Background: Mutations in LMNA gene cause cardiomyopathy, for which mechanistic insights are lacking.
Results: Dusp4 expression is enhanced in hearts with LMNA cardiomyopathy, and its overexpression in mice causes it by activating AKT-mTOR signaling that impairs autophagy.
Conclusions: Dusp4 causes cardiac dysfunction and may contribute to the development of LMNA cardiomyopathy.
Significance: Revealing pathogenic mechanisms of LMNA cardiomyopathy is essential for the development of mechanism-based therapies.
Keywords: Akt, Autophagy, Cardiomyopathy, Dual Specificity Phosphoprotein Phosphatase, ERK, Lamin, mTOR, Signal Transduction, Transgenic Mice
Abstract
Mutations in the lamin A/C gene (LMNA) cause a diverse spectrum of diseases, the most common of which is dilated cardiomyopathy often with skeletal muscular dystrophy. Lamin A and C are fundamental components of the nuclear lamina, a dynamic meshwork of intermediate filaments lining the nuclear envelope inner membrane. Prevailing evidence suggests that the nuclear envelope functions as a signaling node and that abnormality in the nuclear lamina leads to dysregulated signaling pathways that underlie disease pathogenesis. We previously showed that activated ERK1/2 in hearts of a mouse model of LMNA cardiomyopathy (LmnaH222P/H222P mice) contributes to disease, but the complete molecular pathogenesis remains poorly understood. Here we uncover a pathogenic role of dual specificity phosphatase 4 (Dusp4), which is transcriptionally induced by ERK1/2. Dusp4 is highly expressed in the hearts of LmnaH222P/H222P mice, and transgenic mice with cardiac-selective overexpression of Dusp4 display heart dysfunction similar to LMNA cardiomyopathy. In both primary tissue and cell culture models, overexpression of Dusp4 positively regulates AKT-mTOR signaling, resulting in impaired autophagy. These findings identify a pathogenic role of Dusp4 in LMNA cardiomyopathy.
Introduction
A-type lamins (lamin A/C) are a fundamental component of the nuclear lamina, a meshwork of intermediate filaments lining the nuclear envelope inner membrane (1, 2). Although ubiquitously expressed in most mammalian somatic cells, site- and sequence-specific mutations in LMNA lead to tissue-selective diseases, the most common of which affects striated muscle in the form of dilated cardiomyopathy (hereinafter referred to as LMNA cardiomyopathy) (1). Mechanistic insights into how mutant variants of lamin A/C trigger disease pathogenesis are just beginning to be elucidated.
Recent evidence suggests that A-type lamins play a dynamic role in regulating signal transduction, particularly ERK1/2 signaling (1, 3–5). We previously showed that activated ERK1/2 in hearts of LmnaH222P/H222P mice, a mouse model of LMNA cardiomyopathy, contributes to disease (6–8). However, it remains unclear how ERK1/2 mediates pathogenic mechanisms causing dilated cardiomyopathy. We also recently demonstrated that AKT-mammalian target of rapamycin (mTOR)4 signaling is activated in hearts of LmnaH222P/H222P mice (9). Activation of mTOR in hearts of mouse models of LMNA cardiomyopathy (LmnaH222P/H222P and Lmna−/− mice) leads to impaired productive autophagy, and its reactivation by mTOR inhibition by rapamycin and its analogs ameliorates the disease (9, 10). In the current study, we set out to explore pathogenic mechanisms mediated by ERK1/2 activation in the pathogenesis of LMNA cardiomyopathy. We focused on the Dusp family of phosphatases, which catalyze dephosphorylation of tyrosine and serine/threonine residues on MAP kinases, some of which are induced in an ERK1/2-dependent manner, and establish feedback inhibition (11, 12). We found that the nuclear resident ERK1/2-induced phosphatase DUSP4 contributes to the pathogenesis of LMNA cardiomyopathy, linking activated ERK1/2 to AKT-mTOR signaling.
EXPERIMENTAL PROCEDURES
Animals
The Columbia University Medical Center Institutional Animal Care and Use Committee approved all protocols using animals. LmnaH222P/H222P mice were generated and genotyped as described (13). Dusp4 transgenic mice were generated at the Herbert Irving Comprehensive Cancer Center Transgenic Mouse Facility (Columbia University Medical Center). Dusp4 cDNA was subcloned into pPSKII-MHC-GH downstream of the α-myosin heavy chain promoter and upstream of a human growth hormone polyadenylation signal. Minigenes of ∼7.3 kb containing Dusp4 cDNA were microinjected into superovulated CBA/B6F1 fertilized oocytes and transferred to pseudopregnant females. One surviving founder, identified by PCR of extracted DNA from tails using primer sets indicated in supplemental Table S3, was backcrossed to WT B6/CBAF1 to produce male transgenic positive breeders (F1). The male breeders were backcrossed to female WT CBA/B6F1 (F2) to obtain an adequate number of mice for the experiments. Only F2 mice were used in the study. Autophagy was induced in vivo by fasting mice (given only water) for 24 h.
RNAseq and Enrichment Analysis Using DAVID
RNAseq and data analysis were performed at the Herbert Irving Comprehensive Cancer Center Next Generation Sequencing Laboratory (Columbia University Medical Center). Briefly, total RNA from mouse ventricular tissue of 8-week-old Dusp4 Tg+ and Tg− mice (n = 3/group) was extracted using TRIzol (Invitrogen). Double-stranded cDNA from the isolated RNA was generated using TruSeq RNA sample preparation kit v2 followed by library construction with TruSeq single-read cluster kit v3-cBot-HS (Illumina). The resulting cDNA library was sequenced with HiSeq 2000 genome analyzer (Illumina) using 2 × 107 single-ended 100 base pair reads. Sequenced reads were mapped to the mouse reference genome UCSC version mm9 with TopHat version 1.3.3, and differentially expressed genes were assessed using Cuffdiff, a part of the Cufflinks version 1.3.0 package (14). A scatter plot was generated using multiplot function from Genepattern (15).
Differentially expressed genes (as determined by Cuffdiff, q < 0.05) were separated into the up-regulated or down-regulated list. The gene lists were further filtered to eliminate low expressing genes (Cuffdiff expression value <1). The filtered gene lists were separately entered into DAVID to identify enriched Gene Ontology (GO) terms. The resulting functional clusters were filtered based on enrichment score and p value. Classification stringency was set to “high,” which resulted in a more defined GO terms with sizable number of genes grouped into annotated clusters.
Cell Culture and Reagents
C2C12 cells (American Type Culture Collection) were maintained at 37 °C with 5% CO2 and subcultured at ∼60–70% confluency. Unmodified and stable C2C12 cells were cultured in DMEM supplemented with 10% FBS (Invitrogen). Viral packaging cell line 293T cells were maintained in the same media. WT AMPKγ1, AMPKγ1 H150R, WT AMPKα2, and K45R AMPKα2 were obtained from Addgene. Working concentrations of 200 μm 5-aminoimidazole-4-carboxyamide ribonucleoside (AICAR) (Sigma-Aldrich), 100 ng ml−1 EGF (MP Biomedicals), 100 μm H2O2 (Thermo Fisher Scientific), 200 nm insulin (Sigma-Aldrich), and 10 ng ml−1 TGF-β (PeproTech) were prepared from stocks dissolved in water. Working concentrations of 4 μm ionomycin (Sigma-Aldrich) and 20 μm PD98059 (EMD Biosciences) were prepared from stocks dissolved in Me2SO. Glucose uptake was measured using a glucose assay kit (Biovision) according to the manufacturer's instructions. Briefly, 1 × 105 cells were plated onto 10-cm plates, and the media were replaced 8 h later after cell attachment. The cells were incubated for 48 h, after which glucose consumption was calculated by subtracting glucose concentrations in culture media from the original 25 mm glucose concentration in DMEM. Isocitrate dehydrogenase 3 (IDH3) activity converting NAD+ to NADH was measured in extracts from 50 mg of mouse ventricular tissue using the isocitrate dehydrogenase activity assay kit (Biovision) according to the manufacturer's instructions.
RNA Isolation and qPCR
Total RNA was isolated using TRIzol and cDNAs generated from 1 μg primed with a 1:1 ratio of random hexameric primers and oligo(dT) using SuperScript Reverse Transcriptase II (Invitrogen). qPCR was performed on an ABI 7300 real time PCR system (Applied Biosystems) using SYBR green (USB). qPCR primers for Acadl, Atf4, cFos, cJun, Col1a1, Col1a2, Elk4, Fn1, Hprt, Idh3a, Myl7, Myl4, Nppa, Nppb, Pdk4, Pkm2, and Sln have been described elsewhere (6–8, 16, 17). Additional primers were generated using Primer3 and are listed in supplemental Table S3. Hprt mRNA was assessed to ensure equal fidelity in enzymatic reactions and was used as internal control to normalize qPCR results. Fold changes in gene expression were determined by the ΔΔCt method and are presented as fold change over WT controls.
Protein Extraction and Western Blot Analysis
Whole cell extracts were isolated using radioimmunoprecipitation assay buffer (Sigma-Aldrich) with protease inhibitor mixture (Roche Applied Science), 1 mm PMSF, and 1 mm sodium vanadate. 15–30 μg were loaded for SDS-PAGE. The following antibodies were purchased from Cell Signaling Technology: phospho-ACC (antibody 3661), ACC (antibody 3676), phospho-AKT (Ser-473 antibody 4060, Thr-308 antibody 4056), AKT (antibody 4691), phospho-AMPKα (Thr-172 antibody 2535), AMPKα (antibody 2532), phospho-ERK1/2 (antibody 9101), LC3B (antibody 2775), phospho-MEK1/2 (antibody 9121), MEK1/2 (antibody 9122), phospho-mTOR (Ser-2448 antibody 2971), mTOR (antibody 2972), p62/SQSTM1 (antibody 5114), phospho-S6 (Ser-240/244 antibody 2215), S6 (antibody 2217), phospho-ULK1 (Ser-757 antibody 6888), and ULK1 (antibody 8054). The following antibodies were purchased from Santa Cruz Biotechnology: ERK1/2 (sc-94), Dusp4 (sc-1200), lamin A (sc-20680), β-actin (sc-47778), and α-tubulin (sc-12462-R). Quantification of blots was performed with ImageJ (18), normalized to loading controls as indicated, and presented as arbitrary units or fold change over untreated or WT controls.
