig. 1E). These results indicate that HDAC1 inhibits erythroid differentiation and promotes cell proliferation. Because HDAC2 often coexists in HDAC1-containing corepressor complexes, HDAC2 was also knocked down by shRNA-mediated gene silencing (supplemental Fig. S2A). Down-regulation of HDAC2 also significantly affected erythroid differentiation (supplemental Fig. S2B), suggesting that the HDAC1/2-associated complexes are important for differentiation. Interestingly, HDAC2 knockdown had much less of an effect in MEL cell proliferation, although the HDAC2 knockdown was more efficient than that of HDAC1 (supplemental Fig. S2C).
GATA-1- and FOG-1-associated Deacetylase Activities Decrease after Differentiation
GATA-1 is a key transcription factor required for erythroid differentiation. It has been shown that GATA-1 associates with the NuRD or the MeCP1 corepressor complexes that contain HDAC1 and HDAC2 (36, 37). The interaction between GATA-1 and NuRD is mediated through the GATA-1-associated protein FOG-1 (36–39). It appears that the NuRD complex is required for both GATA-1-mediated repression and GATA-1-mediated activation (43). Because HDAC1 deacetylase activity is down-regulated after differentiation, we speculated that GATA-1-associated deacetylase activity of the GATA-1-containing complex is also reduced after differentiation.
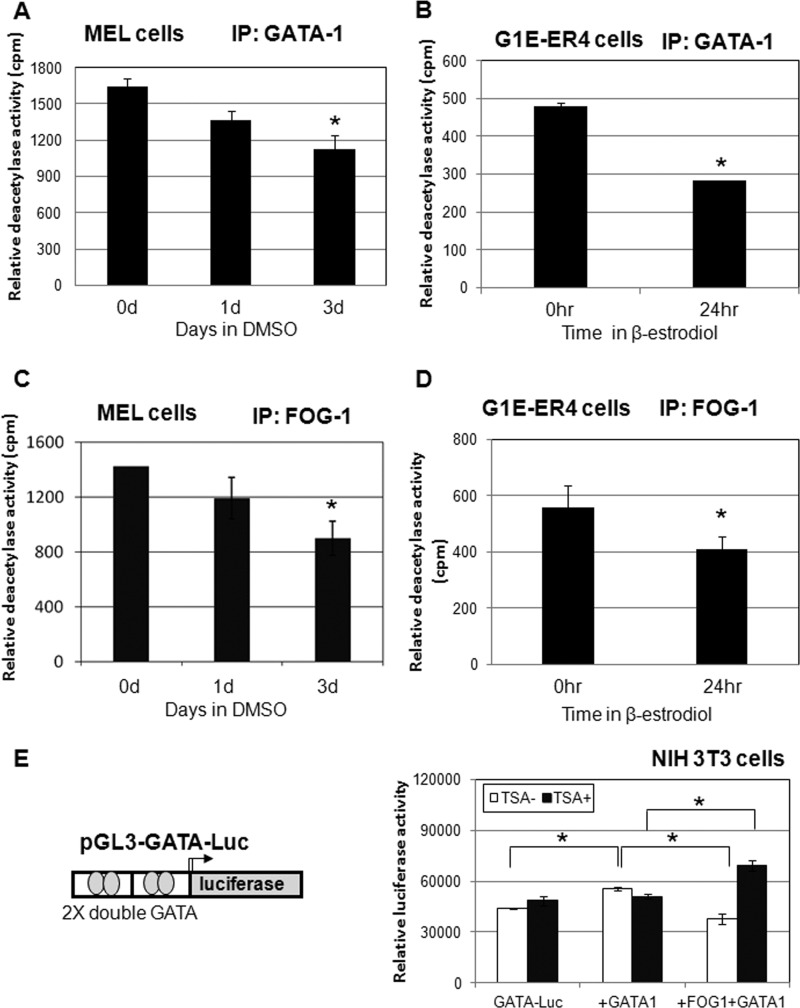
We first examined whether GATA-1-associated HDAC activity changes upon DMSO-induced MEL cell differentiation. Cell lysates were prepared at time intervals following the addition of DMSO to the cell culture and subjected to immunoprecipitation using GATA-1 antibody. The GATA-1-associated HDAC activity declined upon the addition of DMSO (Fig. 2A), although the cellular HDAC1 protein levels remained unchanged (Fig. 1B). We also tested GATA-1-associated deacetylase activity in another erythroid model system, the G1E cells. G1E are GATA-1-null erythroid progenitor cells (46); G1E-ER4 cells were engineered to stably express estrogen-inducible GATA-1 (47, 56). The addition of estrogen led to translocation of GATA-1 into the nucleus and rapid induction of erythroid differentiation. The G1E-ER4 cells were treated with estrogen for 24 h, and whole cell extracts were prepared for immunoprecipitation with GATA-1. GATA-1-associated deacetylase activity was decreased almost 40% after treatment with estrogen (Fig. 2B).
FIGURE 2.
GATA-1-associated deacetylase activity is down-regulated during differentiation of MEL cells. A and B, GATA-1-associated deacetylase activity decreases upon differentiation. Whole cell extracts from MEL cells (A) or G1E-ER4 cells (B) incubated with 1.5% DMSO or 1 μm estradiol for the indicated time periods were subjected to immunoprecipitation (IP) with GATA-1 antibodies. Immunoprecipitates were then assayed for HDAC activity. *, significant difference when compared with 0 d (Student's t test, p < 0.01). C and D, FOG-1-associated deacetylase activity is reduced upon differentiation. Whole cell extracts from MEL or G1E-ER4 cells incubated with 1.5% DMSO or 1 μm estradiol for the indicated time periods were subjected to immunoprecipitation with FOG-1 antibodies. Immunoprecipitates were then assayed for HDAC activity. *, significant difference when compared with 0 d (Student's t test, p < 0.01). E, deacetylase activity is needed for FOG-1 repression of GATA-1-mediated reporter activity. GATA-1 and FOG-1 expression vectors were transfected together with the GATA-1 luciferase reporter in NIH 3T3 cells. The cells were treated with or without TSA overnight before harvest. The luciferase assay was performed 48 h after transfection. *, significant differences (Student's t test, p < 0.01). Data shown are the means ± S.E. of five independent experiments.
Because FOG-1 mediates GATA-1 and NuRD complex interactions, we further tested whether FOG-1-associated deacetylase activity was also reduced after differentiation. Indeed, through immunoprecipitation with FOG-1 antibody, FOG-1-associated deacetylase activity was also reduced after differentiation in both MEL and G1E-ER4 cells (Fig. 2, C and D). These results indicate that although GATA-1 and FOG-1 remain associated with the NuRD corepressor complex after induction of differentiation (37, 43), their associated deacetylase activity is significantly reduced.
