Background: Parkin is recruited to defective mitochondria to promote degradation by an autophagy mechanism (mitophagy).
Results: VDACs specifically interact with Parkin on defective mitochondria and are required for efficient targeting of Parkin to mitochondria and subsequent mitophagy.
Conclusion: VDACs recruit Parkin to defective mitochondria.
Significance: A novel mechanistic aspect of Parkin-dependent mitophagy is proposed that may be relevant to Parkinson disease.
Keywords: Autophagy, Mass Spectrometry (MS), Mitochondria, Parkin, Ubiquitination, VDAC, Mitophagy
Abstract
Mutations in the ubiquitin ligase Parkin and the serine/threonine kinase PINK1 can cause Parkinson disease. Both proteins function in the elimination of defective mitochondria by autophagy. In this process, activation of PINK1 mediates translocation of Parkin from the cytosol to mitochondria by an unknown mechanism. To better understand how Parkin is targeted to defective mitochondria, we purified affinity-tagged Parkin from mitochondria and identified Parkin-associated proteins by mass spectrometry. The three most abundant interacting proteins were the voltage-dependent anion channels 1, 2, and 3 (VDACs 1, 2, and 3), pore-forming proteins in the outer mitochondrial membrane. We demonstrate that Parkin specifically interacts with VDACs when the function of mitochondria is disrupted by treating cells with the proton uncoupler carbonyl cyanide p-chlorophenylhydrazone. In the absence of all three VDACs, the recruitment of Parkin to defective mitochondria and subsequent mitophagy are impaired. Each VDAC is sufficient to support Parkin recruitment and mitophagy, suggesting that VDACs can function redundantly. We hypothesize that VDACs serve as mitochondrial docking sites to recruit Parkin from the cytosol to defective mitochondria.
Introduction
The serine/threonine kinase PINK1 and the RING family ubiquitin ligase Parkin were recently found to be involved in the elimination of defective mitochondria by mitochondrial autophagy (mitophagy) (1, 2). Mitophagy is thought to protect cells from the deleterious effects of accumulating functionally deficient mitochondria and increased production of reactive oxygen species (3, 4). Because mutations in both proteins are associated with familial forms of Parkinson disease, failure to degrade defective mitochondria may underlie the disease in some cases.
Parkin is recruited from the cytosol to mitochondria with a disrupted inner mitochondrial membrane potential (5). Upon targeting to mitochondria, Parkin is activated and ubiquitinates mitochondrial proteins (6, 7, 32). Parkin variants that lack ubiquitin ligase activity can be recruited to mitochondria but are unable to promote mitophagy, suggesting that ubiquitination of mitochondrial proteins by Parkin is essential for mitophagy but not for its recruitment to mitochondria (2). Several mitochondrial ubiquitination targets of Parkin have been identified in mammalian systems and in flies. Among them are mitofusins, proteins promoting fusion of mitochondria (8–13). Ubiquitination and subsequent degradation of mitofusins could prevent fusion of defective mitochondria with intact ones to keep them isolated and facilitate selective degradation (5, 14). Another ubiquitination target of Parkin is Miro1, a protein that links mitochondria to microtubules for plus-end directed movement (15, 16). Phosphorylation of Miro1 by PINK1 is required for ubiquitination by Parkin, leading to degradation of Miro1 and arrest of mitochondrial movement. Finally, one of the three mammalian voltage-dependent anion channels, VDAC1,3 is ubiquitinated by Parkin, whereas VDAC2 is not (17, 18). It has been speculated that ubiquitination of VDAC1 is required for mitophagy, but mouse embryonic fibroblasts (MEFs) lacking both VDAC1 and VDAC3 (VDAC1/3−/− MEFs) eliminate mitochondria as efficiently as control wild-type MEFs (18), compelling further studies to establish a role for VDACs in mitophagy. Parkin-dependent ubiquitination of mitochondrial proteins allows p62/SQSTM1, a ubiquitin and LC3-binding protein that links ubiquitinated proteins and organelles to the downstream autophagy machinery, to bind defective mitochondria (17–19).