Stable Transgene and shRNA Expression in C2C12 Cells
H222P-lamin A point mutant was generated from WT cDNA using a QuikChange II site-directed mutagenesis kit (Stratagene), which replaced an adenine to a cytosine at nucleotide 665. WT lamin A or H222P-lamin A and FLAG coding sequences were cloned into pBabe-puro (Addgene). Murine Dusp4 cDNA was isolated from reverse transcribed C2C12 mRNA and cloned into pBabe-puro. pBabe-retro vectors encoding lamin A, H222P-lamin A, Dusp4, GFP-LC3 (Addgene), and pBabe vector with no insert were co-transfected with pCL-Eco into 293T cells to generate retroviruses. For stable knockdown of Dusp4, we used three independent shRNAs in pLKO.1 lentiviral vector (Open Biosystems) targeting the following sequences: CAATCACTTTGAGGGTCATTA (Sh1), CGAGTACATCGACGCAGTAAA, and GTCTCTTCAGACTGTCCCAAT (data not shown). The lentiviral vectors were co-transfected into 293T cells with the packaging vectors pCMV-dR8.2 dvpr and pCMV-VSV-G (Addgene). DNA sequencing was performed at the DNA sequencing lab at Columbia University Medical Center to confirm all constructs. Virus-infected cells were selected with 2 μg ml−1 puromycin.
Microscopy
Immunofluorescence microscopy was performed on stable C2C12 cells using standard protocols with FLAG antibodies (Sigma-Aldrich). Immunofluorescence images were captured using a LSM 510 confocal laser scanning system (Carl Zeiss) in the Confocal and Specialized Microscopy Core at the Herbert Irving Comprehensive Cancer Center (Columbia University Medical Center) using the 488-nm line of argon laser. For fluorescence microscopy of stable C2C12 cells expressing GFP-LC3B, fixed cells were counterstained with TO-PRO-3 (Invitrogen) and visualized under a DP72 fluorescence microscope (Olympus). Fluorescent images were captured using DP72 basic software package, and the image analysis (puncta count) was performed manually using ImageJ (18).
Transthoracic Echocardiography
Dusp4 transgenic mice were anesthetized with 1–2% isoflurane and placed on a heating pad (37 °C) attached to an electrocardiographic monitor. Echocardiography was performed using Vevo 770 imaging system (VisualSonics) equipped with a 30-MHz transducer. The parameters were measured for at least three cardiac cycles. A “blinded” echocardiographer performed the examinations and interpreted the results.
Cell Viability Assay and Annexin V/Propidium Iodide Staining
Cell viability was determined by trypan blue exclusion assay. Annexin V/propidium iodide staining was performed using the Dead Cell apoptosis kit with annexin V Alexa Fluor® 488 and propidium iodide (Invitrogen) according to the manufacturer's instructions. Fluorescence signals were acquired using FACSCalibur (Becton Dickinson).
Glucose Deprivation and Autophagy Induction
To induce autophagy, stable C2C12 cells cultured in DMEM containing 10% FBS were gradually adapted to 10% bovine calf serum (BCS) (Invitrogen) over a 48-h period. Following adaptation to BCS, the cells were rinsed three times with PBS and incubated with glucose and sodium pyruvate-free DMEM (Invitrogen) with or without 10% BCS for indicated times. Alternatively, C2C12 cells in DMEM with 10% BCS were treated with 100 nm temsirolimus (Sigma-Aldrich) for 6 h. Lysosomal protease inhibition during glucose deprivation was performed by adding 10 μg ml−1 pepstatin and 10 μg ml−1 leupeptin (both Sigma-Aldrich) in glucose- and sodium pyruvate-free DMEM with 10% BCS.
Statistical Analysis
Graphpad (Prism Software) was used to perform statistical analyses. Statistical significance was determined by a two-tailed, unpaired Student's t test with a value of p < 0.05 considered significant. Values with error bars shown in the figures are the means ± standard errors of means. The sample sizes are indicated in the figure legends. Statistically significant enrichment with DAVID was based on the Fisher's exact test p value with p < 0.05 considered significant and enrichment score (negative log of averaged p values of all subclusters).
RESULTS
Dusp4 Is Highly Expressed in Hearts of Mouse Models of LMNA Cardiomyopathy
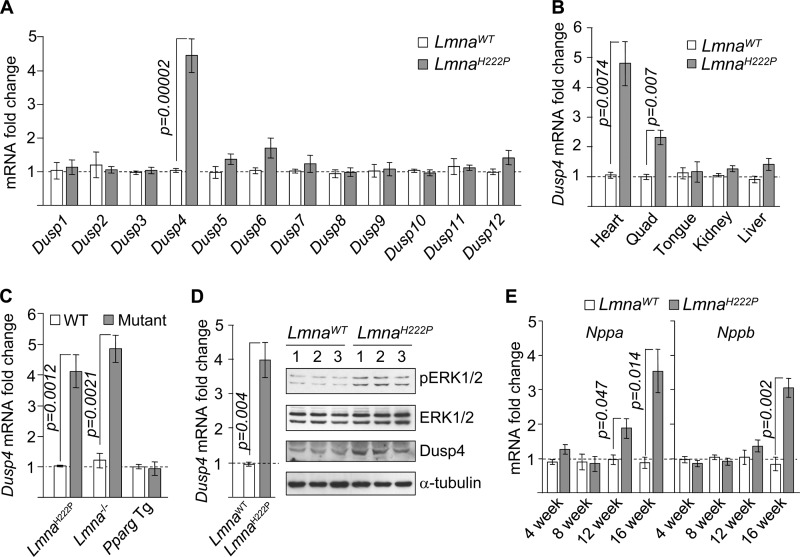
We originally focused on the Dusp family of phosphatases, with the initial intent of understanding the mechanism of ERK1/2 activation. This was based on the observation that activation of MEK1/2, an upstream kinase that phosphorylates ERK1/2, was not observed in hearts of LmnaH222P/H222P mice (data not shown), suggesting a defect in ERK1/2 dephosphorylation. Of 12 Dusp mRNAs assessed by quantitative real time PCR (qPCR), we observed a specific increase of Dusp4 (∼4-fold) in hearts of 16-week-old male LmnaH222P/H222P mice (Fig. 1A). This was contrary to our expectation and suggested that high Dusp4 expression resulted from ERK1/2 activation. Enhanced Dusp4 mRNA expression was observed in cardiac and to a lesser extent (∼2-fold) in quadriceps muscle but not in other tissues examined (Fig. 1B). It was observed in hearts from LmnaH222P/H222P and Lmna−/− mouse models of LMNA cardiomyopathy (13, 19) but not from PPARγ transgenic mice (20) that also develop dilated cardiomyopathy (Fig. 1C). Dusp4 mRNA and the encoded protein were both increased in hearts of 4-week-old LmnaH222P/H222P mice, coincident with ERK1/2 activation (Fig. 1D). Enhanced Dusp4 expression occurred prior to detectable signs of cardiomyopathy (13) and up-regulation of natriuretic factors encoded by Nppa and Nppb (Fig. 1E), which are expressed as a consequence of left ventricular dilatation. Similarly high Dusp4 expression occurred in female LmnaH222P/H222P mice but with delayed kinetics (data not shown), consistent with the later onset of cardiomyopathy relative to male mice (13). Hence, Dusp4 appears to be involved in the development of cardiomyopathy rather than affected as a consequence.
FIGURE 1.
Dusp4 is highly expressed in hearts of LmnaH222P/H222P mice. A, qPCR analysis of Dusp1–Dusp12 mRNA in hearts of 16-week-old LmnaH222P/H222P (LmnaH222P) and WT (LmnaWT) mice (n = 5). B, qPCR analysis of Dusp4 mRNA in various tissues from 16-week-old LmnaH222P/H222P and WT mice (n = 3). C, qPCR analysis of Dusp4 mRNA in hearts from 8-week-old LmnaH222P/H222P mice (n = 3), 6-week-old Lmna−/− mice (n = 3), and 8-week-old PPARγ transgenic (Pparg Tg) mice (n = 4). WT denotes controls for the respective mouse models. D, qPCR analysis of Dusp4 mRNA (left panel) and Western blot (right panel) of phospho-ERK1/2 (pERK1/2), total ERK1/2 (ERK1/2), DUSP4, and α-tubulin in hearts of 4-week-old LmnaH222P/H222P and WT mice (n = 3). The numbers above the blots denote individual samples. E, qPCR analysis of Nppa and Nppb mRNA in hearts of 4–16-week-old LmnaH222P/H222P and WT mice (n = 3).
H222P-Lamin A Induces Dusp4 in Energy-deprived Cells
To dissect the molecular mechanism(s) responsible for enhanced Dusp4 expression in hearts of LmnaH222P/H222P mice, we used C2C12 cell culture model. Although C2C12 cells are myoblasts of murine skeletal muscle origin, they are relatively easy to culture and genetically manipulate and have or can adopt some features of cardiomyocytes (21, 22). We used retroviral transduction to generate cells stably expressing either FLAG-tagged WT (C2-WT) or a variant (C2-H222P) lamin A identical to that in LmnaH222P/H222P mice. These cells stably expressed the respective proteins, but because expression of H222P-lamin A alone did not induce Dusp4 expression or ERK1/2 activation, we tested various conditions to recapitulate the pattern of expression in the hearts of LmnaH222P/H222P mice.
