Background: Regulation of transcriptional activity of heat shock factor-1 (HSF1) is widely thought to be the main point of control for heat shock protein (Hsp) expression.
Results: Glutamine increases Hsf1 gene transcription in a C/EBPβ-dependent manner and up-regulates HSF1 activity.
Conclusion: Glutamine is an activator for both HSF1 expression and transactivation.
Significance: Glutamine-induced HSF1 expression provides a novel mechanistic frame for HSF1-Hsp axis regulation.
Keywords: C/EBP Transcription Factor, Glutamine, Heat Shock Protein, Intestinal Epithelium, Transcription, Heat Shock Factor-1
Abstract
Heat shock transcription factor-1 (HSF1) is the master regulator for cytoprotective heat shock protein (Hsp) expression. It is widely thought that HSF1 expression is non-inducible, and thus the key control point of Hsp expression is regulation of the transactivation activity of HSF1. How HSF1 expression is regulated remains unknown. Herein we demonstrate that glutamine (Gln), a preferred fuel substrate for the gut, enhanced Hsp expression both in rat colonic epithelium in vivo and in cultured non-transformed young adult mouse colonic epithelial cells. This was associated with up-regulation of the transactivation activity of HSF1 via increased HSF1 trimerization, nuclear localization, DNA binding, and relative abundance of activating phosphorylation at Ser-230 of HSF1. More intriguingly, Gln enhanced HSF1 protein and mRNA expression and Hsf1 gene promoter activity. Within the −281/−200 region of the Hsf1 promoter, deletion of the putative CCAAT enhancer-binding protein (C/EBP) binding site abolished the HSF1 response to Gln. C/EBPβ was further shown to bind to this 82-bp sequence both in vitro and in vivo. Gln availability strikingly altered the ratio of C/EBPβ inhibitory and active isoforms, i.e. liver-enriched inhibitory protein and liver-enriched activating protein. Liver-enriched inhibitory protein and liver-enriched activating protein were further shown to be an independent repressor and activator, respectively, for Hsf1 gene transcription, and the relative abundance of these two C/EBPβ isoforms was demonstrated to determine Hsf1 transcription. We show for the first time that Gln not only enhances the transactivation of HSF1 but also induces Hsf1 expression by activating its transcription in a C/EBPβ-dependent manner.
Introduction
During the course of evolution, organisms have developed inherent stress-responsive molecular machinery to dampen proteotoxic stresses they encounter in the environment. This is primarily accomplished by heat shock proteins (Hsp),2 a family of highly conserved proteins. A prominent feature of stressed cells is the increased synthesis of cytoprotective Hsps that aid in the refolding of misfolded peptides and restrain protein aggregation (1). The up-regulation of Hsp gene expression following stress primarily occurs on the transcriptional level (2, 3). Heat shock transcription factor-1 (HSF1) is the master regulator for transcriptional activation of many key Hsp genes, including Hsp70 and Hsp25 (2). Upon activation, HSF1 binds to conserved regulatory sequences known as heat shock elements where it activates Hsp gene expression in response to stress (4–6). It has long been believed that HSF1 is constitutively expressed in most tissues and cell types and that HSF1-regulated increases in Hsp expression occur primarily via posttranslational mechanisms (7). As a result, how HSF1 proceeds through a multistep pathway involving a monomer-to-trimer transition, nuclear accumulation, acquisition of DNA binding ability, and extensive posttranslational modifications has been a primary focus of a large body of research directed at understanding the regulation of Hsp expression (2, 3, 7–9).
However, contrary to this notion, new evidence has recently emerged suggesting that HSF1 expression may not remain constant and can be induced (10–14). Mechanistically, Yang et al. (15) demonstrated that riluzole, a United States Food and Drug Administration-approved drug for the treatment of amyotrophic lateral sclerosis, can increase cellular HSF1 latent monomer content by blunting its turnover. This increased HSF1 content led to a more robust heat shock response during stress (15). Hyperthermia (11, 14), laser therapy (13), and hemorrhagic shock (10) have been reported to up-regulate HSF1 mRNA expression in multiple tissues and cell types, suggesting that regulation of HSF1 expression may occur at a pretranslational level(s). Apart from these artificial chemical and stress-related inducers with inherent toxicities, a physiologically relevant regulation pathway for HSF1 transactivation activity and expression via a substrate normally present in the cell has yet to be identified.
Glutamine (Gln), one of the most functionally versatile amino acids, is involved in a diverse range of physiological processes (16). Gln is released in significant quantities from skeletal muscle stores following stress and injury (16) and thus appears to be a key substrate for cells and tissues following stress and may serve as a signal for activation of the cellular stress response. As the preferred respiratory fuel for the gut epithelium, particularly following stress, Gln has long been studied as a promising agent to preserve intestinal functional/structural integrity and promote intestinal recovery during injury or stress (16, 17). Accumulating evidence has shown that Gln also appears to regulate numerous genes involved in the cellular response to stress (16). Recent data from our group indicate that Gln administration in pharmacologic and clinically relevant doses can safely enhance the key protective Hsp expression in experimental models of inflammatory and infectious injury (18–20). Specifically, Gln-mediated induction of Hsps appears to be vital for Gln-mediated gut protection during various stress and injurious conditions (21–23). Currently, Gln is the only physiologically relevant substrate known to enhance the human heat shock response (24). However, the mechanism(s) by which Gln enhances Hsp expression remains elusive.
Given the essential role of HSF1 in activating Hsp gene transcription and recent evidence indicating that HSF1 expression may be inducible, we hypothesize that Gln up-regulates HSF1 transactivation activity and/or its expression, which ultimately leads to enhanced Hsp expression. Here we report for the first time that Gln not only enhances HSF1 activation but also intriguingly up-regulates actual Hsf1 gene expression per se. Furthermore, we demonstrate that this up-regulation of Hsf1 gene expression occurs at the transcriptional level, and the amino acid-responsive CCAAT enhancer-binding protein (C/EBP) β (25–28) is essential for this Gln-mediated HSF1 response.
EXPERIMENTAL PROCEDURES
Animals
Animal use was approved by the Institutional Animal Care and Use Committee of the University of Colorado. As detailed previously (29), colitis was induced in male Sprague-Dawley rats (250–300 g; Charles River) drinking ad libitum distilled water containing 5% DSS (w/v; molecular weight, 36,000–50,000; MP Biomedicals) for 7 consecutive days. The presence of colitis after the 7-day DSS induction was confirmed by disease activity, histopathology, and markers of mucosal inflammation as described (29). Gln treatment was started immediately before DSS treatment initiation and continued throughout the whole 7-day DSS colitis induction. Gln (0.75 g/(kg·day)) or sham (isovolemic sterile water) was administered to animals by oral gavage. Another set of healthy animals not exposed to DSS served as healthy controls and were randomized into two groups (n = 13/group for Gln or sham treatment). After the 7-day Gln or sham treatment, rats were killed, and the mucosa was gently scraped off to determine Hsp and HSF1 content.
Cell Culture
Conditionally immortalized young adult mouse colonic epithelial cells (YAMCs) were provided by Dr. Robert Whitehead (Vanderbilt University). Rat small intestinal epithelial cell line IEC-6 was obtained from the American Type Culture Collection. Mouse embryonic fibroblasts (MEFs) from wild-type and c/ebpβ−/− mice were a courtesy from Dr. Peter Johnson (National Institutes of Health, Bethesda, MD) (30). YAMCs are a conditionally immortalized mouse colonic intestinal epithelial cell line derived from the Immortomouse, a transgenic animal containing a temperature-sensitive SV40 large T antigen (tsA58) under the control of an IFN-γ-dependent promoter (31). YAMCs were maintained under permissive conditions (33 °C) in RPMI 1640 medium supplemented with 5 units/ml murine IFN-γ (Sigma), 2 mm Gln, 5% fetal bovine serum (FBS; v/v), 1% ITS+ Premix (v/v; BD Biosciences), 50 units/ml penicillin, and 50 μg/ml streptomycin. Under nonpermissive conditions at 37 °C in the absence of IFN-γ, these cells undergo differentiation and develop mature epithelial cell functions and properties. Before being exposed to Gln at varied concentrations, all cells were cultured in IFN-γ-free and antibiotic-free medium under non-permissive conditions of 37 °C for 16 h to induce colonocyte differentiation. During this time, SV40 large T antigen is no longer produced, and any remaining protein misfolds because of the temperature-sensitive mutation (tsA). After the initial 16-h differentiation-induction period, cells were exposed to serum-free, antibiotic-free, and Gln-free RPMI 1640 medium containing 0, 0.5, or 2 mm Gln. Cells were returned to the incubator at 37 °C for 6 h before harvest. To investigate how Gln affects HSF1 mRNA expression kinetics, cells were harvested after 45-, 72-, 90-, and 135-min incubation in Gln at varied concentrations. For heat shock experiments, immediately after being switched to serum and antibiotic-free medium with Gln at varied concentrations, cells were subjected to heat shock treatment by immersion in a water bath at 43 °C for 30 min. Cells were harvested immediately after heat shock or allowed to recover at 37 °C for 1–6 h as needed by specific experiments. IEC-6 cells and MEFs were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 2 mm Gln, 50 mg/liter penicillin, and 50 mg/liter streptomycin. IEC-6 cells were cultured in DMEM supplemented with 5% FBS and 100 units of insulin. MEFs were cultured in DMEM supplemented with 10% FBS. To investigate the effect of Gln on HSF1 expression in IEC-6 and MEF cells, the medium was replaced by serum-free, antibiotic-free, and Gln-free DMEM containing 0, 0.5, or 2 mm Gln. Cells were returned to the incubator at 37 °C for 6 h before harvest. Each experiment repeated four times.
RNA Extraction and Real Time RT-PCR
Total RNA was extracted from YAMCs using an RNeasy kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad) and random hexanucleotide primer. The sense and antisense primers for mouse HSF1 were obtained from Qiagen (QT00172508). Amplicon expression in each sample was normalized to β-actin. After normalization, expression of HSF1 was quantified using a ΔΔCt calculation.
