Background: Although PAK1 regulates cytoskeleton and microtubule dynamics, its role in controlling the functions of MCAK remains unknown.
Results: PAK1 phosphorylates MCAK and thereby regulates both its localization and function.
Conclusion: MCAK is a cognate substrate of PAK1.
Significance: This study provides a novel mechanistic insight into PAK1 regulation of MCAK functions.
Keywords: Cell Signaling, Centriole, Centromeres, Cytoskeleton, Microtubules, MCAK, MT Dynamics, p21-activated Kinase 1
Abstract
Although p21-activated kinase 1 (PAK1) and microtubule (MT) dynamics regulate numerous fundamental processes including cytoskeleton remodeling, directional motility, and mitotic functions, the significance of PAK1 signaling in regulating the functions of MT-destabilizing protein mitotic centromere-associated kinesin (MCAK) remains unknown. Here we found that MCAK is a cognate substrate of PAK1 wherein PAK1 phosphorylates MCAK on serines 192 and 111 both in vivo and in vitro. Furthermore, we found that PAK1 phosphorylation of MCAK on serines 192 and 111 preferentially regulates its microtubule depolymerization activity and localization to centrosomes, respectively, in the mammalian cells.
Introduction
A fine balance of microtubule (MT)3 dynamics between the depolymerized and polymerized phases plays a vital role in numerous fundamental processes including chromosomal segregation and directional cell migration. The MT dynamics is coordinately regulated by MT-stabilizing and -destabilizing proteins that promote or attenuate polymerization, respectively, which in turn modulates resulting functions. For example, MT-destabilizing protein mitotic centromere-associated kinesin (MCAK) plays an important role in mitotic spindle assembly as well as in faithful chromosome segregation (1, 2). MCAK localizes at the spindle poles, centromeres, and kinetochores during mitosis (3, 4). The ability of MCAK to attenuate or accelerate MT depolymerization ensures proper attachment of microtubules to kinetochores. The MCAK activity is tightly regulated by mitotic kinases such as Aurora-A, Aurora-B, Plk1, and Cdk1 (5–12).
The small GTPases such as Rac and cdc42 are major regulators of cytoskeletal dynamics (13) and mediate its effect through p21-activated kinases (PAKs). The group I PAK family members regulate GTPase-dependent cytoskeletal remodeling, either utilizing its catalytic activity or interacting with other proteins (14). PAK1 plays a prominent role in mitosis as well as in actin and microtubule remodeling. For example, PAK1 phosphorylates myosins, a family of actin-based molecular motor proteins (15), and mammalian PAK1 phosphorylates myosin light chain on Ser-19 in neuronal cell, resulting in stabilization of the localized actin network through formation of the GIT1·PIX·Rac·PAK complex (16). PAK1 is also involved in the phosphorylation of proteins that control MT dynamics such as stathmin/oncoprotein 18, which destabilizes MT by binding to tubulin dimers and inhibits tubulin polymerization to promote MT disassembly (17). Phosphorylation of oncoprotein 18 by PAK1 inactivates this protein, resulting in the stabilization of MT at the leading edges of migrating cells. PAK1 phosphorylation of tubulin cofactor B augments the heterodimerization of α/β-tubulins, leading to tubulin polymerization (18). PAK1 can also phosphorylates dynein light chain 1, a component of the cytoplasmic dynein complex, which moves along with MT. PAK1 phosphorylates DLC1 at Ser-88, which in turn affects vesicle formation and trafficking due to its effects on MT (19).
Because both PAK1 and MCAK are localized to centrosomes (4, 12, 20–23) and PAK1 is upstream of other mitotic kinases involved in the regulation of MCAK function, we made an attempt to delineate the role of PAK1 in the regulation of MCAK function. We found that MCAK is a PAK1 substrate and demonstrated that PAK1 phosphorylation of MCAK regulates both its localization and MT depolymerization activity.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
ZR-75 cells were purchased from American Tissue Culture Collection (Manassas, VA). MCF-7/T423E cells are MCF-7 cells stably transfected with a tetracycline-inducible constitutively active PAK1 (PAK1-T423E) where PAK1 expression is induced by the addition of doxycycline (1 μg/ml) (24–27). PAK1 wild-type and PAK1−/− murine embryonic fibroblasts (MEFs) were kindly provided by Dr. Chernoff and were used in our earlier study (28). ZR-75 cells, MCF-7/T423E cells, and PAK1 wild-type and PAK1−/− MEFs were cultured in Dulbecco's modified Eagle's medium (DMEM)/F-12 (Mediatech) supplemented with 10% fetal calf serum (Hyclone). Antibodies against PAK1 (catalogue number A301-259A) and Aurora B (catalogue number A300-431A) were purchased from Bethyl Laboratories (Montgomery, TX). Anti-MCAK antibody (catalogue number NB100-2588) and phospho-MCAK antibody (catalogue number ab74146) were purchased from Novus Biologicals and Abcam, respectively. Antibodies against α-tubulin (catalogue number T9026) and γ-tubulin (catalogue number T6557) were purchased from Sigma. Anti-Aurora-A antibody (catalogue number GTX13824) and anti-PAK2 antibody (catalogue number sc-1872) were obtained from GeneTex (Irvine, CA) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Pericentrin antibody (PRB-432C) was obtained from Berkeley Antibody Co.
Cloning and Site-directed Mutagenesis
Human MCAK full length (NCBI accession number NM_006845.3) and its deletions (amino acids 1–187, 1–255, 188–591, 255–591, and 592–725) were cloned either into GST-tagged vector pGEX-5x-1 (GE Healthcare) or His-tagged vector pcDNA3.1/HisC (Invitrogen) using the Clontech Infusion PCR cloning kit. GFP-MCAK full length was kindly provided by Dr. Satoko Aoki, Tokyo University of Science, Japan (29). Mutations in full-length GST-MCAK and GFP-MCAK constructs were created using a site-directed mutagenesis kit (Stratagene, Cedar Creek, TX).