Next, we investigated whether GATA-1- and FOG-1-mediated repression depends on histone deacetylase activity. We constructed a reporter gene construct consisting of only two double GATA binding sites from the GATA-1 promoter to study the specific effect of GATA-1 in the absence of other cis elements and hematopoietic transcription factors (Fig. 2E). GATA-1 slightly increased the reporter activity in NIH 3T3 cells. The increase of reporter activity by GATA-1 was repressed by FOG-1, conforming that FOG-1 recruits corepressor complexes (36). We then examined whether the histone deacetylase activity is needed for FOG-1-mediated repression. Cells were treated with TSA overnight after transfection. Treatment with TSA relieved the repression mediated by FOG-1. Further, TSA converted FOG-1 from repressor to activator (Fig. 2E). This result suggests that the FOG-1 recruited complex may contribute to GATA-1 activation when the deacetylase activity is suppressed in the complex.
GATA-1-associated HDAC1 Is Acetylated during Differentiation, and Acetylated HDAC1 Promotes Differentiation
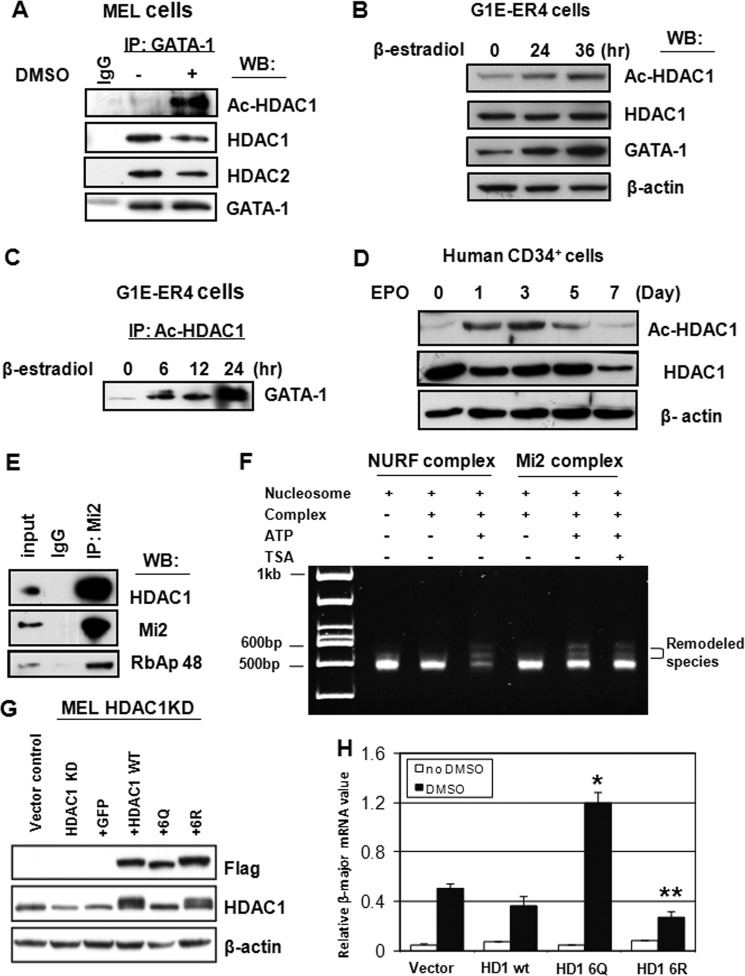
Given that HDAC1 can be acetylated by p300 and that acetylated HDAC1 inhibits the deacetylase activities of HDAC1 and HDAC2 (22, 24), combined with our observation that the GATA-1-associated HDAC complex loses deacetylase activity, whereas cellular HDAC1 protein levels remain unchanged (Fig. 2), we reasoned that GATA-1-associated HDAC1 is acetylated in differentiated cells. To test this hypothesis, MEL cells were induced to differentiate with DMSO. The extracts were then immunoprecipitated with GATA-1 antibodies. The resulting complexes were then subjected to Western blotting with antibodies specific for acetylated HDAC1 (supplemental Fig. S3). GATA-1-associated HDAC1 was acetylated, and HDAC1 acetylation was significantly increased during erythroid differentiation (Fig. 3A). In contrast, GATA-1-associated HDAC1 levels decreased to a much lesser extent. We also tested acetylated HDAC1 levels in G1E-ER4 cells. Accordingly, we found that acetylated HDAC1 levels increased upon differentiation (Fig. 3B), but overall HDAC1 levels remained unchanged (Fig. 3B). Consistent with the results from MEL cells, the association of acetylated HDAC1 and GATA-1 was also markedly increased in G1E-ER4 cells after differentiation (Fig. 3C). Next, we tested whether acetylated HDAC1 can also be induced in human CD34+ hematopoietic stem cells. CD34+ cells can be induced to differentiate along the erythroid lineage by exposure to erythropoietin. Before induction, there were very low levels of acetylated HDAC1 in the cells. After induction of differentiation, acetylated HDAC1 gradually increased and then decreased at later stages of differentiation (Fig. 3D). This result suggests that the dynamic acetylation pattern of HDAC1 may play an important role in erythroid differentiation.
FIGURE 3.
GATA-1-associated HDAC1 is acetylated upon differentiation. A, nuclear extracts from MEL cells treated with or without 1.5% DMSO for 3 days were subjected to immunoprecipitation (IP) with GATA-1 antibodies. The precipitates were analyzed by Western blotting (WB) with antibodies as indicated. B, G1E-ER4 cells were treated with estradiol for the indicated time periods. Whole cell extracts were collected and subjected to Western blot analysis with antibodies as indicated. C, G1R-ER4 cells were treated with estradiol for the indicated time periods. Whole cell extracts were immunoprecipitated with anti-acetylated HDAC1 antibodies and Western blotted with anti-GATA-1 antibodies. D, human CD34+ cells were treated with erythropoietin (EpO) for the indicated time periods. Whole cell extracts were collected and subjected to Western blot analysis with antibodies as indicated. E, preparation of Mi2-associated protein complexes. Nuclear extracts from MEL cells were immunoprecipitated with Mi2 antibodies, and the precipitate was analyzed by Western blot with antibodies as indicated. F, TSA does not affect the NuRD complex nucleosome remodeling activity. A nucleosome remodeling assay was performed with Drosophila NURF or the Mi2 complex with or without ATP or TSA. G, overexpression of FLAG-HDAC1 and mutants in HDAC1 knockdown (KD) MEL cells. Expression levels of FLAG-HDAC1 or mutants were analyzed by Western blot. H, HDAC1 knockdown cells with overexpressed HDAC1 or mutants were treated with DMSO for 3 days, and β-major mRNA levels were measured by RT-PCR. * and ** indicate significant differences when compared with vector control (Student's t test, p < 0.01). Data shown are the means ± S.E. of three independent experiments.