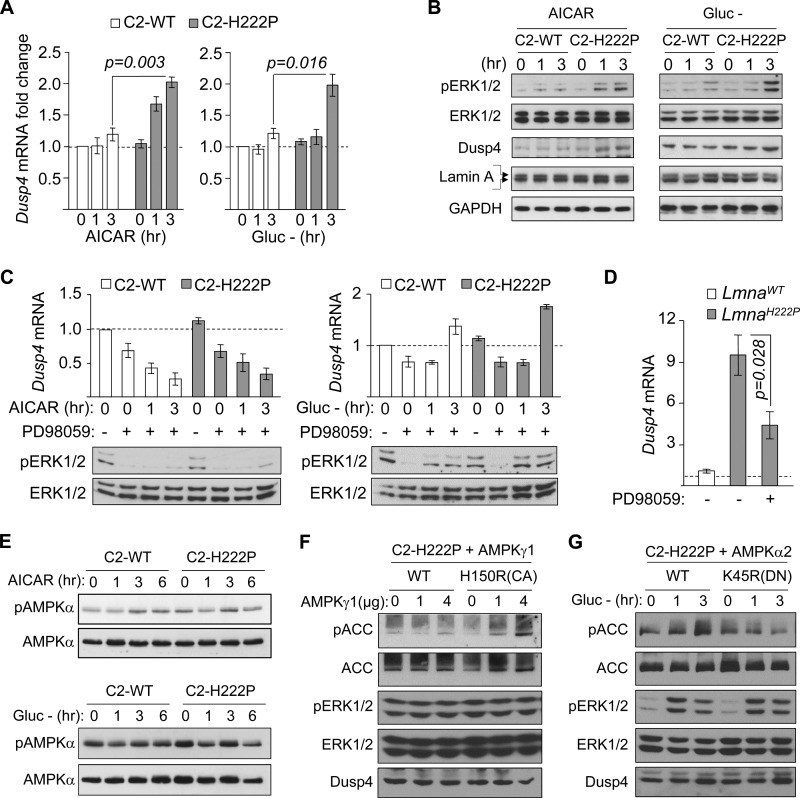
Of several stimuli tested, AICAR and glucose deprivation significantly induced Dusp4 mRNA expression (∼2-fold) in C2-H222P but not C2-WT cells, suggesting that C2C12 cells expressing H222P-lamin A had enhanced Dusp4 mRNA expression under energy deficit (Fig. 2A). To test whether the enhanced Dusp4 expression was correlated with ERK1/2 activation, as in hearts of LmnaH222P/H222P mice, we assessed phosphorylated ERK1/2 levels in C2-WT and C2-H222P cells subjected to AICAR or glucose deprivation. As expected, ERK1/2 phosphorylation was increased in C2-H222P relative to C2-WT cells (Fig. 2B). Responses to AICAR or glucose deprivation were comparable between unmodified C2C12 and cells stably transduced with retrovirus carrying empty vector or C2-WT cells (data not shown). These results demonstrated a correlation between enhanced Dusp4 expression and ERK1/2 activation in energy-deprived C2-H222P, recapitulating the expression pattern in hearts from LmnaH222P/H222P mice. We next asked whether ERK1/2 activation drives the enhanced Dusp4 expression. We inhibited MEK1/2 with PD98059 in C2-WT and C2-H222P cells stimulated by either AICAR or glucose deprivation. In cells treated with AICAR, PD98059 pretreatment decreased ERK1/2 phosphorylation and abrogated Dusp4 mRNA induction (Fig. 2C, left panel). In contrast, cells subjected to glucose deprivation circumvented MEK1/2 inhibition to increase ERK1/2 phosphorylation and Dusp4 mRNA induction (Fig. 2C, right panel). These results suggest that ERK1/2 activity is required for Dusp4 expression. Similar results were obtained with U0126, another MEK1/2 inhibitor (data not shown). We also measured Dusp4 mRNA in the hearts of 16-week-old LmnaH222P/H222P mice treated with 1 mg kg−1 day−1 of PD98059 for 4 weeks, which improves cardiac function (7). Dusp4 mRNA expression was reduced by ∼60% in PD98059-treated mice relative to controls given Me2SO (Fig. 2D). Collectively, these data demonstrated that high cardiac Dusp4 expression in LmnaH222P/H222P mice and energy-deprived C2-H222P was primarily ERK1/2-dependent.
FIGURE 2.
Dusp4 expression is enhanced in response to energy deficit in C2C12 stably expressing H222P-lamin A. A, qPCR analysis of Dusp4 mRNA 0, 1, and 3 h after AICAR treatment (left panel) or glucose deprivation (Gluc −, right panel) in C2C12 cells stably expressing FLAG-tagged WT lamin A (C2-WT) or H222P-lamin A (C2-H222P) (n = 3 experiments). B, Western blot of protein extracts 0, 1, and 3 h after AICAR treatment (left panel) or glucose deprivation (Gluc −; right panel) in C2C12 stably expressing C2-WT or C2-H222P. The blots were probed with antibodies against phospho-ERK1/2 (pERK1/2), total ERK1/2 (ERK1/2), DUSP4, and lamin A. Two bands in the lamin A blot represent the FLAG-tagged and endogenous proteins. A representative blot is shown from five experiments. C, qPCR analysis of Dusp4 mRNA and representative Western blot of pERK1/2 and ERK1/2 in C2-WT and C2-H222P subjected to 30 min of pretreatment with PD98059 followed by PD98059 and AICAR co-treatment (left panel) or glucose deprivation (right panel) for 0, 1, and 3 h. Dusp4 mRNA values are presented as fold change over untreated C2-WT. Untreated and treated with PD98059 are denoted by − and +, respectively (n = 3 experiments). D, qPCR analysis of Dusp4 mRNA in hearts of LmnaH222P/H222P (LmnaH222P) and WT (LmnaWT) mice subjected to 4 weeks of treatment with Me2SO or PD98059. Dusp4 mRNA values are presented as fold change over WT. Treatment with or without PD98059 (Me2SO given in place of PD98059) are denoted by − and +, respectively (n = 4). E, Western blot analysis of phospho-AMPK and AMPK in C2-WT and C2-H222P treated with either AICAR (top panels) or subjected to glucose deprivation (Gluc −; bottom panels) for 0, 1, 3, and 6 h. A representative blot from three experiments is shown. F, Western blot analysis of phospho-ACC, ACC, phospho-ERK1/2, ERK1/2, and DUSP4 in C2-H222P transiently transfected with increasing concentration of WT AMPKγ1 or H150R constitutively active (CA) AMPKγ1 point mutant expression vector. A representative blot from two experiments is shown. G, Western blot analysis of phospho-ACC, ACC, phospho-ERK1/2, ERK1/2, and DUSP4 in C2-H222P transiently transfected with WT AMPKα2 or K45R dominant negative (DN) AMPKα2 point mutant expression vector subjected to 0, 1, and 3 h of glucose deprivation. A representative blot from two experiments is shown.
Given that the enhanced Dusp4 expression was observed in C2-H222P cells only after subjecting them to conditions that activate AMP-activated protein kinase (AMPK), we assessed the phosphorylation status of Thr-172 on the AMPKα subunit and observed that phosphorylation of this site does not significantly increase with either AICAR treatment or glucose deprivation (Fig. 2E). However, given that AMP and AMP-mimetics like AICAR can induce allosteric activation of AMPK without increasing Thr-172 phosphorylation, we further assessed its potential involvement by utilizing constitutively active and dominant negative forms of AMPK. When we expressed constitutively active point mutant AMPKγ1 H150R in C2-H222P cells, ERK1/2 phosphorylation or Dusp4 expression were not enhanced (Fig. 2F). This suggested that AMPK activation in and of itself was not sufficient to drive Dusp4 expression. In addition, when C2-H222P cells expressing dominant negative AMPKα2 K45R were subjected to glucose deprivation, we still observed enhanced ERK1/2 phosphorylation and Dusp4 expression (Fig. 2G). These results suggest that AMPK is not significantly involved in Dusp4 expression in C2-H222P cells after AICAR treatment or under glucose deprivation.
Cardiac-selective Overexpression of Dusp4 Impairs Heart Function
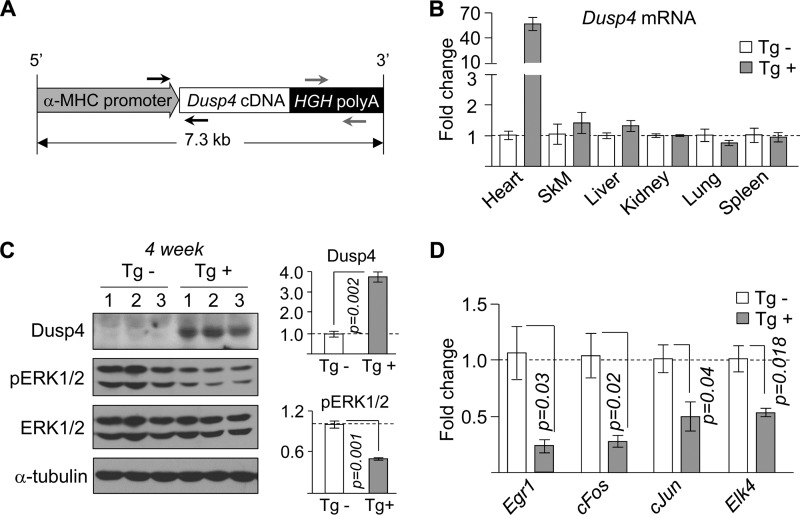
To directly determine whether enhanced cardiac Dusp4 expression contributes to development of cardiomyopathy, we generated a transgenic mouse line with cardiac-selective overexpression (Fig. 3, A and B). Two founders were produced, one of which died at 8 weeks of age. The transgenic mice bred normally, producing 42% male and 58% female (of 131 total mice) offspring with the transgene detected in ∼50%. DUSP4 protein expression was ∼3.7-fold higher in hearts of transgene positive (Tg+) mice compared with transgene negative littermates (Tg−), with a corresponding decrease in phosphorylated ERK1/2 (Fig. 3C) and expression of mRNAs encoded by genes it activates (Fig. 3D). Therefore, the cardiac phenotypes observed in these transgenic mice are the result of Dusp4 expression alone without the influence of activated ERK1/2, which is distinct from the expression profile in hearts of LmnaH222P/H222P mice (in which expression of both Dusp4 and phosphorylated ERK1/2 are enhanced). Overexpression of Dusp4 did not disrupt the normal balance of other Dusp family members, and no significant difference in body weight between Tg+ and Tg− mice was observed (data not shown).