In Vitro Kinase Assay
The in vitro kinase assay was performed either using full-length GST-MCAK fusion protein or its deletion constructs as a substrate and recombinant purified GST-PAK1 or GST-PAK1 kinase-dead as an enzyme. The reaction was carried out in HEPES buffer (50 mm HEPES, 10 mm MgCl2, 2 mm MnCl2, and 1 mm dithiothreitol) containing 100 ng of recombinant purified GST-PAK1 enzyme, 10 μCi of [γ-32P]ATP, and 25 μm cold ATP. GST alone was used as a negative control. GST proteins were purified using glutathione-Sepharose beads following the manufacturer's instructions. The reaction mixture (30 μl) was incubated in a water bath for 30 min at 30 °C, the reaction was stopped by adding 10 μl of 4× SDS loading buffer and separated by SDS-PAGE, and the results were analyzed by autoradiography.
In Vivo Kinase Assay
Cells were grown in 60-mm dishes in medium containing 10% fetal bovine serum and antibiotic-antimycotic solution. After 24 h, cells were transfected with either control vector or T7-PAK1 and the substrate T7-MCAK. We used 1 μg of PAK1 and 1 μg of MCAK plasmid DNA to transfect the cells. After 24 h of transfection, the cells were washed with phosphate-buffered saline (PBS), and serum-free medium was added. After 24 h, the cells were changed to phosphate-free medium with sodium pyruvate and 2% dialyzed serum and labeled with [32P]orthophosphate (0.2 mCi/ml) for 6 h. Whenever required, after labeling, cells were changed to complete medium containing 10% serum and kept for 6 h. The cells were then lysed, immunoprecipitated with the indicated antibodies, and separated by SDS-PAGE, and the results were analyzed by autoradiography.
In Vitro Microtubule Depolymerization Assay
For this assay, various GST-MCAK constructs were used along with pure rhodamine-labeled tubulin, and the assay was carried out as described earlier (12). The microtubule depolymerization assay was performed in a 15-μl volume reaction buffer containing BRB80, 1 mm DTT, 133 μg/ml casein, 1.5 mm MgATP, and 1.5 μm rhodamine-labeled and GMPCPP-stabilized microtubules. Reactions were started with incubation of the kinase and GST-MCAK in the reaction buffer for 10 min without the microtubules, and then the polymerization or depolymerization was initiated by the addition of microtubules. Reactions were allowed to incubate for 5–20 min, and then 2 μl of each reaction was put on a glass slide and sealed under the coverslip. An LSM-710 confocal microscope was used to capture 25 images for each time point. Representative figures from three individual experiments are shown here. For quantitation, the number of microtubules per 71 × 56-μm fields from three individual micrographs for each sample was counted. The segmented line tool of the ImageJ software was used to draw a line across the length of the microtubules to get the exact length of the microtubules. Only microtubules greater than 6 μm were considered for analysis of each sample. Data represents mean ± S.D. of all counted samples.
In Vivo Microtubule Regrowth/Polymerization Assay
The MT regrowth or polymerization assay was carried out as described elsewhere (7, 18, 30–32). ZR-75 cells were grown on coverslips. After 24–36 h of growth, nocodazole treatments for microtubule biogenesis assays were performed. Briefly, cells were treated with 4 μm nocodazole for 30 min, rinsed twice with PBS, and then incubated in medium with 10% serum for 0 and 15 min, respectively, to allow for new microtubule biogenesis. A mouse monoclonal antibody against α-tubulin was used for colocalization of tubulin with PAK1 or MCAK, respectively. Goat anti-mouse Alexa Fluor 555 antibody was used for tubulin, and goat anti-rabbit Alexa Fluor 488 was used for PAK1 or MCAK. For quantitation, 25 cells were scored for each condition, a fraction of cells with different lengths of microtubules were counted using the Zen software in the LSM 710 confocal microscope, and quantification of the microtubules was carried out using ImageJ software.
siRNA Transfection
siRNAs against PAK1 (catalogue number M-003521-03) and Aurora-A (catalogue number L-003545-01) were purchased from Dharmacon RNAi Technologies, and siRNA against MCAK (catalogue number sc-105596) was purchased from Santa Cruz Biotechnology. 100 nmol/liter siRNA (control, PAK1, and Aurora-A) was used to transfect the cells along with 4 μl of Oligofectamine (Invitrogen) in 6-well plates following the manufacturer's protocol. Specific protein knockdown was checked after 48 h of transfection by Western blot analysis.
GST Pulldown Assay
In vitro transcription and translation of PAK1 was performed using a T7 TnT kit (Promega) in which 1 μg of plasmid DNA was translated in the presence of [35S]methionine. The GST pulldown assays were performed by incubating equal amounts of GST and GST-fused full-length proteins and deletion constructs immobilized on glutathione-Sepharose beads (GE Healthcare) with in vitro translated 35S-labeled protein in GST binding buffer (25 mm Tris-HCl, pH 8.0, 50 mm NaCl, 10% glycerol, and 0.1% Nonidet P-40). Bound proteins were isolated by incubating the reaction mixture with rotation for 3 h at 4 °C. After washing the beads with GST binding buffer, the proteins were eluted in 4× SDS loading buffer, separated by SDS-PAGE, and visualized by autoradiography.
Immunoprecipitation and Immunoblotting
Cells were grown in complete medium containing 10% fetal bovine serum and antibiotic-antimycotic solution (Invitrogen). Whenever needed, cells were incubated with 2 mm thymidine (Sigma-Aldrich) for 16 h followed by an 8-h release in complete medium. This procedure was repeated, and after the second release, cells were washed thrice with PBS and incubated in lysis buffer (20 mm HEPES, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 10 mm NaF, 1 mm NaVO4, 0.5% Nonidet P-40, protease inhibitor mixture (Roche Applied Science), and phosphatase inhibitor mixture (Sigma-Aldrich)) for 30 min on ice. The protein concentration was estimated, and an equal concentration of protein was resolved by 10% SDS-PAGE, transferred to nitrocellulose membrane, probed with the appropriate antibodies, and detected using an ECL detection kit. Immunoprecipitation was carried out overnight at 4 °C using 1 μg of antibody/mg of protein. Complexes were collected using protein G beads by incubating with rotation for 4 h at 4 °C. After extensive washing in buffer containing 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, and 1 mm EDTA, proteins were detected as mentioned earlier.