Because GATA-1-associated HDAC1 is within the NuRD complex, the next question that we asked was whether the loss of HDAC1 deacetylase activity affects the nucleosome remodeling activity of the NuRD complex. The NuRD complex was purified from MEL cells with anti-Mi2 antibodies (Fig. 3E). The complex was then subjected to a remodeling assay with or without treatment with the deacetylase inhibitor TSA (Fig. 3F). The results show that histone deacetylase activity was not required for NuRD remodeling activity. Thus, loss of deacetylase activity during erythroid differentiation did not affect NuRD-associated chromatin remodeling activity. This suggests that the remodeling activity may be required for NuRD-mediated activation and that it may positively or negatively influence the recruitment of activators or repressors to chromatin, respectively.
We next reasoned that if down-regulation of HDAC1 deacetylase activity is required for erythroid differentiation, then introduction of the HDAC1 6Q mutant, which mimics acetylated HDAC1 and has defective deacetylase activity (22), should promote differentiation. Conversely, the HDAC1 6R mutant, which mimics active, nonacetylated HDAC1, should inhibit erythroid differentiation and promote cell proliferation. To test these possibilities, we stably expressed siRNA-resistant FLAG-tagged HDAC1, HDAC1 6R, or HDAC1 6Q in HDAC1 knockdown MEL cells using the retroviral vector pOZ (57, 58). The expression levels of exogenous HDAC1 or HDAC1 mutants in the stable clones were comparable with the expression levels of endogenous HDAC1 (Fig. 3G). HDAC1 6Q-associated deacetylase activity was significantly reduced when compared with that of HDAC1 WT or HDAC1 6R in cell extracts (supplemental Fig. S4). HDAC1 6Q increased β-globin expression when compared with the vector control (Fig. 3H). In contrast, HDAC1 6R inhibited MEL cell differentiation. Overexpression of wild type HDAC1 did not significantly affect β-globin expression, suggesting that a portion of wild type HDAC1 is acetylated in differentiated cells. The proliferation rates of these cells were also examined (supplemental Fig. S5). HDAC1 6Q did not affect MEL cell proliferation. However, cells overexpressing wild type HDAC1 or HDAC1 6R promoted cell proliferation. Thus, the data suggest that HDAC1 activity is important for inhibiting erythroid differentiation and promoting proliferation. Down-regulation of HDAC1-associated deacetylase activity is an important step required for the onset of erythroid differentiation programs.
GATA-1 Up-regulates Its Own Promoter Activity through the Recruitment of Acetylated HDAC1
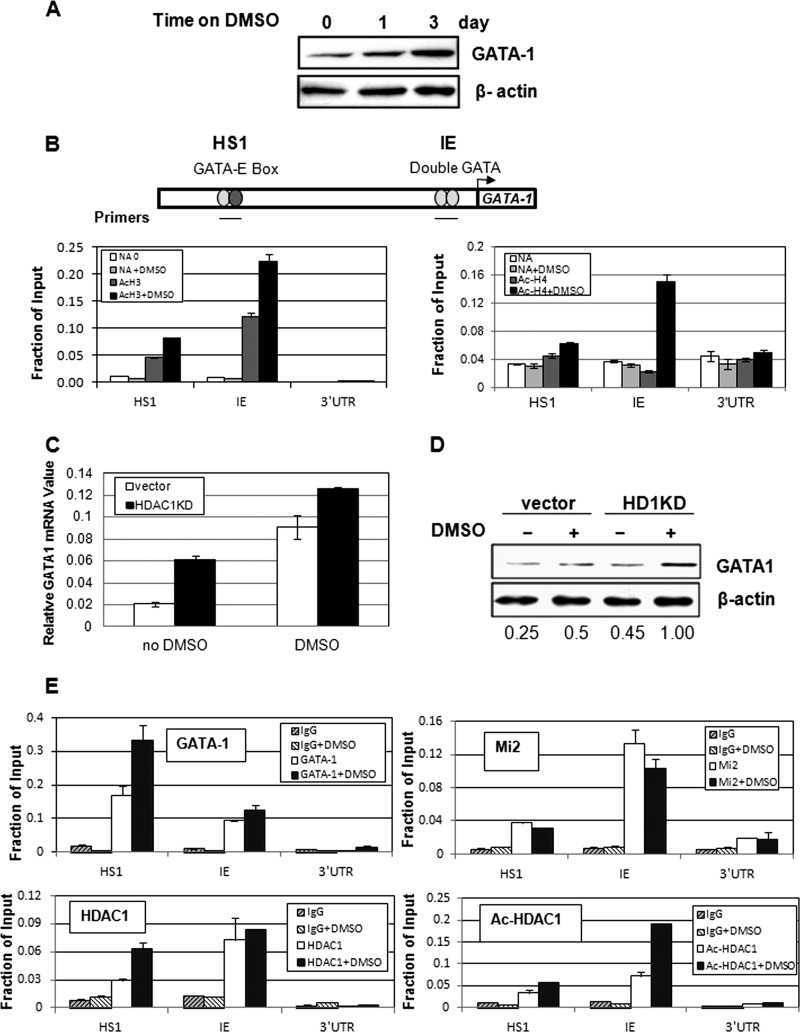
GATA-1 can regulate its own promoter activity through binding to several GATA sites located in its own promoter and enhancer regions in erythrocytes and in megakaryocytes (59). During differentiation of MEL cells, GATA-1 levels increased (Fig. 4A), supporting previous observations demonstrating that GATA-1 promoter activity is auto-regulated (59). The major regulatory regions of the GATA-1 gene locus are located in HS1 and in the hematopoietic specific promoter region (IE) (Fig. 4B). These regions are important for GATA-1-mediated promoter activation (59, 60). We tested the histone acetylation levels at these sites in MEL cells. As reported previously, the HS1 and IE regions were moderately acetylated in MEL cells before induction (61) (Fig. 4B). Upon DMSO-mediated induction, acetylation levels of histone H3 and H4 were significantly increased (Fig. 4B), indicating that the GATA-1 gene locus is activated upon erythroid differentiation. We next tested whether GATA-1 gene activation is regulated by HDAC1. In HDAC1 knockdown MEL cells, both GATA-1 mRNA and protein levels increased (Fig. 4, C and D), suggesting that HDAC1 negatively regulates GATA-1 promoter activity.
