Background: RNF168 promotes chromosomal break localization of 53BP1 and BRCA1; 53BP1 loss rescues homologous recombination (HR) in BRCA1-deficient cells.
Results: RNF168 depletion suppresses HR defects caused by BRCA1 silencing; RNF168 influences HR similarly to 53BP1.
Conclusion: RNF168 is important for HR defects caused by BRCA1 loss.
Significance: Although RNF168 promotes BRCA1 and 53BP1 localization to chromosomal breaks, RNF168 affects HR similarly to 53BP1.
Keywords: BRCA1, Chromosomes, DNA Repair, Genomic Instability, Homologous Recombination, RNF168, Homology-directed Repair, Single Strand Annealing
Abstract
The RING finger nuclear factor RNF168 is required for recruitment of several DNA damage response factors to double strand breaks (DSBs), including 53BP1 and BRCA1. Because 53BP1 and BRCA1 function antagonistically during the DSB repair pathway homologous recombination (HR), the influence of RNF168 on HR has been unclear. We report that RNF168 depletion causes an elevated frequency of two distinct HR pathways (homology-directed repair and single strand annealing), suppresses defects in HR caused by BRCA1 silencing, but does not suppress HR defects caused by disruption of CtIP, RAD50, BRCA2, or RAD51. Furthermore, RNF168-depleted cells can form ionizing radiation-induced foci of the recombinase RAD51 without forming BRCA1 ionizing radiation-induced foci, indicating that this loss of BRCA1 recruitment to DSBs does not reflect a loss of function during HR. Additionally, we find that RNF168 and 53BP1 have a similar influence on HR. We suggest that RNF168 is important for HR defects caused by BRCA1 loss.
Introduction
Mutations in DNA damage response factors are associated with human diseases with distinct pathologies, including cancer predisposition and immune deficiency. For example, inherited mutations in the RING finger nuclear factor RNF168 have been found in patients with decreased antibody maturation via class switch recombination, which involves nonhomologous end joining repair of programmed double strand breaks (DSBs)2 (1, 2).
Consistent with a role in DSB repair, RNF168 is recruited to DSBs, as measured by its formation of ionizing radiation-induced foci (IRIF), and furthermore is important for radioresistance (1, 3, 4). RNF168 is an E3 ubiquitin ligase that is critical for IRIF of several downstream DNA damage response proteins, which notably includes both 53BP1 and the breast and ovarian cancer susceptibility factor BRCA1 (1, 3, 4). However, these two DNA damage response factors appear to act antagonistically during DSB repair by homologous recombination (HR), in that the loss of 53BP1 can rescue HR in BRCA1-deficient cells (5, 6). Thus, determining how RNF168 influences HR, relative to BRCA1 and 53BP1, is important to understand its role during genome maintenance.
HR includes at least two distinct pathways (7, 8). Homology-directed repair (HDR) is a conservative HR pathway that involves strand invasion of a homologous template via the recombinase RAD51, followed by nascent DNA synthesis. In contrast, single strand annealing (SSA) is a nonconservative HR pathway that bridges two homologous segments that flank a DSB, thereby causing a deletion between the homologous segments. Accordingly, a key mechanistic distinction between HDR and SSA is the relative requirement for RAD51, in that RAD51 and its co-factor BRCA2 are required for HDR but inhibit SSA (9, 10). In contrast, several factors that influence DSB end processing mutually affect HDR and SSA. For example, nonhomologous end joining factors that protect DSB ends from processing (e.g., KU70 and XRCC4) inhibit both HDR and SSA (9, 11, 12). Conversely, factors that facilitate end processing (e.g., CtIP, RAD50, and NBS1) promote both HDR and SSA (11–13). Similarly, BRCA1 promotes both HDR and SSA (9) and hence has been postulated to promote end processing during HR (14), although the precise function of BRCA1 during HR remains elusive (12, 15, 16). In contrast, 53BP1 inhibits HDR (17, 18), whereas its effect on SSA has been unclear.
Because RNF168 is critical for recruitment of both BRCA1 and 53BP1 to DSBs (1, 3, 4), which have opposing effects on HR (5, 6), we have sought to characterize the influence of RNF168 during HR. We show that depletion of RNF168 causes an elevated frequency of HDR and SSA and substantially suppresses HR defects (HDR, SSA, and RAD51 IRIF) and anaphase chromosome separation defects caused by BRCA1 silencing. Furthermore, we find that cells depleted of RNF168 can form RAD51 IRIF without forming BRCA1 IRIF. In contrast, depletion of RNF168 does not suppress HR defects in cells deficient in several other HR factors (CtIP, RAD50, BRCA2, and RAD51). Finally, we show that 53BP1 influences HR similarly to RNF168. These findings indicate that RNF168 is important for HR defects caused by BRCA1 silencing.
EXPERIMENTAL PROCEDURES
Cell Lines, siRNAs, and Plasmids
The U2OS reporter cell lines were described previously (19, 20). Linearized reporter plasmids were electroporated into passage immortalized 53BP1−/− MEFs (5), followed by selection using hygromycin for the DR-GFP-hyg version and the other reporters using puromycin, and intact reporters were confirmed for individual clones by Southern blotting (19).
Many siRNAs were from Thermoscientific: siRNF168 (pool of 4, L-007152-00), siRNF168#18 (distinct single not in the pool, D-007152-18), siBRCA1 (D-003461-07) (21), siBRCA1#6 (D-003461-06), siRAD50 (pool of 4, M-005232-01), and nontargeting siCTRL (D-001810-01). Other siRNAs were siBRCA2 (pool of three; Santa Cruz, SC-29825) and a single CtIP siRNA (22). To generate the siRNF168#18-resistant expression vector, silent mutations were introduced into FLAG-RNF168 WT and C19S (23) that was inserted into pCAGGS-BSKX (9). To generate pCAGGS-dn53BP1, a segment of 53BP1 (encoding amino acids Asp-1052 to Pro-1639) was fused downstream from tandem nuclear localization signals and a HA immunotag, which was then inserted into pCAGGS-BSKX (9). The 53BP1 expression vector was described previously (Addgene 19836) (24), as were other plasmids (9). Sequences of siRNAs and primers for RT-PCR analysis are shown in supplemental Fig. S1A.
Repair Assays, Immunoblot, and Cell Cycle Analysis
For RNAi, 1 × 105 cells were plated on top of a mixture of 5 pmol of each siRNA incubated with 1.8 μl of RNAiMAX for 25 min. For experiments with two different siRNAs, siCTRL is added to each of the single siRNA transfections to ensure the same total concentration of siRNA (10 pmol). Two days after siRNA transfection, these cells were co-transfected with siRNA as described above (10 pmol total) with 0.8 μg of I-SceI expression vector (pCBASce) using 3.6 μl of Lipofectamine 2000 (Invitrogen). Some I-SceI transfections also included 0.4 μg of pCAGGS-FLAG-RNF168, 0.4 μg of pCAGGS-dn53BP1, 0.4 μg of pCAGGS-BSKX (EV), 0.4 μg of BARD1-hb202, 0.2 μg of BRC3, or 0.1 μg of RAD51-K133R. For 53BP1−/− experiments, 105 cells were plated 1 day prior to transfection with 0.8 μg of pCBASce along either with 0.8 μg of 53BP1 expression vector (24) or a control empty vector (pCMV/EV) or with a GFP expression vector alone (pCAGGS-NZEGFP) (25), each with 3.6 μl of Lipofectamine 2000 (Invitrogen). Three days after I-SceI transfection, the percentage of GFP+ cells was determined by FACS analysis (CYAN ADP, Dako). For comparison, the repair value for each transfection was divided by the mean repair value for the parallel control transfection (i.e., siCTRL and/or EV). Additionally, absolute GFP+ frequencies are presented in supplemental Table S1. The repair values are the means of at least three independent transfections, error bars reflect the standard deviation, and statistics were performed with the unpaired t test.