Immunofluorescence and Confocal Microscopy
Cells were grown on glass coverslips. After 24 h, nocodazole (10 ng/ml) was added to the medium, and cells were incubated for 16 h. Then the cells were washed three times with PBS, fixed, and permeabilized using 0.5% Triton X-100 for 5 min. Cells were then washed three times with PBS and stained for γ-tubulin, PAK1, phospho-MCAK, pericentrin, and Aurora-A using the appropriate antibodies followed by incubation with secondary antibody conjugated with Alexa Fluor 488 (green; Invitrogen) or Alexa 546 (red; Invitrogen). The coverslips were counterstained with DAPI (blue; Invitrogen) for DNA. Coverslips were mounted in SlowFade mounting medium (Invitrogen) and sealed onto glass slides. Samples were imaged using a laser-scanning confocal microscope (LSM 710) equipped with a 63×/1.4 numeric aperture objective following established methods. Quantitative analysis was conducted using the Zen software. The images were taken at room temperature, and each representative image is at the same cellular level and magnification.
Super-resolution Imaging
The microtubules of PAK1 wild-type and PAK1−/− MEFs were imaged using a super-resolution microscope (Leica Microsystems). The nuclei were stained with DRAQ5 for 5 min. Images of the nuclei are confocal images. The images of the microtubules are super-resolution images. The images were cropped and merged with the images of the nuclei.
RESULTS AND DISCUSSION
PAK1 Regulates MT Dynamics
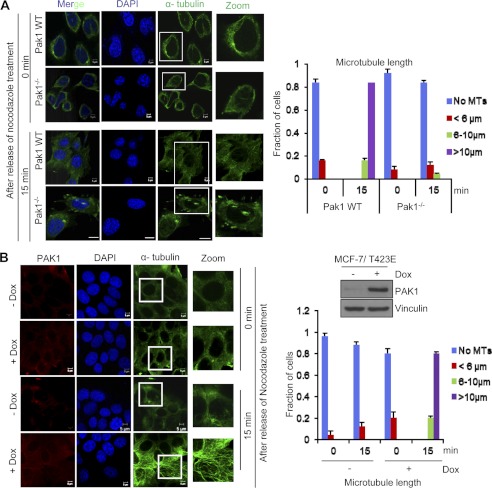
To explore the possible role of PAK1 signaling in MT remodeling, we performed an in vivo MT regrowth/polymerization assay in PAK1−/− and PAK1 wild-type MEFs. We observed decreased MT regrowth/polymerization in the PAK1−/− MEFs as compared with the wild-type PAK1 MEFs due to MT depolymerization activity in the absence of PAK1 (Fig. 1A). Because we observed decreased MT biogenesis under the PAK1-depleted condition in PAK1−/− MEFs, we further analyzed the MT biogenesis/polymerization activity under PAK1 overexpression conditions. For this purpose, we chose an isogenic MCF-7/T423E cell line stably expressing PAK1 wherein the effect of PAK1 hyperactivation could be studied in the same cells under a regulatable expression of catalytically active PAK1 (24–27). We found increased MT regrowth/polymerization under PAK1 overexpression in MCF-7/T423E cells after treatment with doxycycline compared with untreated cells presumably due to defective MT depolymerization activity (Fig. 1B). Together, these results suggest that PAK1 signaling regulates MT depolymerization and associated MT dynamics.
FIGURE 1.
Effect of PAK1 on MT dynamics. A, in vivo MT regrowth/polymerization in PAK1 wild-type (WT) and PAK1−/− MEFs. Cells were treated with nocodazole for 30 min, and after release at time intervals of 0 and 15 min, cells were fixed, stained with the indicated antibodies, and analyzed using confocal microscopy. The right side panel represents the graphical representation of the length of microtubules in each condition in 25 cells, and the y axis represents the fraction of cells representing the different lengths of microtubules scored. B, MCF-7/T423E cells in the presence and absence of doxycycline (Dox; 1 μg/ml) were treated with nocodazole for 30 min, and after release at time intervals of 0 and 15 min, cells were fixed, stained with the indicated antibodies, and the MT regrowth/polymerization was analyzed using confocal microscopy. The right side panel represents the graphical representation of the length of microtubules in each condition in 25 cells, and the y axis represents the fraction of cells with the different lengths of microtubules scored. Western blot analysis for PAK1 is provided to show the expression levels of PAK1 in MCF-7/T423E cells in the presence and absence of doxycycline. Error bars represent S.D. Scale bars = 5 μm.
PAK1 Interacts with the MCAK·Aurora-A·γ-Tubulin Complex
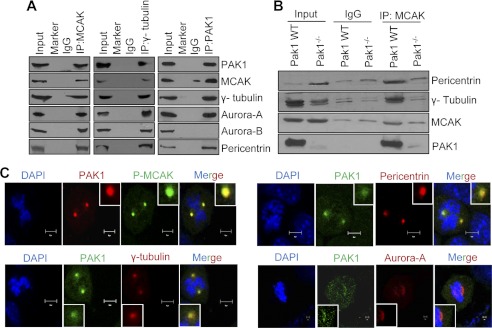
From the published data, it is evident that Auroras play a significant role in MT dynamics by phosphorylating MCAK. PAK1 is an upstream kinase of Auroras, and because of its established role in MT dynamics, we evaluated the association of PAK1 with the endogenous Auroras and MCAK during mitosis. We found that MCAK effectively interacts with PAK1, γ-tubulin, Aurora-A, Aurora-B, and pericentrin, whereas PAK1 interacts with all the above proteins with the exception of Aurora-B in the centrosomes (Fig. 2, A and B). This raises the possibility that Aurora-B and PAK1 may reside in different complexes. Consistent with these findings, we also found evidence of colocalization of the endogenous PAK1 with MCAK, Aurora-A, and γ-tubulin using confocal scanning microscopy (Fig. 2C). These results suggest that PAK1 interacts selectively with MCAK·Aurora-A·γ-tubulin and regulates MT dynamics.
FIGURE 2.