Plasmid Constructs
Mouse Hsp70 promoter (−1041/−1) and 5′ deletion fragments of the mouse Hsf1 promoter (see Fig. 5A) were generated by PCR from genomic mouse DNA using primers and antisense primers containing restriction sites at their 5′-ends, respectively (supplemental Table 1). MluI sites were added to the 5′ terminus of all the forward primers, and a HindIII site and a BlgII site were added to the reverse primer for Hsp70 and Hsf1 promoter constructs, respectively. Amplified fragments were subsequently cloned into the pGL3-Basic reporter vector (Promega) using the corresponding restriction sites. For site-directed mutagenesis by internal deletions or base substitutions (see Fig. 5, B and C), fragments of the 5′ part of the Hsf1 promoter were generated by using MluI-HSF1F281 primer and respective antisense primers listed in supplemental Table 1, and fragments of the 3′ part were generated by using BlgII-HSF1R antisense primer and respective primers listed in supplemental Table 1. The liver-enriched activating protein (LAP) expression plasmid pcDNA-LAP was a gift from Dr. Peter Johnson (32) (NCI-Frederick, National Institutes of Health). The liver-enriched inhibitory protein (LIP) expression plasmid pcDNA-LIP was a generous gift from Dr. J. Friedman (33) (University of Colorado). All the primers used are listed in supplemental Table 1.
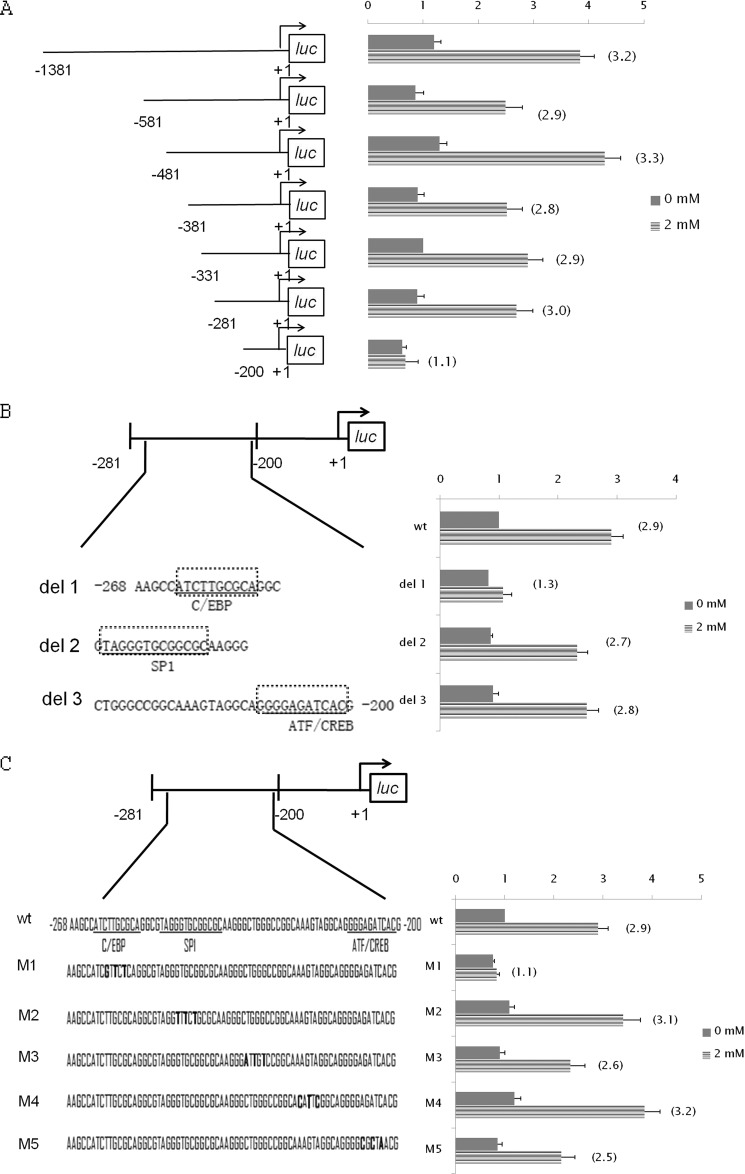
FIGURE 5.
Identification of key cis-element(s) of Hsf1 promoter responsible for Gln-induced activation of Hsf1 transcription. A, effect of Gln on 5′ deletions of the Hsf1 promoter. YAMCs were transiently transfected with luc constructs containing progressive 5′ deletions of the Hsf1 promoter. B, response of −281/−200 Hsf1 promoter bearing various internal deletions of putative transcription factor binding sites to Gln. YAMCs were transiently transfected with luc constructs containing internal deletions of the Hsf1 promoter as indicated by boxes with dotted lines. The putative binding sites for C/EBP, SP1, and activating transcription factor (ATF)/cAMP-response element-binding protein (CREB) sites are underlined. C, response of −281/−200 Hsf1 promoter bearing various base substitution mutations to Gln. YAMCs were transiently transfected with luc constructs containing various base substitution mutations of the −281/−200 region. The bold letters indicate the substituted bases. For both the internal deletion and base substitution analyses, 24 h after transfection, cells were exposed to Gln at 0 or 2 mm for 6 h before harvest for preparation of cell extracts and determination of luc activity. The -fold induction, defined as the ratio of the relative luc activity of cells treated with 2 mm Gln to that of cells treated with 0 mm Gln, is indicated in parentheses to the right of the bars. For A–C, error bars represent S.E.
Western Blotting
M-PER mammalian protein extraction reagent (Pierce) with protease and phosphatase inhibitors (Roche Applied Science) added was used to extract the total protein from rat mucosal tissue and to prepare whole cell extracts of YAMCs and IEC-6 cells. To investigate the intracellular localization and phosphorylation of HSF1, nuclear and cytoplasmic extracts were prepared from YAMCs with the Pierce Nuclear/Cytoplasmic Fractionation kit (Pierce). Lysates were quantified for total protein using the BCA protein assay (Pierce), separated by SDS-PAGE, and transferred to polyvinylidene difluoride (PVDF) membranes. Selected samples were treated with 80 units of calf intestine alkaline phosphatase (Sigma-Aldrich) at 37 °C for 60 min before electrophoresis. The membrane was incubated with a primary antibody against Hsp25 (Stressgen), Hsp70 (Stressgen), β-actin (Santa Cruz Biotechnology), HSF1 (Cell Signaling Technology), Ser(P)-307-HSF1 (Santa Cruz Biotechnology), Ser(P)-230-HSF1 (Santa Cruz Biotechnology), anti-α-tubulin (Calbiochem) as a cytoplasmic marker, or anti-lamin B1 (Santa Cruz Biotechnology) as a nuclear marker. For LIP immunoblotting, an antibody detecting all C/EBPβ isoforms (Thermo catalog number MAI-827) was used, whereas for LAP immunoblotting, an antibody (Cell Signaling Technology catalog number 3087) specifically detecting the p38 and p35 LAPs but not LIP was used. Blots were then stained with the appropriate horseradish peroxidase-conjugated secondary antibody. Reactive bands were detected with enhanced chemiluminescence reagents (Pierce), and exposed with a chemiluminescent darkroom system (UVP). Quantification of images was done by scanning densitometry using LabWorks 4.0 image acquisition and analysis software (UVP). β-Actin was used as a loading control.
Native PAGE
Blue native gel electrophoresis was performed to visualize the oligomerization of HSF1. Whole cell extracts prepared by M-PER extraction reagent were separated by native PAGE using the Novex Bis-Tris gel system according to the manufacturer's specifications (Invitrogen). Native gels were then transferred to PVDF membrane (Millipore), and HSF1 protein was visualized by Western blotting.
Electrophoretic Mobility Shift Assay (EMSA) (Gel Shift) and Supershift
Nuclear extracts were prepared from YAMCs with the Pierce Nuclear Extraction kit (Pierce) and stored at 80 °C until use. Two sets of 3′-end biotin-labeled synthetic complementary oligonucleotides corresponding to consensus binding sequence of C/EBP and HSF binding site (heat shock element (HSE)) were obtained from Signosis. A complementary oligonucleotide covering the −80/−51 region of the AHSG promoter, which has been shown to harbor a proven binding site for C/EBP (34), was obtained by PCR. EMSA was performed using the LightShift Chemiluminescent EMSA kit (Pierce) according to the manufacturer's protocol. Nuclear extracts (5–10 μg of protein) were incubated with 1 μg of poly[d(I-C)], 2 μl of binding buffer (100 mm Tris-HCl, 500 mm KCl, 10 mm dithiothreitol (DTT) (pH 7.5)), and biotin end-labeled oligonucleotide probe in a total volume of 20 μl for 15 min at room temperature. After separation by 5% non-denaturing polyacrylamide gel electrophoresis, the binding reaction mixtures were transferred onto a nylon membrane (Pierce), and the membrane was UV-cross-linked. The biotin-labeled DNA was probed with streptavidin-HRP conjugate for chemiluminescence detection.
Supershift experiments were carried out by incubating nuclear extracts with 1 μl of anti-HSF1 (Cell Signaling Technology), anti-C/EBPβ (Thermo) antibody, or preimmune serum. For competition experiments, 200-fold excess unlabeled HSE competitor oligonucleotides or 100-fold excess unlabeled −281/−200 Hsf1 promoter sequence obtained by PCR was used.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed using the ChIP Assay kit (catalog number 17-295, Upstate/Millipore, Temecula, CA) according to the manufacturer's protocol with modifications. Briefly, the cells were treated with 1% formaldehyde for 5 min at 37 °C, and the cross-linking was stopped by 0.125 m glycine. After three washes with PBS buffer, the cells were sonicated in SDS Lysis Buffer (catalog number 20-163, Upstate/Millipore) on ice to obtain ∼500-bp DNA fragments. The samples were precleared with single-stranded DNA/protein A beads and incubated with 10 μg of rabbit anti-HSF1 (Cell Signaling Technology) or anti-C/EBPβ (Santa Cruz Biotechnology) or 5 μl of rabbit preimmune serum at 4 °C overnight. The samples were eluted, decross-linked at 65 °C overnight, and then treated with 0.1 mg/ml proteinase K. DNAs were purified with a QIAquick PCR product purification kit (catalog number 28104, Qiagen). Enrichment of the DNA was analyzed by PCR, and products were run on an agarose gel or measured by real time PCR as described above using primers listed in supplemental Table 1. A negative control PCR for each immunoprecipitation using IGX1A negative control primer targeting open reading frame-free intergenic DNA (Qiagen) was performed. The relative level of C/EBPβ binding was quantified by correcting for the amount of input DNA and control antibody (preimmune IgG) DNA.