PINK1 is required to target Parkin to defective mitochondria (7). PINK1 is a mitochondrial protein with a single N-terminal transmembrane domain. In intact mitochondria, PINK1 is constitutively degraded upon import into mitochondria, and steady-state levels are very low (20–22). After treating cells with CCCP, a proton uncoupler that dissipates the inner mitochondrial membrane potential, PINK1 becomes stabilized in the outer mitochondrial membrane (20). The kinase activity of PINK1 is required to subsequently target Parkin to mitochondria (17, 20, 23). Several models have been proposed for how PINK1 mediates targeting of Parkin to mitochondria, but the mechanism is not clear. To better understand how Parkin is recruited to defective mitochondria, we identified interaction partners of Parkin on defective mitochondria using immunoprecipitation and mass spectrometry. This approach identified the three VDACs as the predominant mitochondrial-binding partners of Parkin. We show that VDACs are necessary for efficient recruitment of Parkin to defective mitochondria and subsequent mitophagy.
EXPERIMENTAL PROCEDURES
Cell Lines and Plasmids
All cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 0.1 mg/ml streptomycin. To generate 293 cells stably expressing FLAG-Parkin, human Parkin was cloned into a Gateway-adapted pcDNA5-FRT/TO vector (Invitrogen) in-frame with an N-terminal 3×HA-3×FLAG affinity tag (FLAG-Parkin), and integrated into Flp-In T-REx 293 cells using the Flp-In T-Rex mammalian expression system according to the manufacturer's instructions (Invitrogen). Primary VDAC1/3−/− MEFs were a generous gift from William J. Craigen (Baylor College of Medicine, Houston, TX) and immortalized by serial passaging. VDAC1/3−/− MEFs do not express detectable levels of endogenous Parkin. To express N-terminally 3×FLAG-tagged Parkin (human), and C-terminally 2×HA-tagged VDAC1 (human), VDAC2 or VDAC3 (mouse) in these MEFs, the respective cDNAs were cloned into the pMX retroviral vector, sequences were confirmed, and retroviruses for infection of VDAC1/3−/− MEFs were generated by transfection of the pMX plasmids into PlatE packaging cells (24). MEFs stably expressing the transgenes were obtained by subcloning infected cells. Primers and sequence information are available upon request.
siRNA and Real-time PCR
For siRNA experiments, MEFs were transfected with a mix of three VDAC2 siRNAs at a final concentration of 20 nm (Invitrogen, catalogue no. 1320003, oligonucleotide ID MSS247970, GACCAAGUACAAAUGGUGUGAGUAU; MSS247971, GGCUCAUCUAAUACAGACACUGGUA; MSS278681, GAGGAUCAAUUUAUCAGAAAGUAUG) (28) or a control nontargeting siRNA (Invitrogen, catalogue no. 12935-200) using Lipofectamine RNAiMAX (Invitrogen) as described by the manufacturer. Cells were used for further experiments 2 days after transfection. To determine the efficiency of VDAC2 knockdown, RNA was harvested using the RNeasy kit (Qiagen). cDNA was generated with the SuperScript III First Strand cDNA synthesis system using oligo(dT) primers according to the manufacturer's instructions (Invitrogen). Quantitative PCR was performed in triplicate using a SYBR Green mix (Roche Applied Science). Measured transcript levels were normalized to β-actin and compared with the nontargeting control siRNA. Efficiency of VDAC2 knockdown was also analyzed by Western blotting of VDAC1/3−/− MEFs expressing HA-tagged VDAC2.