PAK1 interacts with MCAK at the centrosomes. A, Western blot analysis of PAK1, MCAK, γ-tubulin, Aurora-A, Aurora-B, and pericentrin after immunoprecipitating (IP) with the indicated antibodies in the centrosomal proteins isolated from the synchronized ZR-75 cells. B, Western blot analysis for PAK1, MCAK, pericentrin, and γ-tubulin after immunoprecipitating with MCAK antibody in the centrosomal proteins isolated from the synchronized PAK1 wild-type and PAK1−/− MEFs. C, synchronized ZR-75 cells were released after 6 h and analyzed for the endogenous PAK1 localization with centrosomal proteins pericentrin, phospho-MCAK (P-MCAK), Aurora-A, and γ-tubulin by double immunofluorescence staining. The antibodies specific for the particular proteins were used for the staining. DAPI was used as a nuclear stain. Scale bars = 10 μm.
MCAK Is a Novel PAK1 Substrate
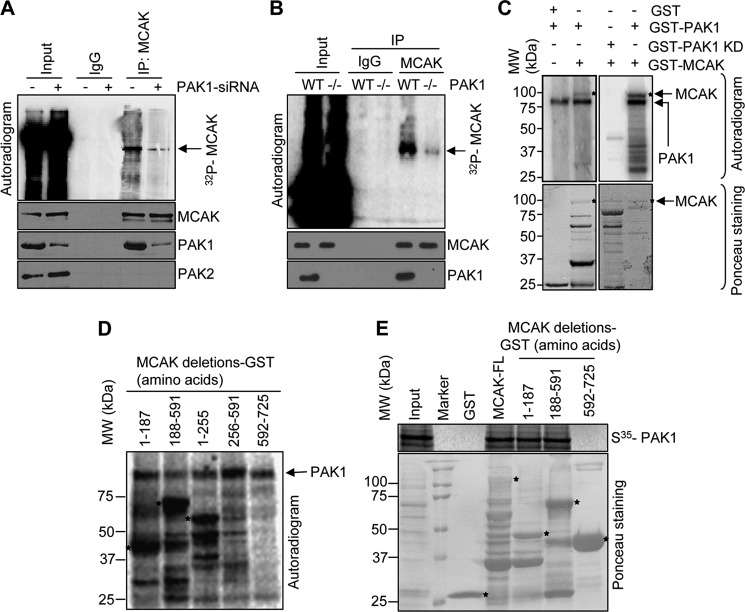
As PAK1 is upstream of Aurora-A kinase and found to be associated with MCAK in the centrosomes (Fig. 2, A–C), we next investigated the ability of PAK1 to phosphorylate MCAK. We found that selective depletion of PAK1 compromises the phosphorylation status of the endogenous MCAK as well as the amount of PAK1 pulled down by MCAK (Fig. 3A). Consistently, we also observed a decreased phosphorylation of MCAK in PAK1−/− MEFs (Fig. 3B) compared with PAK1 wild-type MEFs, providing a clue that MCAK might be a PAK1-interacting substrate. Furthermore, PAK1 enzyme but not kinase-dead enzyme readily phosphorylates the GST-MCAK (Fig. 3C). Results from in vitro phosphorylation studies involving purified PAK1 enzyme and a series of MCAK deletion constructs revealed the presence of a PAK1 phosphorylation site or sites within the N-terminal amino acid 1–187 and 1–255 regions (Fig. 3D and supplemental Fig. 1). In addition, results from GST pulldown assays indicated that 35S-labeled PAK1 interacts with amino acids 1–591 of MCAK (Fig. 3E). Group-based phosphorylation site predicting and scoring platform (GPS) (33) analysis of this region revealed the presence of potential PAK1 phosphorylation sites in MCAK: Ser-95, Ser-111, and Ser-192 (supplemental Fig. 2A).
FIGURE 3.
MCAK is a novel substrate of PAK1. A, in vivo phosphorylation of MCAK after PAK1 knockdown using PAK1-specific siRNA in ZR-75 cells. After treating the cells with PAK1 siRNA for 24 h, they were washed with PBS and incubated in serum-free medium for 24 h. After 24 h, the cells were changed to phosphate-free medium with sodium pyruvate and 2% dialyzed serum and labeled with [32P]orthophosphate (0.2 mCi/ml) for 6 h. After 6 h, cells were changed to complete medium containing 10% serum and kept for 6 h. The cells were then lysed and immunoprecipitated (IP) with the MCAK antibody, separated by SDS-PAGE, transferred onto nitrocellulose membrane, and analyzed by autoradiography. B, in vivo phosphorylation of MCAK in PAK1 wild-type and PAK1−/− MEFs. The assay was carried out as described in A. C, GST alone and MCAK-GST were used in an in vitro kinase assay with recombinant purified PAK1 and PAK1 kinase-dead enzyme. Asterisks in the upper panel represent the MCAK phosphorylation. The lower panel shows the Ponceau staining for the GST and MCAK-GST used in the reaction. Asterisks represent the MCAK-GST band. D, in vitro kinase assay was carried out with deletion constructs of MCAK-GST using recombinant purified PAK1 enzyme (PAK1-GST). Asterisks represent the phosphorylation of MCAK. E, in vitro translated 35S-labeled PAK1 was used to study its binding with GST, MCAK-GST, and its deletion constructs. The extent of binding was measured by signal intensity using autoradiography. The Ponceau-stained blot shows the equal amounts of GST proteins used in the reaction. Asterisks in the lower panel represents deletion constructs of MCAK-GST.