FIGURE 3.
Cardiac-selective overexpression of Dusp4 reduces ERK1/2 signaling. A, schematic diagram of targeting vector used to generate Dusp4 transgenic mice. α-MHC denotes α-myosin heavy chain, and HGH poly(A) denotes human growth hormone polyadenylation signal. The black and gray arrows indicate annealing positions of genotyping primers. B, qPCR analysis of Dusp4 mRNA in various tissues (SkM denotes skeletal muscle) from 4-week-old Dusp4 Tg+ and Tg− littermates (n = 3 samples). C, Western blot of phosphorylated ERK1/2 (pERK1/2), total ERK1/2 (ERK1/2), DUSP4, and α-tubulin in hearts of 4-week-old Dusp4 Tg+ and Tg− littermates (left panel) (n = 3 samples). The numbers above the blots denote individual samples. Quantification of DUSP4 and pERK1/2 normalized to α-tubulin and ERK1/2, respectively, is presented as fold change over WT (right panel). D, qPCR analysis of expression of genes downstream of ERK1/2 in 4-week-old Dusp4 Tg+ and Tg− littermates (n = 3 samples).
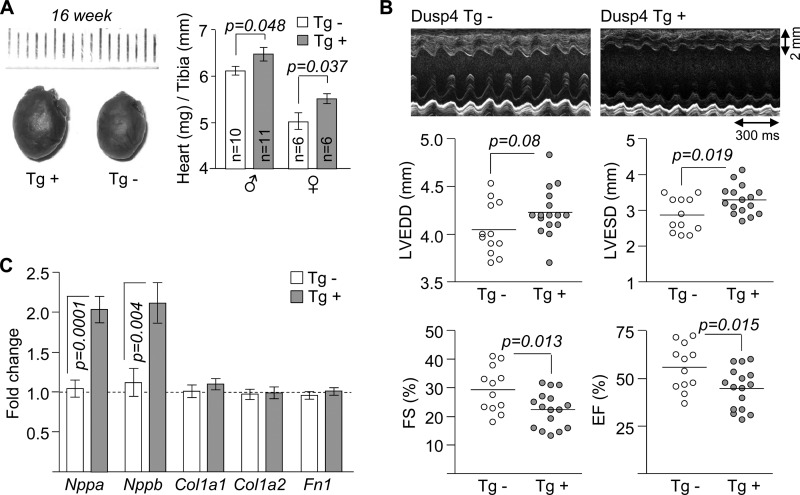
At 16 weeks of age, there was a subtle but statistically significant enlargement of hearts in both male and female Dusp4 Tg+ mice (Fig. 4A) that was not present at 8 weeks (data not shown). Motion mode echocardiographic tracings and data derived from them revealed significantly increased left ventricular end systolic diameters, decreased fractional shortening, and decreased ejection fraction but not increased left ventricular end diastolic diameters (Fig. 4B). These results demonstrated that Dusp4 overexpression caused cardiac dysfunction with an apparent defect in cardiac contractility, which recapitulates some but not all phenotypes in LmnaH222P/H222P mice (13). Hearts from Tg+ mice also exhibited significantly enhanced expression Nppa and Nppb, consistent with left ventricle dilatation, but not Col1a1, Col1a2, and Fn1, encoding proteins involved in fibrosis, a predominant feature of later stage LMNA cardiomyopathy (Fig. 4C).
FIGURE 4.
Cardiac-selective overexpression of Dusp4 impairs heart function. A, representative hearts (left panels) and heart weight to tibia length ratios (right panels) of 16-week-old Dusp4 Tg+ and Tg− littermates. The hash marks above hearts correspond to millimeters. B, representative motion mode echocardiographic tracings (top panels) and graphic representations for left ventricular end diastolic diameter (LVEDD), left ventricular end systolic diameter (LVESD), fractional shortening (FS), and ejection fraction (EF) of 16-week-old Dusp4 Tg+ mice and Tg− littermates (n = 12 mice for Tg− and n = 16 for Tg+). Each circle represents an individual mouse, and the horizontal bars denote means. C, qPCR analysis of Nppa, Nppb, Col1a1, Col1a2, and Fn1 mRNA expression in hearts of 16-week-old Dusp4 Tg+ and Tg− littermates (n = 10 samples).
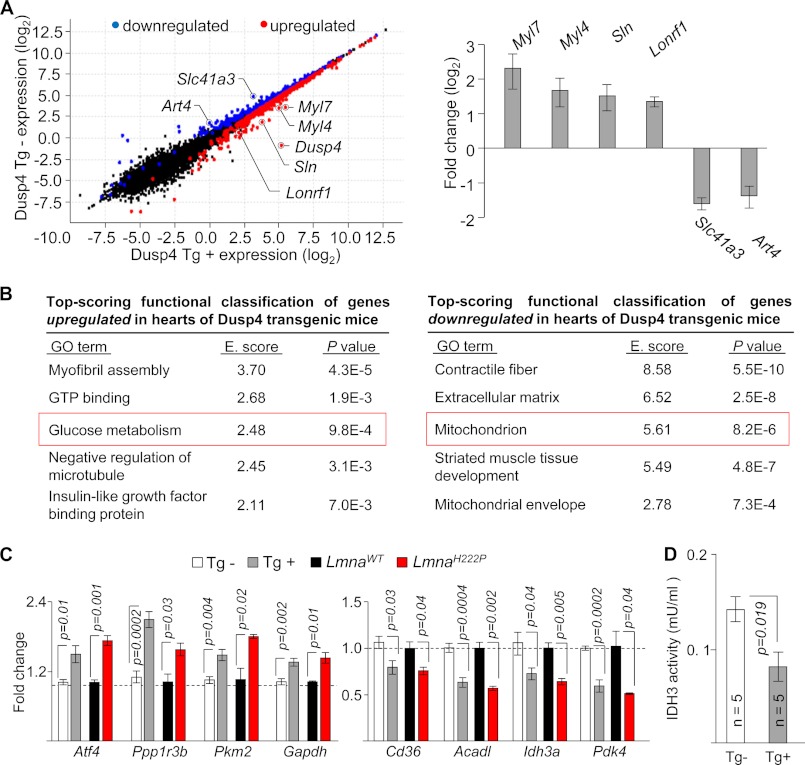
Dusp4 Alters Expression of Genes Involved in Metabolism
To obtain a genome-wide view of the functional impact of heart-selective Dusp4 overexpression, we assessed cardiac transcriptomes of Tg+ and Tg− mice (n = 3/group) by RNAseq. We used ventricular tissue from 8-week-old male mice, which is prior to observable signs of cardiomyopathy, to minimize potential secondary effects of disease. Of 22,310 mapped genes, ∼10,700 genes (cutoff based on fragments/kilobase of exons/million bases mapped > 1) were expressed in heart samples (see supplemental data), consistent with previous RNAseq results using mouse heart tissue (23, 24). At 8 weeks of age, Dusp4 overexpression led to significant (q < 0.05) up-regulation of 410 genes, with Dusp4 and fetal isoforms of myosin light chains among the highest scoring and significant down-regulation of 212 genes (Fig. 5A, left panel). Analysis by qPCR was performed to validate a select number of genes detected by RNAseq (Fig. 5A, right panel). We next explored functional classification of genes with expression that were significantly altered by Dusp4 overexpression. Enrichment analysis using GO terms with up-regulated and down-regulated genes revealed alterations in biological processes relating to cardiac metabolism (Fig. 5B). Among the top scoring GO terms enriched with up-regulated genes included “glucose metabolism” (Fig. 5B, left panel, and supplemental Table S1). Conversely, top scoring GO terms enriched with down-regulated genes included “mitochondrion” and “mitochondrial envelope” (Fig. 5B, right panel, and supplemental Table S2). This suggested that Dusp4 overexpression caused metabolic disturbances in the heart. We therefore examined expression of selected genes listed under “glucose metabolism” and “mitochondrion” by qPCR and confirmed that these were bona fide changes in hearts of Dusp4 Tg+ mice (Fig. 5C). Moreover, these changes were also observed in the hearts of LmnaH222P/H222P mice with high cardiac Dusp4 expression (Fig. 5C). To further confirm the functional consequence of the altered gene expression, we assessed enzymatic activity of IDH3, for which expression of the α subunit was reduced in hearts of Dusp4 Tg+ mice (Fig. 5D). Ventricular tissue isolated from 8-week-old male Dusp4 Tg+ mice exhibited reduced IDH3 activity relative to Tg− littermates. These results collectively revealed abnormalities in cardiac metabolism in Dusp4 transgenic mice.
