Background: The regulatory mechanism underlying the interaction of the Rabex-5 MIU domain with ubiquitinated cargos remains unclear.
Results: Rabex-5 guanine nucleotide exchange factor (GEF) mutants affected interactions of ubiquitinated cargos.
Conclusion: GDP/GTP exchange in the GEF domain controls the MIU domain interactions with the ubiquitinated cargos.
Significance: Rabex-5 GEF activity acts as an intramolecular switch for spatiotemporal trafficking of the ubiquitinated cargos.
Keywords: Allosteric Regulation, Endocytosis, Guanine Nucleotide Exchange Factor (GEF), Rab, Ubiquitination, Ubiquitin-binding Domain
Abstract
Ubiquitin (Ub)-dependent endocytosis of membrane proteins requires precise molecular recognition of ubiquitinated cargo by Ub-binding proteins (UBPs). Many UBPs are often themselves monoubiquitinated, a mechanism referred to as coupled monoubiquitination, which prevents them from binding in trans to the ubiquitinated cargo. However, the spatiotemporal regulatory mechanism underlying the interaction of UBPs with the ubiquitinated cargo, via their Ub-binding domains (UBDs) remains unclear. Previously, we reported the interaction of Rabex-5, a UBP and guanine nucleotide exchange factor (GEF) for Rab5, with ubiquitinated neural cell adhesion molecule L1, via its motif interacting with Ub (MIU) domain. This interaction is critical for the internalization and sorting of the ubiquitinated L1 into endosomal/lysosomal compartments. The present study demonstrated that the interaction of Rabex-5 with Rab5 depends specifically on interaction of the MIU domain with the ubiquitinated L1 to drive its internalization. Notably, impaired GEF mutants and the Rabex-5E213A mutant increased the flexibility of the hinge region in the HB-VPS9 tandem domain, which significantly affected their interactions with the ubiquitinated L1. In addition, GEF mutants increased the catalytic efficiency, which resulted in a reduced interaction with the ubiquitinated L1. Furthermore, the coupled monoubiquitination status of Rabex-5 was found to be significantly associated with interaction of Rabex-5 and the ubiquitinated L1. Collectively, our study reveals a novel mechanism, wherein the GEF activity of Rabex-5 acts as an intramolecular switch orchestrating ubiquitinated cargo-binding activity and coupled monoubiquitination to permit the spatiotemporal dynamic exchange of the ubiquitinated cargos.
Introduction
Ubiquitination-mediated degradation of membrane proteins is crucial to protein quality control and attenuation of the receptor-mediated signaling pathway. Ubiquitination aids these processes by specifying the proteins that should be transported to lysosomes through the multivesicular endosomal pathway (1, 2). Ubiquitinated proteins are sorted into distinct pathways, via their association with several classes of Ub-binding domain (UBD)2-containing proteins, thereby controlling downstream biochemical processes (3–6). A number of endocytic Ub-binding proteins (UBPs) such as Eps15, Epsin, Sts, Hrs, and Rabex-5, a guanine nucleotide exchange factor (GEF) for Rab5-undergo self-monoubiquitination, a phenomenon known as coupled monoubiquitination, which prevents them from binding in trans to the ubiquitinated cargo proteins, thus providing an efficient, intrinsic UBP switch off mechanism (3, 7–10). However, the spatiotemporal control mechanism relating to the ability of these endocytic UBPs to associate and/or dissociate with ubiquitinated cargos during endocytic trafficking has not been elucidated.
UBPs often possess numerous UBDs belonging to different classes (11). Despite this, different UBDs function collectively or independently to recognize a ubiquitinated target. Studies on Rabex-5 provided the first structural insight into the combinatorial recognition of Ub by multiple UBDs in a single protein. Rabex-5 contains 2 independent UBDs, the A20-zinc finger (ZnF) and a motif interacting with Ub (MIU) (Fig. 1A), which binds Ub with higher affinities when compared with other Ub-binding motifs (12). The presence of multiple UBDs allows UBPs to either bind to several mono-Ub molecules in multimonoubiquitinated cargos (4) or to engage with a single mono-Ub through different UBDs (11, 13, 14), thus increasing their binding affinity. However, these domains exhibit different biochemical properties. One such example is that Ub employs the Asp-58 surface residue to associate with the A20-ZnF domain and the Ile-44 patch to bind to the MIU domain (13, 14). The A20-ZnF domain possesses Ub-ligase activity (13–16) and promotes Ras ubiquitination to attenuate the Ras signaling pathway (17). To exert Ub-ligase activity, the A20-ZnF domain binds Ub-charged Ubc5 (an E2 enzyme) in a manner that depends upon the residues involved in Ub binding. In a previous study, we reported that the MIU domain, rather than the A20-ZnF domain, is specifically involved in the trafficking of ubiquitinated cargo into endosomes via a direct interaction (18). Our findings thus suggested that the MIU and A20-ZnF domains are functionally distinct in the Ub signaling pathways.
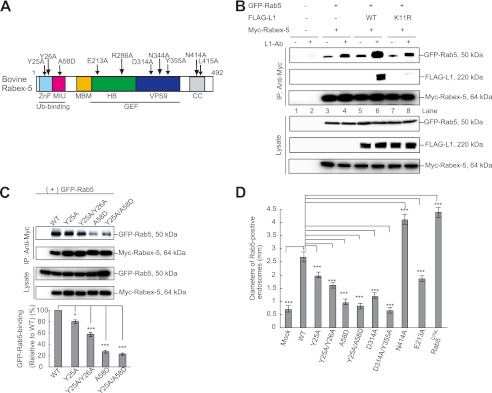
FIGURE 1.
UBDs regulate the Rab5-binding activity of Rabex-5. A, schematic diagram showing the domain structure of bovine Rabex-5. Light blue, ZnF; magenta, MIU; orange, membrane-binding motif (MBM); green, HB; blue, VPS9; and gray, CC domain. The arrows indicate the Rabex-5 mutants used in this study. B, N2a cells co-expressing the indicated plasmids were incubated in the presence or absence of L1-Ab. The cells were then lysed, and immunoblot analysis was performed. To control the functionality of the antibodies, cell lysates were applied (Lysate). C, cells co-expressing GFP-Rab5 with the indicated plasmids were lysed and analyzed by immunoblotting (upper). The bars represent the relative densitometric value of GFP-Rab5 to Rabex-5WT (lower). The data are mean ± S.E. in triplicate. *, p < 0.05; ***, p < 0.001. D, the diameters of the largest GFP-Rab5-labeled endosomes in 30 N2a cells expressing Rabex-5WT, its mutants, and Rab5Q79L were measured: the graph shows the mean and calculated S.E. ***, p < 0.001.
Rabex-5 originally identified as a GEF for Rab5 (19), is a small GTPase that is associated with the early endosomal membrane and is involved in the regulation of early endosome fusion, as well as endocytosis (20–22). The recruitment of the Rabaptin5·Rabex-5 complex to early endosomal membranes containing Rab5-GTP has been proposed as a positive-feedback mechanism that results in the local nucleotide exchange of Rab5 in restricted areas of the endosomal membrane (19, 23). Rabex-5 has been reported to get translocated to the plasma membrane upon ligand stimulation with either the EGF or L1 antibody (Ab) (14, 18) and induce the production of Rab5-positive enlarged vacuoles due to the increment of the activated form of Rab5 (18). These findings suggest a plausible mechanism of ubiquitinated cargo trafficking, which is similar to that of the vacuolar protein sorting 9 protein (Vps9p), a yeast homolog of Rabex-5, wherein cross-talk occurs between the GEF activity of the VPS9 domain, and the Ub-binding activity, which involves the coupling of Ub conjugation to the ER degradation (CUE) domain (24). However, the underlying mechanism integrating the Ub-binding activity, as well as the GEF activity for Rab5 in Rabex-5, has not yet been resolved.