Plasmid Transfection and Promoter Activity Assays
For transfections, cells were seeded at low density (25–30% confluence). After 24 h of culture, the medium was replaced under permissive conditions but with no antibiotics, and cells were grown to 70% confluence. Transfections were then performed using the Lipofectamine 2000 (Invitrogen) transfection reagent using 0.5 μg of DNA/culture and following the manufacturer's recommendations. 24 h after transfection, cells were shifted to non-permissive conditions for 16 h before use in an experiment. In certain experiments as specified, pcDNA-LAP or pcDNA-LIP plasmids and the Hsf1 promoter-luciferase (p1381/luc) construct were co-transfected into YAMCs or c/ebpβ−/− MEFs using the Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's protocol. 0.125 μg of pRL-SV40-Renilla (Promega) was co-transfected for correction of transfection efficiency. Cells were lysed in 120 μl of 1× Passive Lysis Buffer, and Dual-Luciferase reporter assays were performed according to the manufacturer's directions (Promega). An amount of 20 μl of lysate was used for measurement in a luminometer (Monolight 3010, BD Biosciences). Each transfection was repeated three times. The results were reported as relative luciferase activity, which represents a ratio of luciferase firefly activity/Renilla activity. The data represent the mean ± S.E. of three different experiments repeated in triplicate.
Statistical Analysis
Data are expressed as mean ± S.E. Treatment differences were tested using one-way analysis of variance followed by Tukey's test or Bonferroni posttest (SPSS 12.0, SPSS Inc., Chicago, IL) as specified. Otherwise for two-group mean comparison, unpaired two-tailed Student's t test for independent samples was used. A probability of 0.05 was accepted as being statistically significant.
RESULTS
Gln Enhances Hsp Gene Expression and Hsp70 Promoter-driven Reporter Gene Expression
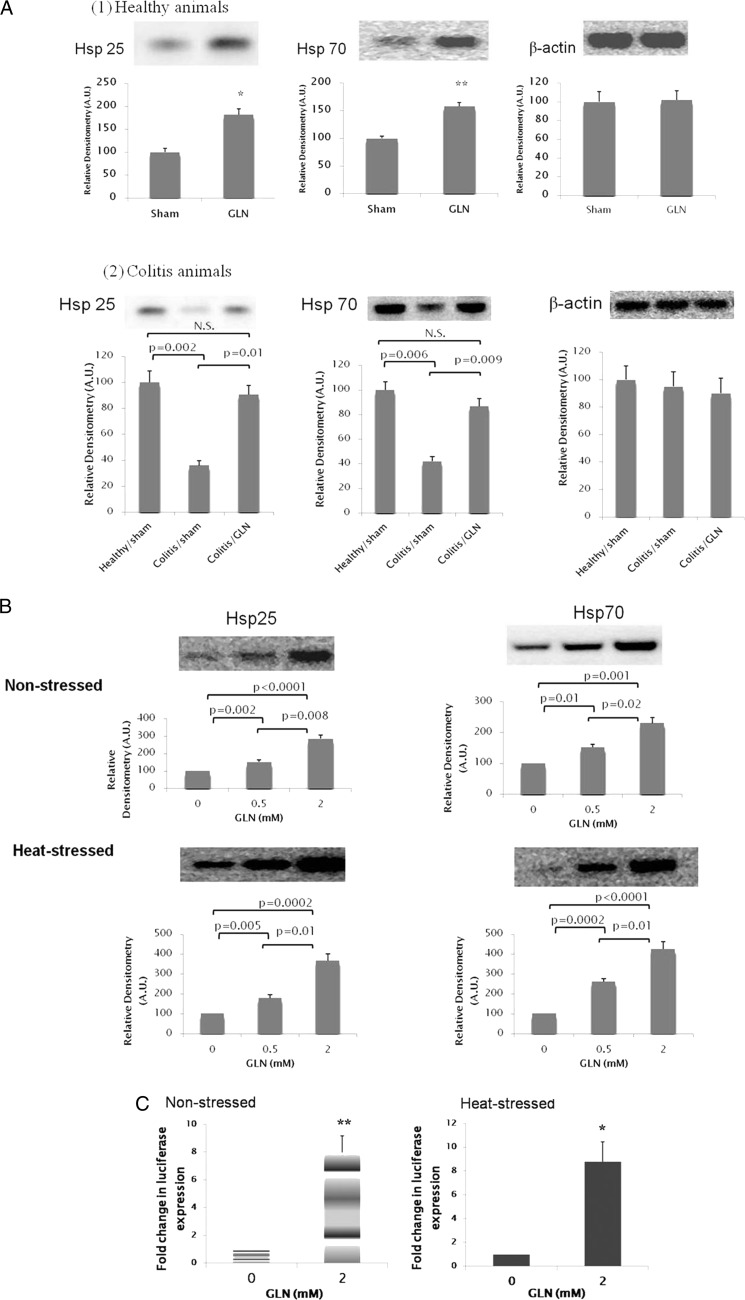
As shown in Fig. 1, bolus Gln treatment increased basal expression of Hsp25 and Hsp70 in the colonic epithelium of healthy animals (Fig. 1A, panel 1). In animals developing DSS-induced colitis (as confirmed with clinical assessment, histological grading, and markers of inflammation and apoptosis as we reported previously (29)), there was a significant decrease in Hsp25 and Hsp70 expression in colonic epithelium as compared with healthy animals. This decreased Hsp expression was corrected with Gln administration (Fig. 1A, panel 2). In non-stressed YAMCs (non-transformed murine colonic epithelial cells) treated with Gln at 0, 0.5, and 2 mm, basal expression of Hsp25 and Hsp70 increased with Gln supplementation in a dose-dependent manner. Gln administration also led to a dose-dependent increase of Hsp expression in YAMCs following heat shock at 43 °C for 30 min (Fig. 1B).
FIGURE 1.
Gln up-regulates colonic Hsp25 and Hsp70 expression. A, Gln treatment up-regulates colonic mucosal Hsp25 and Hsp70 protein expression in healthy rats (panel 1) and rats with colitis (panel 2). Rats received DSS + Gln (0.75 g/(kg·day)) or DSS + sham (isovolemic sterile water) treatment for 7 consecutive days (Days 0–7), and accumulation of stress-inducible Hsp (Hsp25 and Hsp70) in colonic mucosa was examined by Western blot at Day 7. Healthy rats not exposed to DSS treatment received Gln or sham treatment only (n = 13/treatment group). For panel 1, *, p = 0.01; **, p = 0.008 compared with sham by Student's t test; for panel 2, Hsp abundance in mucosal tissue of healthy animals receiving sham treatment was used as a reference. Results are expressed as relative densitometry as detailed under “Experimental Procedures.” Means are compared via post hoc Tukey's test. N.S., not significant. B, Gln enhances total cellular Hsp25 and Hsp70 protein expression in YAMCs. Non-stressed YAMCs (non-transformed murine colonic epithelial cells) were treated with Gln at various concentrations for 6 h before harvest. In the heat-stressed condition, cells were heat-shocked (43 °C for 30 min) immediately after being shifted to media with varied Gln concentrations followed by a 2-h recovery. The image shown is representative of four individual experiments (n = 4). Results are expressed as relative densitometry as detailed under “Experimental Procedures” (n = 4). Means are compared via post hoc Tukey's test. C, Gln enhanced Hsp70 promoter-driven luciferase gene expression. YAMCs were transfected with the distal 1035-bp mouse Hsp70 promoter fused into a luciferase reporter plasmid. In non-stressed condition, cells were exposed to Gln at varied concentrations for 6 h at 37 °C. In the heat-stressed condition, cells were heat-shocked (43 °C for 30 min) immediately after being shifted to media with varied Gln concentrations and then returned to 37 °C for 6 h before harvest. Gln increased Hsp70 promoter-driven luciferase expression in both non-stressed (left panel) and heat-stressed (right panel) cells. Data are expressed as -fold change in luciferase expression over 0 mm Gln in the non-stressed (left) or heat-stressed (right) condition (n = 4). *, p = 0.006; **, p = 0.002 by Student's t test. For A–D, error bars represent S.E. A.U., arbitrary units.
To investigate whether Gln can up-regulate Hsp gene promoter activity and thus lead to transcriptional activation of the Hsp genes, we studied the Hsp70 promoter-driven reporter gene expression pattern in cells exposed to Gln. Incubation with Gln for 6 h led to an 8.0-fold (p < 0.01) increase in basal expression of Hsp70-luciferase expression. In heat-stressed cells, Gln at 2 mm resulted in an 8.8-fold increase (p < 0.01) in Hsp70-luciferase expression as compared with cells treated with 0 mm Gln (Fig. 1C).
Gln Enhances HSF1 Spontaneous and Heat-induced Trimerization
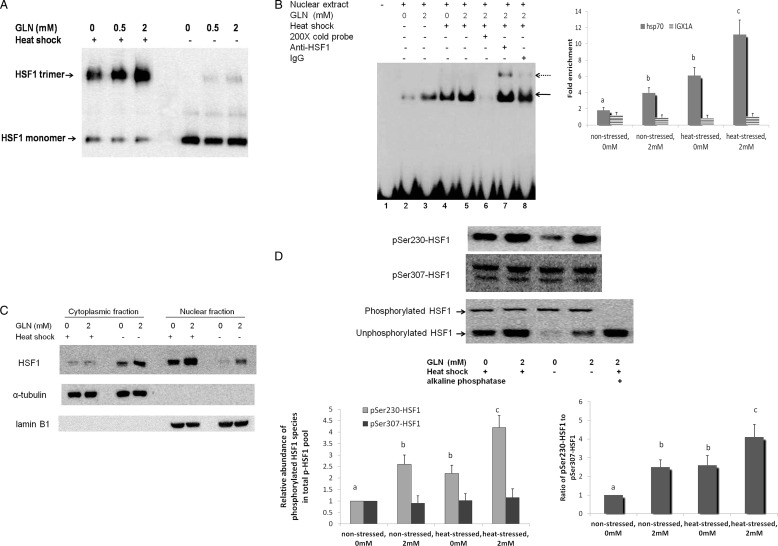
As we showed that Gln can transcriptionally activate Hsp gene expression, we further examined whether Gln affected one or more of the key events essential for the transactivation activity of HSF1. Trimerization of latent monomeric HSF1 is the initial step of HSF1 activation, which enables DNA binding. Non-reducing native polyacrylamide electrophoresis was used to visualize oligomerized HSF1. In non-stressed cells, Gln at 0.5 and 2 mm resulted in a significant spontaneous trimerization of HSF1. When cells were subjected to heat shock, HSF1 began to transition from its monomeric form to its trimeric form even in the absence of Gln. Gln further enhanced HSF1 trimerization after heat shock injury in a dose-dependent manner (Fig. 2A).