FIGURE 5.
Dusp4 overexpression alters metabolic gene expression. A, scatter plot (left panel) of expression level per genes (based on Cuffdiff analysis) from RNAseq results of 8-week-old hearts from Dusp4 Tg+ and Tg− mice. Genes showing expression of 0 were filtered out to enable plotting. Blue and red circles denote down-regulated and up-regulated genes, respectively, in Dusp4 Tg+ mice relative to Tg− littermates. Black circles denote genes with no significant difference. qPCR validation of Myl7, Myl4, Sln, Lonrf1, Slc41a3, and Art4 mRNA expression is shown on the right. The bar graph represents fold change over transgenic negative controls (n = 6 samples). B, top scoring functional classification of genes up-regulated (left panel) or down-regulated (right panel) in hearts of Dusp4 Tg+ mice using DAVID. GO terms with higher enrichment score (E. score) indicate more significant enrichment. The red boxes highlight GO terms selected for further analysis. C, qPCR validation of mRNA expression of genes associated with “glucose metabolism” that are up-regulated (left panel) and genes associated with “mitochondrion” that are down-regulated (right panel) in hearts of 8-week-old Dusp4 Tg+ mice (n = 6/group), 16-week-old LmnaH222P/H222P mice (n = 3/group), and their respective controls. D, IDH3 enzymatic activity in ventricular tissue isolated from 8-week-old male Dusp4 Tg+ and Tg− mice. mU denotes milliunit in which one unit of IDH3 is the amount of enzyme that will generate 1.0 μmol of NADH/min at pH 8 at 37 °C.
Dusp4 Overexpression Activates AKT-mTOR and Impairs Autophagy in the Heart
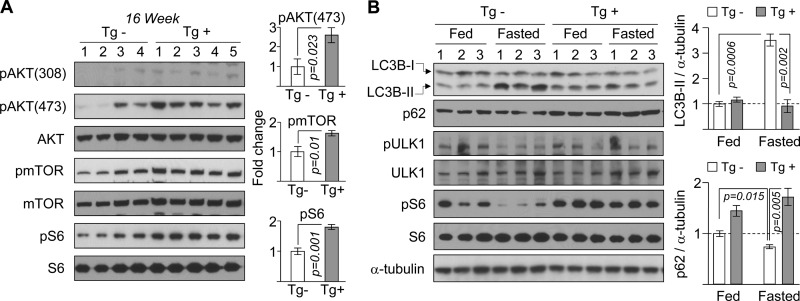
We then sought an explanation for the altered expression of genes in metabolism in hearts of LmnaH222P/H222P and Dusp4 transgenic mice. Given that the AKT-mTOR signaling pathway, which plays a central role in regulating metabolism, is activated in the hearts of LmnaH222P/H222P mice (9, 10), we hypothesized that similar activation occurred in hearts of Dusp4 transgenic mice. Indeed, although some variability was observed in Dusp4 Tg− controls, we observed statistically significant enhancement in the expression of phosphorylated AKT in hearts of Dusp4 Tg+ mice (Fig. 6A). Enhanced phosphorylation of mTOR and its downstream target S6 ribosomal protein was also observed (Fig. 6A). Therefore, Dusp4 overexpression correlated with activated AKT-mTOR signaling in both LmnaH222P/H222P and Dusp4 Tg+ mice. A pathogenic consequence of activated AKT-mTOR in hearts of LmnaH222P/H222P mice is impaired autophagy (9), which is a lysosome-dependent recycling of damaged/cytoplasmic cargo, enclosed in double-membrane vesicles termed autophagosomes. Given the identical signaling profile in hearts of Dusp4 transgenic mice, we examined autophagy in hearts of these mice fasted for 24 h. This was achieved by assessing levels of lipidated LC3B (LC3B-II) produced during autophagosome formation and expression of p62, an autophagosome-associated protein that is degraded with autophagocytized cargo (25, 26). In hearts of Tg− mice, fasting resulted in a significant increase in LC3B-II and a decrease in p62 levels (Fig. 6B), indicative of enhanced autophagy. In contrast, no significant differences were observed in LC3B-II and p62 levels between fed and fasted Tg+ mice, demonstrating impaired autophagy, a phenotype consistent with that observed in the LmnaH222P/H222P mice (9). Likewise, whereas the 24-h fasting reduced expression of phosphorylated serine 757 ULK1 and phosphorylated S6 in Tg− controls, no significant changes in the phosphorylation status of these proteins were observed in the Tg+ mice (Fig. 6B), further implicating involvement of mTOR in inhibiting autophagy.
FIGURE 6.
Dusp4 overexpression activates AKT-mTOR and impairs autophagy in Dusp4 Tg+ mice. A, Western blot of phosphothreonine AKT (pAKT(308)), phosphoserine AKT (pAKT(473)), AKT, phospho-mTOR (pmTOR), mTOR, phospho-S6 (pS6), and S6 in hearts of 16-week-old Dusp4 Tg+ and Tg− mice (left panel). The numbers above the blots denote individual heart samples. Quantification of phospho-proteins, normalized to their respective total proteins, are presented as fold change over Tg− controls (right panel) (n = 4 for Tg− and n = 5 for Tg+). B, Western blot of LC3B, p62, phosphoserine 757 ULK1 (pULK1), ULK1, phospho-S6 (pS6), S6, and α-tubulin in hearts of 16-week-old Dusp4 Tg+ mice and Tg− littermates fasted for 24 h (Fasted) or fed ad libitum (Fed) (top right panel). The numbers above the blots denote individual heart samples. LC3B-I denotes nonlipidated, and LCB-II denotes lipidated LC3B. LC3B-II and p62 levels are presented as fold change over fed mice (bottom right panel) (n = 3).
Dusp4 Overexpression Activates AKT-mTOR and Impairs Autophagy in C2C12 Myoblasts
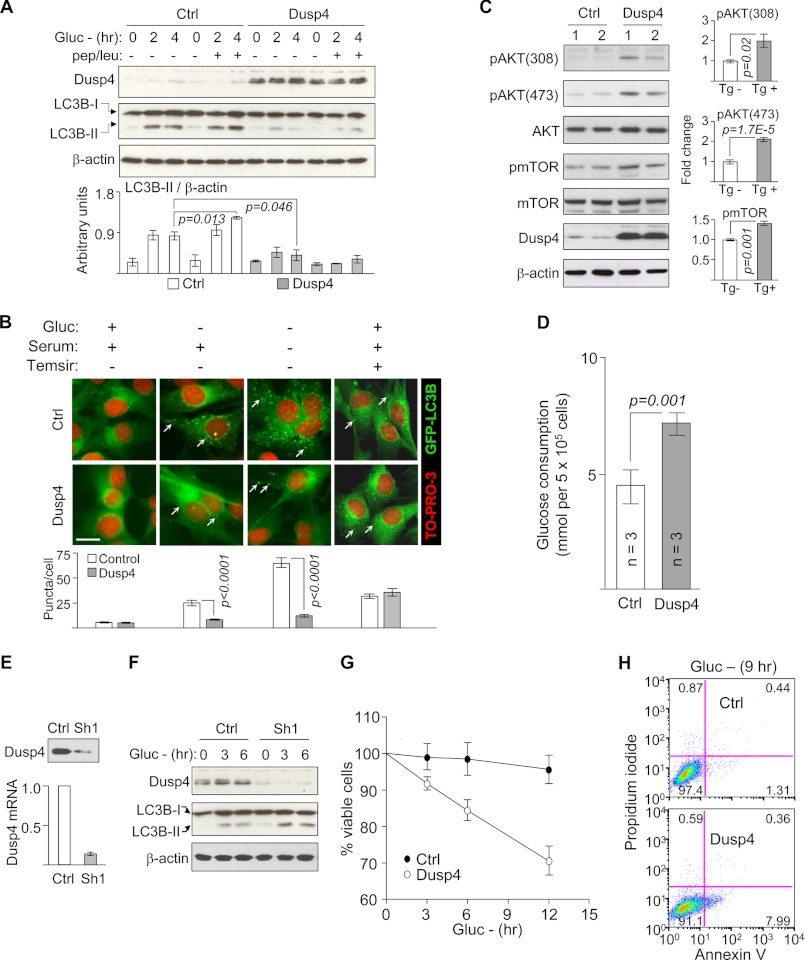
To validate the effects of Dusp4 on autophagy in cultured cells, we generated C2C12 cells stably overexpressing the protein. Autophagy induction by glucose deprivation enhanced LC3B-II expression in control cells containing empty vector but not in cells expressing Dusp4 (Fig. 7A). To confirm that this resulted from impaired autophagosome formation rather than excessive degradation, we treated cells with lysosomal protease inhibitors pepstatin and leupeptin during glucose deprivation. In glucose-deprived control cells, protease inhibitors further enhanced LC3B-II levels (Fig. 7A), demonstrating autophagosome accumulation. In contrast, they had negligible effects in glucose-deprived Dusp4-overexpressing cells, suggesting impaired autophagosome formation. As an alternative approach, we generated C2C12 cells co-expressing Dusp4 and GFP-tagged LC3B. Autophagy induction elicits relocalization of GFP-LC3B from diffusely cytoplasmic to punctate structures indicative of LC3B-II incorporation into autophagosomes (27). Under basal conditions, few GFP-LC3B puncta were observed in control and Dusp4-overexpressing C2C12 cells (Fig. 7B). Upon deprivation of either glucose or glucose and serum, there were significantly fewer puncta in Dusp4-overexpressing cells compared with controls, confirming the negative effect of Dusp4 on autophagy. Because impaired autophagy in hearts of LmnaH222P/H222P mice can be reversed by inhibiting mTOR (9), we assessed whether this is the case in Dusp4-overexpressing C2C12 cells. We treated Dusp4-overexpressing cells with the rapamycin analog temsirolimus and observed significantly increased formation of GFP-LC3B puncta in both control and Dusp4-overexpressing cells (Fig. 7B). This demonstrated that autophagic responses were intact in Dusp4-overexpressing cells and could be activated if mTOR is inhibited. This is consistent with our observations in LmnaH222P/H222P and Dusp4 Tg+ mice and confirms activated mTOR as the culprit in impaired autophagy. Indeed, Dusp4 overexpression alone was sufficient to enhance AKT and mTOR phosphorylation in C2C12 cells relative to controls (Fig. 7C). To further confirm activation of AKT, we measured glucose uptake in Dusp4-overexpressing cells, which is regulated by AKT (28). Glucose consumption was increased in Dusp4-overexpressing cells relative to controls (Fig. 7D), demonstrating an additional functional consequence of AKT activation. These results confirmed that Dusp4 overexpression impairs autophagy by activating AKT-mTOR signaling. In a reciprocal experiment, stable knockdown of Dusp4 in C2C12 cells exhibited enhanced autophagy under glucose deprivation (Fig. 7, E and F).