For immunoblot analysis, protein was extracted 2 days after transfection using 20 mm Tris, pH 8, 100 mm NaCl, 1 mm EDTA, 0.5% IGEPAL, 1 mm DTT, and Roche protease inhibitor mixture and eight freeze/thaw cycles. Equal amounts of protein (Bio-Rad protein assay) were loaded for each immunoblot. The blots were probed with the following antibodies for detection by ECL (GE/Amersham Biosciences): BRCA1 (Abcam, ab16780), RNF168 (Millipore, 06-1130), actin (Sigma, A2066), 53BP1 (Abcam, ab36823), GAPDH (Abcam, ab9482), and HRP-conjugated secondary antibodies (Santa Cruz Biotechnology, sc-2004 and sc-2005).
For cell cycle analysis, 2 days after siRNA treatment, the cells were incubated with bromodeoxyuridine (BrdU, 10 mm) for 1 h, fixed in 70% ethanol, and stained with FITC-conjugated anti-BrdU antibody (BD Biosciences, 51–33284X) and propidium iodide (Sigma). The values are the means of three independent transfections, and statistics were performed as for the repair assays.
Anaphase and IRIF Analysis, Including Microscope Image Acquisition
For anaphase and IRIF analysis, cells were plated on chamber slides the day after siRNA or plasmid transfection. For anaphase analysis, following 2 days of culturing on the slides, the cells were fixed in 2% paraformaldehyde followed by immunostaining with anti-PICH antibody (Abnova H00054821-D01P) and counterstaining by DAPI in Vectashield Mounting Medium (Vector Laboratories). Images were acquired using an AX-70 microscope (Olympus) equipped with a 40× NA 0.75 UPlanFl objective, with Retiga Exi camera (QImaging) and ImagePro software (MediaCybernetics). For each siRNA treatment condition, >100 anaphases were accumulated from several independent transfections (n ≥ 3). Anaphase bridges were scored as continuous DAPI staining between anaphase chromosomes. Subsequently, localization of PICH at anaphase bridges was scored. Statistics were performed with Fisher's exact test.
To examine IRIF, following 1 day of culturing on the slides, the cells were treated with IR (Mark 1 irradiator Cs137) and allowed to recover prior to fixation in 2% paraformaldehyde. For RAD51 and BRCA1 IRIF, pre-extraction (25 mm Hepes, 50 mm NaCl, 1 mm EDTA, 3 mm MgCl2, 300 mm sucrose, and 0.5% Triton X-100) was performed before fixation in 2% paraformaldehyde. Antibodies used for immunofluorescence: anti-Rad51 (Santa Cruz, H-92), anti-RPA (Calbiochem, RPA34-20 and Ab-3), anti-53BP1 (Abcam, ab36823), anti-BRCA1 (Santa Cruz, D-9), and anti-HA immunotag (Bethyl Laboratories, A190–107F). Subsequently, the slides were stained with Alexa Fluor 568 goat anti-rabbit IgG and/or Alexa Fluor 488 goat anti-mouse IgG (Invitrogen), and DAPI in Vectashield mounting medium (Vector Laboratories). Images were acquired as for the anaphase analysis.
Regarding quantification of IRIF, for RAD51 IRIF, >150 cells were scored for the presence of ≥5 RAD51 foci to determine the frequency of RAD51 IRIF+ cells. RAD51 IRIF frequencies represent the means of three independent transfections, and statistics were performed using the unpaired t test. For the RAD51 and BRCA1 co-staining experiment, RAD51 IRIF+ cells were secondarily scored as containing <5 BRCA1 foci (IRIF−), to determine the frequency of RAD51 IRIF+/BRCA1 IRIF− cells. For the RPA IRIF experiment, >200 cells for each siRNA treatment, accumulated from two independent transfections, were scored for the number of RPA IRIF. The statistics for the RAD51/BRCA1 co-staining, and RPA staining, were performed with Fisher's exact test.
RESULTS
RNF168 Inhibits Two Distinct HR Pathways: HDR and SSA
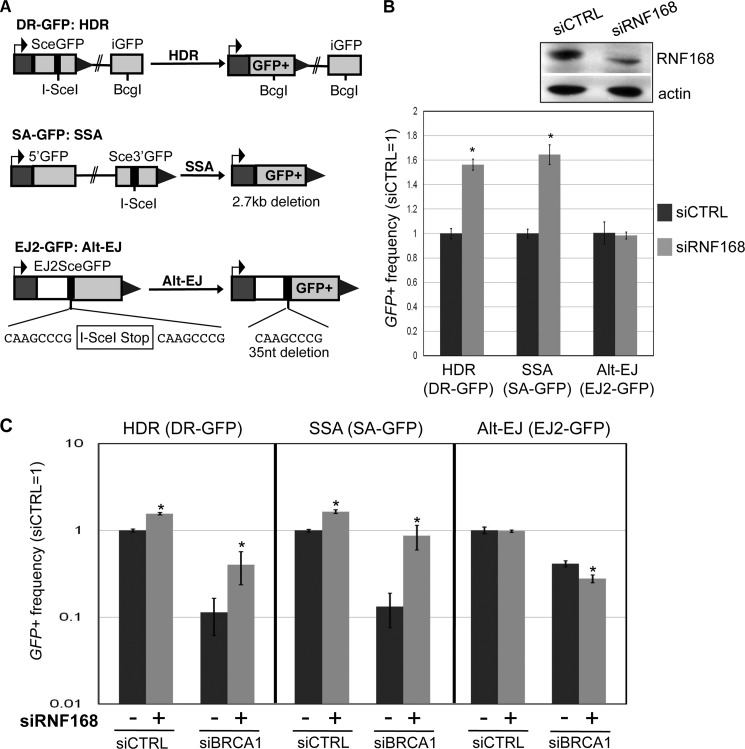
We examined the influence of RNF168 during distinct DSB repair events, using a panel of previously described GFP-based reporters that contain a recognition site for the rare-cutting endonuclease I-SceI (Fig. 1A) (22). With DR-GFP, GFP+ is restored via HDR of an I-SceI-induced DSB that uses a downstream homologous template. With SA-GFP, GFP+ is restored via SSA between two repeats that flank an I-SceI-induced DSB, which causes a 2.7-kb deletion. Notably, GFP+ cells in SA-GFP could also potentially arise via HDR with crossing over, although such HDR events are rare in mitotic cells (26, 27) and thus do not likely significantly contribute to this assay (9). In addition to HR, we have examined alternative end joining (Alt-EJ), which is similar to SSA but using shorter stretches of homology that are closer to DSB ends. Such Alt-EJ repair events do not require the classical nonhomologous end joining factors that mediate V(D)J recombination (28–30). To examine Alt-EJ, we used EJ2-GFP, in which GFP+ is restored by repair that bridges 8 nucleotides of microhomology flanking the I-SceI-induced DSB, which causes a 35-bp deletion (22).
FIGURE 1.