FIGURE 2.
Gln availability affects the key events essential for acquisition of HSF1 for its transactivation activity. A, Gln enhances HSF1 spontaneous and heat-induced trimerization. HSF1 trimerization in YAMCs was examined by non-denaturing gel electrophoresis in the non-stressed state or 1 h after heat shock (43 °C for 30 min). Gln increases the accumulation of activated trimeric form of HSF1 in both conditions. B, Gln enhances binding of HSF1 to HSE. Nuclear extracts from YAMCs were obtained in the non-stressed state or immediately after heat shock treatment (43 °C for 30 min), and binding of HSF1 in YAMC nuclear extracts to HSE was examined by EMSA (left panel). Biotin-labeled HSE was used as the probe. Non-biotin-labeled HSE at a 200-fold molar excess relative to the probe was used as the competitor. The HSF1-HSE complex is indicated by a solid arrow. Supershifts are indicated by a dotted arrow. HSF1 binding to HSE in vivo was further determined by quantitative ChIP assay (right panel). Primers for IGX1A were used as a negative control in all immunoprecipitations (n = 3). The experimental conditions that do not share the same letter are significantly different from each other for Hsp70 promoter DNA enrichment (0 or 2 mm) as determined by post hoc Tukey's test (p < 0.05). DNA enrichment for IGX1A is not significantly different between the various conditions. C, Gln enhances HSF1 nuclear localization. HSF1 abundance was analyzed via Western blot in cytoplasmic and nuclear fractions of YAMCs obtained in the non-stressed state or 1 h after heat shock treatment (43 °C for 30 min). α-Tubulin and lamin B1 are markers for the purity of cytoplasmic and nuclear fractions, respectively. D, Gln increased the relative abundance of activating phosphorylation of nuclear HSF1 at Ser-230 but did not affect that of the repressing phosphorylation of nuclear HSF1 at Ser-307. Nuclear extracts of YAMCs obtained in the non-stressed state or 1 h after heat shock treatment (43 °C for 30 min) were immunoblotted with anti-Ser(P)-230-HSF1, anti-Ser(P)-307-HSF1, and anti-HSF1 antibodies. The higher molecular weight band in the lower blot represents global nuclear phosphorylated HSF1 (p-HSF1), which disappeared with addition of alkaline phosphatase. The experimental conditions that do not share the same letter are significantly different from each other (post hoc Tukey's test, p < 0.05). The relative abundance of Ser(P)-307-HSF1 is not significantly different between the various conditions. For B and D, error bars represent S.E.
Gln Enhances Binding of HSF1 to Heat Shock Element
Trimerized HSF1 is capable of binding to the HSE of Hsp gene promoters. The HSE is composed of multiple inverted repeats of the pentanucleotide motif NGAAN. We examined the binding of HSF1 to HSE in nuclear extracts using EMSA (Fig. 2B, left panel). The specificity of HSF1-HSE binding was confirmed with the disappearance of the HSE-protein complex when an excess of unlabeled HSE probes were competing with the labeled probes for HSF1 binding (lane 6). The HSF1 binding to HSE was further identified with a supershift assay when the extracts were preincubated with HSF1 antibody (lane 7), whereas nonspecific IgG was not able to supershift the complex (lane 8). Heat shock treatment led to initial HSF1-HSE complex formation even in the absence of Gln (lane 4 versus lane 2). However, consistent with our finding of Gln-mediated increases in HSF1 trimerization in non-stressed cells, our data show a small, yet significant increase in HSF1-HSE binding following treatment with 2 mm Gln versus 0 mm Gln. In heat-stressed cells, 2 mm Gln further potentiated heat-induced HSF1-HSE binding. Using a quantitative ChIP assay (Fig. 2B, right panel), we further confirmed that Gln enhanced the binding of HSF1 to HSE under both unstressed and heat-stressed conditions in vivo.
Gln Enhances HSF1 Nuclear Localization and Relative Abundance of Activating Phosphorylation of HSF1 at Ser-230
Another key feature associated with HSF1 activation is increased nuclear localization (35, 36). The abundance of HSF1 was assessed in isolated nuclear and cytoplasmic fractions, respectively. Gln increased HSF1 nuclear abundance in non-stressed cells. Heat stress resulted in initial nuclear translocation of HSF1, and Gln (2 mm) further increased heat-induced HSF1 nuclear translocation (Fig. 2C).
The conversion of latent HSF1 monomers to their trimerized form is required for HSF1-DNA binding to occur. However, DNA binding of HSF1 is not necessarily sufficient for transcriptional activation of Hsp gene expression. Key posttranslational modifications are also required for the transactivation activity of HSF1 (37–39). Phosphorylation of HSF1 at specific serine residues may serve a stimulatory or inhibitory role in the transactivation activity of HSF1. Of these phosphorylation events, phosphorylation of Ser-230 is essential for transactivation of HSF1 and enhanced Hsp expression (40), whereas Ser-307 phosphorylation has been shown to serve as a key repression mechanism for the activity of HSF1 (37, 41, 42). The molar ratio between the activating and repressing sites is suggested to be a key determinant of the magnitude of the transcriptional activity of HSF1 (40). Hence, we examined the relative abundance of Ser-203 and Ser-303 phosphorylations of HSF1 in total HSF1 phosphopeptides. As shown in Fig. 2D, Gln increased the relative abundance of activating phosphorylation of nuclear HSF1 at Ser-230 in the total phosphorylated HSF1 pool but did not affect the relative abundance of repressing phosphorylation of nuclear HSF1 at Ser-307, which shifted the ratio of these two phosphorylated species toward the activating phosphorylation at Ser-230.
Gln Enhances HSF1 Expression as a Transcription Factor Per Se
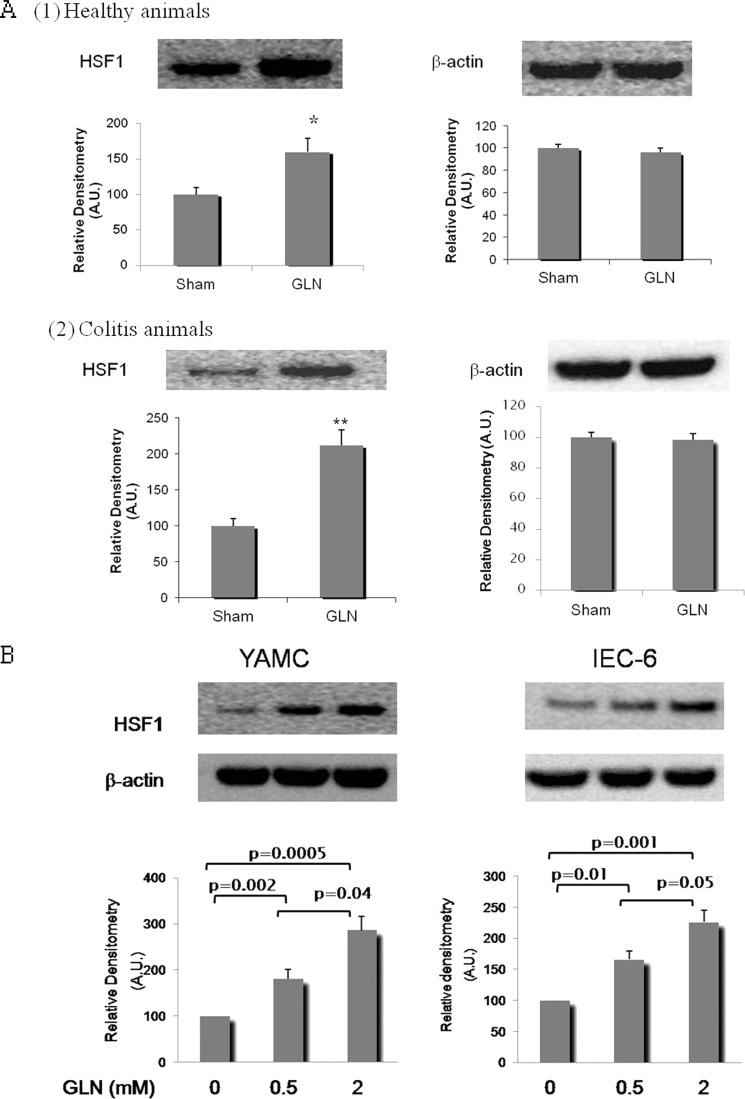
As shown in Fig. 3A, administration of Gln at 0.75 g/(kg·day), a non-toxic Gln dose commonly used to protect against experimental organ injury (18, 20, 29), increased HSF1 protein abundance in colonic epithelium by 61% in healthy animals and 112% in animals with colitis. Consistent with the in vivo findings, there was a dose-dependent increase of total YAMC cellular HSF1 abundance when cells were incubated with Gln for 6 h at 37 °C (Fig. 3B). To confirm that this effect was not limited to one cell line, this effect of Gln on HSF1 expression was also demonstrated in a rat small intestinal epithelial cell line (IEC-6 cells) (Fig. 3B).
FIGURE 3.