FIGURE 7.
Dusp4 overexpression activates AKT-mTOR and impairs autophagy in C2C12 cells. A, representative Western blot of DUSP4, LC3B, and β-actin in C2C12 stably overexpressing Dusp4 deprived of glucose (Gluc −) for 0, 2, and 4 h or co-treated with pepstatin and leupeptin (pep/leu) (top panel). Ctrl denotes control: C2C12 transduced with empty vector. Untreated and treated with pepstatin/leupeptin are indicated by − and +, respectively. Quantification of LC3B-II is presented in arbitrary units (bottom panel; n = 3 experiments). B, fluorescent micrograph of GFP-LC3B co-expressed in Dusp4-overexpressing or control C2C12 subjected to 9 h of glucose deprivation, 6 h of glucose + serum deprivation, or 6 h of temsirolimus (Temsir) (top panel). White arrows show GFP-LC3B puncta. Removal and addition of glucose, serum, or temsirolimus in culture media are denoted by − and +, respectively. The nuclei were counterstained with TO-PRO-3. Scale bar, 20 μm. Enumerated GFP-LC3B puncta/cell (bottom) (n = 60 cells/condition from three experiments). C, Western blot of pAKT (S473, T308), AKT, pmTOR, mTOR, DUSP4, and β-actin in control and Dusp4-overexpressing C2C12. The numbers above the blots denote individual preparations. Representative blots are shown from two experiments. D, glucose uptake measurements in control and Dusp4-overexpressing C2C12 after 48 h in culture. E, representative Western blot (top panel) and qPCR (bottom panel) of C2C12 with stable knockdown of Dusp4. Ctrl and Sh1 denote C2C12 stably transduced with lentivirus carrying empty vector and shRNA that targets Dusp4, respectively. Dusp4 mRNA values are presented as fold change relative to control (n = 3 experiments). F, Western blot of DUSP4, LC3B-II, and β-actin in C2C12 control or expressing Sh1 deprived of glucose (Gluc −) for 0, 3, and 6 h. LC3B-I denotes nonlipidated LC3B. A representative blot is shown from three experiments. G, cell viability analysis on control and Dusp4-overexpressing cells deprived of glucose (Gluc −) for 0, 3, 6, and 12 h). The data are presented as percentages of viable cells at just prior to glucose deprivation (n = 5 for each time point). H, annexin V/propidium iodide staining of control and Dusp4-overexpressing cells deprived of glucose (Gluc −) for 9 h. The numbers in each quadrant represent percentages of cells. Representative flow cytometry analysis is shown from three experiments.
Given the impaired autophagy in Dusp4-overexpressing cells, we assessed whether it had pathogenic effects. Following glucose deprivation, there were less viable Dusp4-overexpressing cells compared with control cells (Fig. 7G), consistent with the well established notion that autophagy protects cells from energy deficit (25). Because cells that are unable to induce autophagy default to apoptosis (29), we reasoned that enhanced apoptosis may explain the reduced viability observed in glucose-deprived Dusp4-overexpressing cells. Indeed, annexin V/propidium iodide staining revealed that a higher percentage of Dusp4-overexpressing cells undergo apoptosis under glucose deprivation than control cells (Fig. 7H) despite the anti-apoptotic AKT signaling. These results collectively demonstrated a pathogenic consequence of Dusp4 overexpression in cells.
AKT Activation and Impaired Autophagy Is Predominantly Mediated by Dusp4
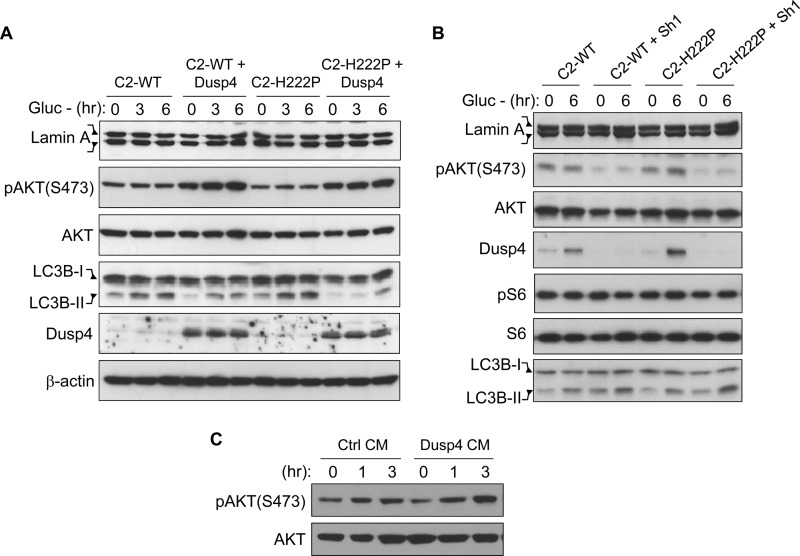
We next sought to determine whether co-expressing H222P-lamin A with Dusp4 would further activate AKT and exacerbate autophagy inhibition. We found that neither WT nor H222P-lamin A expression alone had observable effects on AKT activation and autophagy induction in glucose-deprived cells despite the ∼2-fold induction of Dusp4 in H222P-lamin A-expressing cells (Fig. 8A). In contrast, co-expression with Dusp4 activated AKT and impaired autophagy to similar levels in cells expressing either WT or H222P-lamin A. These results supported the hypothesis that Dusp4 is the main driving force behind AKT activation and autophagy impairment and that its expression level is important in mediating these effects. In a reciprocal experiment, we performed Dusp4 knockdown in C2-WT and C2-H222P cells and measured AKT activation and autophagy induction under glucose deprivation (Fig. 8B). Dusp4 knockdown decreased AKT activation and enhanced autophagy to similar levels in both C2-WT and C2-H222P cells, whereas no significant changes in phosphorylated S6 levels were observed (Fig. 8B). The S6 results were surprising given that an mTOR-containing complex mTORC1 activates p70 S6 kinase, the upstream activator of S6 (30), and may be due to the immortalized nature of C2C12 cells. Nonetheless, these results further suggested that AKT-mTOR activation and pathogenic effects of impaired autophagy are primarily mediated through Dusp4.
FIGURE 8.
AKT activation and impaired autophagy is predominantly mediated by Dusp4 via soluble factor(s). A, Western blot of lamin A, pAKT(S473), AKT, LC3B-II, DUSP4, and β-actin in C2C12 stably co-expressing either WT or H222P-lamin A with Dusp4 subjected to glucose deprivation (Gluc −) for 0, 3, and 6 h. Two bands in the lamin A blot represent the FLAG-tagged and endogenous proteins. LC3B-I denotes nonlipidated LC3B. A representative blot is shown from three experiments. B, Western blot of lamin A, pAKT(S473), AKT, DUSP4, phospho-S6, S6, and LC3B-II in C2-WT and C2-H222P with stable knockdown of DUSP4 deprived of glucose for 0 and 6 h. Two bands in the lamin A blot represent the FLAG-tagged and endogenous proteins. LC3B-I denotes nonlipidated LC3B. A representative blot is shown from three experiments. C, Western blot of pAKT(S473) and AKT in control (Ctrl) C2C12 that were treated with conditioned medium (CM) from either control or Dusp4-overexpressing C2C12 (Dusp4) for 0, 1, and 3 h. A representative blot is shown from three experiments.
Dusp4-mediated AKT Activation Is Mediated in Part through Soluble Factor(s)
To begin to dissect the mechanism how Dusp4 activates AKT, we assessed whether this effect was mediated by soluble factor(s) produced by Dusp4-overexpressing cell. We treated control C2C12 cells with conditioned medium from either Dusp4-overexpressing cells or control cells (Fig. 8C). Cells treated with conditioned media from Dusp4-overexpressing cells had higher expression of phosphorylated AKT compared with those treated with conditioned media from control cells (Fig. 8C). Cell doubling time between Dusp4-overexpressing and control cells were comparable (data not shown), suggesting that potential differences in depletion of growth factors in the FBS that were supplemented to the culture media are not responsible for the difference in AKT activation. This suggested that Dusp4-mediated AKT activation occurs in part via soluble factors produced by Dusp4-overexpressing cells.