Mass Spectrometry
HEK293 cells stably expressing FLAG-Parkin or control cells not expressing FLAG-Parkin were grown to 60–80% confluence in 15-cm plates, treated for 1 h with 10 μm CCCP, and then washed with PBS yielding 2 ml of packed cells. All subsequent steps were done at 4 °C. Cells were disrupted with a Dounce homogenizer in 10 mm HEPES (pH 7.7), 1 mm EGTA, 250 mm sucrose, 7 mm β-mercaptoethanol, and protease inhibitors (0.5 mm PMSF, 10 μg/ml leupeptin, 5 μg/ml chymostatin, 3 μg/ml elastatinal, 1 μg/ml pepstatin A). Cell lysates were centrifuged for 15 min at 1,000 × g to pellet nuclei and cell debris, and the supernatant was centrifuged for 15 min at 10,000 × g to pellet mitochondria. After washing the mitochondrial membrane pellet twice in 10 mm HEPES (pH 7.7), 1 mm EGTA, 250 mm sucrose, it was solubilized in buffer A (100 mm HEPES (pH 7.7), 250 mm KCl, 2 mm MgCl2, 2 mm EDTA, 1 mm NaF, 10% glycerol, 1% digitonin, 7 mm β-mercaptoethanol, and protease inhibitors) and centrifuged for 1 h at 45 K in a Ti70 rotor (200,000 × g; Beckman). The supernatant was immunoprecipitated with anti-FLAG beads (M2 monoclonal antibody; Sigma), and bound material was eluted with 3×FLAG peptide (0.25 mg/ml; Sigma). Mass spectrometry analysis was performed as previously described (25). Proteins were considered candidate Parkin-interacting proteins if they were identified in the Parkin immunoprecipitations but not in control immunoprecipitations from Flp-In-293 cells. NSAF values were calculated according to Florens et al. (26).
Immunoprecipitations
For confirming the association of VDACs with FLAG-Parkin, 293 cells stably expressing FLAG-Parkin were incubated for 1 h with 10 μm CCCP, washed with PBS, and disrupted in buffer A with a Dounce homogenizer on ice. After centrifugation for 1 h at 45 K in a TLA-45 rotor (125,000 × g; Beckman), the supernatant was immunoprecipitated with anti-FLAG beads. Proteins were resolved on 10% SDS-polyacrylamide gels, transferred onto PVDF membranes, and detected with an anti-FLAG (M2, Sigma), anti-VDAC (mouse monoclonal 20B12AF2; MitoSciences) and anti-GAPDH (GT239; GeneTex) antibodies by enhanced chemifluorescence. The anti-VDAC antibody was raised against full-length human VDAC1 and recognizes VDAC1 and VDAC3 but not VDAC2 (supplemental Fig. S1). Ubiquitination of HA-VDACs was determined in VDAC1/3−/− MEFs by immunoprecipitation with anti-HA beads (Sigma) and Western blotting with anti-ubiquitin antibodies (P4D1; Covance).
Immunofluorescence Microscopy
For immunostaining, cells were grown on coverslips, washed in PBS, and fixed for 10 min at room temperature in 1× PBS containing 4% paraformaldehyde. Cells were then permeabilized by incubation with 1× PBS containing 0.5% Triton X-100 for 5 min at room temperature, transferred into 1× PBS with 0.2% Tween 20 (PBS/Tween), and incubated for 30 min in blocking buffer (5% goat serum, 0.2% fish skin gelatin, 0.2% Tween in 1× PBS). Primary antibody incubations were performed for 30 min at room temperature in blocking solution, and cells were washed three times in PBS/Tween and incubated with Alexa Fluor 488 and Texas Red-labeled secondary antibodies in blocking buffer for 30 min. Cells were washed with PBS/Tween and mounted in Prolong Gold (Invitrogen). Antibodies used for immunostaining are: anti-FLAG (M2, 1:1000, mouse monoclonal; Sigma), anti-Tom20 (1:100, rabbit polyclonal, Santa Cruz FL-283), and rabbit or mouse anti-HA antibodies. Images were acquired on a Zeiss M1 fluorescence microscope with 40× and 63× Plan-Apochromat objectives and the MetaMorph software (Molecular Devices, Sunnyvale, CA). Image contrast and brightness were adjusted using Adobe Photoshop. To compute statistical significance for multiple comparisons, we used one-way ANOVA and the Tukey-Kramer method (StatPlus, AnalystSoft Inc.).