To investigate the molecular mechanism that integrates the Ub-binding and GEF activities of Rabex-5, we employed the Rabex-5-mediated endocytic pathway of the ubiquitinated neuronal cell adhesion molecule L1 as the model system (18) and assessed the functional impact of UBDs on GEF activity and vice versa. We found that Ub binding-deficient Rabex-5 mutants significantly diminish the interaction with Rab5. Intriguingly, we found that the GEF activity of Rabex-5 plays a critical role in the regulation of the interaction with the ubi quitinated L1. Furthermore, we showed a significant association between the coupled monoubiquitination and the interaction of Rabex-5 with the ubiquitinated L1. Taken together, our data provide compelling evidence for the spatiotemporal regulation of the ubiquitinated cargo trafficking by integrating the Ub-binding activity, as well as the GEF activity of Rabex-5.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and DNA Constructs
Mouse neuroblastoma N2a cells and human embryonic kidney 293T (HEK293T) cells were cultured and transfected by using Lipofectamine 2000 (Invitrogen) as described previously (18). The human Rab5a construct was amplified using PCR from cDNA and subcloned into an EGFP-N1 vector. The constructs for the FLAG-tagged L1 mutants and GFP-Rab5 mutants were as described previously (18). pCI-neo (Promega, Madison, WI, USA), encoding a full-length Myc-tagged bovine Rabex-5, was generously provided by Dr. J. S. Bonifacino (Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health). Myc-tagged Rabex-5 mutants were generated using a QuikChange Site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions, as were primers containing the corresponding mutant sequences. The constructs were checked by sequencing.
Immunoprecipitation, Western Blotting, and Subcellular Fractionation
Immunoprecipitation and Western blotting were performed as described previously (18). Briefly, N2a cells incubated with anti-L1 polyclonal antibody or anti-mouse L1 monoclonal antibody (5 μg/106 cells) were lysed in lysis buffer containing protease inhibitors for 60 min on ice. The cell lysates were clarified by centrifugation prior to separation by SDS-PAGE. The resolved proteins were electrotransferred to PVDF membranes and then blocked in TBST containing 5% nonfat milk. The membranes were probed with primary antibodies, followed by the addition of horseradish peroxidase-conjugated secondary antibody (Pierce). Specific proteins were visualized with enhanced chemiluminescence detection reagents (Pierce). Results were quantified using the Quantity One 1-D analysis software (Bio-Rad). For immunoprecipitation, the clarified cell lysates were incubated with the primary antibodies and protein G-Sepharose (GE Healthcare) overnight at 4 °C. The beads were washed extensively with lysis buffer and solubilized in SDS sample loading buffer. Subcellular fractionation was conducted as described previously (18). Cells were homogenized using a Dounce homogenizer and centrifuged at 850 × g for 10 min to remove nuclei and cell debris, and postnuclear supernatants were subjected to ultracentrifugation at 200,000 × g for 10 min in a Himac CS120GXL centrifuge (Hitachi, Tokyo, Japan) to separate the membrane (pellet) and cytosolic (supernatant) fractions. Antibodies purchased were FLAG (M2; Sigma), Myc (9E10 hybridoma; Roche Applied Science), hemagglutinin (HA; Covance Research Products, Berkeley, CA), Ub (Dako Cytomation Denmark A/S, Glostrup, Denmark), GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA), L1 (goat polyclonal IgG; Santa Cruz Biotechnology), and Rab5 (Cell Signaling Technology, Danvers, MA). Affinity purified polyclonal anti-Rabex-5, anti-Rab5, and anti-L1 antibodies were raised using a C-terminal fragment of bovine Rabex-5 (residues 426–481), full-length human Rab5a, and full-length rat L1 as immunogens, respectively.
Biotinylation Assay for Endocytosis and Recycling
Cells pretreated with cycloheximide (10 μg/ml) and leupeptin (0.3 mm) were washed with ice-cold PBS and biotinylated by incubating with 300 μg/ml of EZ-Link-Sulfo-NHS-SS-Biotin (Pierce) for 30 min at 4 °C, as described previously (18). To measure recycling of the endocytosed proteins, the cells were incubated at 37 °C for various times in normal medium and then washed with the stripping solution, as described previously (25). Cell extracts were prepared, and cell debris was removed by centrifugation at 14,000 × g for 20 min. Clarified cell extracts were precipitated using streptavidin and immobilized on agarose beads at 4 °C for 2 h. After 5 washes with cell lysis buffer, the bound proteins were removed using SDS sample buffer.
Immunofluorescent Microscopy
Immunocytochemistry was performed as described previously (18). At 48 h after transfection, the cells were rinsed with PBS, fixed in 4% formaldehyde for 30 min, permeabilized with 0.3% Triton X-100 in PBS for 30 min, and then blocked with goat serum. Following incubation with the primary antibodies and further with Alexa Fluor 488-, Alexa Fluor 563-, and Alexa Fluor 633-conjugated secondary antibodies (Molecular Probes, Eugene, OR), the coverslips were mounted with fluorescent mounting medium (Vectashield, Vector, CA). Labeled cells were visualized using a 1X71 fluorescence microscope (Olympus, Tokyo, Japan) under a ×60 oil immersion objective lens. Quantification of surface and/or intracellular fluorescence intensities of L1 was performed with MetaMorph imaging software (Universal Imaging Corp., PA) using an arbitrary threshold. Laser-scanning confocal microscopy was performed using an Olympus FLUOVIEW FV1000 confocal laser scanning microscope equipped with a ×63 oil immersion objective lens. In at least 3 independent experiments, 30 cells were photographed and analyzed for each construct.
Investigation of the Conformational Changes in the Mutants by Molecular Dynamics Simulations
On the basis of the crystal structure of the wild-type human Rabex-5 helix bundle (HB)-VPS9 tandem domain (PDB code 2OT3, chain A) (26), E212A, N343A, and E212A/N343A mutants were modeled using an MOE program (Chemical Computing Group, Montreal, Canada). The system was solvated in a periodic cubic box filled with TIP3P water molecules with an Amber ff99 force field (27). Before the molecular dynamics (MD) simulation, the modeled structure was optimized using the AMBER7 package (28). During the MD simulation, the system was warmed to 300 K for 20 ps in the NVT ensemble, and the simulation was continued at 300 K for 2.0 ns with a 500-ps time step in the NVT ensemble using the AMBER7 package. After the system had reached a sufficiently equilibrated state, the last 200 conformations were sampled at 1-ps intervals for structural analysis. The fluctuations in the atomic positions were calculated as B-factors.
Statistical Analyses
All experiments were repeated at least three times, and the values represent mean ± S.E. Student's t test was used to calculate statistical significance. *, **, and *** represent p < 0.05, p < 0.01, and p < 0.001, respectively. The error bars denote the S.E.
RESULTS
The Recruitment of Rab5 to Rabex-5 Depends on the Interaction of Rabex-5 with the Ubiquitinated L1
Previously, we showed that the dephosphorylation and ubiquitination of L1, which are followed by its association with the MIU domain of Rabex-5, enhance the internalization and sorting of the ubiquitinated L1 into lysosomes (18). Penengo et al. (14) reported that Rabex-5 was translocated to the plasma membrane upon EGF ligand stimulation. We found that Rabex-5 enlarged the early endosomal compartments upon stimulation with L1 ligands (18). On the basis of these results, we hypothesized that the interaction of the MIU domain of Rabex-5 with ubiquitinated cargos on the plasma membrane might trigger the recruitment of Rab5 to its GEF domain to stimulate endocytosis of the ubiquitinated cargos.
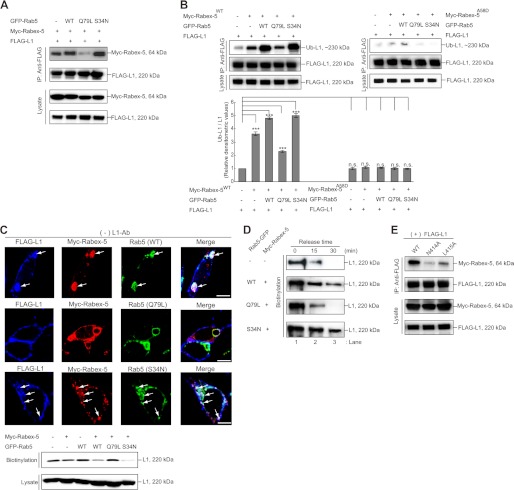
To determine whether the interaction of Rabex-5 with ubiquitinated L1 leads to the recruitment of Rab5 to Rabex-5 upon L1-Ab stimulation, we transfected FLAG-tagged L1WT and a FLAG-tagged L1K11R, a ubiquitination-deficient mutant (18) separately, in combination with GFP-Rab5 and Myc-tagged Rabex-5 into N2a cells expressing L1 endogenously (Fig. 1B). Subsequently, we incubated the transfected N2a cells separately in the presence and absence of L1-Ab, respectively, which mimics L1-L1 homophilic binding (18).