FIGURE 4.
HDAC1 is recruited to the GATA-1 promoter to repress GATA-1 transcription. A, GATA-1 protein levels increase after MEL cell differentiation. Nuclear extracts from DMSO-treated MEL cells were analyzed by Western blotting for GATA-1 protein levels. B, the GATA-1 promoter is acetylated upon DMSO induction. MEL cells treated with DMSO were subjected to ChIP against Ac-H3 or Ac-H4 antibodies. The resulting precipitated DNA was analyzed by real time PCR. Primer locations are indicated as solid lines. C, vector control and HDAC1 knockdown (KD) MEL cells were treated with DMSO for 1 day. GATA-1 mRNA levels were determined by real time RT-PCR. Results are the means ± S.E. of three independent experiments. D, vector control and HDAC1 knockdown cells were treated with DMSO for 1 day. GATA-1 protein levels were examined by Western blotting. The protein band intensities were quantified using the 8900 imaging system (Alpha Innotech) and normalized to actin. E, acetylated HDAC1 is recruited to the GATA-1 promoter after differentiation. MEL cells treated with DMSO were subjected to ChIP with antibodies against GATA-1, Mi2, HDAC1, and acetylated HDAC1, as indicated. The resulting precipitated DNA was analyzed by real time PCR. Primer locations are indicated as solid lines. For ChIP assays, the results of a representative experiment of three independent experiments are shown. Data are means ± S.E. of three real time PCR results.
We further studied the recruitment of GATA-1 and the NuRD complex to the GATA-1 promoter during DMSO-induced erythroid differentiation. Mi2 consistently bound to the HS1 and IE regions of the GATA-1 locus before and after induction (Fig. 4E). HDAC1 binding did not change at the IE region and increased at the HS1 region after DMSO induction. It is not clear why HDAC1 binding increased at the HS1 region after DMSO induction. However, acetylated HDAC1 significantly increased after DMSO induction at both sites (Fig. 4E), which may cause reduction of total deacetylase activity at these regions, resulting in gene activation.
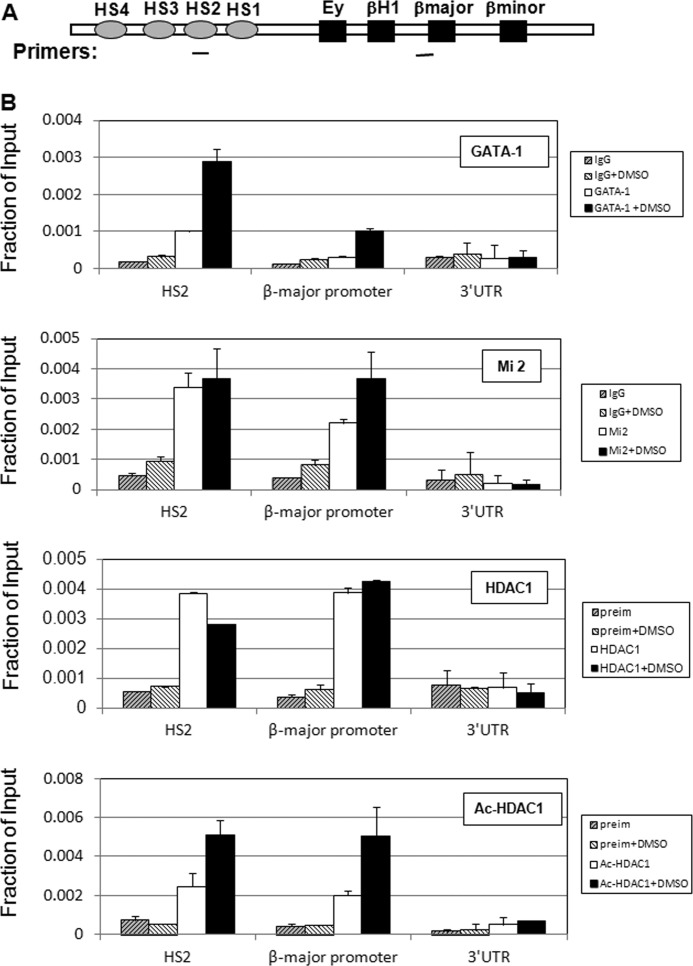
Acetylated HDAC1 Is Located at the β-Globin Promoter and the Locus Control Region after Erythroid Differentiation
In MEL cells, expression of adult β-globin can be greatly induced upon treatment with DMSO, which results from binding of erythroid-specific and non-erythroid-specific transcription factors and cofactors to the locus control region and the β-major promoter (53–55, 62). Given that acetylated HDAC1 is recruited to the GATA-1 promoter in association with the NuRD complex, we asked whether acetylated HDAC1 can also be recruited to the β-globin locus in a similar manner. Chromatin immunoprecipitation experiments were performed with MEL cells with or without the addition of DMSO. As reported previously, GATA-1 is recruited to both the locus control region HS2 and the β-major promoter region of the β-globin gene after DMSO treatment (63) (Fig. 5A). Consistent with the observation at the GATA-1 locus, Mi2 was present at both regions before induction. The level of binding was increased at the promoter region and remained unchanged in the HS2 region (Fig. 5B), suggesting that NuRD may act as coactivator at the promoter region after induction (43). HDAC1 strongly associated with both regions before induction of differentiation and remained strongly bound at the β-major promoter region after differentiation. The binding at the HS2 region was slightly decreased after induction (Fig. 5B). Importantly, the recruitment of acetylated HDAC1 to both HS2 and the β-major promoter was significantly increased after DMSO treatment (Fig. 5B), indicating that the overall deacetylase activity at HS2 and the promoter region may be reduced after DMSO-induced erythroid differentiation. The reduction of deacetylase activity at these regulatory regions may lead to the activation of β-globin transcription.
FIGURE 5.
Acetylated HDAC1 is recruited to the β-globin locus after differentiation. A, schematic representation of the mouse β-globin gene locus. Locations of PCR primer sets are underlined. B, MEL cells treated with DMSO were subjected to ChIP with antibodies against GATA-1, Mi2, HDAC1, and acetylated HDAC1, as indicated. The resulting precipitated DNA was analyzed by real time PCR with primers as indicated. For ChIP assays, the results of a representative experiment of three independent experiments are shown. Data are means ± S.E. of three real time PCR results. preim, preimmunoserum from rabbit.