RESULTS
VDACs Are Major Interaction Partners of Parkin on Defective Mitochondria
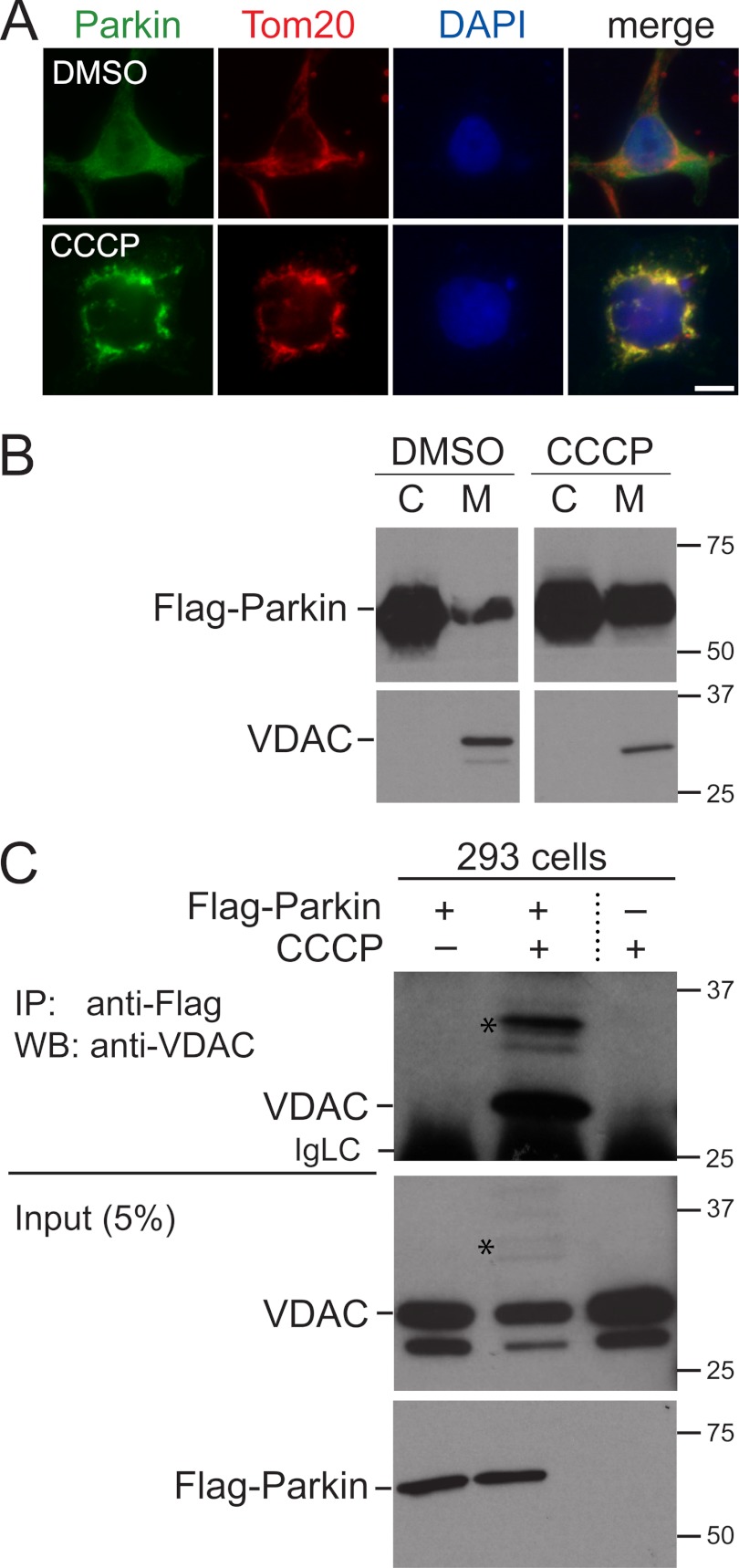
To understand how Parkin is recruited to defective mitochondria, we sought to identify mitochondrial proteins that interact with Parkin in response to mitochondrial damage using affinity purification of FLAG-tagged Parkin and mass spectrometry. We generated 293 cells stably expressing FLAG-tagged Parkin and treated them for 1 h with CCCP. As previously described, under these conditions, some Parkin translocates from the cytosol to mitochondria and co-localizes with the mitochondrial marker protein Tom20 (Fig. 1A) (5). To increase the likelihood of detecting mitochondrial interaction partners of Parkin, we obtained a subcellular membrane fraction enriched in mitochondria. As previously shown, the amount of Parkin in this fraction increases after treatment of cells with CCCP for 1 h (Fig. 1B) (5). This membrane fraction was solubilized and subjected to immunoprecipitation with anti-FLAG antibodies, and Parkin-associated proteins were identified by mass spectrometry (two-dimensional LC-MS/MS, MudPIT).