Incubation of cells co-expressing Myc-tagged Rabex-5 and GFP-Rab5 with L1-Ab slightly enhanced the interaction of Myc-tagged Rabex-5 with GFP-Rab5 by 14.3 ± 1.9% (Fig. 1B, lane 3 versus lane 4, n = 3, p < 0.05), implying that this enhancement might be due to the L1-Ab-dependent ubiquitination of the endogenous L1. Notably, the interaction of Myc-tagged Rabex-5 with GFP-Rab5 in cells co-expressing FLAG-tagged L1WT was found to be significantly increased by 82.4 ± 2.8% (Fig. 1B, lane 5 versus lane 6, n = 3, p < 0.01). However, the interaction of Myc-tagged Rabex-5 with GFP-Rab5 in cells expressing FLAG-tagged L1K11R was comparable with that in the cells co-expressing Myc-tagged Rabex-5 and GFP-Rab5 (14.1 ± 1.8%, Fig. 1B, lane 7 versus lane 8, n = 3, p < 0.05).
Notably, FLAG-tagged L1WT was found to coimmunoprecipitate with Myc-tagged Rabex-5, upon the incubation with L1-Ab (Fig. 1B, lane 6), indicating the formation of a ubiquitinated L1·Rabex-5·Rab5 complex. These results suggest that L1-L1 homophilic binding results in an increased interaction between Rabex-5 and Rab5, which in turn depends specifically on the interaction of the ubiquitinated L1 with Rabex-5.
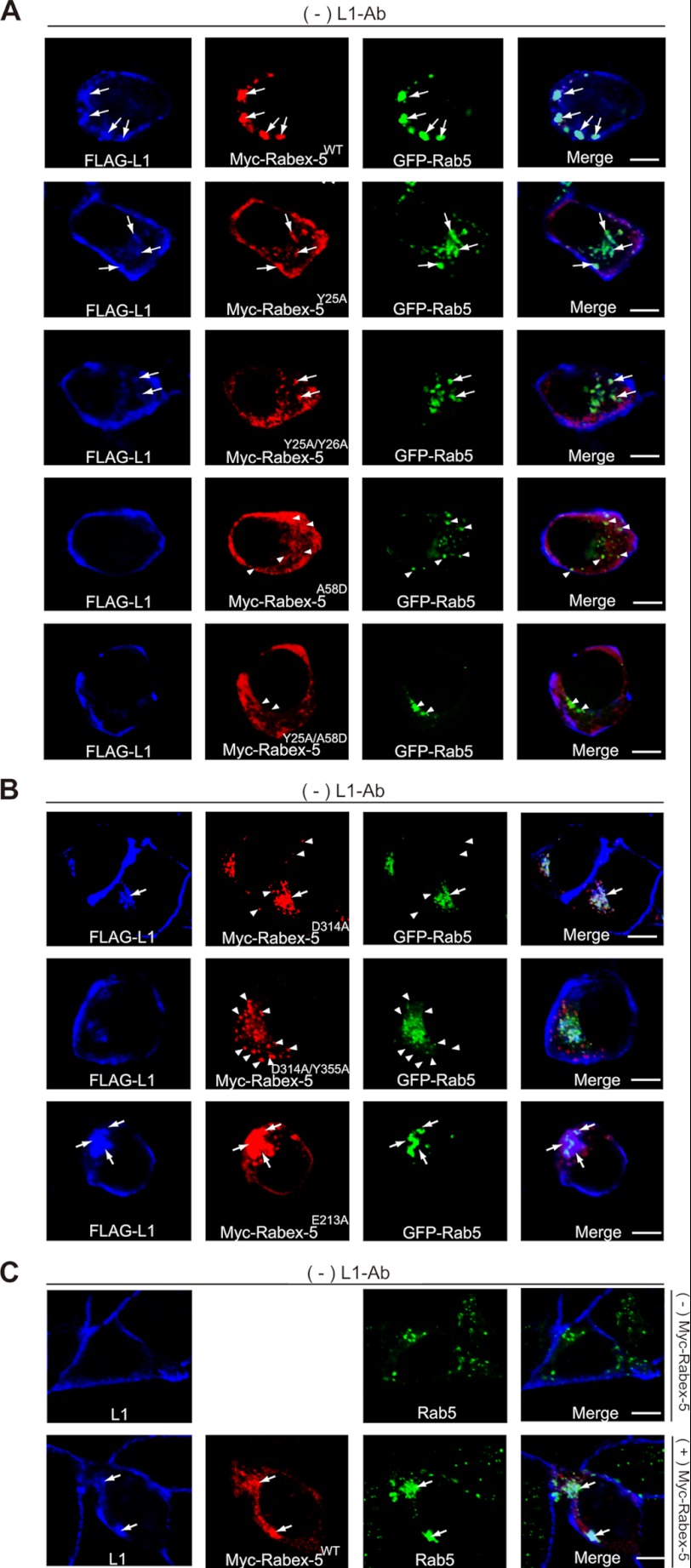
To further investigate the functional role of the enhanced recruitment of Rab5 to Rabex-5 due to the interaction of the ubiquitinated L1 with UBDs of Rabex-5, we assessed the endocytosis of L1, Rab5-binding, and determined the size of Rab5-positive early endosomes by using Rabex-5 mutants with impaired UBDs. Rabex-5 mutants with impaired UBDs, such as Rabex-5Y25A, Rabex-5Y25A/Y26A, Rabex-5A58D, and Rabex-5Y25A/A58D (Fig. 1A) (13–15), displayed the following properties: (i) reduction of Rab5-binding activity by 21.2 ± 4.6, 45.3 ± 4.1, 76.1 ± 3.5, and 79.5 ± 3.2% (n = 3), respectively, compared with Rabex-5WT (Fig. 1C); (ii) reduction of L1 accumulation on endosomes due to their overexpression (Fig. 2A, arrows), consistent with previous data (18). In particular, the accumulation of L1 in cells expressing Rabex-5A58D or Rabex-5Y25A/A58D was not detected, which is comparable with that of cells that lacked the overexpressing of Rabex-5WT (Fig. 2C, upper); (iii) partial co-localization of Rabex-5Y25A and Rabex-5Y25A/Y26A with L1 and Rab5-positive endosomes (Fig. 2A, arrows) was the third property. However, most Rabex-5A58D and Rabex-5Y25A/A58D showed no such co-localization with Rab5-positive endosomes (Fig. 2A, arrowheads) and were found to be distributed throughout the cytoplasmic region. These observations are supported by a previous finding that showed that membrane targeting of Rabex-5 is mediated by the association of its UBDs with ubiquitinated membrane proteins (10); and (iv) reduction in the diameter of Rab5-positive early endosomes by 24.2 ± 3.5, 41.1 ± 3.8, 66.2 ± 2.5, and 71.4 ± 2.2% (n = 30), respectively, when compared with Rabex-5WT (Fig. 1D).
FIGURE 2.
Impact of Rabex-5 mutants on the subcellular distributions of L1 and Rab5. A, N2a cells co-expressing the indicated plasmids in the absence of L1-Ab were stained with FLAG-tagged L1 (blue), Myc-tagged Rabex-5 (red), and GFP-Rab5 (green). The arrows indicate the accumulation of L1 on Rabex-5-positive endosomes. The arrowheads indicate that Rab5-positive endosomes do not co-localize with Rabex-5 UBDs mutants. Scale bar, 5 μm. B, N2a cells transfected with the indicated plasmids in the absence of L1-Ab were stained with FLAG-tagged L1 (blue), Myc-tagged Rabex-5 (red), and GFP-Rab5 (green). The arrows indicate the accumulation of L1 on Rabex-5-positive endosomes. The arrowheads indicate that Rabex-5 GEF mutants do not co-localize with Rab5-positive endosomes. Scale bar, 5 μm. C, N2a cells in the absence of L1-Ab were stained with endogenous L1 (blue) and endogenous Rab5 (green) (upper). N2a cells transfected with Myc-tagged Rabex-5 were stained with endogenous L1 (blue), Myc-tagged Rabex-5 (red), and endogenous Rab5 (green) (lower). Note that overexpression of Myc-tagged Rabex-5 induced the accumulation of endogenous L1 and enlarged the endogenous Rab5-positive endosomes. Scale bar, 5 μm.