RNF168 inhibits HDR and SSA, and depletion of RNF168 can suppress the defects in these HR events caused by BRCA1 silencing. A, shown are three reporters with an I-SceI recognition site, along with each respective DSB repair event that restores GFP+: HDR for DR-GFP, SSA for SA-GFP, and Alt-EJ for EJ2-GFP. B, depletion of RNF168 causes an increase in HDR and SSA, but not Alt-EJ. U2OS reporter cell lines were transfected with siRNF168 (pool of four siRNA sequences) prior to co-transfection of siRNF168 and an expression vector for I-SceI. Shown are the frequencies of GFP+ cells for each reporter cell line, relative to parallel transfections with a nontargeting siRNA (siCTRL). *, p < 0.0001 (n ≥ 3). Also shown are immunoblot signals for RNF168 and actin for siRNF168- and siCTRL-treated samples. C, depletion of RNF168 suppresses the HDR and SSA defects caused by BRCA1 silencing. U2OS reporter cell lines were treated with siCTRL or siBRCA1, along with (+) or without (−) siRNF168. For all (−) siRNF168 treatments, siCTRL was added to ensure equivalent concentrations of total siRNA. Repair was then analyzed as in B. Shown are the frequencies of GFP+ cells for the HDR, SSA, and Alt-EJ reporter cell lines for each siRNA treatment, relative to siCTRL. *, p < 0.0001 for HDR and SSA, p = 0.0025 for Alt-EJ, versus the (−) parallel siRNF168 condition (n ≥ 3).
To determine the effect of RNAi depletion of RNF168 on these DSB repair events, we transfected siRNAs into human osteosarcoma (U2OS) cell lines containing these reporters (19) and subsequently expressed I-SceI. After these transfections, we determined the percentage of GFP+ cells using FACS analysis, which provides the frequency of the repair event marked by restoration of a GFP+ cassette. Depletion of RNF168 via siRNF168 (pool of four siRNAs), relative to a nontargeting siCTRL, was confirmed by immunoblotting (2-fold; Fig. 1B) and quantitative RT-PCR (supplemental Fig. S1B). From these experiments, we found that siRNF168 treatment caused an increase in HDR and SSA (both 1.6-fold) but had no significant effect on Alt-EJ (Fig. 1B).
Depletion of RNF168 Suppresses HR Defects Caused by BRCA1 Silencing
The above findings indicate that RNF168 is important to inhibit HR; however RNF168 is critical for recruiting the HR factor BRCA1 to DSBs, as measured by IRIF analysis (1, 3, 4). To reconcile these findings, we hypothesized that BRCA1 recruitment to DSBs is not required for HR in RNF168-depleted cells. A corollary of this hypothesis is that HR defects caused by BRCA1 loss can be suppressed by RNF168 depletion. To test this notion, we examined repair in U2OS cells with RNAi-mediated silencing of RNF168, BRCA1, or both BRCA1 and RNF168 (Fig. 1C and supplemental Fig. S2). We confirmed efficient reduction in RNA levels by quantitative RT-PCR analysis of cells treated with siRNF168, siBRCA1, both siBRCA1 and siRNF168, and siCTRL (supplemental Fig. S1B). From these experiments, we found that siBRCA1 treatment caused a substantial decrease in HDR and SSA (Fig. 1C and supplemental Fig. S2), which is consistent with previous results with Brca1 disruption (9). Importantly, we found that cells treated with both siBRCA1 and siRNF168 showed a striking increase in HDR and SSA, compared with siBRCA1-treated cells (Fig. 1C and supplemental Fig. S2; 3.6- and 7-fold, respectively). In contrast, we found that siBRCA1 treatment caused a moderate decrease in Alt-EJ, which was not rescued by siRNF168 treatment (Fig. 1C).
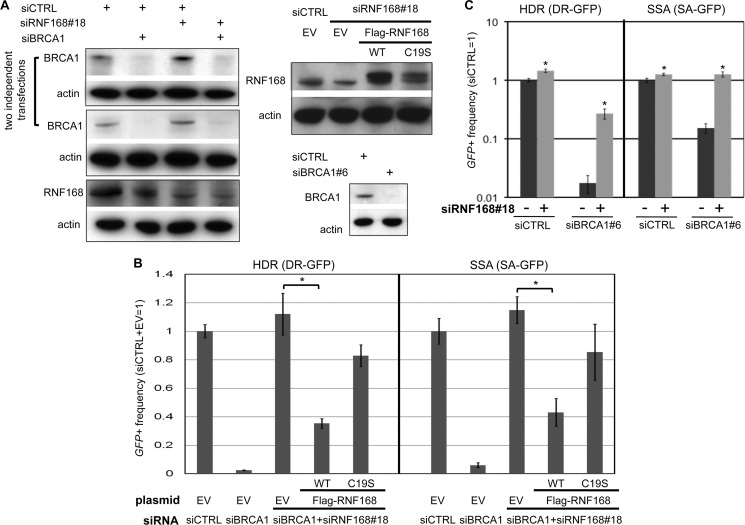
To confirm these findings, we utilized a distinct siRNA sequence to deplete RNF168 (siRNF168#18) and performed complementation analysis using an expression vector for FLAG-RNF168 with silent mutations to evade siRNF168#18. We evaluated expression vectors for WT RNF168, as well as a mutant form (C19S) that is predicted to disrupt the RING domain (1). Using immunoblot analysis, we confirmed expression of FLAG-RNF168, efficient depletion of BRCA1 in the cells treated with siBRCA1 alone and both siBRCA1 and siRNF168#18, and efficient depletion of RNF168 (4-fold; modestly greater than with the siRNF168 pool of four distinct siRNAs) in the cells treated with siRNF168#18 alone and both siBRCA1 and siRNF168#18 (Fig. 2A). Also from this analysis, we note that the C19S mutant of RNF168 is expressed at a lower level than WT, which is consistent with previous findings that RING domain mutants of RNF168 show lower expression (1). In addition, we note that siRNF168#18-treated cells show a minor increase in BRCA1 protein levels (Fig. 2A) but nevertheless show a marked defect in BRCA1 IRIF formation (Fig. 3A).
FIGURE 2.
Transient RNF168 expression inhibits HR in cells co-depleted of BRCA1 and endogenous RNF168. A, immunoblot analysis showing RNAi depletion of BRCA1 and RNF168, and expression of FLAG-RNF168. Shown are immunoblot signals for BRCA1, RNF168, and actin for cells treated with siRNAs used in B and C. Also shown (upper right) are immunoblot signals for RNF168 and actin for cells after co-transfection with siRNA along with the expression vectors that are used in B for FLAG-RNF168 WT, FLAG-RNF168 C19S (a RING domain mutant), or control EV. B, depletion of RNF168 by a distinct single siRNA (#18) suppresses HR defects in siBRCA1-treated cells, which is inhibited by transient expression of RNF168. U2OS reporter cell lines were examined as in Fig. 1C, except using siRNF168#18 and including siRNF168#18-resistant expression vectors for FLAG-RNF168 WT, FLAG-RNF168 C19S, or control EV during the I-SceI transfection. Shown are the frequencies of GFP+ cells for the HDR and SSA reporter cell lines and each siRNA treatment, relative to siCTRL and EV. *, p < 0.0001 (n = 3). C, repair analysis using a distinct siBRCA1. U2OS reporter cell lines were examined as in Fig. 1C, using siBRCA1#6 and siRNF168#18. Shown are the frequencies of GFP+ cells for the HDR and SSA reporter cell lines for each siRNA treatment, relative to siCTRL. *, p ≤ 0.0001 (n = 6).