Gln enhances HSF1 expression per se. A, Gln treatment up-regulates colonic mucosal HSF1 protein expression in healthy rats (panel 1) and rats with colitis (panel 2). Rats received DSS + Gln (0.75 g/(kg·day)) or DSS + sham (isovolemic sterile water) treatment for 7 consecutive days (Days 0–7), and accumulation of HSF1 in colonic mucosa was examined by Western blot at Day 7. Healthy rats not exposed to DSS treatment received Gln or sham treatment only (n = 13/treatment group). Results are expressed as relative densitometry as detailed under “Experimental Procedures.” *, p = 0.02; **, p = 0.009 compared with sham by Student's t test. B, Gln up-regulates HSF1 protein expression in YAMCs and IEC-6 cells. YAMCs or IEC-6 cells were treated with Gln at various concentrations for 6 h before harvest. The image shown is representative of four individual experiments. Results are expressed as relative densitometry as detailed under “Experimental Procedures” (n = 4). Means are compared via post hoc Tukey's test. For A and B, error bars represent S.E. A.U., arbitrary units.
Gln Transcriptionally Activates Hsf1 Gene Expression
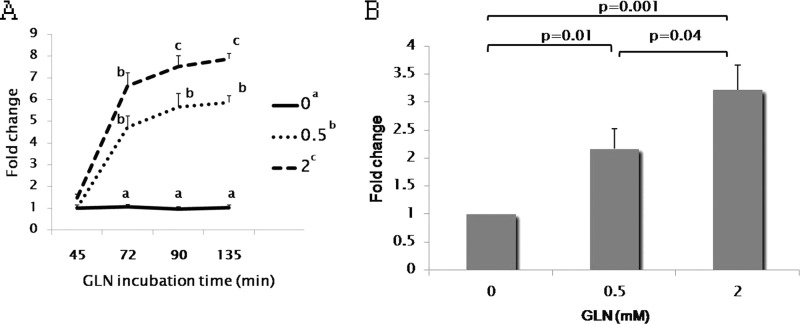
Utilizing RT-PCR, we demonstrated that Gln incubation at 37 °C (non-stress conditions) increases the HSF1 mRNA level in YAMCs. This Gln-mediated increase in HSF1 mRNA expression occurred as early as 72 min after the initiation of Gln treatment (Fig. 4A). This effect on HSF1 mRNA expression kinetics suggests that modulation of HSF1 expression by Gln occurs at a transcriptional level.
FIGURE 4.
Gln transcriptionally activates HSF1 gene expression. A, Gln enhanced HSF1 mRNA expression in YAMCs. Cells were harvested for total RNA at 45, 72, 90, and 135 min after exposure to Gln, and HSF1 mRNA abundance was determined by real time RT-PCR. Results are expressed as -fold changes over the HSF1 mRNA level at 0 mm Gln after 45-min incubation. In the figure key, Gln treatments at varied concentrations that do not share a common letter are different (0 versus 0.5 mm, p < 0.0001; 0.5 versus 2 mm, p = 0.005; 0 versus 2 mm, p < 0.0001; post hoc Tukey's test). The mean HSF1 mRNA levels were compared at the indicated time points between various Gln concentrations, and the means at a certain time point that do not share a common letter are significantly different (p < 0.05, Bonferroni posttests). B, Gln enhances Hsf1 gene promoter activity. A 1381-bp sequence upstream of the murine Hsf1 gene translation start site was fused to a luciferase reporter plasmid, and the construct was transiently transfected into YAMCs. Sixteen hours after transfection, cells were exposed to Gln at various concentrations for 6 h before harvest for preparation of cell extracts and determination of luc activity. Data are expressed as -fold change in luciferase expression over 0 mm Gln (n = 3). Means are compared via post hoc Tukey's test. For A and B, error bars represent S.E.
To further confirm this, we analyzed Hsf1 gene promoter activity following exposure to different Gln concentrations. We fused a 1381-bp sequence upstream of the murine Hsf1 gene translation start site to a luciferase reporter plasmid and transiently transfected the construct into YAMCs. As shown in Fig. 4B, Hsf1 promoter-driven luciferase expression was increased with Gln supplementation in a dose-dependent manner. Gln at 2 mm led to 3.2-fold increase in Hsf1 gene promoter activity over basal conditions (without supplemental Gln), indicating that Gln transcriptionally activates Hsf1 gene expression by increasing Hsf1 gene promoter activity.
Key cis-Element(s) of Hsf1 Promoter Responsible for Gln-induced Activation of Hsf1 Transcription
To identify the key cis-element(s) involved in Gln-induced Hsf1 transcription activation, luciferase constructs bearing the Hsf1 promoter deleted to a variety of lengths (p1381/luc to p200/luc) were initially utilized to identify the minimal promoter length required for Gln-induced transcriptional response. These constructs were transiently transfected into YAMCs, and the response to Gln was determined by luciferase assay. As shown in Fig. 5A, deletion constructs down to p281/luc did not have a significant effect on Gln inducibility, whereas the deletion construct p200/luc attenuated the Gln response, suggesting that the key positive cis-element responsible for the Gln response is present in the Hsf1 promoter between bp −281 and −200.
Using the TRANSFAC transcription factor database, we found that the Hsf1 promoter sequence at −263/−254 exhibits similarity with a binding site for C/EBP, the sequence at −249/−237 has similarity to a binding site for SP1, and the sequence at −211/−202 has similarity to a binding site for activating transcription factor/cAMP-response element-binding protein. To determine the involvement of each site in the Gln-mediated response, we transfected the cells with a series of mutant constructs bearing internal deletions of the three putative binding sites within the −281/−200 sequence and analyzed the luciferase activity of these constructs. As shown in Fig. 5B, none of these deletion mutants had a significant effect on basal promoter activity. However, deletion of the putative C/EBP site (del 1) abolished Gln-mediated transcriptional activation, whereas neither del 2 nor del 3 had significant effects on either basal activity or Gln inducibility. To confirm the finding derived from the internal deletion analysis, we further scanned the −281/−200 sequence with a series of base substitution mutant constructs designated M1 to M5 (Fig. 5C). Consistent with the internal deletion analysis, the mutation of C/EBP binding site (M1) resulted in a loss of the Gln response, whereas the other mutations had minimal effects on the Gln-mediated response. Taken together, these results suggest that the putative −263/−254 C/EBP site is essential to Gln-mediated activation of Hsf1 transcription.
C/EBPβ Mediates Gln-induced Activation of Hsf1 Transcription
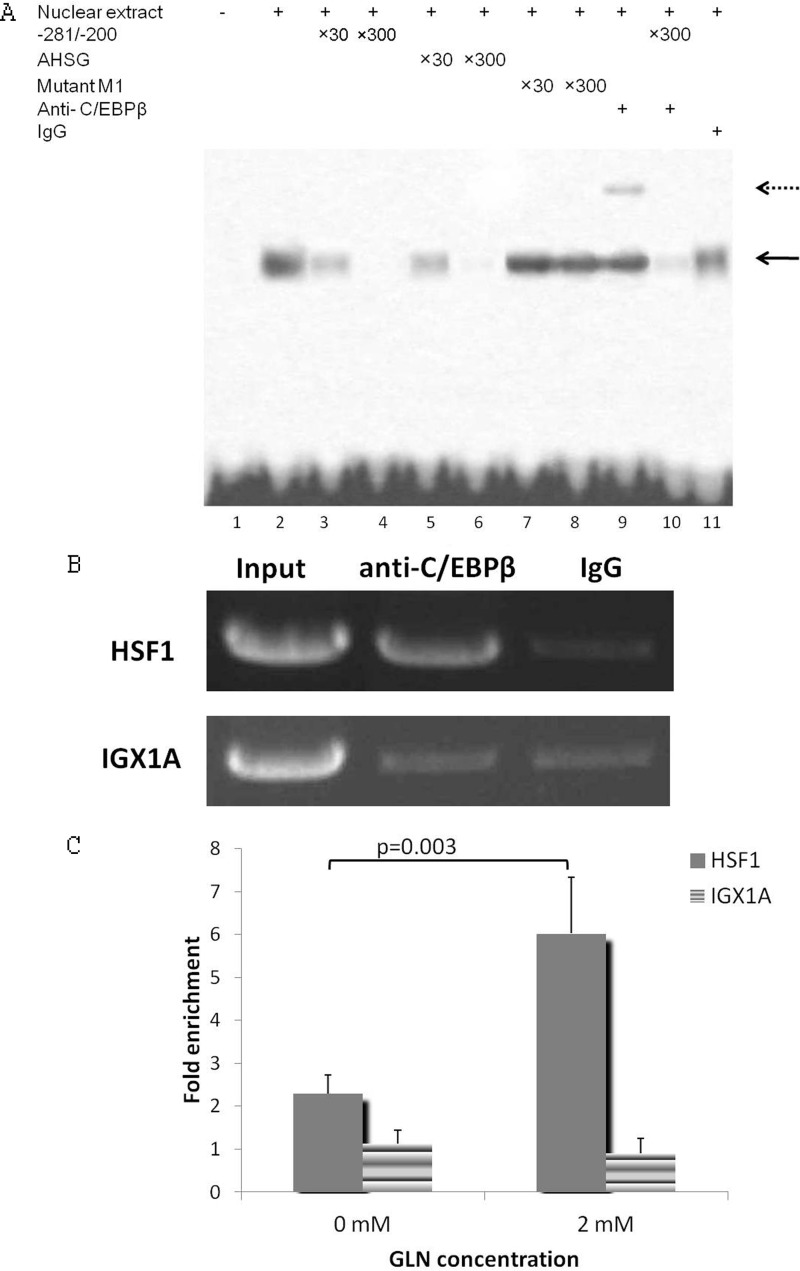
EMSA was performed to prove there is actual in vitro binding of C/EBP to the −281/−200 sequence (Fig. 6A). A biotin-labeled proven C/EBP binding sequence was used as a probe. The shifts were competed out by the −281/−200 sequence (Fig. 6A, lanes 3 and 4), a high affinity C/EBP binding site found in the AHSG promoter (Fig. 6A, lanes 5 and 6), but not by the mutant M1 sequence (Fig. 6A, lanes 7 and 8). Therefore, the −263/−255 sequence has been identified as a C/EBP binding site. Furthermore, the competition experiments with varied amounts of excess competitor demonstrate that the C/EBP proteins have an affinity for the −263/−255 binding site of Hsf1 promoter comparable with that for the high affinity site in AHSG.
FIGURE 6.