Notably, the MIU domain, rather than the A20-ZnF domain, affected Rab5-binding activity and reduced the size of Rab5-positive endosomes significantly, thus strengthening the hypothesis that the interaction of Rab5 with Rabex-5 further depends on interaction of the MIU domain with the ubiquitinated L1. Taken together, these findings highlight one of the different biochemical properties of these UBDs. Consequently, these results further led to the investigation of the mechanism by which Rabex-5 orchestrates between the sequential binding with the ubiquitinated L1 and Rab5, via the MIU and HB-VPS9 domains prior to the internalization of the ubiquitinated L1.
The HB-VPS9 Tandem Domain Regulates the Interaction of the MIU Domain with the Ubiquitinated L1
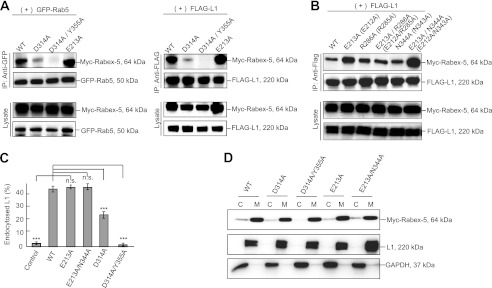
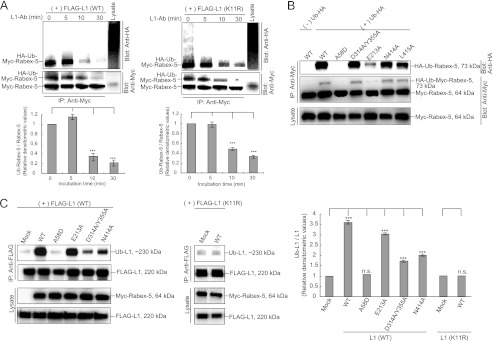
To determine whether the HB-VPS9 domain modulates interaction of the MIU domain with the ubiquitinated L1, we assessed the interaction of Rabex-5D314A (Fig. 1A), a GEF activity-impaired Rabex-5 mutant (26), and FLAG-tagged L1 or Rab5S34N, a GDP-locked form of Rab5 (29). Intriguingly, interactions of the Rabex-5D314A mutant with Rab5 and L1 were found to be significantly decreased by 58.3 ± 3.5 and 62.1 ± 5.5%, respectively (Fig. 3A, n = 3, p < 0.01), and the Rabex-5D314A mutant was partially co-localized with L1 and Rab5-positive endosomes (Fig. 2B, arrows). Furthermore, Rabex-5D314A/Y355A, a GEF activity impaired Rabex-5 double mutant (26), impaired the interactions with L1 and Rab5 (Fig. 3A) and was found to be predominantly localized on the peripheral endosomes (Fig. 2B, arrowheads), where it is not co-localized with the internalized L1. Expectedly, Rabex-5D314A and Rabex-5D314A/Y355A significantly reduced the size (in diameter) of Rab5-positive endosomes (Fig. 1D), despite the targeting the GEF mutants to the membrane fraction (Fig. 3D). These results are in agreement with a previous observation where the GEF activity of Rabex-5 was identified to be indispensable for its recruitment to early endosomes (10).
FIGURE 3.
Analysis of Rabex-5 GEF mutants. A, the cells were co-transfected with the indicated plasmids. Cell lysates were immunoprecipitated and analyzed by immunoblotting. B, cells co-expressing FLAG-tagged L1 with the indicated Myc-tagged Rabex-5 mutants were immunoprecipitated and analyzed by immunoblotting. The parentheses indicate human amino acid residues. C, fluorescence intensities were quantified, and the percentage of perinuclear to total fluorescence was plotted (right). ***, p < 0.001; n.s., not significant. D, immunoblot showing the membrane and cytosol distribution in N2a cells expressing the indicated plasmids and endogenous L1 and GAPDH, as described above. C, cytosol; M, membrane.
Although Rabex-5WT expression led to an increased accumulation of L1 on endosomes (Fig. 2, A and C, arrows), the expressions of Rabex-5D314A and Rabex-5D314A/Y355A resulted in a comparatively lesser accumulation (Fig. 2B). Expectedly, the percentage of intracellular to total fluorescence intensity of L1 in cells expressing Rabex-5D314A and Rabex-5D314A/Y355A was significantly decreased by 55.2 ± 3.5 and 97.2 ± 1.7% (n = 3), respectively, when compared with Rabex-5WT (Fig. 3C). These results suggest 2 possibilities: the allosteric regulation of stability by the GEF domain for the formation of the ubiquitinated L1·Rabex-5·Rab5 complex or the decreased binding to the ubiquitinated L1 due to the mislocalization of GEF mutants.
To exclude the latter possibility, we searched for another GEF mutant whose subcellular localization was comparable with that of Rabex-5WT. Although it has been reported that mutation of Glu-213 prevents solubility (26), we observed that Myc-tagged Rabex-5E213A significantly increased its binding to Rab5 and L1 by 31.5 ± 4.1% (Fig. 3A, right panels, n = 3, p < 0.01) and 42.3 ± 2.9% (Fig. 3A, left panels, n = 3, p < 0.01), respectively. In addition, Rabex-5E213A expression also resulted in the accumulation of L1 on endosomes (Fig. 2B, arrows).
Glu-213, which is located at the interface between the HB and VPS9 domains, has been shown to play a critical role in the structural stability of these domains (26). Therefore, we hypothesized that conformational changes in the HB-VPS9 tandem domain caused by Rabex-5E213A might be involved in regulation of the interaction with ubiquitinated L1 and/or its structural stability through the MIU domain.
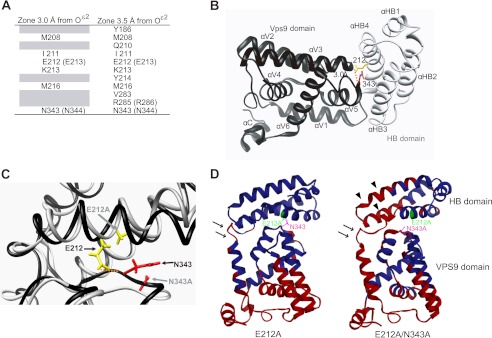
This prompted us to identify the amino acid residues within hydrogen bond distance of the Oϵ2 atom of Glu-212 on the basis of the crystal structure of the human Rabex-5 HB-VPS9 tandem domain (26). The distance between the Oϵ2 atom of Glu-212 and the amide nitrogen of Asn-343 in human Rabex-5 was the closest (2.99 Å) among all the atom pairs (Fig. 4, A and B). Among the identified residues, we examined the impact of the following mutants on the interaction with FLAG-tagged L1 by co-immunoprecipitation: Myc-tagged Rabex-5E213A, Rabex-5R286A, Rabex-5E213A/R286A, Rabex-5N344A, and Rabex-5E213A/N344A (Fig. 1A). Strikingly, the interaction of the ubiquitinated L1 with Myc-tagged Rabex-5E213A/N344A increased most significantly by 92.1 ± 4.3% (Fig. 3B, n = 3, p < 0.01), compared with that of Myc-tagged Rabex-5WT, leading to the accumulation of L1 on endosomes (data not shown). Thus, our findings proved that the interface between the HB and VPS9 domains play a critical role in regulating the structural stability of the interaction between Rabex-5 and the ubiquitinated cargos.
FIGURE 4.