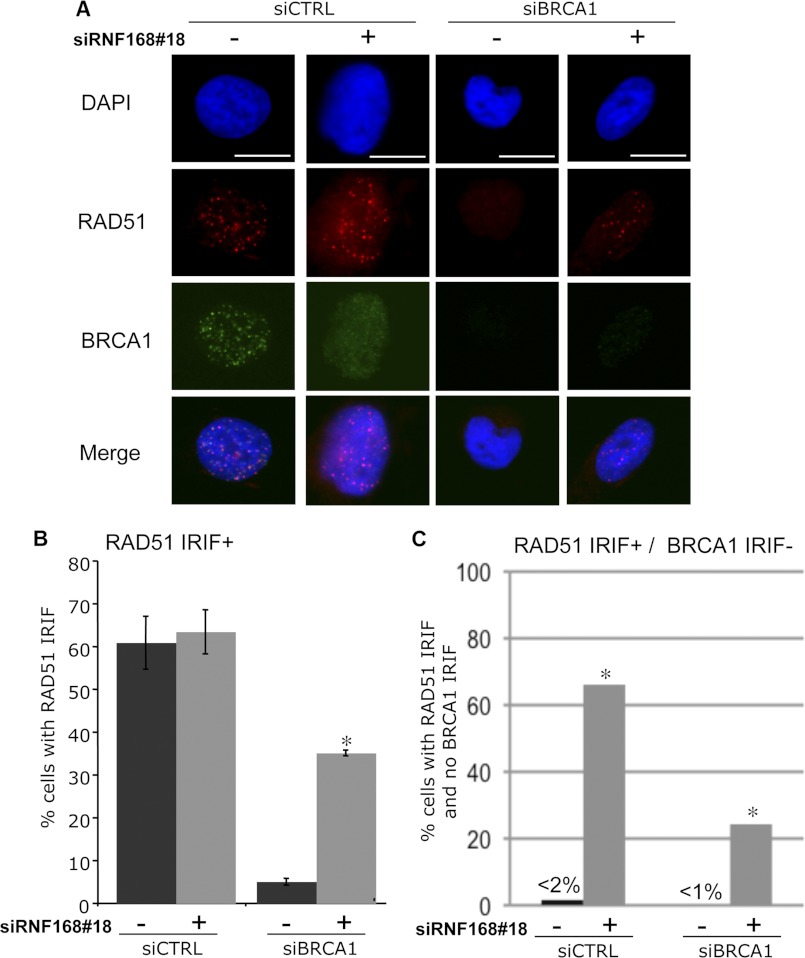
FIGURE 3.
RNF168-depleted cells show a diminished requirement for BRCA1 in formation of RAD51 IRIF. U2OS cells were treated with siRNAs as in Fig. 2, cultured for 2 days, treated with 6 Gy of IR (Cs137), and allowed to recover for 4 h prior to pre-extraction, fixation, and immunostaining with RAD51 antibodies, or co-staining with antibodies against RAD51 and BRCA1. Cells with ≥5 RAD51 foci were scored as RAD51 IRIF+, and cells with <5 BRCA1 foci were scored as BRCA1 IRIF−. A, shown are DAPI, RAD51 immunostaining, and BRCA1 immunostaining signals for an siCTRL cell that is RAD51 IRIF+/BRCA1 IRIF+, an siBRCA1 cell that is RAD51 IRIF−/BRCA1 IRIF−, and siRNF168#18 and both siBRCA1 and siRNF168#18 cells that are RAD51 IRIF+/BRCA1 IRIF−. Bars, 10 μm. B, depletion of RNF168 suppresses the RAD51 IRIF defect caused by BRCA1 silencing. Shown are the frequencies of cells with ≥5 RAD51 foci (RAD51 IRIF+) for each of the siRNA treatments shown in A. *, p < 0.0001 versus the parallel (−) siRNF168 condition (n ≥ 3). C, RNF168-depleted cells can form RAD51 IRIF without BRCA1 IRIF, which is uncommon in control cells. Shown are the frequencies of cells with ≥5 RAD51 foci and <5 BRCA1 foci (RAD51 IRIF+/BRCA1 IRIF−) for each of the siRNA treatments shown in A. *, p < 0.0001 versus the (−) parallel siRNF168 condition using Fisher's exact test.
Using this distinct RNF168 siRNA (#18), along with complementation via FLAG-RNF168, we examined effects of RNF168 depletion on the frequency of HR in cells with BRCA1 silencing. Similar to results shown above (Fig. 1C), we found that cells treated with siBRCA1 and siRNF168#18 show a significant increase in HDR and SSA, relative to siBRCA1-treated cells (Fig. 2B), which we also observe using a distinct siRNA targeting BRCA1 (siBRCA1#6; Fig. 2, A and C). Furthermore, we found that transient expression of FLAG-RNF168 WT substantially inhibited HDR and SSA in cells treated with siBRCA1 and siRNF168#18 (Fig. 2B; 3.2- and 2.6-fold, respectively). In contrast, expression of the C19S mutant form of FLAG-RNF168 had only a minor effect on HDR and SSA in these cells (Fig. 2B; 1.3-fold decrease), indicating that an intact WT RING domain is important for substantial inhibition of HR. Altogether, these results indicate that depletion of RNF168 can suppress HR defects caused by BRCA1 silencing.
RNF168-depleted Cells Show a Diminished Requirement for BRCA1 for Forming RAD51 IRIF
BRCA1 is important for recruitment of the central HR factor RAD51 to DSBs, based on IRIF analysis (31, 32). Accordingly, based on our above findings with the HR reporter assays, we predicted that RNF168 depletion would enable formation of RAD51 IRIF in a manner that is relatively independent of BRCA1. We examined RAD51 IRIF in cells treated with siCTRL, siBRCA1, siRNF168#18, and both siBRCA1 and siRNF168#18, following treatment with 6 Gy of IR and 4 h of recovery (Fig. 3, A and B). In this experiment, siBRCA1-treated cells showed a much lower frequency of cells with RAD51 IRIF, compared with both siCTRL- and siRNF168#18-treated cells (12-fold lower, p < 0.0001), as expected (31, 32). However, cells treated with both siBRCA1 and siRNF168#18 showed a significantly greater frequency of RAD51 IRIF, compared with siBRCA1-treated cells (7-fold higher, p < 0.0001). Thus, RNF168 depletion appears to diminish the requirement for BRCA1 for formation of RAD51 IRIF.
Based on this above findings, along with recent reports showing that RNF168-depleted cells can form RAD51 IRIF (33, 34), we predicted that RNF168-depleted cells should be able to form RAD51 IRIF without BRCA1 IRIF. We tested this hypothesis using immunofluorescence co-staining with RAD51 and BRCA1 antibodies, using the same siRNA and IR treatments as above (Fig. 3, A and C). We then determined the frequency of cells that were positive for RAD51 IRIF but lacked BRCA1 IRIF (RAD51 IRIF+/BRCA1 IRIF− cells). Consistent with previous studies showing a significant association between RAD51 and BRCA1 in recruitment to DSBs (32, 35), siCTRL- and siBRCA1-treated samples showed a very low frequency of RAD51 IRIF+/BRCA1 IRIF− cells (<2%). In contrast, cells treated with siRNF168#18 alone and both siBRCA1 and siRNF168#18 showed a substantial frequency of RAD51 IRIF+/BRCA1 IRIF− cells (66 and 24%, respectively). These findings support the notion that RNF168-depleted cells show a diminished requirement for BRCA1 for formation of RAD51 IRIF.
RNF168 Depletion Suppresses the RPA IRIF and Anaphase Chromosome Separation Defects Caused by BRCA1 Silencing
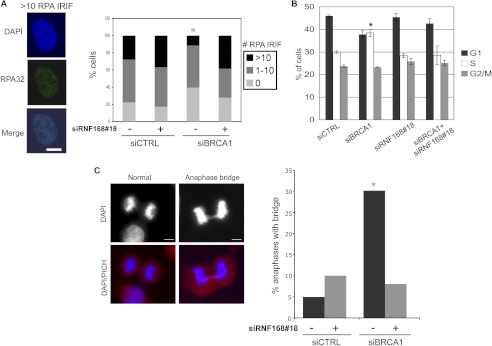
In addition to RAD51 recruitment, another step of HR is recruitment of the ssDNA-binding factor replication protein A (RPA) to DSBs, which is likely mediated by end resection that forms ssDNA (14, 36). Such end resection is predicted to be required for RAD51 nucleoprotein filament formation during HDR and also for strand annealing during SSA (12). BRCA1 has been shown to promote RPA IRIF, although the effects are more modest than for RAD51 IRIF (14). Consistent with this notion, we found that the fraction of cells with >10 RPA IRIF (RPA32 subunit) was modestly reduced in siBRCA1-treated cells compared with siCTRL cells (2.4-fold, p < 0.0001; Fig. 4A). In contrast, the fraction of cells with >10 RPA IRIF for cells treated with both siBRCA1 and siRNF168#18 was not reduced compared with siCTRL cells and accordingly was significantly higher than siBRCA1-treated cells (p < 0.0001; Fig. 4A). These findings indicate that depletion of RNF168 can also suppress the modest RPA IRIF defect caused by BRCA1 silencing.