C/EBPβ binds to the −281/−200 sequence of Hsf1 promoter. A, C/EBPβ can bind to the −281/−200 Gln-response element in vitro as determined by gel mobility shift assays. Biotin-labeled oligos with the canonical c/EBP consensus sequence were used as probes. The wild-type −281/−200 area of Hsf1 promoter, the C/EBP consensus sequence in the AHSG promoter (−80/−51), or the mutated M1 −281/−200 sequence was used as a competitor at a 30- or 300-fold molar excess relative to the probe. The DNA-protein complex is indicated by a solid arrow. Supershifts, which are indicated by a dotted arrow, were done with specific anti-C/EBPβ antibody. B, ChIP assay to show that C/EBPβ binds to the −281/−200 Gln-response element in vivo. Sheared chromatin (average size, 500 bp) from cells treated with 2 mm Gln was immunoprecipitated using 10 μg of anti-C/EBPβ (Santa Cruz Biotechnology) or 5 μl of rabbit preimmune serum. Primers for IGX1A were used as a negative control in all immunoprecipitations. Immunoprecipitated DNA was analyzed by PCR, and products were run on an agarose gel. The image shown is representative of four separate experiments. C, Gln increases C/EBPβ binding to the Hsf1 promoter at −281/−200 as determined by ChIP assay. Immunoprecipitated DNA was analyzed by real time PCR. Gln at 2 mm significantly enhanced Hsf1 promoter DNA (−281/−200) enrichment compared with 0 mm (p < 0.05). DNA enrichment for IGX1A is not significantly different between the different Gln concentrations (n = 3). Means are compared via Student's t test. Error bars represent S.E.
Because C/EBPβ has been reported to play a key regulatory role in amino acid-mediated transcription activation (27, 28), supershifts were further performed to identify whether C/EBPβ can bind to the −281/−200 sequence. First, we confirmed that anti-C/EBPβ antibody can supershift the DNA-protein complex as shown in Fig. 6A, lane 9. When using the −281/−200 sequence as a competitor for the biotin-labeled C/EBPβ consensus sequence, the biotin-labeled supershift was removed, suggesting that C/EBPβ can indeed bind to the −281/−200 sequence.
To confirm the binding of C/EBPβ to the Hsf1 promoter at −281/−200 in vivo, ChIP analysis was performed on pooled chromatin samples prepared from the YAMCs. A −281/−200 sense and antisense primer pair containing the C/EBP binding site showed a much stronger PCR signal in anti-C/EBPβ-precipitated samples than IgG-precipitated samples (Fig. 6B), whereas amplifications were equivalent for anti-C/EBPβ- and IgG-precipitated samples when using the negative control primers for IGX1A gene. This confirms that C/EBPβ binds to the Hsf1 promoter at −281/−200 in vivo. We further examined the induction of C/EBPβ binding to the Hsf1 promoter in response to Gln by quantifying immunoprecipitated DNA using quantitative PCR. Notably, C/EBPβ binding to Hsf1 promoter was induced by 2.6-fold by Gln (Fig. 6C).
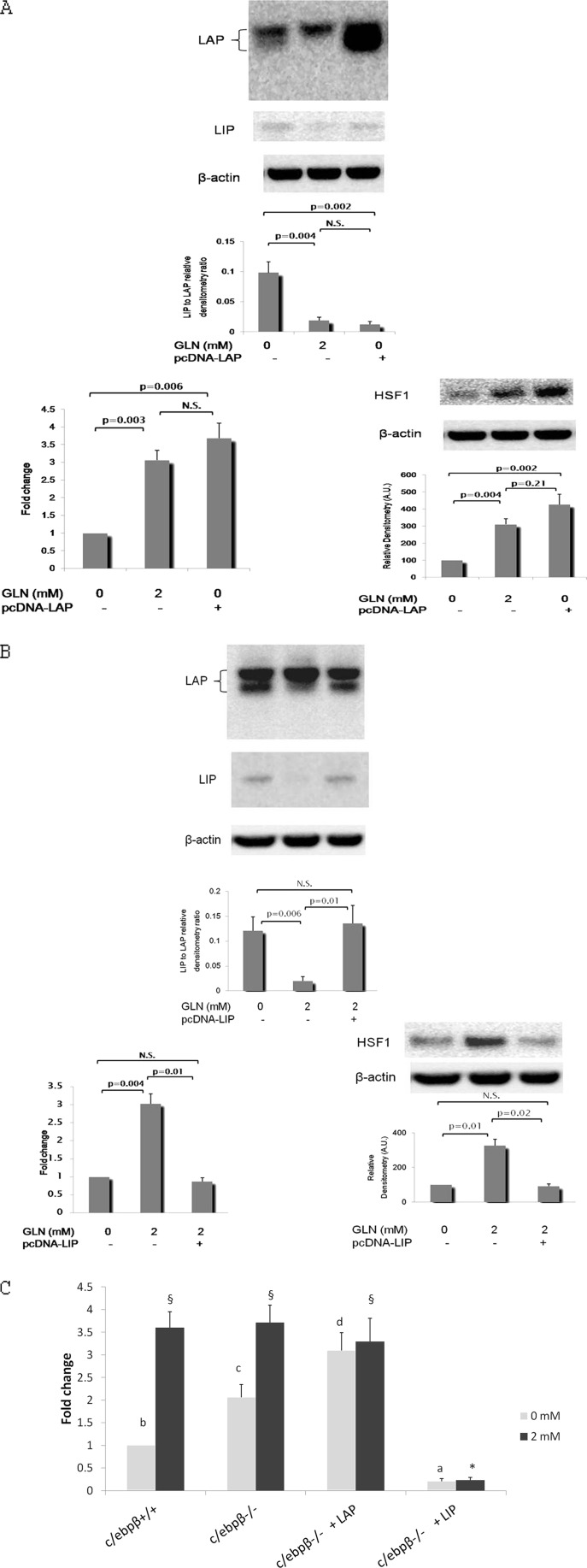
As we identified that the C/EBP binding site is a key cis-element involved in Gln-mediated Hsf1 transcription activation and confirmed that C/EBPβ can bind to the −281/−200 sequence, we further attempted to identify a potential role of C/EBPβ in the Gln-mediated Hsf1 response. Transcription of the intronless c/ebpβ gene results in a single mRNA. LAP, the active isoform of C/EBPβ, and liver-enriched inhibitory protein (LIP), the inhibitory dominant-negative isoform of C/EBPβ, are the primary products from translation of this single C/EBPβ mRNA via alternative start sites. In cells transfected with expression plasmid vector alone, Gln deprivation led to increases in both LIP and LAP expression, but the LIP expression increase was much greater, leading to a significantly increased LIP:LAP ratio (Fig. 7, A and B, upper panels).
FIGURE 7.
C/EBPβ mediates Gln-induced activation of Hsf1 transcription via LIP/LAP ratio. A, effects of overexpression of C/EBPβ LAP on the Gln-mediated HSF1 response. YAMCs were transfected with expression plasmids bearing LAP cDNA resulting in LAP overexpression (top). B, effects of overexpression of C/EBPβ LIP on the Gln-mediated HSF1 response. YAMCs were transfected with expression plasmids bearing LIP cDNA resulting in LIP overexpression (top). For A and B, Hsf1 promoter activity (lower left) and HSF1 protein expression (lower right) were determined in cells transfected with and without 1381-bp Hsf1 promoter-luc plasmids, respectively. C, effects of individual C/EBPβ LAP and LIP isoforms on Hsf1 promoter activity in response to Gln. LIP and LAP cDNA expression plasmids were co-transfected in c/ebpβ−/− MEF cells with Hsf1 promoter-luc reporter plasmids. The experimental conditions that do not share the same letter or symbol are significantly different from each other at the same Gln concentration (0 or 2 mm) as determined by Bonferroni posttests (p < 0.05). For promoter assays, data are expressed as -fold change in luciferase expression over pcDNA vector-transfected YAMCs exposed to 0 mm Gln (A and B) or c/ebpβ+/+ MEFs at 0 mm Gln (C) (n = 4). Means are compared via post hoc Tukey's test. N.S., not significant. The image shown is representative of four separate experiments. For A–C, error bars represent S.E.
We next examined Hsf1 transcription in LAP- or LIP-overexpressing cells transfected with expression plasmids bearing LAP or LIP cDNA. Overexpression of LAP strikingly increased Hsf1 promoter activity and derepressed HSF1 expression in the absence of Gln (Fig. 7A, lower panels), whereas overexpression of LIP led to deactivated Hsf1 promoter activity and HSF1 protein expression even when Gln was supplied at 2 mm (Fig. 7B, lower panels).
To determine the individual effect of LAP and LIP on the Gln-mediated HSF1 response independently of one another, cDNA expression plasmids of each isoform were co-transfected into c/ebpβ−/− MEF cells with Hsf1 promoter-luciferase reporter plasmids. Irrespective of Gln abundance, LAP strikingly enhanced Hsf1 promoter activity, whereas LIP affected the Hsf1 promoter in the opposite manner (Fig. 7C).
Taken together, our data demonstrate that LAP serves as an activating trans-factor for Hsf1 transcription, whereas LIP inhibits Hsf1 transcription, and furthermore, the relative abundance of LIP and LAP appears to determine HSF1 expression. Gln availability strikingly altered the LIP:LAP ratio, which seems to be responsible for the Gln-mediated activation of Hsf1 transcription.
DISCUSSION
Enhanced Hsp expression has the potential to be beneficial in numerous clinical settings including sepsis, shock, tissue ischemia/reperfusion injury, and autoimmune/inflammatory diseases. As a result, interventions aimed at enhancing the heat shock response could lead to improved outcome in clinical illness and injury. Presently, laboratory methods utilizing chemical or gene therapy-based mechanisms to enhance Hsp expression still have limited or no clinical relevance because of their inherent toxicities (43, 44). Thus, the search has continued for a safe pharmacologic or metabolic agent to induce Hsps (43). A significant amount of data from our previous work and that of others demonstrates that Gln is able to enhance Hsp expression and further confer protection against a variety of stress and injury conditions at cellular, tissue, and organismal levels (18–20). Given the long standing safety profile of Gln in clinical use (24, 45, 46), Gln has the potential to emerge as a clinically relevant enhancer of the heat shock response in illness and injury. Here we show that Gln has the ability to enhance Hsp expression by acting on the HSF1-Hsp axis. Our data provide the first evidence demonstrating that Gln not only activates multiple steps in the HSF1 transactivation pathway but also up-regulates actual HSF1 expression. Another novel finding of our study is that the enhancement of HSF1 expression by Gln, at least in part, occurs at the transcriptional level.