Structural modeling of the human Rabex-5 HB-VPS9 tandem domain. A, amino acid residues in human Rabex-5 located within 3 or 3.5 Å from the Oϵ2 atom of Glu-212 in the HB-VPS9 tandem domain. The parentheses indicate the bovine amino acid residues. B, ribbon diagram of the human HB (gray) and VPS9 domains (black) (PDB code 2OT3). Glu-212 (E213 in bovine) and Asn-343 (N344 in bovine) are highlighted as stick models in yellow and red, respectively. A dotted orange line indicates hydrogen bonds. C, structural comparison of human Rabex-5WT (black) and the Rabex-5E212A/N343A mutant (gray) in the HB-VPS9 tandem domains. The structure of the Rabex-5E212A/N343A mutant was calculated by MD simulation. A dotted orange line indicates hydrogen bonds. D, comparisons of the atomic fluctuations (B-factor) of the human Rabex-5 mutants with Rabex-5WT. Amino acid residues whose fluctuations are higher than that of the Rabex-5WT are shown in red. The arrows and arrowheads indicate the increment of fluctuations of the hinge region between the HB and VPS9 domains and the HB domain of Rabex-5E212A/N343A mutant, respectively.
Furthermore, to analyze the stability, as well as the dynamic properties of the interface between the HB and VPS9 domains of Rabex-5E212A and Rabex-5E212A/N343A (human Rabex-5 numbering), we performed MD simulations. The MD simulations revealed that the distances between the Cα atoms of Glu-212 and Asn-343 in human Rabex-5E212A (11.0 ± 0.4 Å) and Rabex-5E212A/N343A (10.1 ± 0.7 Å) mutants increased significantly by 45 and 35%, respectively, when compared with that between the Oϵ2 atom of Glu-212 and amide nitrogen of Asn-343 in Rabex-5WT (7.5 ± 0.5 Å) (Fig. 4C). Despite the increase in the flexibilities of the VPS9 domain and the hinge region between αHB4 and αV1 of Rabex-5E212A and Rabex-5E212A/N343A increased (Fig. 4D, arrows), the flexibility of the HB domain of Rabex-5E212A/N343A was further promoted, in comparison to Rabex-5E212A (Fig. 4D, arrowheads). Corroborating our findings, a study by Delprato et al. (26) demonstrated the role of the HB domain in allosteric regulation, which has also been observed in RIN1 (30). Collectively, these structural and functional interpretations lend further support to the hypothesis that the interface between the HB and VPS9 domains aids in modulating the interaction between the MIU domain and the ubiquitinated L1.
Activation of Rabex-5 GEF Activity Weakens the Interaction of Rabex-5 with the Ubiquitinated L1
Next, we sought to determine the location and mechanism underlying the dissociation of the GDP-Rab5·Rabex-5·ubiquitinated L1 complex, which results in the release of the ubiquitinated L1 following internalization. On the basis of our findings that the HB-VPS9 tandem domain regulates interaction with the ubiquitinated L1, we proposed that the dissociation of ubiquitinated cargos from Rabex-5·Rab5 complexes is induced by a conformational change of the HB-VPS9 tandem domain during the GDP/GTP exchange reaction (23, 31).
To test this hypothesis, we co-expressed GFP-Rab5S34N or GFP-Rab5Q79L, a constitutively active Rab5 mutant (29), with Myc-tagged Rabex-5 and examined the effect of these GFP-Rab5 mutants on the interaction of Myc-tagged Rabex-5 with FLAG-tagged L1. Surprisingly, the interaction of Myc-tagged Rabex-5 with FLAG-tagged L1 was found to be significantly increased in the presence of GFP-Rab5WT or GFP-Rab5S34N (Fig. 5A), thereby resulting in a significant increase in the accumulation of ubiquitinated L1 by 5.2 ± 0.2-fold (Fig. 5B, n = 3, p < 0.01). Indeed, co-expression of Myc-tagged Rabex-5 with the GFP-Rab5WT or GFP-Rab5S34N mutants led to the accumulation of L1·Rabex-5·Rab5 complexes on endosomes (Fig. 5C, upper panels, arrows), thus resulting in depletion of biotinylated L1 from the plasma membrane (Fig. 5C, lower panels). In contrast, co-expression of Myc-tagged Rabex-5 with Rab5Q79L diminished the interaction of Rabex-5 with the ubiquitinated L1 (Fig. 5A), resulting in the decreased accumulation of the ubiquitinated L1 (Fig. 5B) on endosomes (Fig. 5C, upper), without altering L1 levels on the cell surface (Fig. 5C, lower). The attenuated impact of Rab5Q79L expression on Rabex-5-mediated L1 accumulation provided 2 possibilities: the lack of the substrate GDP-Rab5, which is essential for internalization of Rabex-5 in cells expressing Rab5Q79L, might result in a reduced interaction with the ubiquitinated L1, and alternatively, the enhancement of L1 recycling regulated by Rab5Q79L overexpression might occur between the endosomes and plasma membrane.
FIGURE 5.
Activation of Rabex-5 GEF activity decreased ubiquitinated L1 binding. A, N2a cells expressing the indicated plasmids were lysed and analyzed by immunoblotting. B, the accumulation of ubiquitinated L1 was examined by immunoprecipitation (IP) and analyzed by immunoblotting (upper). The bars represent the relative densitometric value of Ub-L1/L1 (lower). Note that a compatible amount of the ubiquitinated L1 in cells only expressing FLAG-tagged L1 was detected in cells co-expressing with Myc-tagged Rabex-5A58D. The data are mean ± S.E. in triplicate. ***, p < 0.001. C, N2a cells transfected with the indicated plasmids were stained with FLAG-tagged L1 (blue), Myc-tagged Rabex-5 (red), and GFP-Rab5 (green). The arrows indicate L1 accumulation on Rabex-5-positive endosomes (upper). Scale bar, 5 μm. N2a cells expressing the indicated plasmids were labeled with biotin and precipitated with streptavidin-agarose (lower). The amount of L1 on the surface was detected by anti-L1 antibodies. D, L1 recycling was measured. The internalized L1 (lane 1) after glutathione stripping of cells disappeared from the internal pools in mock cells and cells co-expressing GFP-Rab5Q79L within 15 min (lanes 2 and 3), whereas cells co-expressing GFP-Rab5WT and GFP-Rab5S34N exhibited the delayed disappearance of L1. The relative densitometric value of L1 divided by the incubation time was calculated as the release rate from endosomes (in the text). E, N2a cells expressing the indicated plasmids were lysed and analyzed by immunoblotting.
To examine these possibilities, we directly measured the release rate of the internalized L1 from endosomes in cells co-expressing Myc-tagged Rabex-5 with GFP-Rab5 mutants (Fig. 5D). Cells co-expressing Myc-tagged Rabex-5 with GFP-Rab5WT or GFP-Rab534N demonstrated a significant delay in exiting and/or releasing of the internalized L1 from endosomes (1.5 ± 0.2%/min and 1.3 ± 0.3%/min, respectively), whereas cells co-expressing Myc-tagged Rabex-5 with GFP-Rab5Q79L showed a comparable rate with that of the control (2.6 ± 0.3%/min) (Fig. 5D), indicating that activation of the Rabex-5 GEF activity is required for reduced interaction with the ubiquitinated L1 on early endosomes. Therefore, increased accumulation of the ubiquitinated L1 on endosomes might be due to the limited rate of GDP-Rab5 binding and their release from the HB-VPS9 tandem domain (26, 32).
To further validate this finding, we tested Myc-tagged Rabex-5N414A and Rabex-5L415A (Fig. 1A), constitutively activated GEF mutants that increased catalytic efficiency (kcat/Km), where Asn-414 and Leu-415 are located in an autoinhibitory region that overlaps with the binding site for Rabaptin-5 (33). Indeed, Myc-tagged Rabex-5N414A significantly increased the size of Rab5-positive endosomes, comparable with that of Rab5Q79L-induced endosomes (Fig. 1D). Notably, the interaction of these mutants with the ubiquitinated L1 decreased significantly in both mutants: 58.2 ± 2.8 and 53.5 ± 2.2%, respectively (Fig. 5E, n = 3, p < 0.01), compared with Rabex-5WT. Furthermore, Myc-tagged Rabex-5N414A exhibited a reduced accumulation of the ubiquitinated L1 (Fig. 6C). Therefore, our results highlight that the activation of Rabex-5 GEF activity on early endosomes is required to diminish its interaction with the ubiquitinated L1, which in turn aids in its release from Rabex-5·Rab5 complexes on endosomes and further stimulates the homotypic early endosome fusion simultaneously.
FIGURE 6.