FIGURE 4.
Depletion of RNF168 suppresses RPA IRIF and anaphase chromosome separation defects caused by BRCA1 silencing. Prior to examining RPA IRIF, cell cycle, and anaphases, U2OS cells were treated with siCTRL or siBRCA1, along with (+) or without (−) siRNF168#18, as in Fig. 2. A, depletion of RNF168 suppresses the decrease in RPA IRIF caused by siBRCA1 treatment. Cells 2 days after transfection were treated with 10 Gy of IR (Cs137) and allowed to recover for 4 h prior to fixation and immunostaining with RPA (RPA32 subunit) antibodies. Shown is a representative siCTRL-treated cell with >10 RPA IRIF, and the percentage of cells with 0, 1–10, and >10 RPA IRIF for each siRNA treatment. *, p < 0.0001 for number of cells >10 RPA IRIF versus all other siRNA treatments using Fisher's exact test (n > 200 accumulated from two independent transfections). B, treatment with siBRCA1 causes a modest shift of G1 to S phase cells. Shown are the percentages of cells in G1, S, and G2/M for each siRNA condition, based on co-staining for BrdU and propidium iodide. *, p = 0.0008 versus siCTRL (n ≥ 3). C, depletion of RNF168 suppresses the anaphase bridges caused by siBRCA1 treatment. Shown are representative images of a normal anaphase, and an anaphase bridge co-stained with PICH and DAPI from siBRCA1-treated cells. Bars, 10 μm. Shown is the percentage of bridges in late anaphase for each siRNA treatment, scored using DAPI staining. This frequency was determined from analyzing over 100 anaphases for each siRNA treatment, which were accumulated from several independent transfections (n ≥ 3). *, p < 0.0006 versus all other siRNA treatments using Fisher's exact test.
We also examined the effects of siRNF168#18 and/or siBRCA1 treatment on cell cycle and mitosis. With cell cycle profiles (Fig. 4B and supplemental Fig. S3), we found that siBRCA1-treated cells showed a modest shift from G1 phase to S phase (38% S), as compared with the other siRNA treatments (28–30% S). Next, because HR-deficient cells show anaphase chromosome separation defects (37), we tested whether siRNF168#18 treatment could suppress such defects in siBRCA1-treated cells. For this, we examined a marker of incomplete chromosome separation: anaphase bridges that are detected by the DNA dye DAPI (Fig. 4C). We found that the frequency of anaphase bridges was higher in cells treated with siBRCA1 versus those treated with siCTRL, siRNF168#18 alone, and both siBRCA1 and siRNF168#18 (>3-fold, p < 0.0006; Fig. 4C). Notably, we also found that the vast majority (80%; Fig. 4C) of anaphase bridges induced by siBRCA1 treatment contained PICH, which is a factor that associates with anaphase abnormalities caused by HR deficiency, replication stress, or decatenation stress (37, 38). In summary, these results indicate that RNF168 is important for the HR and anaphase chromosome separation defects caused by BRCA1 silencing.
Depletion of RNF168 Suppresses HR Defects Caused by a Dominant Negative Peptide of BARD1, but Not by Disruption of Several Other HR Factors
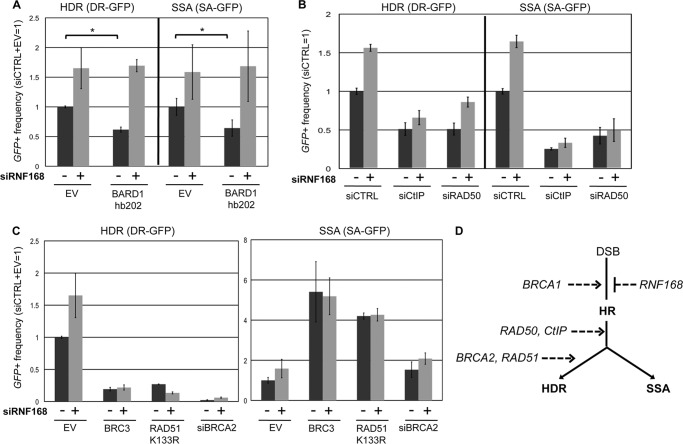
As an independent approach to examine the influence of RNF168 on HR in BRCA1-deficient cells, we disrupted BARD1, which binds to BRCA1 to form a heterodimer complex with E3 ubiquitin ligase activity (39, 40). Specifically, we expressed a BARD1 dominant negative peptide that has been shown to disrupt HR: BARD1-hb202, which contains the N-terminal 202 residues of BARD1 that includes its RING finger domain but lacks the Ankyrin and BRCT domains (39). We co-transfected expression vectors for I-SceI and BARD1-hb202 in the U2OS reporter cells and found that BARD1-hb202 caused a reduction in HDR and SSA (1.6-fold; Fig. 5A). However, in siRNF168-treated cells, BARD1-hb202 had no effect on repair, in that HDR and SSA frequencies were even higher than the siCTRL cells (Fig. 5A). These findings indicate that RNF168 depletion can suppress HR defects caused by a dominant negative peptide fragment of BARD1.
FIGURE 5.
Depletion of RNF168 suppresses HR defects caused by a dominant negative peptide of BARD1, but not by disruption of CtIP, RAD50, BRCA2, or RAD51. To examine effects of RNF168 depletion on repair in cells disrupted for several HR factors, U2OS reporter cell lines were treated with siRNAs or expression vectors for dominant negative peptides, along with (+) or without (−) siRNF168. Shown are the frequencies of GFP+ cells for the HDR and SSA reporter cell lines and each siRNA treatment, relative to siCTRL/siCTRL+EV = 1. A, expression of a BARD1 dominant negative peptide (BARD1 hb202) inhibits HR, but not in siRNF168-treated cells. *, p ≤ 0.0013 (n ≥ 3). B, cells treated with siCTIP or siRAD50 show a decreased frequency of HR, and siRNF168 does not restore HR in these cells. C, cells expressing dominant negative peptides of BRCA2 or RAD51 (BRC3 or RAD51-K133R, respectively) or treated with siBRCA2, show a decreased frequency of HDR and an increased frequency of SSA; furthermore, siRNF168 does not restore HDR in these cells. D, shown is a model whereby RNF168 acts antagonistically to BRCA1 during the regulation of HR, which is distinct from the execution of HR that requires CtIP, RAD50, BRCA2, and RAD51.