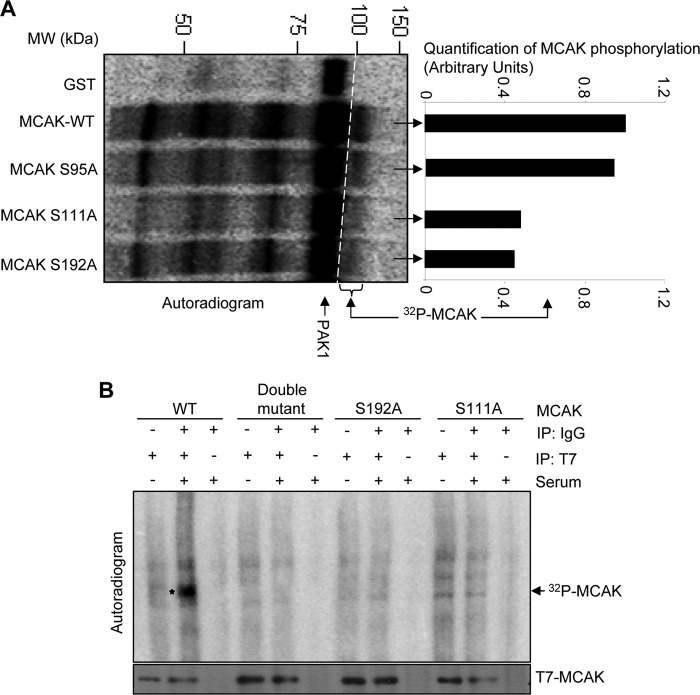
We next individually mutated these serine residues to alanine in the full-length GST-MCAK and evaluated the phosphorylation of MCAK mutants in vitro. We noticed an inability of PAK1 to phosphorylate MCAK-S111A and -S192A mutants as compared with the wild-type MCAK (Fig. 4A and supplemental Fig. 2B), suggesting that MCAK contains two PAK1 phosphorylation sites, Ser-192 and Ser-111. To validate the PAK1 phosphorylation of MCAK in a physiologically relevant setting, we cotransfected ZR-75 breast cancer cells with T7-MCAK or T7-MCAK-S192A, T7-MCAK-S111A, or T7-MCAK-S192A/S111A mutant, and in vivo phosphorylation assays involving metabolic labeling of cells with labeled orthophosphoric acid were performed. Consistent with in vitro studies, we noticed a substantial reduction in the phosphorylation of MCAK-S192A and -S111A as well as the MCAK-S111A/S192A double mutant as compared with MCAK (Fig. 4B). From the earlier studies, it is evident that serum induces PAK1 activity (34, 35); accordingly, we found increased phosphorylation of MCAK in the presence of serum (Fig. 4B). These findings suggest that MCAK is a cognate substrate of PAK1 and that Ser-111 and Ser-192 are the functionally active PAK1 phosphorylation sites in MCAK.
FIGURE 4.
PAK1 phosphorylates MCAK at positions Ser-111 and Ser-192. A, in vitro kinase assay with MCAK-GST and its mutants using recombinant purified PAK1 enzyme (PAK1-GST). Phosphorylated MCAK was visualized by autoradiography. The right side panel represents the quantification analysis of MCAK phosphorylation using ImageJ software. B, in vivo kinase assay was carried out after transfecting the ZR-75 cells with either control vector, T7-PAK1, T7-MCAK, or its mutants. After transfection, cells were labeled with [32P]orthophosphate for 6 h in the presence and absence of serum. Cell lysates were then prepared, immunoprecipitated (IP) with the indicated antibodies, separated by SDS-PAGE, transferred onto nitrocellulose membrane, and visualized by autoradiography.
PAK1 Phosphorylation of MCAK Inhibits Its MT Depolymerization Activity
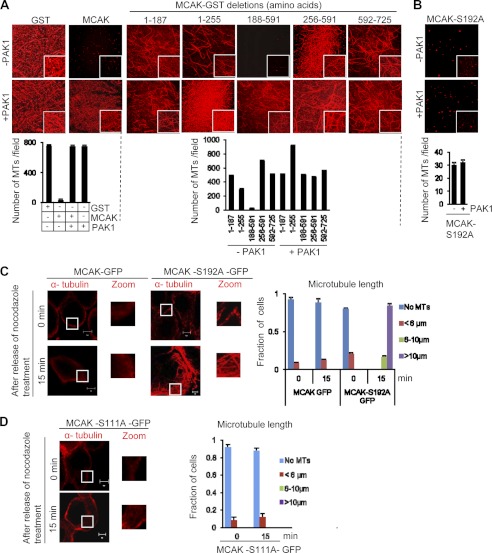
Because both PAK1 and MCAK participate in MT remodeling, we explored whether PAK1 could regulate the MT depolymerization activity of MCAK. We incubated rhodamine-labeled, GMPCPP-stabilized MT with PAK1 and MCAK for 5 min and visualized its polymerization status by immunofluorescence. As expected, incubation of GMPCPP-stabilized MT with MCAK depolymerized the MT. Surprisingly, inclusion of PAK1 completely inhibited the ability of MCAK to depolymerize MT, revealing for the first time the ability of PAK1 to regulate the depolymerization activity of MCAK (Fig. 5A). To identify the region of MCAK responsible for the noticed inhibitory effect of PAK1 on MT depolymerization, we used MCAK deletion mutants encompassing various domains and identified the minimal catalytic domain (amino acids 188–591) containing depolymerization activity, which could be completely inhibited by PAK1 (Fig. 5A) presumably due to phosphorylation of MCAK. Computational GPS analysis of the region encompassing amino acids 188–591 revealed the presence of only one PAK1 phosphorylation site, Ser-192.
FIGURE 5.
PAK1 phosphorylation of MCAK at position Ser-192 regulates its MT depolymerization activity. A, in vitro MT depolymerization activity of MCAK-GST and its deletion constructs in the presence of recombinant purified PAK1 enzyme (PAK1-GST). Rhodamine-labeled, GMPCPP-stabilized microtubules were incubated with MCAK-GST and its deletion constructs in the presence and absence of recombinant purified PAK1 enzyme for 5 min, and images were taken using confocal microscopy. The lower panel represents the quantification of in vitro MT depolymerization activity. For quantification, images were captured using the LSM-710 confocal microscope, and 25 images were taken for each time point. Representative figures from three individual experiments are shown here. For quantification, the number of microtubules per 71 × 56-μm fields from three individual micrographs for each sample were counted. The segmented line tool of the ImageJ software was used to draw a line across the length of the microtubules to get the exact length of the microtubules. Only microtubules greater than 6 μm were considered for analysis for each sample. Data represent mean ± S.D. of those samples. B, in vitro MT depolymerization activity of MCAK-S192A in the presence and absence of recombinant purified PAK1 enzyme. Rhodamine-labeled, GMPCPP-stabilized microtubules were incubated with MCAK-S192A-GST in the presence and absence of recombinant purified PAK1 for 5 min, and images were taken using confocal microscopy. The lower panel represents the quantification of in vitro MT depolymerization activity. The graph represents the number of microtubules per field in each condition. Error bars represent the mean of the S.D. C and D, in vivo MT depolymerization analysis using MCAK-GFP and its phosphorylation site mutants in mitotic ZR-75 cells. ZR-75 cells were transfected with MCAK-GFP, MCAK-S192A-GFP, or MCAK-S111A-GFP and treated with nocodazole (4 μg/ml). After 0- and 15-min release from nocodazole, cells were fixed, stained with the indicated antibodies, and analyzed using confocal microscopy. Microtubule length was scored in >25 cells per experiment. The graph shows the average of three independent experiments. Error bars represent S.D. Scale bars = 5 μm.