Determination of monoubiquitinated Rabex-5 levels. A, N2a cells co-expressing Myc-tagged Rabex-5 with HA-tagged Ub (left) or N2a cells co-expressing Myc-tagged Rabex-5, FLAG-tagged L1K11R, and HA-tagged Ub (right) were stimulated with L1-Ab. Immunoprecipitation (IP) and immunoblotting were performed as indicated (uppers). The mean amount of Ub-Rabex-5 normalized to Ub-Rabex-5 in the absence of L1-Ab is presented in the graphs (lower). The data are mean ± S.E. in triplicate. ***, p < 0.001. B, N2a cells were co-transfected with the indicated plasmids. Immunoprecipitation and immunoblotting were performed as indicated. C, the accumulation of ubiquitinated L1 was examined by immunoprecipitation and analyzed by immunoblotting (left). The bars represent the relative densitometric value of Ub-L1/L1 (right). The data are mean ± S.E. in triplicate. ***, p < 0.001.
The Interaction of the Ubiquitinated L1 with Rabex-5 Is Correlated with the Coupled Monoubiquitination of Rabex-5
Because Rabex-5 undergoes UBD dependent-coupled monoubiquitination (10, 13–15), it is plausible that this coupled monoubiquitination might prevent it from binding in trans to the ubiquitinated L1. To address this possibility, we determined the amount of monoubiquitinated Myc-tagged Rabex-5 in cells co-expressing FLAG-tagged L1WT or FLAG-tagged L1K11R upon incubation with L1-Ab. Previously, we showed that incubation with L1-Ab led to the ubiquitination of L1 and facilitated the interaction of ubiquitinated L1 with Rabex-5 (18). Strikingly, the level of monoubiquitinated Myc-tagged Rabex-5 was significantly decreased in cells co-expressing FLAG-tagged L1WT or FLAG-tagged L1K11R depending on the incubation time with L1-Ab (Fig. 6A). This finding suggests that the L1-Ab stimulation-dependent de-ubiquitination of Rabex-5 may release an intramolecular “cis” interaction to permit a functional “trans” interaction of the MIU domain with the ubiquitinated L1. In addition, the level of monoubiquitinated Myc-tagged Rabex-5 in cells co-expressing L1WT was comparable with that in the cells co-expressing FLAG-tagged L1K11R (Fig. 6A). This finding suggests that an unknown mechanism underlying the L1-L1 homophilic interactions, via the extracellular and/or intracellular domain, with the exception of the ubiquitinated site of L1, could be involved in stimulation of de-ubiquitination of Rabex-5.
To further investigate the relationship between the coupled monoubiquitination of Rabex-5 and the interaction of Rabex-5 with the ubiquitinated L1, we determined the monoubiquitination level of the Rabex-5 mutants tested in this study by co-expressing them with HA-tagged Ub. Consistent with previous data (10, 15), the co-expression of Rabex-5 with HA-tagged Ub resulted in the monoubiquitination of Rabex-5, whereas co-expression of Rabex-5A58D with Ub-HA did not induce Rabex-5 monoubiquitination (Fig. 6B), indicating the MIU-dependent coupled monoubiquitination of Rabex-5. The level of Myc-tagged Rabex-5E213A monoubiquitination, which substantially increases interaction with the ubiquitinated L1 (Fig. 3A), was found to be decreased when compared with that of wild-type Rabex-5 (Fig. 6B). Notably, the levels of monoubiquitination of a subset of the impaired GEF mutants and constitutively activated GEF mutants of Rabex-5 were found to be reduced, when compared with that of wild-type Rabex-5 (Fig. 6B). Corroborating these findings, accumulation of the ubiquitinated L1 was detected in cells expressing Myc-tagged Rabex-5WT and Rabex-5E213A but not in cells expressing the impaired GEF mutants or constitutively activated GEF mutants (Fig. 6C). Taken together, these results indicate that the interaction of Rabex-5 with the ubiquitinated L1 correlates with the coupled monoubiquitination status of Rabex-5.
DISCUSSION
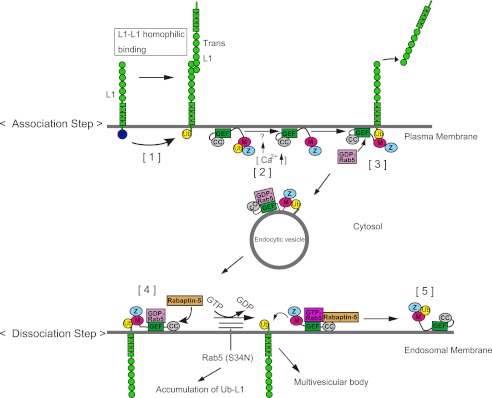
On the basis of the findings from our previous study (18) and this study, we proposed a model describing the mechanisms underlying L1 ligand-induced L1 ubiquitination on the plasma membrane followed by Rabex-5-mediated internalization, and the pre-determined release of the ubiquitinated L1 from Rabex-5, destined for lysosomal degradation via early endosomes (Fig. 7). Previously, we reported that L1-L1 homophilic binding leads to dephosphorylation at the tyrosine-based sorting motif to recruit AP-2 and ubiquitination at the cytoplasmic domain of L1 (Fig. 7, step 1) (18). Upon L1-L1 homophilic binding, coupled monoubiquitination of Rabex-5, presumably in a “switched off” state in which its MIU domain interacts with the mono-Ub moiety in cis, undergoes de-ubiquitination to convert Rabex-5 to a “switched on” state in which the MIU domain is positioned for trans interactions (Fig. 7, step 2). Subsequently, Rabex-5 interacts with the ubiquitinated L1 on the plasma membrane via the MIU domain, followed by Rab5 recruitment, which in turn triggers internalization of the ubiquitinated L1 from the plasma membrane (Fig. 7, step 3). The ubiquitinated L1·Rabex-5·GDP-Rab5 complex on the endocytic vesicle is then transported to the early endosomal membrane.
FIGURE 7.
A proposed mechanism for the spatiotemporal regulation of Rabex-5 in the endocytic trafficking of ubiquitinated L1. M, motif-interacting with Ub; Z, ZnF. Step 1, Src kinase is involved in the phosphorylation of a tyrosine-based motif, thereby regulating recruitment of the AP-2 protein. However, the dephosphorylation of this motif upon L1-L1 homophilic interactions results in the recruitment of AP-2, causing the clathrin-mediated endocytosis of L1 from the plasma membrane. The dephosphorylation of L1 also triggers the recruitment of an unidentified E3-ligase to ubiquitinate L1 on the plasma membrane. Step 2, the coupled monoubiquitination of Rabex-5, presumably in a switched off state, in which the MIU domain interacts with the mono-Ub moiety in cis, undergoes deubiquitination by an unidentified deubiquitinase upon L1 ligand stimulation, which might cause a Ca2+ influx. Then, monoubiquitinated Rabex-5 undergoes deubiquitination, converting Rabex-5 to a switched on state in which the MIU domain is positioned for trans interactions. Step 3, the interaction of ubiquitinated L1 with Rabex-5 recruits Rab5, which in turn triggers the endocytosis of ubiquitinated L1 from the plasma membrane. Next, the ubiquitinated L1·Rabex-5·GDP-Rab5 complex in an endocytic vesicle is transported to the endosomal membrane. Step 4, Rabex-5 turns on the GEF activity of the HB-VPS9 tandem domain by recruiting Rabaptin-5 to endosomes. Step 5, the conversion of GDP-Rab5 to GTP-Rab5 catalyzed by the GEF domain releases the Rabex-5·GTP-Rab5·Rabaptin-5 complex from the ubiquitinated L1 on the endosomes. Releasing the ubiquitinated L1 from the MIU domain allows this domain to interact with the mono-Ub moiety in cis and to return to the coupled monoubiquitinated Rabex-5 for another cycle.
Upon its arrival to the early endosomes, Rabex-5 turns on the GEF activity of the HB-VPS9 tandem domain by recruitment of Rabaptin-5 to endosomes (33, 34). The exchange of GDP/GTP catalyzed by the HB-VPS9 tandem domain facilitates release of the ubiquitinated L1 from the Rabex-5·GTP-Rab5·Rabaptin-5 complex. Consequently, the activated form of Rab5, GTP-Rab5, stimulates homotypic early endosomal fusion events on the endosomes (Fig. 7, step 4). Releasing the ubiquitinated L1 from the MIU domain allows this domain to interact with the mono-Ub moiety in cis and return to the coupled monoubiquitinated Rabex-5 for another cycle (Fig. 7, step 5).