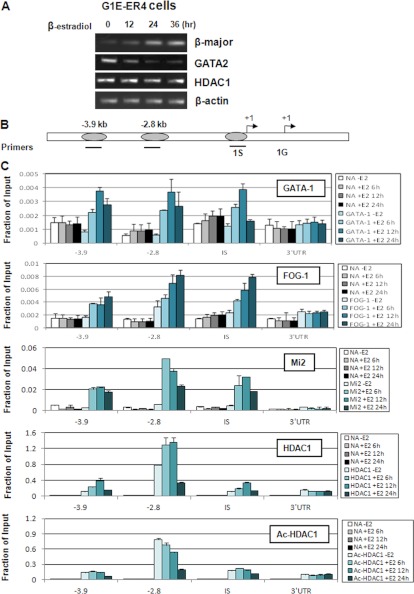
GATA-2 Levels Were Down-regulated by GATA-1 through Recruitment of Active HDAC1 in G1E-ER4 Cells
Because the recruitment of acetylated HDAC1 to gene promoters led to transcriptional activation, we asked whether acetylated HDAC1 recruitment is decreased at genes that are repressed by GATA-1. The GATA-2 gene is known to be targeted by GATA-1-mediated repression. Because there is very low expression of GATA-2 in MEL cells, we used G1E-ER4 cells to study GATA-2 gene regulation. In G1E-ER4 cells, the addition of estradiol led to translocation of GATA-1 into the nucleus and rapid induction of erythroid differentiation (Fig. 6A) (33). Accordingly, GATA-2 expression was decreased upon induction by estrogen (33, 48) (Fig. 6A). We investigated whether the recruitment of HDAC1 was involved in GATA-2 gene repression. GATA-1-mediated repression involves the binding of GATA-1 to GATA motifs located at the −3.9 kb, −2.8 kb, and IS promoter region of the GATA-2 gene ((41, 64, 65) (Fig. 6B)). Before induction, there was low to no binding of GATA-1 and FOG-1 (Fig. 6C). There was low binding of HDAC1 except at the −2.8 region. Interestingly, acetylated HDAC1 binding at this region was high, indicating that although HDAC1 was present at this region of the GATA-2 locus before induction, the deacetylase activity was low. Mi2 binding before induction was also very low at all regions, suggesting that HDAC1 may be recruited to the GATA-2 locus without association of the NuRD complex and independent from GATA-1. Upon induction by estradiol, GATA-1 binding increased, and so did binding of FOG-1, Mi2, and HDAC1, suggesting that GATA-1 recruits the NuRD corepressor complex to the GATA-2 locus through FOG-1. Consistent with the recruitment of the GATA-1-associated NuRD complex, acetylated HDAC1 levels were decreased. This result agrees with previous studies showing that after induction, histone H3 and H4 levels at the GATA-2 gene locus were reduced and expression of GATA-2 was repressed in differentiated erythroid cells (41). Interestingly, after 24 h of induction, GATA-1, Mi2, and HDAC1 binding was significantly reduced, suggesting that GATA-1 and the NuRD complex dissociated from the regulatory elements. It is not clear why FOG-1 binding remained high. One possibility is that FOG-1 may be recruited through GATA-1-independent mechanisms.
FIGURE 6.
Increasing levels of HDAC1 and decreasing levels of acetylated HDAC1 are recruited to the GATA-2 locus after induction of differentiation in G1E-ER4 cells. A, G1E-ER4 cells were treated with estradiol for the indicated time periods. Total RNA was extracted, and RT-PCR was performed with primers for the indicated genes. B, schematic representation of the mouse GATA-2 locus. Locations of the PCR primer sets are underlined. C, G1E-ER4 cells treated with estradiol were subjected to ChIP with antibodies against GATA-1, FOG-1, Mi2, HDAC1, or acetylated HDAC1. The resulting precipitated DNA was analyzed by real time PCR with the primers indicated. The results of a representative experiment of three independent experiments are shown. Data are means ± S.E. of three real time PCR results. NA, no antibody control.
DISCUSSION
In this study, we showed that HDAC1 deacetylase activity was down-regulated during erythroid differentiation and that knockdown of HDAC1 promoted erythroid differentiation. The down-regulation of HDAC1 deacetylase activity was not due to the decrease of protein levels but through the acetylation of HDAC1. We showed previously that the acetyltransferase p300 can acetylate HDAC1 and that acetylated HDAC1 has no deacetylase activity (22). Further, acetylated HDAC1 also inhibits the deacetylase activity of HDAC2 through dimerization with HDAC2 (24). Thus, during erythroid differentiation, HDAC1 acetylation results in loss of histone deacetylase activity of HDAC1-containing corepressor complexes.
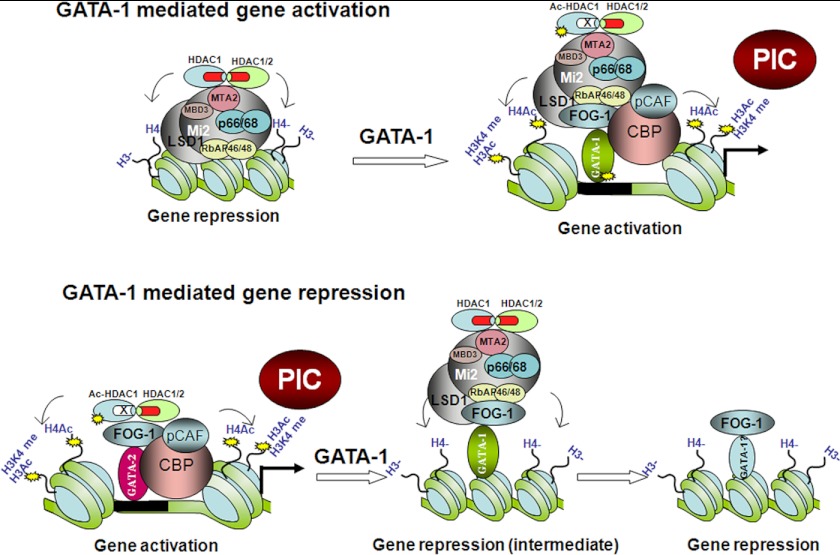
It has been reported that GATA-1 can recruit NuRD corepressor complexes through FOG-1 to target gene promoters and repress transcription (36–39). Interestingly, the NuRD complex was also required for the activation of GATA-1-directed transcription (43). In this study, we further studied the dynamic recruitment of the NuRD complex during gene activation and repression. Upon erythroid differentiation, the β-globin and the GATA-1 genes are activated, whereas the expression of the GATA-2 gene is repressed. We found that the NuRD complex was recruited to the β-globin and GATA-1 gene loci in undifferentiated cells. After differentiation, HDAC1 was acetylated, which extinguished deacetylase activities of the NuRD complex recruited by GATA-1. HDAC1 may be acetylated by p300/CBP during gene activation as it was shown that p300/CBP is recruited to GATA-1-activated promoters (34, 56, 66). Thus, during erythroid differentiation, p300/CBP may play multiple roles in GATA-1-mediated gene activation. First, it can acetylate histone tails to establish an active chromatin structure; second, it can acetylate GATA-1 to enhance GATA-1 binding to chromatin; and third, it can acetylate HDAC1 to suppress its deacetylase activity within the NuRD corepressor complex to keep chromatin acetylated (Fig. 7).