Enhanced Transactivation Activity of HSF1 by Gln
The main points of control in the regulation of HSF1 are widely considered to be the key steps leading to HSF1 acquiring its transactivating capacity, leading to induction of the Hsp response (2, 3, 7–9). As a result, we first focused on determining whether Gln affects HSF1 trimerization, nuclear localization, nuclear binding of HSF1 to the HSE, and phosphorylation of nuclear HSF1. We demonstrate here that Gln enhanced HSF1 nuclear accumulation, a prominent feature of HSF1 activation (36), in both non-stressed and heat-stressed conditions. Furthermore, spontaneous and heat-induced HSF1 trimerization was increased by Gln administration. This effect of Gln was associated with an increase in HSF1 DNA binding ability in both non-stressed and heat shock conditions. HSF1 trimerization and DNA binding alone are insufficient for transcriptional induction of Hsp genes without adequate posttranslational modifications via specific phosphorylation (37, 40). In this study, for the first time, we show that Gln administration can enhance the relative abundance of activating phosphorylation of HSF1 at Ser-230 while not affecting that of repressing phosphorylation at Ser-307, which overall favors HSF1 transcriptional activation (40).
Collectively, our data are supported by a previous study by Morrison et al. (22), who showed that Gln treatment also increased HSF1 nuclear localization and HSE binding in non-stressed fibroblasts and further increased HSF1-HSE binding and HSF1 global phosphorylation after heat shock (22). In a similar study, Ropeleski et al. (47) demonstrated that Gln increased heat-induced HSF1-HSE binding in small intestinal epithelial IEC-18 cells, but they were not able to detect a concurrent Gln-dependent difference in HSF1 trimerization immediately after heat shock. Consistently, we did not observe any difference in HSF1 trimerization with Gln treatment immediately after heat shock at 43 °C for 30 min (supplemental Fig. 1). However, a significant Gln-dependent increase in HSF1 trimerization was observed 1 h after heat shock (Fig. 2A). Furthermore, Gln treatment increased the duration of HSF1 trimerization (supplemental Fig. 1). Therefore, the effects of Gln on HSF1 activation may also be attributed to a more sustainable HSF1 activation state. This appears to be translated into a more robust Hsp induction following heat shock. This is supported by the finding of Ropeleski et al. (47) that Gln treatment did increase the duration of HSF1-HSE binding following heat shock and further increased Hsp mRNA and protein expression and Hsp70 promoter-driven reporter expression at 2 and 6 h, respectively, after heat shock (47).
Transcriptional Activation of Hsf1 Gene by Gln
Here, for the first time, we show that Gln induces Hsf1 gene transcription with two lines of evidence: Gln increases HSF1 mRNA expression and further increases Hsf1 gene promoter activity. We have mapped the key cis-element responsible for the induction of Hsf1 gene activation by Gln to the promoter fragment at −267/−248. This region exhibits a similarity to the C/EBP consensus sequence. Based on this, we proceeded to demonstrate that C/EBPβ binds to this sequence both in vitro and in vivo.
Limited information is available concerning the amino acid control of gene expression in mammalian cells. Amino acid-mediated transcriptional regulation has been reported for a small number of genes such as asparagine synthetase (48), C/EBP-homologous protein (CHOP) (49), and collagen Iα (50). To date, the amino acid-response elements, which are cis-elements within the promoter of a gene that mediate transcriptional regulation by an amino acid, have only been identified in asparagine synthetase and CHOP genes (51–54). The sequences of the asparagine synthetase and CHOP amino acid-response element regions show similarity with the specific binding sites of the C/EBP family transcription factors (55). Members of C/EBP proteins (C/EBPα–C/EBPζ) have been found to play a critical role in regulating numerous cellular responses including cellular growth, differentiation, and energy metabolism (56). Of particular interest, C/EBPβ has been reported to be involved in amino acid-trigged transcription activation (27, 28). Here we also show that C/EBPβ appears to be the key trans-factor mediating the ability of Gln to activate Hsf1 transcription.
We demonstrate that the ratio of C/EBPβ inhibitory and active isoforms (LIP:LAP) serves as an independent factor mediating the Gln-induced HSF1 response. The cellular ratio of LIP and LAP has been shown to be a vital determinant of gene expression regulation (57, 58). We show in this study that the relative abundance of LAP and LIP, which is regulated by Gln supply, appears to be the key control machinery in mediating the Hsf1 transcription response to Gln. In the c/ebpβ−/− background, individual LAP and LIP expression leads to induced and suppressed Hsf1 transcription, respectively, irrespective of Gln supply. Furthermore, in YAMCs with a c/ebpβ+/+ background, we confirmed that overexpression of LAP derepresses Hsf1 transcription during Gln deprivation, whereas overexpression of LIP deactivates Hsf1 transcription in the presence of a sufficient Gln supply. This suggests that LAP and LIP act as an independent activator and repressor for Hsf1 transcription. Consistent with our finding, Siu et al. (59) also showed that LAP and LIP appear to act as an activator and repressor for transcription of asparagine synthetase, another nutrient-responsive gene, as overexpression of LAP increased asparagine synthetase gene transcription in HepG2 hepatoma cells, whereas LIP blocked enhanced asparagine synthetase transcription following amino acid or glucose deprivation.
In summary, our data demonstrate that Gln not only acts as an inducer of Hsf1 gene activation but also intriguingly can act as a competent inducer of HSF1 expression by activating its transcription. The relative abundance of LIP and LAP seems to serve as a tuning mechanism linking Gln availability to Hsf1 transcription. Here we report a dual control machinery in mobilizing HSF1 that ultimately leads to a robust induction of the heat shock response in the gut. Our data suggest that Gln can potentially be developed as an independent clinically viable inducer of expression of the transcription factor HSF1 and the heat shock response.
Supplementary Material
Acknowledgments
We thank R. Whitehead and J. Friedman for cells and plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM078312 (to P. E. W.). This work was also supported by a start-up grant from the Department of Anesthesiology, University of Colorado (to H. X.).

This article contains supplemental Fig. 1 and Table 1.
- Hsp
- heat shock protein
- C/EBP
- CCAAT enhancer-binding protein
- CHOP
- C/EBP-homologous protein
- HSE
- heat shock element
- HSF1
- heat shock transcription factor-1
- IEC
- intestinal epithelial cell
- LAP
- liver-enriched activating protein
- LIP
- liver-enriched inhibitory protein
- MEF
- mouse embryonic fibroblast
- YAMC
- young adult mouse colonic epithelial cell
- DSS
- dextran sulfate sodium
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- AHSG
- α2-Heremans-Schmid glycoprotein
- luc
- luciferase.
REFERENCES
- 1. Ritossa F. (1996) Discovery of the heat shock response. Cell Stress Chaperones 1, 97–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anckar J., Sistonen L. (2007) Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv. Exp. Med. Biol. 594, 78–88 [DOI] [PubMed] [Google Scholar]
- 3. Pirkkala L., Nykänen P., Sistonen L. (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15, 1118–1131 [DOI] [PubMed] [Google Scholar]
- 4. Zimarino V., Wu C. (1987) Induction of sequence-specific binding of Drosophila heat shock activator protein without protein synthesis. Nature 327, 727–730 [DOI] [PubMed] [Google Scholar]
- 5. Sorger P. K., Lewis M. J., Pelham H. R. (1987) Heat shock factor is regulated differently in yeast and HeLa cells. Nature 329, 81–84 [DOI] [PubMed] [Google Scholar]
- 6. Larson J. S., Schuetz T. J., Kingston R. E. (1988) Activation in vitro of sequence-specific DNA binding by a human regulatory factor. Nature 335, 372–375 [DOI] [PubMed] [Google Scholar]
- 7. Wu C. (1995) Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11, 441–469 [DOI] [PubMed] [Google Scholar]
- 8. Morimoto R. I. (1993) Cells in stress: transcriptional activation of heat shock genes. Science 259, 1409–1410 [DOI] [PubMed] [Google Scholar]
- 9. Voellmy R. (1994) Transduction of the stress signal and mechanisms of transcriptional regulation of heat shock/stress protein gene expression in higher eukaryotes. Crit. Rev. Eukaryot. Gene Expr. 4, 357–401 [PubMed] [Google Scholar]
- 10. Sundar S. V., Li Y. Y., Rollwagen F. M., Maheshwari R. K. (2005) Hemorrhagic shock induces differential gene expression and apoptosis in mouse liver. Biochem. Biophys. Res. Commun. 332, 688–696 [DOI] [PubMed] [Google Scholar]
- 11. Sonna L. A., Hawkins L., Lissauer M. E., Maldeis P., Towns M., Johnson S. B., Moore R., Singh I. S., Cowan M. J., Hasday J. D. (2010) Core temperature correlates with expression of selected stress and immunomodulatory genes in febrile patients with sepsis and noninfectious SIRS. Cell Stress Chaperones 15, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Metzler B., Abia R., Ahmad M., Wernig F., Pachinger O., Hu Y., Xu Q. (2003) Activation of heat shock transcription factor 1 in atherosclerosis. Am. J. Pathol. 162, 1669–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou J. D., Luo C. Q., Xie H. Q., Nie X. M., Zhao Y. Z., Wang S. H., Xu Y., Pokharel P. B., Xu D. (2008) Increased expression of heat shock protein 70 and heat shock factor 1 in chronic dermal ulcer tissues treated with laser-aided therapy. Chin. Med. J. 121, 1269–1273 [PubMed] [Google Scholar]
- 14. Ding X. Z., Smallridge R. C., Galloway R. J., Kiang J. G. (1996) Rapid assay of HSF1 and HSF2 gene expression by RT-PCR. Mol. Cell. Biochem. 158, 189–192 [DOI] [PubMed] [Google Scholar]