Cross-talk between the Ub-binding Activity of the MIU Domain and GEF Activity for Rab5
Although some previous studies using Vps9p, the yeast homolog of Rabex-5, suggested the possible involvement of cross-talk between the Ub-binding activity of the CUE domain and the GEF activity of the VPS9 domain in the ubiquitinated cargo trafficking pathway (24, 35), the underlying mechanism has not been demonstrated directly. Here, we provided direct evidence showing that the ubiquitinated cargo-binding activity of the MIU domain and the GEF activity for Rab5 are regulated reciprocally in a spatiotemporal manner during the ubiquitinated L1 endocytic trafficking pathway.
First, we showed that Rabex-5 mutants with Ub-binding deficiency significantly diminished interactions with Rab5, resulting in the reduced diameter of Rab5-positive endosomes. Between the 2 UBDs in Rabex-5, the MIU domain predominantly affects interaction with Rab5. The MIU domain, not the A20-ZnF domain, is directly involved in interaction with the ubiquitinated L1 (18) and the A20-ZnF domain exhibits Ub ligase activity via binding to Ub-charged Ubc5 (13–15). Given our findings and previous reports, the interaction of Rabex-5 with ubiquitinated cargo, via the MIU domain might recruit Rab5 to the GEF domain of Rabex-5 to drive the ubiquitinated cargo from the plasma membrane. Importantly, we also showed that this recruitment of Rab5 to the GEF domain of Rabex-5 depends upon interaction of the MIU domain with the ubiquitinated L1, because the interaction of Rab5 with Rabex-5 in cells expressing L1K11R, a Ub-deficient mutant, was not affected upon incubation with L1-Ab. These data confirm that interaction of the ubiquitinated L1 with Rabex-5 occurs prior to interaction of Rabex-5 with Rab5.
Second, we demonstrated that interaction of the HB-VPS9 tandem domain of Rabex-5 with Rab5 is crucial to interaction of the MIU domain with the ubiquitinated L1. Although Zhu et al. (36) identified the minimum motif required for Rabex-5 targeting the early endosomes, our data revealed that the GEF mutants, which have a diminished interaction with Rab5, showed a little co-localization with Rab5-positive endosomes. These results are in agreement with a previous study that demonstrated that Rabex-5 GEF activity is indispensable for its recruitment to endosomes (10). Therefore, our results provide an alternative explanation for the decreased ubiquitinated cargo-binding activity of the GEF mutants. As these mutants could not be properly targeted to the Rab5-positive early endosomal compartments, they showed a weak interaction with the ubiquitinated L1.
Despite potential effects of the mutation at Glu-213 on misfolding, stability, and solubility of Rabex-5 (26), we showed that Rabex-5E213A and Rabex-5E213A/N344A could be properly targeted to the early endosomes via their interaction with Rab5, and that they significantly modulated the interaction with the ubiquitinated L1 by manipulating the flexibility of the hinge region between HB and VPS9 domains. This implies that the interaction of Rabex-5 with Rab5 might facilitate the stability of the complexes with the ubiquitinated cargo. Furthermore, our findings lend support to a previous hypothesis that the RIN proteins contain Ras association domains that facilitate allosteric regulation of Rab5 exchange activity by GTP bound Ras (30, 37). The HB-VPS9 tandem domains of Rabex-5 and RIN1 have equivalently high exchange activity for Rab5 and Rab21 but relatively weak activity for Rab22 (26). However, a recent study by Faesen et al. (38) reported that the Ub-binding activity of the Ub-specific protease USP7/HAUSP is enhanced allosterically by the metabolic enzyme guanosine 5′-monophosphate synthetase. Therefore, further structural and functional experiments investigating the allosteric effect of the HB-VPS9 tandem domain on the ubiquitinated cargo-binding activity of the MIU domain are warranted. In addition, further experiments for assessing whether the mutation at Glu-213 affects folding, stability, or solubility of full-length of Rabex-5 protein are required.
An exciting possibility raised by our findings is that activation of the GEF domain is involved in the reduced interaction between the MIU domain and the ubiquitinated L1 to release the ubiquitinated cargo on early endosomes. Therefore, it is thought that the release of GDP-Rab5 should be the rate-limiting step. Importantly, the GDP/GTP exchange activity of Rabex-5 is suppressed by the autoinhibitory element that overlaps the binding site for the multivalent effector Rabaptin-5 (33). This autoinhibition can be partially reversed by the mutation of conserved residues on the nonpolar face of the predicted amphipathic helix or by assembly of the complex with Rabaptin-5 (33). Indeed, we showed that the constitutively active GEF mutants Rabex-5N414A and Rabex-5L415A exhibited considerably less ubiquitinated L1-binding activity compared with Rabex-5WT. Collectively, the sequential reaction, starting with GDP-Rab5 binding and progressing to the conversion of GDP-Rab5 to GTP-Rab5 catalyzed by the HB-VPS9 tandem domain, potentially mediates proper Ub-cargo membrane trafficking events depending on the subcellular localization.
Coupled Monoubiquitination Is Involved in the Inactivation of the Ubiquitinated Cargo-binding Activity of the MIU Domain
There is little direct evidence describing the spatiotemporal regulation of UBD-dependent coupled monoubiquitination during the ubiquitinated cargo sorting, although it has been proposed that a number of endocytic UBPs undergo UBD-dependent coupled monoubiquitination in cis that prevents them from binding in trans to the ubiquitinated cargo proteins (3). A study by Chen et al. (39) reported that depolarization-dependent Ca2+ influx induces a rapid and a general decrease of the ubiquitinated state of synaptic proteins, including monoubiquitinated proteins. Consistent with this finding, the present study showed that the amount of monoubiquitinated Rabex-5 significantly decreased upon stimulation with L1 ligands, which in turn opens a calcium influx pathway in the growth cones of rat sensory neurons without altering the membrane voltage (40, 41). However, additional experiments are needed to measure the calcium influx in N2a cells upon L1 ligand stimulation. These data directly prove that coupled monoubiquitinated UBPs are tightly regulated by extracellular signal transduction events to promote the trafficking of ubiquitinated cargo from the plasma membrane.
Finally, our study established a correlation between the interaction of Rabex-5 with the ubiquitinated L1 and the coupled monoubiquitination status of Rabex-5. Therefore, we proved that the MIU domain is not only involved directly in the trafficking of ubiquitinated cargo but also plays a key role in the coupled monoubiquitination of Rabex-5 to inactivate its ubiquitinated cargo-binding activity. However, our results differ from a previous study by Keren-Kaplan et al. (42), which reported that ubiquitination occurs on the Vps9-GEF domain and that it does not affect the GEF activity in vitro. This could be attributed to the different types of UBDs present in human Rabex-5 and yeast Vps9 proteins. Therefore, future studies should consider these apparent differences and include equivalent biochemical assays for Rabex-5.
In conclusion, we have demonstrated that Rabex-5 can orchestrate an on-time association/dissociation of the ubiquitinated L1 via a combination of Ub-binding, GEF activity, and a coupled monoubiquitination status. Efforts are underway to elucidate the molecular machinery underlying the deubiquitination and the coupled monoubiquitination cycles of Rabex-5 upon stimulation by extracellular signal transduction events.
Acknowledgments
We thank Dr. V. Lemmon (University of Miami School of Medicine) for discussions on L1 biology. Measurement and data analyses were performed at the Kagawa School, Tokushima Bunri University.
This work was supported by a Grant-in-aid for Young Scientists (B) (20790085) from the Japan Society for the Promotion of Science, the Uehara Memorial Foundation, an individual research allowance from Doshisha University (to Y. A.), and a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2011-0023402) (to S. L.).
- UBD
- ubiquitin-binding domain
- Ub
- ubiquitin
- MIU
- motif interacting with Ub
- ZnF
- zinc finger
- GEF
- guanine nucleotide exchange factor
- HB
- helix bundle
- VPS9
- vacuolar protein sorting 9
- UBP
- Ub-binding protein
- MD
- molecular dynamics.