FIGURE 7.
The role of the NuRD complex in GATA-1-mediated gene activation and repression. Upper, a model for GATA-1-mediated activation. The NuRD corepressor complex associates with GATA-1-regulated inactive genes in the absence of GATA-1. After induction of differentiation, GATA-1 is recruited to its targeted sites and subsequently recruits p300/CBP and p300/CBP-associated factor (PCAF) coactivators. These coactivators in turn acetylate histones, GATA-1 and HDAC1, resulting in gene activation. PIC, preinitiation complex. Lower, a model for GATA-1-mediated repression. Before differentiation, GATA-2 recruits coactivators and HDACs to its promoter/enhancer region. p300 acetylates HDAC1, resulting in the loss of HDAC1 deacetylase activity and gene activation. After differentiation, GATA-1 displaces GATA-2 and recruits the NuRD complex for an intermediate stage of repression. At later stages of differentiation, the GATA-2 gene is permanently repressed, and the NuRD complex dissociates from the GATA-2 gene.
In contrast, at the GATA-2 locus, the binding of the NuRD complex was low in undifferentiated cells; however, HDAC1 was present and was highly acetylated. The presence of acetylated HDAC1 correlated with the recruitment of p300/CBP, histone H3 and H4 acetylation, and gene activation at this stage. After differentiation, p300/CBP dissociated from the GATA-2 locus (41), and recruitment of acetylated HDAC1 decreased as well. It is not clear whether HDAC1 is recruited by FOG-1 because FOG-1 binding was found to be low at this stage. After induction of erythroid differentiation, GATA-1, FOG-1, and the NuRD complex were recruited to the promoter. Recruitment of HDAC1 also increased during differentiation. In contrast, acetylated HDAC1 recruitment decreased; this indicates that FOG-1 recruits the NuRD complex with high deacetylase activity to the GATA-2 promoter to repress GATA-2 transcription in differentiated cells. Interestingly, at later stages of erythroid differentiation, recruitment of GATA-1 and the NuRD complex was reduced at the GATA-2 locus (Figs. 6C and 7). This agrees with the observation that permanently silenced genes have low occupancy of histone acetyltransferases and HDACs (67). In contrast, FOG-1 still remained bound at the GATA-2 enhancer and promoter elements, an observation that requires further investigation.
It was shown previously that the NuRD-containing MeCP complex is present at inactive genes during erythroid differentiation (37, 68). MeCP is recruited to repressed chromatin through its interaction with MBD2, a methyl DNA-binding protein that recognizes methylated 5-cytosine-phosphoguanine islands. It has been shown that MBD2 is not present in active chromatin where NuRD binds (37, 68). Because both the MeCP and the NuRD complexes can be recruited to the promoters by interaction with FOG-1, it remains to be investigated why the MeCP complex is not present at active promoters, whereas NuRD remains bound. It will be interesting in future studies to examine whether acetylation of HDAC1 plays a role in the recruitment of the MeCP complex.
Our results also showed that NuRD remodeling activity was not dependent on deacetylase activity. This agrees with previous observations showing that Mi2 is needed for GATA-1-mediated gene activation (43). It was also reported that chromatin remodeling complexes may be required for gene activation during erythroid differentiation (63, 69–71). Thus, the NuRD complex may act as corepressor and coactivator depending on the gene context. During gene activation, HDAC1 in the NuRD complex is acetylated by p300/CBP, which results in the loss of deacetylase activity. The gene-activating activity of NuRD is likely mediated by its nucleosome remodeling activity. During the repression phase, the NuRD complex can deacetylate histones and remodel chromatin into a repressive structure (Fig. 7).
Supplementary Material
Acknowledgments
We thank Dr. Gerd Blobel for the FOG-1 expression plasmid; Dr. Emery Bresnick for β-globin and GATA-2 reporter constructs; and Dr. Mitchell Weiss for G1E and G1E-ER4 cells.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL095674 (to Y. Q.); R01 HL091929, R01091929-01A1S1-the American Recovery and Reinvestment Act Administrative Supplement, and R01 HL 090589 (to S. H.); and R01 DK 83389 and R01 DK 52356 (to J. B.). This work was also supported by a grant from the Florida Bankhead Coley Research Foundation (to Y. Q.).

This article contains supplemental Figs. S1–S5.
- HDAC
- histone deacetylase
- MEL
- murine erythroleukemia
- TSA
- trichostatin A
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- DMSO
- dimethyl sulfoxide
- MeCP
- Methyl 5′-cytosine-phosphoguanine binding protein.