- 15. Yang J., Bridges K., Chen K. Y., Liu A. Y. (2008) Riluzole increases the amount of latent HSF1 for an amplified heat shock response and cytoprotection. PLoS One 3, e2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Curi R., Lagranha C. J., Doi S. Q., Sellitti D. F., Procopio J., Pithon-Curi T. C., Corless M., Newsholme P. (2005) Molecular mechanisms of glutamine action. J. Cell Physiol. 204, 392–401 [DOI] [PubMed] [Google Scholar]
- 17. Jeppesen P. B., Mortensen P. B. (2002) Enhancing bowel adaptation in short bowel syndrome. Curr. Gastroenterol. Rep. 4, 338–347 [DOI] [PubMed] [Google Scholar]
- 18. Wischmeyer P. E., Kahana M., Wolfson R., Ren H., Musch M. M., Chang E. B. (2001) Glutamine induces heat shock protein and protects against endotoxin shock in the rat. J. Appl. Physiol. 90, 2403–2410 [DOI] [PubMed] [Google Scholar]
- 19. Xue H., Sawyer M. B., Field C. J., Dieleman L. A., Baracos V. E. (2007) Nutritional modulation of antitumor efficacy and diarrhea toxicity related to irinotecan chemotherapy in rats bearing the ward colon tumor. Clin. Cancer Res. 13, 7146–7154 [DOI] [PubMed] [Google Scholar]
- 20. Xue H., Sawyer M. B., Field C. J., Dieleman L. A., Murray D., Baracos V. E. (2008) Bolus oral glutamine protects rats against CPT-11-induced diarrhea and differentially activates cytoprotective mechanisms in host intestine but not tumor. J. Nutr. 138, 740–746 [DOI] [PubMed] [Google Scholar]
- 21. Singleton K. D., Wischmeyer P. E. (2007) Glutamine's protection against sepsis and lung injury is dependent on heat shock protein 70 expression. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R1839–R1845 [DOI] [PubMed] [Google Scholar]
- 22. Morrison A. L., Dinges M., Singleton K. D., Odoms K., Wong H. R., Wischmeyer P. E. (2006) Glutamine's protection against cellular injury is dependent on heat shock factor-1. Am. J. Physiol. Cell Physiol. 290, C1625–C1632 [DOI] [PubMed] [Google Scholar]
- 23. Ren H., Musch M. W., Kojima K., Boone D., Ma A., Chang E. B. (2001) Short-chain fatty acids induce intestinal epithelial heat shock protein 25 expression in rats and IEC 18 cells. Gastroenterology 121, 631–639 [DOI] [PubMed] [Google Scholar]
- 24. Ziegler T. R., Ogden L. G., Singleton K. D., Luo M., Fernandez-Estivariz C., Griffith D. P., Galloway J. R., Wischmeyer P. E. (2005) Parenteral glutamine increases serum heat shock protein 70 in critically ill patients. Intensive Care Med. 31, 1079–1086 [DOI] [PubMed] [Google Scholar]
- 25. Wheelhouse N. M., Stubbs A. K., Lomax M. A., MacRae J. C., Hazlerigg D. G. (1999) Growth hormone and amino acid supply interact synergistically to control insulin-like growth factor-I production and gene expression in cultured ovine hepatocytes. J. Endocrinol. 163, 353–361 [DOI] [PubMed] [Google Scholar]
- 26. Straus D. S., Marten N. W., Hayden J. M., Burke E. J. (1994) Protein restriction specifically decreases the abundance of serum albumin and transthyretin nuclear transcripts in rat liver. J. Nutr. 124, 1041–1051 [DOI] [PubMed] [Google Scholar]
- 27. Chen H., Pan Y. X., Dudenhausen E. E., Kilberg M. S. (2004) Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. J. Biol. Chem. 279, 50829–50839 [DOI] [PubMed] [Google Scholar]
- 28. Thiaville M. M., Dudenhausen E. E., Zhong C., Pan Y. X., Kilberg M. S. (2008) Deprivation of protein or amino acid induces C/EBPβ synthesis and binding to amino acid response elements, but its action is not an absolute requirement for enhanced transcription. Biochem. J. 410, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xue H., Sufit A. J., Wischmeyer P. E. (2011) Glutamine therapy improves outcome of in vitro and in vivo experimental colitis models. JPEN J. Parenter. Enteral. Nutr. 35, 188–197 [DOI] [PubMed] [Google Scholar]
- 30. Sterneck E., Tessarollo L., Johnson P. F. (1997) An essential role for C/EBPβ in female reproduction. Genes Dev. 11, 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whitehead R. H., VanEeden P. E., Noble M. D., Ataliotis P., Jat P. S. (1993) Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 90, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y., Bevilacqua E., Chiribau C. B., Majumder M., Wang C., Croniger C. M., Snider M. D., Johnson P. F., Hatzoglou M. (2008) Differential control of the CCAAT/enhancer-binding protein β (C/EBPβ) products liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP) and the regulation of gene expression during the response to endoplasmic reticulum stress. J. Biol. Chem. 283, 22443–22456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shao J., Qiao L., Janssen R. C., Pagliassotti M., Friedman J. E. (2005) Chronic hyperglycemia enhances PEPCK gene expression and hepatocellular glucose production via elevated liver activating protein/liver inhibitory protein ratio. Diabetes 54, 976–984 [DOI] [PubMed] [Google Scholar]
- 34. Banine F., Gangneux C., Lebreton J. P., Frebourg T., Salier J. P. (1998) Structural and functional analysis of the 5′-transcription control region for the human α2-HS glycoprotein gene. Biochim. Biophys. Acta 1398, 1–8 [DOI] [PubMed] [Google Scholar]
- 35. Anckar J., Sistonen L. (2011) Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80, 1089–1115 [DOI] [PubMed] [Google Scholar]
- 36. Vujanac M., Fenaroli A., Zimarino V. (2005) Constitutive nuclear import and stress-regulated nucleocytoplasmic shuttling of mammalian heat-shock factor 1. Traffic 6, 214–229 [DOI] [PubMed] [Google Scholar]
- 37. Kline M. P., Morimoto R. I. (1997) Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol. Cell. Biol. 17, 2107–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hietakangas V., Ahlskog J. K., Jakobsson A. M., Hellesuo M., Sahlberg N. M., Holmberg C. I., Mikhailov A., Palvimo J. J., Pirkkala L., Sistonen L. (2003) Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 23, 2953–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R. I. (2009) Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Holmberg C. I., Hietakangas V., Mikhailov A., Rantanen J. O., Kallio M., Meinander A., Hellman J., Morrice N., MacKintosh C., Morimoto R. I., Eriksson J. E., Sistonen L. (2001) Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 20, 3800–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Knauf U., Newton E. M., Kyriakis J., Kingston R. E. (1996) Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev. 10, 2782–2793 [DOI] [PubMed] [Google Scholar]
- 42. Chu B., Soncin F., Price B. D., Stevenson M. A., Calderwood S. K. (1996) Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J. Biol. Chem. 271, 30847–30857 [DOI] [PubMed] [Google Scholar]
- 43. De Maio A. (1999) Heat shock proteins: facts, thoughts, and dreams. Shock 11, 1–12 [DOI] [PubMed] [Google Scholar]
- 44. Weiss Y. G., Maloyan A., Tazelaar J., Raj N., Deutschman C. S. (2002) Adenoviral transfer of HSP-70 into pulmonary epithelium ameliorates experimental acute respiratory distress syndrome. J. Clin. Investig. 110, 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hegazi R. A., Wischmeyer P. E. (2011) Clinical review: optimizing enteral nutrition for critically ill patients—a simple data-driven formula. Crit. Care 15, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wischmeyer P. (2011) Nutritional pharmacology in surgery and critical care: 'you must unlearn what you have learned'. Curr. Opin. Anaesthesiol. 24, 381–388 [DOI] [PubMed] [Google Scholar]
- 47. Ropeleski M. J., Riehm J., Baer K. A., Musch M. W., Chang E. B. (2005) Anti-apoptotic effects of l-glutamine-mediated transcriptional modulation of the heat shock protein 72 during heat shock. Gastroenterology 129, 170–184 [DOI] [PubMed] [Google Scholar]
- 48. Gong S. S., Guerrini L., Basilico C. (1991) Regulation of asparagine synthetase gene expression by amino acid starvation. Mol. Cell. Biol. 11, 6059–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bruhat A., Jousse C., Wang X. Z., Ron D., Ferrara M., Fafournoux P. (1997) Amino acid limitation induces expression of CHOP, a CCAAT/enhancer binding protein-related gene, at both transcriptional and post-transcriptional levels. J. Biol. Chem. 272, 17588–17593 [DOI] [PubMed] [Google Scholar]
- 50. Bellon G., Chaqour B., Wegrowski Y., Monboisse J. C., Borel J. P. (1995) Glutamine increases collagen gene transcription in cultured human fibroblasts. Biochim. Biophys. Acta 1268, 311–323 [DOI] [PubMed] [Google Scholar]
- 51. Barbosa-Tessmann I. P., Chen C., Zhong C., Siu F., Schuster S. M., Nick H. S., Kilberg M. S. (2000) Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 275, 26976–26985 [DOI] [PubMed] [Google Scholar]
- 52. Bruhat A., Jousse C., Carraro V., Reimold A. M., Ferrara M., Fafournoux P. (2000) Amino acids control mammalian gene transcription: activating transcription factor 2 is essential for the amino acid responsiveness of the CHOP promoter. Mol. Cell. Biol. 20, 7192–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guerrini L., Gong S. S., Mangasarian K., Basilico C. (1993) Cis- and trans-acting elements involved in amino acid regulation of asparagine synthetase gene expression. Mol. Cell. Biol. 13, 3202–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Siu F., Bain P. J., LeBlanc-Chaffin R., Chen H., Kilberg M. S. (2002) ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 277, 24120–24127 [DOI] [PubMed] [Google Scholar]
- 55. Fafournoux P., Bruhat A., Jousse C. (2000) Amino acid regulation of gene expression. Biochem. J. 351, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ramji D. P., Foka P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Raught B., Liao W. S., Rosen J. M. (1995) Developmentally and hormonally regulated CCAAT/enhancer-binding protein isoforms influence β-casein gene expression. Mol. Endocrinol. 9, 1223–1232 [DOI] [PubMed] [Google Scholar]
- 58. Gingras A. C., Raught B., Sonenberg N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963 [DOI] [PubMed] [Google Scholar]
- 59. Siu F., Chen C., Zhong C., Kilberg M. S. (2001) CCAAT/enhancer-binding protein-β is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 276, 48100–48107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.