DISCUSSION
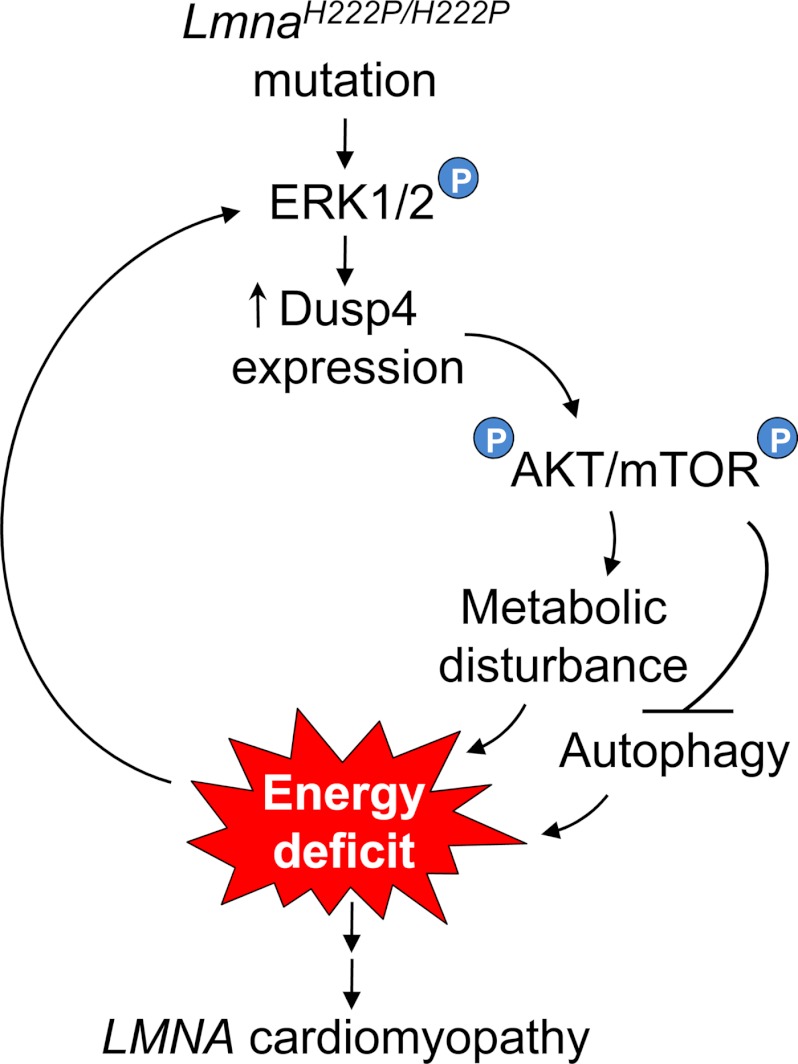
Our findings support a novel mechanism involved in the pathogenesis of LMNA cardiomyopathy (Fig. 9). In this model, LMNA mutation elicits ERK1/2-mediated Dusp4 expression and AKT activation, resulting in abnormal cardiac metabolism. Reduced IDH3 activity in heart may prevent sufficient generation of ATPs from acetyl CoA derived from fatty acid metabolism. Because normal adult hearts preferentially rely on β-oxidation of fatty acids for up to 80% of ATP (31, 32), this may result in cardiomyocyte energy deficit. AKT also activates mTOR, which inhibits autophagy and would exacerbate this energy deficit. Therefore, energy deficit in the heart caused by abnormalities in metabolism and impaired autophagy to compensate for it would perpetuate a self-reinforcing cycle of energy deficit that could over time contribute to the development of cardiomyopathy. Metabolic disturbances including reduced circulating glucose levels and blood chemistries indicative of fasting observed in several murine LMNA cardiomyopathy models (13, 33–35) support our hypothesis, but this has yet to be documented in human patients.
FIGURE 9.
Schematic diagram of proposed model of LMNA cardiomyopathy pathogenesis. The circled P represents phosphorylation. See text for details.
We invariably observed ERK1/2 activation driving enhanced Dusp4 expression in the hearts of LmnaH222P/H222P mice. Although this establishes the basis for enhanced Dusp4 expression, it also presents an apparent paradox, given that Dusp4 can establish feedback inhibition of ERK1/2 signaling (11). Hence, ERK1/2-dependent Dusp4 expression should ultimately be a self-limiting process. However, not only is ERK1/2 activation and Dusp4 expression sustained, but phosphorylated ERK1/2 is actually enriched in nuclei of cardiomyocytes from LmnaH222P/H222P mice (6). An attractive explanation is that A-type lamins function as a molecular scaffold for interaction between ERK1/2 and DUSP4 similar to their role facilitating interactions between ERK1/2 and c-Fos (5). This may be disrupted by alterations in A-type lamins that cause cardiomyopathy.
Enhanced Dusp4 expression has been observed in human hearts with idiopathic dilated cardiomyopathy (36). Reanalysis of microarray data deposited in the Gene Expression Omnibus revealed that Dusp4 mRNA expression was significantly enhanced in spontaneous hypertensive rats with heart failure (GDS3661) (37). Moreover, Dusp4 mRNA expression was enhanced in hearts with isoproterenol-induced hypertrophic cardiomyopathy but not in mice with exercise-induced cardiac hypertrophy (GDS3596) (38), further linking Dusp4 expression to pathological remodeling of the heart. In contrast, cardiac overexpression of Dusp6 does not cause cardiomyopathy but generates increased susceptibility to heart failure after prolonged pathogenic stimuli, such as aortic constriction (39). Because DUSP6 is an ERK1/2-specific phosphatase primarily localized to the cytoplasm, as opposed to DUSP4 that is predominantly nuclear, this difference in their subcellular localization may explain the phenotypic differences. Although our data demonstrate that Dusp4 overexpression is sufficient to cause cardiomyopathy, further studies are necessary to definitively demonstrate that it is an absolute requirement for the development of LMNA cardiomyopathy.
Aberrant activation of ERK1/2 and AKT signaling has been documented in dilated failing hearts from human subjects and in mouse models of various forms of cardiomyopathies (40–47). Compounds that inhibit these pathways are effective in ameliorating cardiomyopathy in LmnaH222P/H222P mice (9, 48). Agents that target ERK1/2 and AKT-mTOR pathways currently being developed to treat cancers may therefore be utilized to treat patients with LMNA cardiomyopathy and provide a therapeutic option for a disease with no current specific treatment option.
Supplementary Material
Acknowledgments
We are grateful to Dr. Gisèle Bonne (Institut de Myologie, Paris, France) for providing LmnaH222P/H222P mice, Dr. Colin L. Stewart (Institute of Medical Biology, Singapore) for providing Lmna−/− mice, and Dr. Ira J. Goldberg (Columbia University, New York) for providing pPSKII-MHC-GH plasmid and PPARγ transgenic mice.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AR048997. This work was also supported by Muscular Dystrophy Association Grant MDA172222 (to H. J. W.). The authors have reported a direct financial conflict of interest: H. J. W. and A. M. are inventors on a pending patent application (PCT/US09/42614) on methods for treating and/or preventing cardiomyopathies by ERK and JNK inhibition filed by the trustees of Columbia University. H. J. W. and J. C. C. are inventors on a provisional patent application on the use of rapamycin and rapamycin analogs for the treatment of dilated cardiomyopathies filed by the trustees of Columbia University.

This article contains supplemental data and Tables S1–S3.
- mTOR
- mammalian target of rapamycin
- Dusp
- dual specificity phosphatase
- AICAR
- 5-aminoimidazole-4-carboxyamide ribonucleoside
- IDH3
- isocitrate dehydrogenase 3
- qPCR
- quantitative real time PCR
- AMPK
- AMP-activated protein kinase
- RNAseq
- RNA sequencing
- GO
- Gene Ontology
- BCS
- bovine calf serum.