REFERENCES
- 1. Cress W. D., Seto E. (2000) Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184, 1–16 [DOI] [PubMed] [Google Scholar]
- 2. Cho Y., Griswold A., Campbell C., Min K. T. (2005) Individual histone deacetylases in Drosophila modulate transcription of distinct genes. Genomics 86, 606–617 [DOI] [PubMed] [Google Scholar]
- 3. Foglietti C., Filocamo G., Cundari E., De Rinaldis E., Lahm A., Cortese R., Steinkühler C. (2006) Dissecting the biological functions of Drosophila histone deacetylases by RNA interference and transcriptional profiling. J. Biol. Chem. 281, 17968–17976 [DOI] [PubMed] [Google Scholar]
- 4. Glozak M. A., Seto E. (2007) Histone deacetylases and cancer. Oncogene 26, 5420–5432 [DOI] [PubMed] [Google Scholar]
- 5. Glaser K. B., Li J., Staver M. J., Wei R. Q., Albert D. H., Davidsen S. K. (2003) Role of class I and class II histone deacetylases in carcinoma cells using siRNA. Biochem. Biophys. Res. Commun. 310, 529–536 [DOI] [PubMed] [Google Scholar]
- 6. Haberland M., Johnson A., Mokalled M. H., Montgomery R. L., Olson E. N. (2009) Genetic dissection of histone deacetylase requirement in tumor cells. Proc. Natl. Acad. Sci. U.S.A. 106, 7751–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inoue S., Mai A., Dyer M. J., Cohen G. M. (2006) Inhibition of histone deacetylase class I but not class II is critical for the sensitization of leukemic cells to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 66, 6785–6792 [DOI] [PubMed] [Google Scholar]
- 8. Lagger G., Doetzlhofer A., Schuettengruber B., Haidweger E., Simboeck E., Tischler J., Chiocca S., Suske G., Rotheneder H., Wintersberger E., Seiser C. (2003) The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol. Cell. Biol. 23, 2669–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Siegel D., Hussein M., Belani C., Robert F., Galanis E., Richon V. M., Garcia-Vargas J., Sanz-Rodriguez C., Rizvi S. (2009) Vorinostat in solid and hematologic malignancies. J. Hematol. Oncol. 2, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Y., LeRoy G., Seelig H. P., Lane W. S., Reinberg D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 [DOI] [PubMed] [Google Scholar]
- 11. Wade P. A., Jones P. L., Vermaak D., Wolffe A. P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8, 843–846 [DOI] [PubMed] [Google Scholar]
- 12. Fatemi M., Wade P. A. (2006) MBD family proteins: reading the epigenetic code. J. Cell Sci. 119, 3033–3037 [DOI] [PubMed] [Google Scholar]
- 13. Feng Q., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Zhang Y. (2002) Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol. Cell. Biol. 22, 536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng Q., Zhang Y. (2001) The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 15, 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ng H. H., Zhang Y., Hendrich B., Johnson C. A., Turner B. M., Erdjument-Bromage H., Tempst P., Reinberg D., Bird A. (1999) MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23, 58–61 [DOI] [PubMed] [Google Scholar]
- 16. Ahringer J. (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16, 351–356 [DOI] [PubMed] [Google Scholar]
- 17. Hutchins A. S., Mullen A. C., Lee H. W., Sykes K. J., High F. A., Hendrich B. D., Bird A. P., Reiner S. L. (2002) Gene silencing quantitatively controls the function of a developmental trans-activator. Mol. Cell 10, 81–91 [DOI] [PubMed] [Google Scholar]
- 18. Kehle J., Beuchle D., Treuheit S., Christen B., Kennison J. A., Bienz M., Müller J. (1998) dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282, 1897–1900 [DOI] [PubMed] [Google Scholar]
- 19. Williams C. J., Naito T., Arco P. G., Seavitt J. R., Cashman S. M., De Souza B., Qi X., Keables P., Von Andrian U. H., Georgopoulos K. (2004) The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity 20, 719–733 [DOI] [PubMed] [Google Scholar]
- 20. Brunmeir R., Lagger S., Seiser C. (2009) Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int. J. Dev. Biol. 53, 275–289 [DOI] [PubMed] [Google Scholar]
- 21. Oh Y. M., Kwon Y. E., Kim J. M., Bae S. J., Lee B. K., Yoo S. J., Chung C. H., Deshaies R. J., Seol J. H. (2009) Chfr is linked to tumour metastasis through the down-regulation of HDAC1. Nat. Cell Biol. 11, 295–302 [DOI] [PubMed] [Google Scholar]
- 22. Qiu Y., Zhao Y., Becker M., John S., Parekh B. S., Huang S., Hendarwanto A., Martinez E. D., Chen Y., Lu H., Adkins N. L., Stavreva D. A., Wiench M., Georgel P. T., Schiltz R. L., Hager G. L. (2006) HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol. Cell 22, 669–679 [DOI] [PubMed] [Google Scholar]
- 23. Qiu Y., Stavreva D. A., Luo Y., Indrawan A., Chang M., Hager G. L. (2011) Dynamic interaction of HDAC1 with a glucocorticoid receptor-regulated gene is modulated by the activity state of the promoter. J. Biol. Chem. 286, 7641–7647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo Y., Jian W., Stavreva D., Fu X., Hager G., Bungert J., Huang S., Qiu Y. (2009) Trans-regulation of histone deacetylase activities through acetylation. J. Biol. Chem. 284, 34901–34910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crispino J. D. (2005) GATA1 in normal and malignant hematopoiesis. Semin. Cell Dev. Biol. 16, 137–147 [DOI] [PubMed] [Google Scholar]
- 26. Weiss M. J., Orkin S. H. (1995) GATA transcription factors: key regulators of hematopoiesis. Exp. Hematol. 23, 99–107 [PubMed] [Google Scholar]
- 27. Migliaccio A. R., Rana R. A., Vannucchi A. M., Manzoli F. A. (2005) Role of GATA-1 in normal and neoplastic hemopoiesis. Ann. N.Y. Acad. Sci. 1044, 142–158 [DOI] [PubMed] [Google Scholar]
- 28. Fujiwara T., O'Geen H., Keles S., Blahnik K., Linnemann A. K., Kang Y. A., Choi K., Farnham P. J., Bresnick E. H. (2009) Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36, 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu M., Riva L., Xie H., Schindler Y., Moran T. B., Cheng Y., Yu D., Hardison R., Weiss M. J., Orkin S. H., Bernstein B. E., Fraenkel E., Cantor A. B. (2009) Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell 36, 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu W., Cheng Y., Keller C. A., Ernst J., Kumar S. A., Mishra T., Morrissey C., Dorman C. M., Chen K. B., Drautz D., Giardine B., Shibata Y., Song L., Pimkin M., Crawford G. E., Furey T. S., Kellis M., Miller W., Taylor J., Schuster S. C., Zhang Y., Chiaromonte F., Blobel G. A., Weiss M. J., Hardison R. C. (2011) Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 21, 1659–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evans T., Felsenfeld G. (1989) The erythroid-specific transcription factor Eryf1: a new finger protein. Cell 58, 877–885 [DOI] [PubMed] [Google Scholar]
- 32. Horak C. E., Mahajan M. C., Luscombe N. M., Gerstein M., Weissman S. M., Snyder M. (2002) GATA-1 binding sites mapped in the β-globin locus by using mammalian chIp-chip analysis. Proc. Natl. Acad. Sci. U.S.A. 99, 2924–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., Weiss M. J. (2004) Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104, 3136–3147 [DOI] [PubMed] [Google Scholar]
- 34. Blobel G. A., Nakajima T., Eckner R., Montminy M., Orkin S. H. (1998) CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. U.S.A. 95, 2061–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cantor A. B., Orkin S. H. (2002) Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21, 3368–3376 [DOI] [PubMed] [Google Scholar]