We then tested whether the suppression of HR defects via RNF168 depletion is specific to BRCA1/BARD1-deficient cells, by examining cells disrupted for other HR factors. Specifically, we investigated the effect of RNF168 on HR in U2OS cells with disruption of the end resection factors CtIP and RAD50, as well as the strand exchange factor RAD51 and its co-factor BRCA2 (7). We first examined cells treated with siRNAs targeting CtIP and RAD50, each of which causes a reduction in HDR and SSA (19, 22). We found that adding siRNF168 to siCtIP- and siRAD50-treated cells caused an equivalent or lesser fold increase on HR, as compared with siCTRL-treated cells (Fig. 5B). Thus, siRNF168 treatment did not rescue HR in siCtIP- and siRAD50-treated cells. Next, we disrupted BRCA2 and RAD51 using two previously described dominant negative peptides that inhibit HDR: a short fragment of BRCA2 (BRC3) and a RAD51 mutant (RAD51-K133R) that is deficient in ATP hydrolysis (9). We also disrupted BRCA2 using siRNA (siBRCA2). We found that BRC3, RAD51-K133R, and siBRCA2 each caused a substantial decrease in HDR and an increase in SSA and that HDR was not rescued in any of these cells by siRNF168 treatment (Fig. 5C). These findings indicate that depletion of RNF168 does not suppress HR defects caused by disruption of CtIP, RAD50, BRCA2, or RAD51 (Fig. 5D).
RNF168 and 53BP1 Have a Similar Influence on HR
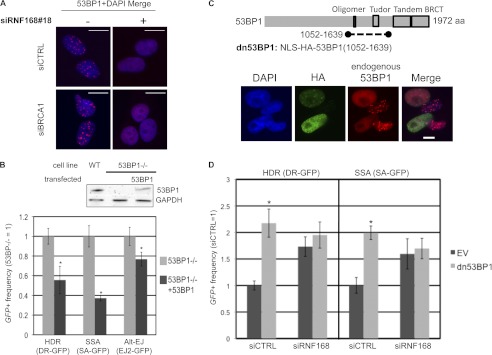
Finally, because RNF168 is critical for recruitment of 53BP1 to DSBs (1, 3, 4), we considered that RNF168 and 53BP1 might have similar effects on HR. To begin with, we examined the formation of 53BP1 IRIF in cells depleted of RNF168 and/or BRCA1. Consistent with previous studies of RNF168-deficient cells (1, 3, 4), we find that siRNF168#18 treatment abolishes 53BP1 IRIF, which we also observe using combined siBRCA1 and siRNF168#18 treatment but not siBRCA1 treatment alone (Fig. 6A).
FIGURE 6.
53BP1 inhibits HR similarly to RNF168. A, RNF168 is required for 53BP1 IRIF. Cells were treated with siRNAs and 10 Gy of IR (Cs137) as in Fig. 4A and allowed to recover 4 h prior to fixation and immunostaining with 53BP1 antibodies. Shown are 53BP1 immunostaining signals from representative nuclei from each siRNA treatment. B, 53BP1 inhibits HDR and SSA, and to a lesser extent Alt-EJ. The reporters shown in Fig. 1A were integrated separately into 53BP1−/− MEFs. These cell lines were transfected with an expression vector for I-SceI, along with an expression vector for 53BP1 or a control EV. Shown are the frequencies of GFP+ cells relative to parallel 53BP1−/− samples, normalized to transfection efficiency via a parallel transfection with a GFP expression vector. *, p < 0.0001 (n ≥ 6). Also shown are immunoblot signals for 53BP1 and GAPDH for WT MEFs, 53BP1−/− MEFs, and 53BP1−/− MEFs following transfection with the 53BP1 expression vector. C, expression of a fragment of 53BP1 (dn53BP1) causes a dominant negative disruption of 53BP1 IRIF. Shown is a diagram of 53BP1 and the dn53BP1 fragment that is fused downstream from tandem nuclear localization signals (NLS) and the HA immunotag. U2OS cells were transfected with an expression vector for dn53BP1, cultured for 2 days, treated with 6 Gy of IR (Cs137), and allowed to recover for 4 h prior to fixation and immunostaining. Shown are HA and 53BP1 immunostaining signals for four representative nuclei. The 53BP1 antibody only detects the endogenous protein, because it is directed against the N terminus, which is absent from dn53BP1. D, the peptide dn53BP1 only causes an increase in HDR and SSA in RNF168-proficient cells. U2OS reporter cells were transfected with siCTRL or siRNF168 2 days prior to co-transfection of the respective siRNA and an expression vector for I-SceI, along with an expression vector for dn53BP1 or EV. Shown are the frequencies of GFP+ cells for the HDR and SSA reporter cell lines for each siRNA and expression vector treatment, relative to siCTRL and EV. *, p < 0.0001 (n ≥ 6).
We then considered whether 53BP1 might influence HR similarly to RNF168. Previous studies of 53BP1 support this prediction, in that disruption of 53BP1 can suppress HR defects caused by BRCA1 loss (5, 6). Furthermore, a dominant negative fragment of 53BP1 has been shown to cause elevated HDR (17), whereas the influence of 53BP1 on SSA has been unclear. However, in contrast to these previous findings with 53BP1, another study showed that HDR of an extrachromosomal plasmid was not affected by 53BP1 loss in mouse embryonic fibroblasts (MEFs) (41). We sought to use chromosomal substrates for HR to clarify whether genetic loss of 53BP1 affects HR similarly to our findings with RNF168 depletion. For this, we integrated the DR-GFP, SA-GFP, and EJ2-GFP reporters (Fig. 1A) into immortalized 53BP1−/− MEFs. We then expressed I-SceI with or without co-expression of human 53BP1 for complementation and subsequently examined the frequency of repair. From these experiments (Fig. 6B), we found that expression of 53BP1 in 53BP1−/− MEFs caused a significant decrease in HDR and SSA (1.8- and 2.7-fold, respectively) and to a lesser extent Alt-EJ (1.3-fold). These results indicate that both RNF168 and 53BP1 are important to inhibit two distinct HR pathways (HDR and SSA) and hence show a similar influence on HR.
Because RNF168 is required for 53BP1 IRIF and because these factors affect HR similarly, we hypothesized that disruption of 53BP1 would fail to cause an increase in HR in RNF168-depleted cells. For this analysis, we disrupted 53BP1 using a dominant negative peptide fragment of 53BP1 (dn53BP1), which contains tandem nuclear localization signals and an HA immunotag fused to amino acids 1052–1639 of 53BP1 (Fig. 6C). This fragment of 53BP1 includes both the oligomerization and Tudor domains, which are important for 53BP1 recruitment to DSBs (42–44). We determined that expressing dn53BP1 disrupts 53BP1 recruitment to DSBs, in that 53BP1 IRIF were abolished in cells that are positive for the HA immunotag of dn53BP1, where endogenous 53BP1 was detected using an antibody directed against an N-terminal fragment that is absent in dn53BP1 (Fig. 6C). We then tested whether dn53BP1 may affect repair only in RNF168-proficient cells, using the U2OS reporter assays. From these experiments, we found that transient co-expression of dn53BP1 with the I-SceI endonuclease caused a significant increase in HDR and SSA in siCTRL cells (2-fold) but importantly did not affect HR in siRNF168-treated cells (Fig. 6D). These findings further support the notion that RNF168 and 53BP1 have a similar influence on HR and also indicate that these factors may function in the same pathway to inhibit HR.
DISCUSSION
RNF168 plays an important role in DNA damage response signaling and radioresistance, as well as immune system function (1, 2). However, the role of RNF168 during a major pathway of DSB repair, HR, has been unclear, particularly because RNF168 promotes the recruitment of both 53BP1 and BRCA1 to DSBs (1, 3, 4), which have antagonistic effects on HR (5, 6). We have examined the influence of RNF168 on HR in cells with or without BRCA1 silencing using several methods: quantifying the frequency of HR repair using DSB reporter assays, examining IRIF formation of the RAD51 recombinase that is critical for HR (45), and quantifying the frequency of anaphase abnormalities that can be caused by HR deficiency (37). With these approaches, we present evidence that RNF168 inhibits HR of reporter assays and that RNF168 is important for HR defects caused by BRCA1 silencing.