REFERENCES
- 1. Dauer W. T., Worman H. J. (2009) The nuclear envelope as a signaling node in development and disease. Dev. Cell 17, 626–638 [DOI] [PubMed] [Google Scholar]
- 2. Lin F., Worman H. J. (1993) Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 268, 16321–16326 [PubMed] [Google Scholar]
- 3. Hernandez L., Roux K. J., Wong E. S., Mounkes L. C., Mutalif R., Navasankari R., Rai B., Cool S., Jeong J. W., Wang H., Lee H. S., Kozlov S., Grunert M., Keeble T., Jones C. M., Meta M. D., Young S. G., Daar I. O., Burke B., Perantoni A. O., Stewart C. L. (2010) Functional coupling between the extracellular matrix and nuclear lamina by Wnt signaling in progeria. Dev. Cell 19, 413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scaffidi P., Misteli T. (2008) Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat. Cell Biol. 10, 452–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. González J. M., Navarro-Puche A., Casar B., Crespo P., Andrés V. (2008) Fast regulation of AP-1 activity through interaction of lamin A/C, ERK1/2, and c-Fos at the nuclear envelope. J. Cell Biol. 183, 653–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muchir A., Pavlidis P., Decostre V., Herron A. J., Arimura T., Bonne G., Worman H. J. (2007) Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J. Clin. Invest. 117, 1282–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muchir A., Shan J., Bonne G., Lehnart S. E., Worman H. J. (2009) Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum. Mol. Genet. 18, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu W., Muchir A., Shan J., Bonne G., Worman H. J. (2011) Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Circulation 123, 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi J. C., Muchir A., Wu W., Iwata S., Homma S., Morrow J. P., Worman H. J. (2012) Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci. Transl. Med. 4, 144ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramos F. J., Chen S. C., Garelick M. G., Dai D. F., Liao C. Y., Schreiber K. H., MacKay V. L., An E. H., Strong R., Ladiges W. C., Rabinovitch P. S., Kaeberlein M., Kennedy B. K. (2012) Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci. Transl. Med. 4, 144ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brondello J. M., Brunet A., Pouysségur J., McKenzie F. R. (1997) The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J. Biol. Chem. 272, 1368–1376 [DOI] [PubMed] [Google Scholar]
- 12. Jeffrey K. L., Camps M., Rommel C., Mackay C. R. (2007) Targeting dual-specificity phosphatases. Manipulating MAP kinase signalling and immune responses. Nat. Rev. Drug Discov. 6, 391–403 [DOI] [PubMed] [Google Scholar]
- 13. Arimura T., Helbling-Leclerc A., Massart C., Varnous S., Niel F., Lacène E., Fromes Y., Toussaint M., Mura A. M., Keller D. I., Amthor H., Isnard R., Malissen M., Schwartz K., Bonne G. (2005) Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet. 14, 155–169 [DOI] [PubMed] [Google Scholar]
- 14. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J. P. (2006) GenePattern 2.0. Nat. Genet. 38, 500–501 [DOI] [PubMed] [Google Scholar]
- 16. Blanchet E., Annicotte J. S., Lagarrigue S., Aguilar V., Clapé C., Chavey C., Fritz V., Casas F., Apparailly F., Auwerx J., Fajas L. (2011) E2F transcription factor-1 regulates oxidative metabolism. Nat. Cell Biol. 13, 1146–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mauro C., Leow S. C., Anso E., Rocha S., Thotakura A. K., Tornatore L., Moretti M., De Smaele E., Beg A. A., Tergaonkar V., Chandel N. S., Franzoso G. (2011) NF-κB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat. Cell Biol. 13, 1272–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abramoff M. D., Magalhaes P. J., Ram S. J. (2004) Image Processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 19. Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C. L., Burke B. (1999) Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147, 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Son N. H., Park T. S., Yamashita H., Yokoyama M., Huggins L. A., Okajima K., Homma S., Szabolcs M. J., Huang L. S., Goldberg I. J. (2007) Cardiomyocyte expression of PPARγ leads to cardiac dysfunction in mice. J. Clin. Invest. 117, 2791–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McMahon D. K., Anderson P. A., Nassar R., Bunting J. B., Saba Z., Oakeley A. E., Malouf N. N. (1994) C2C12 cells. Biophysical, biochemical, and immunocytochemical properties. Am. J. Physiol. 266, C1795–1802 [DOI] [PubMed] [Google Scholar]
- 22. Zebedin E., Mille M., Speiser M., Zarrabi T., Sandtner W., Latzenhofer B., Todt H., Hilber K. (2007) C2C12 skeletal muscle cells adopt cardiac-like sodium current properties in a cardiac cell environment. Am. J. Physiol. Heart Circ. Physiol. 292, H439–H450 [DOI] [PubMed] [Google Scholar]
- 23. Ramsköld D., Wang E. T., Burge C. B., Sandberg R. (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 5, e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakabe N. J., Aneas I., Shen T., Shokri L., Park S. Y., Bulyk M. L., Evans S. M., Nobrega M. A. (2012) Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum. Mol. Genet. 21, 2194–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kroemer G., Mariño G., Levine B. (2010) Autophagy and the integrated stress response. Mol. Cell 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 27. Mizushima N., Yoshimori T., Levine B. (2010) Methods in mammalian autophagy research. Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Plas D. R., Thompson C. B. (2005) Akt-dependent transformation. There is more to growth than just surviving. Oncogene 24, 7435–7442 [DOI] [PubMed] [Google Scholar]
- 29. Boya P., González-Polo R. A., Casares N., Perfettini J. L., Dessen P., Larochette N., Métivier D., Meley D., Souquere S., Yoshimori T., Pierron G., Codogno P., Kroemer G. (2005) Inhibition of macroautophagy triggers apoptosis. Mol. Cell Biol. 25, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sengupta S., Peterson T. R., Sabatini D. M. (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bing R. J., Siegel A., Ungar I., Gilbert M. (1954) Metabolism of the human heart. II. Studies on fat, ketone and amino acid metabolism. Am. J. Med. 16, 504–515 [DOI] [PubMed] [Google Scholar]
- 32. Opie L. H. (1968) Metabolism of the heart in health and disease. I. Am. Heart J. 76, 685–698 [DOI] [PubMed] [Google Scholar]
- 33. Bertrand A. T., Renou L., Papadopoulos A., Beuvin M., Lacène E., Massart C., Ottolenghi C., Decostre V., Maron S., Schlossarek S., Cattin M. E., Carrier L., Malissen M., Arimura T., Bonne G. (2012) DelK32-lamin A/C has abnormal location and induces incomplete tissue maturation and severe metabolic defects leading to premature death. Hum. Mol. Genet. 21, 1037–1048 [DOI] [PubMed] [Google Scholar]
- 34. Cutler D. A., Sullivan T., Marcus-Samuels B., Stewart C. L., Reitman M. L. (2002) Characterization of adiposity and metabolism in Lmna-deficient mice. Biochem. Biophys. Res. Commun. 291, 522–527 [DOI] [PubMed] [Google Scholar]
- 35. Kubben N., Voncken J. W., Konings G., van Weeghel M., van den Hoogenhof M. M., Gijbels M., van Erk A., Schoonderwoerd K., van den Bosch B., Dahlmans V., Calis C., Houten S. M., Misteli T., Pinto Y. M. (2011) Post-natal myogenic and adipogenic developmental. Defects and metabolic impairment upon loss of A-type lamins. Nucleus 2, 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Communal C., Colucci W. S., Remondino A., Sawyer D. B., Port J. D., Wichman S. E., Bristow M. R., Singh K. (2002) Reciprocal modulation of mitogen-activated protein kinases and mitogen-activated protein kinase phosphatase 1 and 2 in failing human myocardium. J. Card. Fail. 8, 86–92 [DOI] [PubMed] [Google Scholar]
- 37. Brooks W. W., Shen S. S., Conrad C. H., Goldstein R. H., Bing O. H. (2010) Transition from compensated hypertrophy to systolic heart failure in the spontaneously hypertensive rat. Structure, function, and transcript analysis. Genomics 95, 84–92 [DOI] [PubMed] [Google Scholar]
- 38. Galindo C. L., Skinner M. A., Errami M., Olson L. D., Watson D. A., Li J., McCormick J. F., McIver L. J., Kumar N. M., Pham T. Q., Garner H. R. (2009) Transcriptional profile of isoproterenol-induced cardiomyopathy and comparison to exercise-induced cardiac hypertrophy and human cardiac failure. BMC Physiol. 9, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Purcell N. H., Wilkins B. J., York A., Saba-El-Leil M. K., Meloche S., Robbins J., Molkentin J. D. (2007) Genetic inhibition of cardiac ERK1/2 promotes stress-induced apoptosis and heart failure but has no effect on hypertrophy in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 14074–14079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baba H. A., Stypmann J., Grabellus F., Kirchhof P., Sokoll A., Schäfers M., Takeda A., Wilhelm M. J., Scheld H. H., Takeda N., Breithardt G., Levkau B. (2003) Dynamic regulation of MEK/Erks and Akt/GSK-3beta in human end-stage heart failure after left ventricular mechanical support. Myocardial mechanotransduction-sensitivity as a possible molecular mechanism. Cardiovasc. Res. 59, 390–399 [DOI] [PubMed] [Google Scholar]
- 41. Haq S., Choukroun G., Lim H., Tymitz K. M., del Monte F., Gwathmey J., Grazette L., Michael A., Hajjar R., Force T., Molkentin J. D. (2001) Differential activation of signal transduction pathways in human hearts with hypertrophy versus advanced heart failure. Circulation 103, 670–677 [DOI] [PubMed] [Google Scholar]
- 42. Karoor V., Vatner S. F., Takagi G., Yang G., Thaisz J., Sadoshima J., Vatner D. E. (2004) Propranolol prevents enhanced stress signaling in Gsα cardiomyopathy. Potential mechanism for β-blockade in heart failure. J. Mol. Cell Cardiol. 36, 305–312 [DOI] [PubMed] [Google Scholar]
- 43. Luckey S. W., Walker L. A., Smyth T., Mansoori J., Messmer-Kratzsch A., Rosenzweig A., Olson E. N., Leinwand L. A. (2009) The role of Akt/GSK-3β signaling in familial hypertrophic cardiomyopathy. J. Mol. Cell Cardiol. 46, 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marin T. M., Keith K., Davies B., Conner D. A., Guha P., Kalaitzidis D., Wu X., Lauriol J., Wang B., Bauer M., Bronson R., Franchini K. G., Neel B. G., Kontaridis M. I. (2011) Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J. Clin. Invest. 121, 1026–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Princen F., Bard E., Sheikh F., Zhang S. S., Wang J., Zago W. M., Wu D., Trelles R. D., Bailly-Maitre B., Kahn C. R., Chen Y., Reed J. C., Tong G. G., Mercola M., Chen J., Feng G. S. (2009) Deletion of Shp2 tyrosine phosphatase in muscle leads to dilated cardiomyopathy, insulin resistance, and premature death. Mol. Cell Biol. 29, 378–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Semple D., Smith K., Bhandari S., Seymour A. M. (2011) Uremic cardiomyopathy and insulin resistance. A critical role for Akt? J. Am. Soc. Nephrol. 22, 207–215 [DOI] [PubMed] [Google Scholar]
- 47. Takeishi Y., Huang Q., Abe J., Che W., Lee J. D., Kawakatsu H., Hoit B. D., Berk B. C., Walsh R. A. (2002) Activation of mitogen-activated protein kinases and p90 ribosomal S6 kinase in failing human hearts with dilated cardiomyopathy. Cardiovasc. Res. 53, 131–137 [DOI] [PubMed] [Google Scholar]
- 48. Muchir A., Reilly S. A., Wu W., Iwata S., Homma S., Bonne G., Worman H. J. (2012) Treatment with selumetinib preserves cardiac function and improves survival in cardiomyopathy caused by mutation in the lamin A/C gene. Cardiovasc. Res. 93, 311–319 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.