- 36. Hong W., Nakazawa M., Chen Y. Y., Kori R., Vakoc C. R., Rakowski C., Blobel G. A. (2005) FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 24, 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodriguez P., Bonte E., Krijgsveld J., Kolodziej K. E., Guyot B., Heck A. J., Vyas P., de Boer E., Grosveld F., Strouboulis J. (2005) GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24, 2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harju-Baker S., Costa F. C., Fedosyuk H., Neades R., Peterson K. R. (2008) Silencing of Aγ-globin gene expression during adult definitive erythropoiesis mediated by GATA-1-FOG-1-Mi2 complex binding at the −566 GATA site. Mol. Cell. Biol. 28, 3101–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Snow J. W., Orkin S. H. (2009) Translational isoforms of FOG1 regulate GATA1-interacting complexes. J. Biol. Chem. 284, 29310–29319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Crispino J. D., Lodish M. B., MacKay J. P., Orkin S. H. (1999) Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell 3, 219–228 [DOI] [PubMed] [Google Scholar]
- 41. Grass J. A., Boyer M. E., Pal S., Wu J., Weiss M. J., Bresnick E. H. (2003) GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. U.S.A. 100, 8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rylski M., Welch J. J., Chen Y. Y., Letting D. L., Diehl J. A., Chodosh L. A., Blobel G. A., Weiss M. J. (2003) GATA-1-mediated proliferation arrest during erythroid maturation. Mol. Cell. Biol. 23, 5031–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miccio A., Wang Y., Hong W., Gregory G. D., Wang H., Yu X., Choi J. K., Shelat S., Tong W., Poncz M., Blobel G. A. (2010) NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 29, 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Letting D. L., Chen Y. Y., Rakowski C., Reedy S., Blobel G. A. (2004) Context-dependent regulation of GATA-1 by friend of GATA-1. Proc. Natl. Acad. Sci. U.S.A. 101, 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang S., Brandt S. J. (2000) mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol. Cell. Biol. 20, 2248–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weiss M. J., Yu C., Orkin S. H. (1997) Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 17, 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gregory T., Yu C., Ma A., Orkin S. H., Blobel G. A., Weiss M. J. (1999) GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood 94, 87–96 [PubMed] [Google Scholar]
- 48. Pal S., Cantor A. B., Johnson K. D., Moran T. B., Boyer M. E., Orkin S. H., Bresnick E. H. (2004) Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc. Natl. Acad. Sci. U.S.A. 101, 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu X., Li X., Valverde K., Fu X., Noguchi C., Qiu Y., Huang S. (2009) LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 106, 10141–10146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang S., Qiu Y., Shi Y., Xu Z., Brandt S. J. (2000) P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. EMBO J. 19, 6792–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hamiche A., Xiao H. (2004) Methods for analysis of nucleosome sliding by Drosophila NURF. Methods Enzymol. 377, 353–363 [DOI] [PubMed] [Google Scholar]
- 52. Kuendgen A., Gattermann N. (2007) Valproic acid for the treatment of myeloid malignancies. Cancer 110, 943–954 [DOI] [PubMed] [Google Scholar]
- 53. Friend C., Scher W., Holland J. G., Sato T. (1971) Hemoglobin synthesis in murine virus-induced leukemic cells in vitro: stimulation of erythroid differentiation by dimethyl sulfoxide. Proc. Natl. Acad. Sci. U.S.A. 68, 378–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sawado T., Igarashi K., Groudine M. (2001) Activation of β-major globin gene transcription is associated with recruitment of NF-E2 to the β-globin LCR and gene promoter. Proc. Natl. Acad. Sci. U.S.A. 98, 10226–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tsiftsoglou A. S., Wong W. (1985) Molecular and cellular mechanisms of leukemic hemopoietic cell differentiation: an analysis of the Friend system. Anticancer Res. 5, 81–99 [PubMed] [Google Scholar]
- 56. Letting D. L., Rakowski C., Weiss M. J., Blobel G. A. (2003) Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 23, 1334–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nakatani Y., Ogryzko V. (2003) Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370, 430–444 [DOI] [PubMed] [Google Scholar]
- 58. Humphrey G. W., Wang Y., Russanova V. R., Hirai T., Qin J., Nakatani Y., Howard B. H. (2001) Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276, 6817–6824 [DOI] [PubMed] [Google Scholar]
- 59. Kobayashi M., Yamamoto M. (2007) Regulation of GATA1 gene expression. J. Biochem. 142, 1–10 [DOI] [PubMed] [Google Scholar]
- 60. Tsai S. F., Strauss E., Orkin S. H. (1991) Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 5, 919–931 [DOI] [PubMed] [Google Scholar]
- 61. Valverde-Garduno V., Guyot B., Anguita E., Hamlett I., Porcher C., Vyas P. (2004) Differences in the chromatin structure and cis-element organization of the human and mouse GATA1 loci: implications for cis-element identification. Blood 104, 3106–3116 [DOI] [PubMed] [Google Scholar]
- 62. Mahajan M. C., Karmakar S., Weissman S. M. (2007) Control of β-globin genes. J. Cell. Biochem. 102, 801–810 [DOI] [PubMed] [Google Scholar]
- 63. Kim S. I., Bresnick E. H. (2007) Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene 26, 6777–6794 [DOI] [PubMed] [Google Scholar]
- 64. Grass J. A., Jing H., Kim S. I., Martowicz M. L., Pal S., Blobel G. A., Bresnick E. H. (2006) Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol. Cell. Biol. 26, 7056–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Martowicz M. L., Grass J. A., Boyer M. E., Guend H., Bresnick E. H. (2005) Dynamic GATA factor interplay at a multicomponent regulatory region of the GATA-2 locus. J. Biol. Chem. 280, 1724–1732 [DOI] [PubMed] [Google Scholar]
- 66. Lee H. Y., Johnson K. D., Fujiwara T., Boyer M. E., Kim S. I., Bresnick E. H. (2009) Controlling hematopoiesis through sumoylation-dependent regulation of a GATA factor. Mol. Cell 36, 984–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang Z., Zang C., Cui K., Schones D. E., Barski A., Peng W., Zhao K. (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kransdorf E. P., Wang S. Z., Zhu S. Z., Langston T. B., Rupon J. W., Ginder G. D. (2006) MBD2 is a critical component of a methyl cytosine-binding protein complex isolated from primary erythroid cells. Blood 108, 2836–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim S. I., Bultman S. J., Jing H., Blobel G. A., Bresnick E. H. (2007) Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol. Cell. Biol. 27, 4551–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim S. I., Bultman S. J., Kiefer C. M., Dean A., Bresnick E. H. (2009) BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl. Acad. Sci. U.S.A. 106, 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mahajan M. C., Narlikar G. J., Boyapaty G., Kingston R. E., Weissman S. M. (2005) Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the β-globin locus control region. Proc. Natl. Acad. Sci. U.S.A. 102, 15012–15017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.