To validate the functional significance of the presumed PAK1 phosphorylation site, we next showed the inability of PAK1 to inhibit the MT depolymerization activity of phosphorylation-deficient MCAK-S192A mutant (Fig. 5B). Such an effect was not observed with the MCAK-S95A mutant (supplemental Fig. 3A), suggesting that active PAK1-mediated phosphorylation of MCAK on Ser-192 might be required to inhibit the depolymerization activity of MCAK by PAK1 signaling.
To investigate the significance of MCAK Ser-192 upon MT depolymerization in vivo, we transfected ZR-75 cells with GFP-MCAK, MCAK-S192A-GFP, or MCAK-S111A-GFP, and cells were treated with nocodazole to completely depolymerize the microtubules. Cells were fixed after allowing microtubules to regrow for 15 min. We noticed an increased microtubule polymerization/regrowth in the cells transfected with MCAK-S192A compared with the cells expressing either MCAK, MCAK-S111A, or MCAK-S95A (Fig. 5, C and D, and supplemental Fig. 3B). These findings suggested that PAK1 phosphorylation of MCAK at Ser-192 contributes to MT dynamics. However, because results from MCAK-S192A and MCAK-S111A suggest that the phosphorylation of both serine residues might be trans-regulated, it remains possible that the intact Ser-111 site may not be phosphorylated in MCAK-S192A. Therefore, we suggest that MCAK Ser-192 preferentially regulates MT dynamics with a potential, yet to be demonstrated, hidden contribution of the Ser-111 site.
Furthermore, we did not find any difference in the MT depolymerization between control and Aurora-A siRNA-treated ZR-75 cells (supplemental Fig. 4), suggesting the involvement of upstream activated kinases such as PAK1 in the regulation of MT depolymerization activity. Because PAK1 is upstream of Aurora-A (20) and MCAK Ser-192 could be phosphorylated by both PAK1 (this study) and Aurora-A (36, 37), the lack of any effect of Aurora-A knockdown on MT depolymerization might be due to phosphorylation of MCAK Ser-192 by PAK1 as the sole kinase in the absence of Aurora-A.
PAK1 Phosphorylation of MCAK at Ser-111 Impacts Its Centrosomal Localization
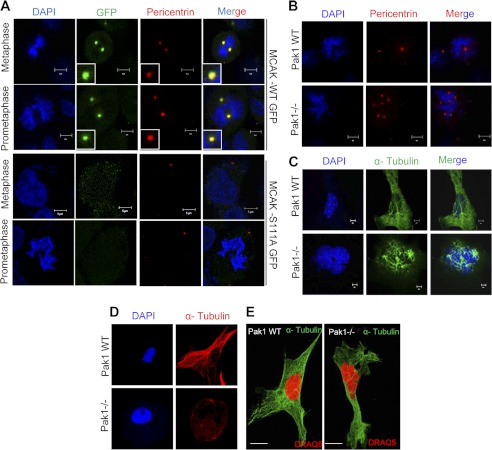
Because PAK1 also phosphorylates MCAK on Ser-111 (Fig. 4 and supplemental Fig. 2), we next evaluated the significance of PAK1 phosphorylation of MCAK at Ser-111 for its centrosomal targeting in human cells. ZR-75 cells transfected with GFP-MCAK, GFP-MCAK-S192A, GFP-MCAK-S111A (but with an intact Ser-192), and GFP-MCAK-S95A were subjected to double thymidine block and then allowed to progress through mitosis for centrosomal localization studies. We found localization of MCAK, MCAK-S192A and -S95A but not MCAK-S111A to the centrosomes along with centrosomal marker protein pericentrin (Fig. 6A and supplemental Fig. 5). Furthermore, we found evidence of defective centrioles in the PAK1−/− MEFs as compared with the wild-type PAK1 MEFs presumably due to lack of Ser-111 phosphorylation in MCAK by PAK1 (Fig. 6B and supplemental Fig. 6A) in addition to any hidden contribution of the Ser-192 site, which might be trans-regulated by Ser-111.
FIGURE 6.
PAK1 phosphorylation of MCAK Ser-111 regulates its centrosomal localization. A, ZR-75 cells transfected with GFP, MCAK-GFP, and MCAK-S111A-GFP were synchronized with double thymidine (2 nm) to the mitotic stage, and centrosomal localization of MCAK was analyzed using confocal microscopy. Anti-pericentrin antibody was used to stain for centrosomes, and DAPI was used as the counterstain. B, abnormal centrioles in PAK1−/− MEFs compared with PAK1 wild-type MEFs. PAK1 wild-type and PAK1−/− MEFs were stained for pericentrin (red), and nuclei were stained with DAPI (blue). C, multiple spindle formation observed in PAK1−/− MEFs compared with PAK1 wild-type MEFs. The PAK1 wild-type and PAK1−/− cells were stained with α-tubulin (green), and nuclei were stained with DAPI. D, long microtubules in PAK1 wild-type MEFs compared with PAK1−/− MEFs. PAK1 wild-type and PAK1−/− MEFs were stained for α-tubulin (red), and nuclei were stained with DAPI (blue). E, multinucleated cells combined with defective MT dynamics were observed in PAK1−/− MEFs compared with PAK1 wild-type MEFs using super-resolution microscopy. The nuclei were stained with DRAQ5 (red). The cells were stained with α-tubulin (green). Scale bars = 5 μm.
Because MCAK is phosphorylated by PAK1 (Fig. 3, A–D) and PAK1−/− MEFs contain easily detectable levels of PAK2 and PAK3 (28), the defect in the centrioles is mainly due to the lack of PAK1. Accordingly, we noticed evidence of abnormal spindles with short microtubules as well as multinucleated cells in the PAK1−/− MEFs as compared with the wild-type PAK1 MEFs (Fig. 6, C, D, and E, and supplemental Fig. 6, B–D). Because such phenotypic effects are normally associated with abnormalities associated with centrosomal proteins (38–40), these findings suggest an inherent role of PAK1 phosphorylation of Ser-111 in MCAK for its centrosomal targeting.