We have also determined that 53BP1, a factor that we have confirmed requires RNF168 for recruitment to DSBs, appears to influence HR similarly to RNF168, in that both factors inhibit HDR and SSA and are important for HR defects caused by BRCA1 loss (5, 6). Furthermore, we have found that disruption of 53BP1 function via a dominant negative peptide only causes a significant increase in HDR and SSA in RNF168-proficient cells, which is consistent with the notion that these factors function in the same pathway to inhibit HR. Apart from HR, RNF168 and 53BP1 promote class switch recombination during antibody maturation (41, 46, 47) and hence appear to share common functions in promoting long range DNA end joining during class switch recombination, as well as being important for HR defects caused by BRCA1 loss. Thus, whereas RNF168 is important for IRIF of both 53BP1 and BRCA1, we suggest that the role of RNF168 during HR is similar to that of 53BP1.
Accordingly, our findings support a relatively unconventional interpretation for the reduction in BRCA1 recruitment to DSBs in RNF168-depleted cells (1, 3, 4). Namely, this reduction in BRCA1 recruitment to DSBs does not appear to reflect a loss of function during HR, because RNF168-depleted cells are proficient at HR and furthermore can form RAD51 IRIF efficiently without forming BRCA1 IRIF. Additionally, RNF168 depletion can suppress the HR and RAD51 IRIF defects caused by BRCA1 silencing. Notably, depletion of another factor important for BRCA1 recruitment to DSBs, the ubiquitin-binding factor RAP80, appears to cause not a loss of BRCA1 function but rather a deregulation of BRCA1 function during HR (48–51). In summary, these findings underscore the notion that reduced BRCA1 recruitment to DSBs does not always reflect a loss of function during HR.
As well, our findings provide a distinct contrast from a recent report showing that cells co-depleted of RNF168, BRCA1, and 53BP1 exhibit a decrease in RAD51 IRIF, compared with cells co-depleted of BRCA1 and 53BP1 (34). Although this result indicates that RNF168 may promote RAD51 IRIF in cells deficient in both BRCA1 and 53BP1, importantly, cells co-depleted of BRCA1 and RNF168 were not examined without simultaneous 53BP1 depletion (34). Accordingly, this recent report did not address the possibility that cells depleted of RNF168 and BRCA1 could exhibit a higher frequency of cells with RAD51 IRIF, compared with BRCA1-depleted cells, which is the result we have shown here. Furthermore, using a number of assays for HR, we find that RNF168 depletion significantly suppresses the HR defect caused by BRCA1 silencing, which supports a substantial role for RNF168 in inhibiting HR in cells with BRCA1 loss.
In contrast to the suppression of HR defects caused by BRCA1 silencing or a dominant negative peptide of BARD1, RNF168 depletion did not rescue HR defects caused by disruption CtIP or RAD50, which are factors critical for the end resection step of HR (12). RNF168 depletion also did not rescue HR defects caused by disruption of BRCA2 or RAD51, which are factors that mediate strand invasion subsequent to end resection (7). Accordingly, these findings indicate that RNF168 and BRCA1 act antagonistically during a step of HR that is distinct from the steps of HR promoted by RAD51, BRCA2, CtIP, and RAD50 (Fig. 5D).
Considering possible mechanisms, RNF168 and BRCA1 could be important for regulating the extent of end processing and/or chromatin remodeling during DSB repair. Consistent with this mechanism, we find that RNF168 does not inhibit Alt-EJ, which likely requires less end processing and minimal chromatin remodeling, relative to HR. Also, we examined localization of the ssDNA-binding factor RPA into IRIF, which is likely mediated by end processing (12, 14, 36), and found that RNF168 depletion can suppress the modest decrease in RPA IRIF caused by BRCA1 silencing. Notably, in considering mechanism, BRCA1 depletion does not cause an obvious effect on RNF168 localization to DSBs (1, 3), which is inconsistent with direct regulation of RNF168 via BRCA1. In summary, given that cells co-depleted of BRCA1 and RNF168 appear to show efficient HR, we postulate that BRCA1 and RNF168 influence the regulation of HR, in contrast to other factors that appear essential to execute HR (e.g., RAD51, BRCA2, CtIP, and RAD50) (Fig. 5D).
Several breast and ovarian carcinomas show BRCA1 deficiency, involving germ line mutations in BRCA1, as well as gene silencing of BRCA1 by promoter methylation and targeting by microRNAs (52–55). Furthermore, genetic disruption of just one allele of BRCA1 has been shown to cause genomic instability in human epithelial cells (56). Thus, characterizing the factors that influence HR in cells with reduced expression of BRCA1 is important for understanding cancer etiology and improving treatment. Along these lines, our findings indicate that RNF168 is important for HR deficiency in cells with BRCA1 silencing. Accordingly, we speculate that evaluating RNF168 may be important to diagnose HR deficiency in tumors with BRCA1 silencing and that RNF168 may be a therapeutic target to restore HR in cells with BRCA1 loss, as has been postulated for 53BP1 (5, 6).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant RO1CA120954 (to J. M. S.). This work was also supported by the Marsha Rivkin Center for Ovarian Cancer Research and by Regents of the University of California Breast Cancer Research Program Grant 181B-0046 (to J. M. S.).

This article contains supplemental Table S1 and Figs. S1–S3.
- DSB
- double strand break
- Alt-EJ
- alternative end joining
- EV
- empty vector
- Gy
- Gray
- HDR
- homology-directed repair
- HR
- homologous recombination
- IR
- ionizing radiation
- IRIF
- ionizing radiation-induced foci
- PICH
- Plk1-interacting checkpoint helicase
- siCTRL
- nontargeting siRNA
- SSA
- single strand annealing
- E3
- ubiquitin-protein isopeptide ligase
- RPA
- replication protein A.