REFERENCES
- 1. Mukhopadhyay D., Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 2. Raiborg C., Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 3. Hicke L., Schubert H. L., Hill C. P. (2005) Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 4. Hurley J. H., Lee S., Prag G. (2006) Ubiquitin-binding domains. Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Traub L. M., Lukacs G. L. (2007) Decoding ubiquitin sorting signals for clathrin-dependent endocytosis by CLASPs. J. Cell Sci. 120, 543–553 [DOI] [PubMed] [Google Scholar]
- 6. Dikic I., Wakatsuki S., Walters K. J. (2009) Ubiquitin-binding domains. From structures to functions. Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 28, 451–455 [DOI] [PubMed] [Google Scholar]
- 8. Woelk T., Oldrini B., Maspero E., Confalonieri S., Cavallaro E., Di Fiore P. P., Polo S. (2006) Molecular mechanisms of coupled monoubiquitination. Nat. Cell. Biol. 8, 1246–1254 [DOI] [PubMed] [Google Scholar]
- 9. Hoeller D., Crosetto N., Blagoev B., Raiborg C., Tikkanen R., Wagner S., Kowanetz K., Breitling R., Mann M., Stenmark H., Dikic I. (2006) Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell. Biol. 8, 163–169 [DOI] [PubMed] [Google Scholar]
- 10. Mattera R., Bonifacino J. S. (2008) Ubiquitin binding and conjugation regulate the recruitment of Rabex-5 to early endosomes. EMBO J. 27, 2484–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harper J. W., Schulman B. A. (2006) Structural complexity in ubiquitin recognition. Cell 124, 1133–1136 [DOI] [PubMed] [Google Scholar]
- 12. Raiborg C., Slagsvold T., Stenmark H. (2006) A new side to ubiquitin. Trends Biochem. Sci. 31, 541–544 [DOI] [PubMed] [Google Scholar]
- 13. Lee S., Tsai Y. C., Mattera R., Smith W. J., Kostelansky M. S., Weissman A. M., Bonifacino J. S., Hurley J. H. (2006) Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat. Struct. Mol. Biol. 13, 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Penengo L., Mapelli M., Murachelli A. G., Confalonieri S., Magri L., Musacchio A., Di Fiore P. P., Polo S., Schneider T. R. (2006) Crystal structure of the ubiquitin binding domains of Rabex-5 reveals two modes of interaction with ubiquitin. Cell 124, 1183–1195 [DOI] [PubMed] [Google Scholar]
- 15. Mattera R., Tsai Y. C., Weissman A. M., Bonifacino J. S. (2006) The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J. Biol. Chem. 281, 6874–6883 [DOI] [PubMed] [Google Scholar]
- 16. Kalesnikoff J., Rios E. J., Chen C. C., Alejandro Barbieri M., Tsai M., Tam S. Y., Galli S. J. (2007) Roles of RabGEF1/Rabex-5 domains in regulating Fcϵ RI surface expression and Fcϵ RI-dependent responses in mast cells. Blood 109, 5308–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan H., Jahanshahi M., Horvath E. A., Liu H. Y., Pfleger C. M. (2010) Rabex-5 ubiquitin ligase activity restricts Ras signaling to establish pathway homeostasis in Drosophila. Curr. Biol. 20, 1378–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aikawa Y. (2012) Rabex-5 protein regulates the endocytic trafficking pathway of ubiquitinated neural cell adhesion molecule l1. J. Biol. Chem. 287, 32312–32323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horiuchi H., Lippé R., McBride H. M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., Zerial M. (1997) A novel Rab5 GDP/GTP exchange to effector complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 19, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 20. Gorvel J. P., Chavrier P., Zerial M., Gruenberg J. (1991) rab5 controls early endosome fusion in vitro. Cell 64, 915–925 [DOI] [PubMed] [Google Scholar]
- 21. Bucci C., Parton R. G., Mather I. H., Stunnenberg H., Simons K., Hoflack B., Zerial M. (1992) The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70, 715–728 [DOI] [PubMed] [Google Scholar]
- 22. Li G., Barbieri M. A., Colombo M. I., Stahl P. D. (1994) Structural features of the GTP-binding defective Rab5 mutants required for their inhibitory activity on endocytosis. J. Biol. Chem. 269, 14631–14635 [PubMed] [Google Scholar]
- 23. Zerial M., McBride H. (2001) Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 24. Donaldson K. M., Yin H., Gekakis N., Supek F., Joazeiro C. A. (2003) Ubiquitin signals protein trafficking via interaction with a novel ubiquitin binding domain in the membrane fusion regulator, Vps9p. Curr. Biol. 13, 258–262 [DOI] [PubMed] [Google Scholar]
- 25. Le T. L., Yap A. S., Stow J. L. (1999) Recycling of E-cadherin. A potential mechanism for regulating cadherin dynamics. J. Cell Biol. 146, 219–232 [PMC free article] [PubMed] [Google Scholar]
- 26. Delprato A., Merithew E., Lambright D. G. (2004) Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell 118, 607–617 [DOI] [PubMed] [Google Scholar]
- 27. Wang J., Cieplak P., Kollman P. A. (2000) How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21, 1049–1074 [Google Scholar]
- 28. Pearlman D. A., Case D. A., Caldwell J. W., Ross W. S., Cheatham T. E., DeBolt S., Ferguson D., Seibel G., Kollman P. (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 91, 1–41 [Google Scholar]
- 29. Stenmark H., Parton R. G., Steele-Mortimer O., Lütcke A., Gruenberg J., Zerial M. (1994) Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13, 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tall G. G., Barbieri M. A., Stahl P. D., Horazdovsky B. F. (2001) Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell 1, 73–82 [DOI] [PubMed] [Google Scholar]
- 31. Ali B. R., Seabra M. C. (2005) Targeting of Rab GTPases to cellular membranes. Biochem. Soc. Trans. 33, 652–656 [DOI] [PubMed] [Google Scholar]
- 32. Esters H., Alexandrov K., Constantinescu A. T., Goody R. S., Scheidig A. J. (2001) Vps9, Rabex-5, and DSS4. Proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J. Mol. Biol. 29, 141–156 [DOI] [PubMed] [Google Scholar]
- 33. Delprato A., Lambright D. G. (2007) Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat. Struct. Mol. Biol. 14, 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lippé R., Miaczynska M., Rybin V., Runge A., Zerial M. (2001) Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol. Biol. Cell 12, 2219–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carney D. S., Davies B. A., Horazdovsky B. F. (2006) Vps9 domain-containing proteins. Activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 16, 27–35 [DOI] [PubMed] [Google Scholar]
- 36. Zhu H., Liang Z., Li G. (2009) Rabex-5 is a Rab22 effector and mediates a Rab22-Rab5 signaling cascade in endocytosis. Mol. Biol. Cell 20, 4720–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han L., Wong D., Dhaka A., Afar D., White M., Xie W., Herschman H., Witte O., Colicelli J. (1997) Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc. Natl. Acad. Sci. U.S.A. 94, 4954–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Faesen A. C., Dirac A. M., Shanmugham A., Ovaa H., Perrakis A., Sixma T. K. (2011) Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol. Cell 7, 147–159 [DOI] [PubMed] [Google Scholar]
- 39. Chen H., Polo S., Di Fiore P. P., De Camilli P. V. (2003) Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc. Natl. Acad. Sci. U.S.A. 100, 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Williams E. J., Doherty P., Turner G., Reid R. A., Hemperly J. J., Walsh F. S. (1992) Calcium influx into neurons can solely account for cell contact-dependent neurite outgrowth stimulated by transfected L1. J. Cell Biol. 119, 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Archer F. R., Doherty P., Collins D., Bolsover S. R. (1999) CAMs and FGF cause a local submembrane calcium signal promoting axon outgrowth without a rise in bulk calcium concentration. Eur. J. Neurosci. 11, 3565–3573 [DOI] [PubMed] [Google Scholar]
- 42. Keren-Kaplan T., Attali I., Motamedchaboki K., Davis B. A., Tanner N., Reshef Y., Laudon E., Kolot M., Levin-Kravets O., Kleifeld O., Glickman M., Horazdovsky B. F., Wolf D. A., Praq G. (2011) Synthetic biology approach to reconstituting the ubiquitylation cascade in bacteria. EMBO J. 11, 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]