We identified MCAK as a novel PAK1-interacting substrate. MCAK is a potent MT depolymerizer and can track tips of polymerizing MT. Its ability to tip track depends on phosphorylation, which may regulate the association of MCAK with the MT lattice as well as with other interacting proteins. Several other proteins orchestrate the regulation of microtubule dynamics. Many of these proteins are destabilizing proteins that are important for inhibiting the MT polymerization and inducing disassembly. During mitosis, MCAK is tightly regulated by kinases and can either abrogate or accelerate the MT depolymerizing activity. The absence or inhibition of MCAK in cells or from Xenopus egg extracts causes abnormal spindles with long MTs as well as improperly attached chromosomes that lead to mis-segregation.
Here we show that PAK1 can interact and phosphorylate MCAK in a physiologically relevant setting. The PAK1-mediated phosphorylation events at Ser-111 and Ser-192 localized MCAK to centrosomes and inhibited its MT depolymerization activity, respectively. In Fig. 4B, we observed that both the single mutants of MCAK (S111A and S192A) are as effective as the double mutant of MCAK in abolishing the in vivo phosphorylation of MCAK, raising the possibility of interdependence in the Ser-111 and Ser-192 phosphorylation. Ser-192 of MCAK is also phosphorylated by another kinase, Aurora-A, to attenuate its MT depolymerization. The noted PAK1-regulated MCAK function may be independent of Aurora-A activity or it may be in coordination with Aurora-A where PAK1 phosphorylates Aurora-A (20), which in turn phosphorylates MCAK (6) to regulate MT dynamics during mitosis (supplemental Fig. 7). It is known that PAK1 plays an instrumental role in the phosphorylation and targeting of Aurora-A kinase to the centrosomes where Aurora-A kinase is needed for the centrosomal duplication and separation during mitosis (20, 26).
Earlier studies suggest that MCAK phosphorylation in the N-terminal region by mitotic kinases like Aurora-A and Aurora-B targets MCAK to spindle poles, centromeres, kinetochores, and chromosome arms (5, 6, 10, 12). As PAK1 is phosphorylating the N-terminal region of MCAK at position Ser-111 similarly to Auroras, we do speculate that PAK1 is involved in the localization of MCAK to centromeres and spindle poles during mitosis. Furthermore, depletion of PAK1 considerably increased MT depolymerization, whereas overexpression of PAK1 diminished the ability of MCAK to remove tubulin subunits from the microtubule end. The presence of MCAK in the same complex as PAK1, γ-tubulin, Aurora-A, and pericentrin suggests that PAK1 interacts with MCAK at the centrosomes during mitosis, and these functions could be regulated by two independent kinases. In brief, the findings presented here have identified MCAK as a novel PAK1-interacting substrate and revealed the exclusivity of phosphorylation-site specific functions of MCAK by the same kinase. Based on these findings, we propose that PAK1 phosphorylation of MCAK at positions Ser-192 and Ser-111 regulates MT dynamics and centrosomal targeting of MCAK, respectively.
Supplementary Material
Acknowledgments
We thank the members of our laboratory for insightful discussions and Poonam R. Molli for in vitro kinase assay.
This work was supported, in whole or in part, by National Institutes of Health Grant CA90970 (to R. K.).

This article contains supplemental Figs. 1–7.
- MT
- microtubule
- PAK
- p21-activated kinase
- MCAK
- mitotic centromere-associated kinesin
- MEF
- murine embryonic fibroblast
- GMPCPP
- guanosine 5′-(α,β-methylene)triphosphate
- GPS
- group-based phosphorylation site predicting and scoring platform
- PIX
- PAK-interacting exchange factor.
REFERENCES
- 1. Manning A. L., Ganem N. J., Bakhoum S. F., Wagenbach M., Wordeman L., Compton D. A. (2007) The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell 18, 2970–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maney T., Wagenbach M., Wordeman L. (2001) Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J. Biol. Chem. 276, 34753–34758 [DOI] [PubMed] [Google Scholar]
- 3. Maney T., Hunter A. W., Wagenbach M., Wordeman L. (1998) Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142, 787–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wordeman L., Mitchison T. J. (1995) Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128, 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang X., Ems-McClung S. C., Walczak C. E. (2008) Aurora-A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol. Biol. Cell 19, 2752–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang X., Lan W., Ems-McClung S. C., Stukenberg P. T., Walczak C. E. (2007) Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol. Biol. Cell 18, 3264–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang L., Shao H., Huang Y., Yan F., Chu Y., Hou H., Zhu M., Fu C., Aikhionbare F., Fang G., Ding X., Yao X. (2011) PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J. Biol. Chem. 286, 3033–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanhaji M., Friel C. T., Kreis N. N., Krämer A., Martin C., Howard J., Strebhardt K., Yuan J. (2010) Functional and spatial regulation of mitotic centromere-associated kinesin by cyclin-dependent kinase 1. Mol. Cell. Biol. 30, 2594–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosasco-Nitcher S. E., Lan W., Khorasanizadeh S., Stukenberg P. T. (2008) Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science 319, 469–472 [DOI] [PubMed] [Google Scholar]
- 10. Andrews P. D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J. R. (2004) Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 6, 253–268 [DOI] [PubMed] [Google Scholar]
- 11. Ohi R., Burbank K., Liu Q., Mitchison T. J. (2007) Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr. Biol. 17, 953–959 [DOI] [PubMed] [Google Scholar]
- 12. Lan W., Zhang X., Kline-Smith S. L., Rosasco S. E., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Walczak C. E., Stukenberg P. T. (2004) Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14, 273–286 [DOI] [PubMed] [Google Scholar]