REFERENCES
- 1. Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P. J., Stankovic T., Taylor A. M., Durocher D. (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 2. Devgan S. S., Sanal O., Doil C., Nakamura K., Nahas S. A., Pettijohn K., Bartek J., Lukas C., Lukas J., Gatti R. A. (2011) Homozygous deficiency of ubiquitin-ligase ring-finger protein RNF168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell Death Differ. 18, 1500–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 4. Pinato S., Scandiuzzi C., Arnaudo N., Citterio E., Gaudino G., Penengo L. (2009) RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol. 10, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bunting S. F., Callén E., Wong N., Chen H. T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., Xu X., Deng C. X., Finkel T., Nussenzweig M., Stark J. M., Nussenzweig A. (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouwman P., Aly A., Escandell J. M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., Haffty B. G., Tommiska J., Blomqvist C., Drapkin R., Adams D. J., Nevanlinna H., Bartek J., Tarsounas M., Ganesan S., Jonkers J. (2010) 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moynahan M. E., Jasin M. (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hartlerode A. J., Scully R. (2009) Mechanisms of double-strand break repair in somatic mammalian cells. Biochem. J. 423, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stark J. M., Pierce A. J., Oh J., Pastink A., Jasin M. (2004) Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 24, 9305–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tutt A., Bertwistle D., Valentine J., Gabriel A., Swift S., Ross G., Griffin C., Thacker J., Ashworth A. (2001) Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20, 4704–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bennardo N., Gunn A., Cheng A., Hasty P., Stark J. M. (2009) Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 5, e1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Symington L. S., Gautier J. (2011) Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 [DOI] [PubMed] [Google Scholar]
- 13. Yang Y. G., Saidi A., Frappart P. O., Min W., Barrucand C., Dumon-Jones V., Michelon J., Herceg Z., Wang Z. Q. (2006) Conditional deletion of Nbs1 in murine cells reveals its role in branching repair pathways of DNA double-strand breaks. EMBO J. 25, 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlegel B. P., Jodelka F. M., Nunez R. (2006) BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 66, 5181–5189 [DOI] [PubMed] [Google Scholar]
- 15. Roy R., Chun J., Powell S. N. (2012) BRCA1 and BRCA2. Different roles in a common pathway of genome protection. Nat. Rev. Cancer 12, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huen M. S., Sy S. M., Chen J. (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 11, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F. W., Chen J., Jackson S. P., Scully R. (2007) Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell 28, 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bunting S. F., Callén E., Kozak M. L., Kim J. M., Wong N., López-Contreras A. J., Ludwig T., Baer R., Faryabi R. B., Malhowski A., Chen H. T., Fernandez-Capetillo O., D'Andrea A., Nussenzweig A. (2012) BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell 46, 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gunn A., Bennardo N., Cheng A., Stark J. M. (2011) Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J. Biol. Chem. 286, 42470–42482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gunn A., Stark J. M. (2012) I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920, 379–391 [DOI] [PubMed] [Google Scholar]
- 21. Hu Y., Ghosh S., Amleh A., Yue W., Lu Y., Katz A., Li R. (2005) Modulation of aromatase expression by BRCA1. A possible link to tissue-specific tumor suppression. Oncogene 24, 8343–8348 [DOI] [PubMed] [Google Scholar]
- 22. Bennardo N., Cheng A., Huang N., Stark J. M. (2008) Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lilley C. E., Chaurushiya M. S., Boutell C., Landry S., Suh J., Panier S., Everett R. D., Stewart G. S., Durocher D., Weitzman M. D. (2010) A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 29, 943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dimitrova N., Chen Y. C., Spector D. L., de Lange T. (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pierce A. J., Johnson R. D., Thompson L. H., Jasin M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. LaRocque J. R., Stark J. M., Oh J., Bojilova E., Yusa K., Horie K., Takeda J., Jasin M. (2011) Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 108, 11971–11976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson R. D., Jasin M. (2000) Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19, 3398–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guirouilh-Barbat J., Huck S., Bertrand P., Pirzio L., Desmaze C., Sabatier L., Lopez B. S. (2004) Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14, 611–623 [DOI] [PubMed] [Google Scholar]
- 29. Haber J. E. (2008) Alternative endings. Proc. Natl. Acad. Sci. U.S.A. 105, 405–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu C., Mills K. D., Ferguson D. O., Lee C., Manis J., Fleming J., Gao Y., Morton C. C., Alt F. W. (2002) Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109, 811–821 [DOI] [PubMed] [Google Scholar]
- 31. Bhattacharyya A., Ear U. S., Koller B. H., Weichselbaum R. R., Bishop D. K. (2000) The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275, 23899–23903 [DOI] [PubMed] [Google Scholar]
- 32. Scully R., Livingston D. M. (2000) In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408, 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sy S. M., Jiang J., Dong S. S., Lok G. T., Wu J., Cai H., Yeung E. S., Huang J., Chen J., Deng Y., Huen M. S. (2011) Critical roles of ring finger protein RNF8 in replication stress responses. J. Biol. Chem. 286, 22355–22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakada S., Yonamine R. M., Matsuo K. (2012) RNF8 regulates assembly of RAD51 at DNA double-strand breaks in the absence of BRCA1 and 53BP1. Cancer Res. 72, 4974–4983 [DOI] [PubMed] [Google Scholar]
- 35. Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D. M. (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88, 265–275 [DOI] [PubMed] [Google Scholar]
- 36. Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S. P. (2007) Human CtIP promotes DNA end resection. Nature 450, 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laulier C., Cheng A., Stark J. M. (2011) The relative efficiency of homology-directed repair has distinct effects on proper anaphase chromosome separation. Nucleic Acids Res. 39, 5935–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan K. L., Hickson I. D. (2011) New insights into the formation and resolution of ultra-fine anaphase bridges. Semin. Cell Dev. Biol. 22, 906–912 [DOI] [PubMed] [Google Scholar]
- 39. Westermark U. K., Reyngold M., Olshen A. B., Baer R., Jasin M., Moynahan M. E. (2003) BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol. Cell. Biol. 23, 7926–7936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Joukov V., Chen J., Fox E. A., Green J. B., Livingston D. M. (2001) Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. U.S.A. 98, 12078–12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ward I. M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J. S., Cascalho M., Chen L., Nussenzweig A., Livak F., Nussenzweig M. C., Chen J. (2004) 53BP1 is required for class switch recombination. J. Cell Biol. 165, 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Iwabuchi K., Basu B. P., Kysela B., Kurihara T., Shibata M., Guan D., Cao Y., Hamada T., Imamura K., Jeggo P. A., Date T., Doherty A. J. (2003) Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J. Biol. Chem. 278, 36487–36495 [DOI] [PubMed] [Google Scholar]
- 43. Ward I., Kim J. E., Minn K., Chini C. C., Mer G., Chen J. (2006) The tandem BRCT domain of 53BP1 is not required for its repair function. J. Biol. Chem. 281, 38472–38477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bothmer A., Robbiani D. F., Di Virgilio M., Bunting S. F., Klein I. A., Feldhahn N., Barlow J., Chen H. T., Bosque D., Callen E., Nussenzweig A., Nussenzweig M. C. (2011) Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell 42, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holthausen J. T., Wyman C., Kanaar R. (2010) Regulation of DNA strand exchange in homologous recombination. DNA Repair 9, 1264–1272 [DOI] [PubMed] [Google Scholar]
- 46. Bohgaki T., Bohgaki M., Cardoso R., Panier S., Zeegers D., Li L., Stewart G. S., Sanchez O., Hande M. P., Durocher D., Hakem A., Hakem R. (2011) Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet. 7, e1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ramachandran S., Chahwan R., Nepal R. M., Frieder D., Panier S., Roa S., Zaheen A., Durocher D., Scharff M. D., Martin A. (2010) The RNF8/RNF168 ubiquitin ligase cascade facilitates class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 107, 809–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu J., Liu C., Chen J., Yu X. (2012) RAP80 protein is important for genomic stability and is required for stabilizing BRCA1-A complex at DNA damage sites in vivo. J. Biol. Chem. 287, 22919–22926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu Y., Scully R., Sobhian B., Xie A., Shestakova E., Livingston D. M. (2011) RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 25, 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Coleman K. A., Greenberg R. A. (2011) The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J. Biol. Chem. 286, 13669–13680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang B., Matsuoka S., Ballif B. A., Zhang D., Smogorzewska A., Gygi S. P., Elledge S. J. (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Walsh T., Casadei S., Lee M. K., Pennil C. C., Nord A. S., Thornton A. M., Roeb W., Agnew K. J., Stray S. M., Wickramanayake A., Norquist B., Pennington K. P., Garcia R. L., King M. C., Swisher E. M. (2011) Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 108, 18032–18037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Turner N., Tutt A., Ashworth A. (2004) Hallmarks of “BRCAness” in sporadic cancers. Nat. Rev. Cancer 4, 814–819 [DOI] [PubMed] [Google Scholar]
- 55. Moskwa P., Buffa F. M., Pan Y., Panchakshari R., Gottipati P., Muschel R. J., Beech J., Kulshrestha R., Abdelmohsen K., Weinstock D. M., Gorospe M., Harris A. L., Helleday T., Chowdhury D. (2011) miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 41, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Konishi H., Mohseni M., Tamaki A., Garay J. P., Croessmann S., Karnan S., Ota A., Wong H. Y., Konishi Y., Karakas B., Tahir K., Abukhdeir A. M., Gustin J. P., Cidado J., Wang G. M., Cosgrove D., Cochran R., Jelovac D., Higgins M. J., Arena S., Hawkins L., Lauring J., Gross A. L., Heaphy C. M., Hosokawa Y., Gabrielson E., Meeker A. K., Visvanathan K., Argani P., Bachman K. E., Park B. H. (2011) Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 17773–17778 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.