- 13. Bokoch G. M. (2003) Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743–781 [DOI] [PubMed] [Google Scholar]
- 14. Eswaran J., Soundararajan M., Knapp S. (2009) Targeting group II PAKs in cancer and metastasis. Cancer Metastasis Rev. 28, 209–217 [DOI] [PubMed] [Google Scholar]
- 15. Brzeska H., Szczepanowska J., Matsumura F., Korn E. D. (2004) Rac-induced increase of phosphorylation of myosin regulatory light chain in HeLa cells. Cell Motil. Cytoskeleton 58, 186–199 [DOI] [PubMed] [Google Scholar]
- 16. Zhang H., Webb D. J., Asmussen H., Niu S., Horwitz A. F. (2005) A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J. Neurosci. 25, 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wittmann T., Bokoch G. M., Waterman-Storer C. M. (2004) Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. J. Biol. Chem. 279, 6196–6203 [DOI] [PubMed] [Google Scholar]
- 18. Vadlamudi R. K., Barnes C. J., Rayala S., Li F., Balasenthil S., Marcus S., Goodson H. V., Sahin A. A., Kumar R. (2005) p21-activated kinase 1 regulates microtubule dynamics by phosphorylating tubulin cofactor B. Mol. Cell. Biol. 25, 3726–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vadlamudi R. K., Li F., Barnes C. J., Bagheri-Yarmand R., Kumar R. (2004) p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 5, 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao Z. S., Lim J. P., Ng Y. W., Lim L., Manser E. (2005) The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol. Cell 20, 237–249 [DOI] [PubMed] [Google Scholar]
- 21. Sanhaji M., Friel C. T., Wordeman L., Louwen F., Yuan J. (2011) Mitotic centromere-associated kinesin (MCAK): a potential cancer drug target. Oncotarget 2, 935–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kinoshita K., Noetzel T. L., Pelletier L., Mechtler K., Drechsel D. N., Schwager A., Lee M., Raff J. W., Hyman A. A. (2005) Aurora-A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol. 170, 1047–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Domnitz S. B., Wagenbach M., Decarreau J., Wordeman L. (2012) MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochores attachment. J. Cell Biol. 197, 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bagheri-Yarmand R., Vadlamudi R. K., Wang R. A., Mendelsohn J., Kumar R. (2000) Vascular endothelial growth factor up-regulation via p21-activated kinase-1 signaling regulates heregulin-β1-mediated angiogenesis. J. Biol. Chem. 275, 39451–39457 [DOI] [PubMed] [Google Scholar]
- 25. Balasenthil S., Sahin A. A., Barnes C. J., Wang R. A., Pestell R. G., Vadlamudi R. K., Kumar R. (2004) p21-activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells. J. Biol. Chem. 279, 1422–1428 [DOI] [PubMed] [Google Scholar]
- 26. Li F., Adam L., Vadlamudi R. K., Zhou H., Sen S., Chernoff J., Mandal M., Kumar R. (2002) p21-activated kinase 1 interacts with and phosphorylates histone H3 in breast cancer cells. EMBO Rep. 3, 767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vadlamudi R. K., Adam L., Wang R. A., Mandal M., Nguyen D., Sahin A., Chernoff J., Hung M. C., Kumar R. (2000) Regulatable expression of p21-activated kinase-1 promotes anchorage-independent growth and abnormal organization of mitotic spindles in human epithelial breast cancer cells. J. Biol. Chem. 275, 36238–36244 [DOI] [PubMed] [Google Scholar]
- 28. Molli P. R., Li D. Q., Bagheri-Yarmand R., Pakala S. B., Katayama H., Sen S., Iyer J., Chernoff J., Tsai M. Y., Nair S. S., Kumar R. (2010) Arpc1b, a centrosomal protein, is both an activator and substrate of Aurora-A. J. Cell Biol. 190, 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aoki S., Ohta K., Yamazaki T., Sugawara F., Sakaguchi K. (2005) Mammalian mitotic centromere-associated kinesin (MCAK): a new molecular target of sulfoquinovosylacylglycerols novel antitumor and immunosuppressive agents. FEBS J. 272, 2132–2140 [DOI] [PubMed] [Google Scholar]
- 30. Waterman-Storer C. M., Worthylake R. A., Liu B. P., Burridge K., Salmon E. D. (1999) Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat. Cell Biol. 1, 45–50 [DOI] [PubMed] [Google Scholar]
- 31. Cau J., Faure S., Comps M., Delsert C., Morin N. (2001) A novel p21-activated kinase binds the actin and microtubule networks and induces microtubule stabilization. J. Cell Biol. 155, 1029–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meunier S., Vernos I. (2011) K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat. Cell Biol. 13, 1406–1414 [DOI] [PubMed] [Google Scholar]
- 33. Zhou F. F., Xue Y., Chen G. L., Yao X. (2004) GPS: a novel group-based phosphorylation predicting and scoring method. Biochem. Biophys. Res. Commun. 325, 1443–1448 [DOI] [PubMed] [Google Scholar]
- 34. Charvet C., Auberger P., Tartare-Deckert S., Bernard A., Deckert M. (2002) Vav1couples T cell receptor to serum responsible factor-dependent transcription via a MEK dependent pathway. J. Biol. Chem. 277, 15376–15384 [DOI] [PubMed] [Google Scholar]
- 35. Senger D. L., Tudan C., Guiot M. C., Mazzoni I. E., Molenkamp G., LeBlanc R., Antel J., Olivier A., Snipes G. J., Kaplan D. R. (2002) Suppression of Rac activity induces apoptosis of human glioma cells but not normal human astrocytes. Cancer Res. 62, 2131–2140 [PubMed] [Google Scholar]
- 36. Tanenbaum M. E., Medema R. H., Akhmanova A. (2011) Regulation of localization and activity of the microtubule depolymerase MCAK. Bioarchitecture 1, 80–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Katayama H., Sasai K., Kloc M., Brinkley B. R., Sen S. (2008) Aurora kinase-A regulates kinetochore/chromatin associated microtubule assembly in human cells. Cell Cycle 7, 2691–2704 [DOI] [PubMed] [Google Scholar]
- 38. Meraldi P., Honda R., Nigg E. A. (2002) Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 21, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shu H. B., Joshi H. C. (1995) γ-Tubulin can both nucleate microtubule assembly and self-assemble into novel tubular structures in mammalian cells. J. Cell Biol. 130, 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Purohit A., Tynan S. H., Vallee R., Doxsey S. J. (1999) Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 147, 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.