FIGURE 1.
Parkin is recruited to mitochondria in response to CCCP treatment and associates with VDAC. A, 293 cells stably expressing FLAG-Parkin were treated for 1 h with 10 μm CCCP or DMSO as a control and immunostained for FLAG-Parkin and the mitochondrial marker Tom20. FLAG-Parkin is normally diffusely localized throughout the cell but is enriched at mitochondria after CCCP treatment. Scale bar, 20 μm. B, as in A except that cells were subjected to subcellular fractionation to generate a cytosol fraction (C) and a membrane fraction enriched in mitochondria (M). FLAG-Parkin and VDACs were detected by Western blotting. Molecular masses are indicated in kDa. C, 293 cells expressing FLAG-Parkin and control 293 cells were treated for 1 h with CCCP or DMSO as a control. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibodies, and associated VDACs were detected with anti-VDAC antibodies by Western blotting (WB). VDACs specifically co-immunoprecipitate with FLAG-Parkin when cells are treated with CCCP. 5% of the lysate used for the immunoprecipitations was loaded for the input. Stars indicate higher molecular mass bands that likely represent covalently modified VDAC. The anti-VDAC antibody was raised against full-length human VDAC1 and cross-reacts with VDAC3 (supplemental Fig. S1B). The minor band below the one marked VDAC may represent VDAC3, which migrates faster than VDAC1 (supplemental Fig. S1B). IgLC indicates the immunoglobulin light chain of the anti-FLAG antibodies.
By far the most prominent proteins detected by mass spectrometry were the three porins VDAC1, 2, and 3 (Table 1). VDACs form large channels in the outer mitochondrial membrane through which ions and metabolites are exchanged between the cytosol and mitochondria. In addition to this role, VDACs interact with a number of other proteins under certain cellular conditions, for example, with hexokinase and proteins of the Bcl-2 family (27). Because mitochondrial proteins serving as putative docking sites for Parkin on mitochondria would be predicted to be among the most abundantly detected proteins interacting with Parkin, VDACs were attractive candidates for this function. We confirmed that Parkin interacts with VDACs by co-immunoprecipitation experiments with total lysates of cells treated for 1 h with CCCP. In these experiments, VDACs co-immunoprecipitate with FLAG-tagged Parkin when cells were treated with CCCP, but not in control cells treated with DMSO (Fig. 1C). Importantly, the overall abundance of FLAG-Parkin and VDACs in these lysates is similar in cells treated with CCCP or DMSO, indicating that FLAG-Parkin does not associate with VDACs in a nonspecific manner during the immunoprecipitation procedure (Fig. 1C). We estimate that approximately 5% of total VDACs co-immunoprecipitate with FLAG-Parkin under these conditions. In addition, we performed these co-immunoprecipitation experiments starting with mitochondrial membrane fractions and obtained similar results (data not shown). So far, we have been unable to co-immunoprecipitate endogenous Parkin with VDACs in 293 cells.
TABLE 1.
Identification of mitochondrial proteins associated with Parkin by mass spectrometry
FLAG-Parkin was expressed in 293 cells and immunoprecipitated from a mitochondrial fraction of cells treated for 1 h with 10 μm CCCP. Shown are the three most abundantly detected proteins (VDACs 1, 2, and 3). Also shown are Miro1 and 2 and two mitofusins (Mfn1 and 2), as well as Parkin itself. The number of unique peptides, number of spectra (reflecting the fact that some peptides are identified more than once in an analysis), and percent sequence coverage for each protein are indicated. These Parkin-associated proteins were not detected in control immunoprecipitations from 293 cells not expressing FLAG-Parkin (data not shown).
Other Parkin-associated proteins identified by mass spectrometry included Miro1 and Miro2 and the two mitofusins (Mfn1, Mfn2), albeit lower abundance than the VDACs (Table 1). Both Miro1 and mitofusins were recently found to be ubiquitination targets of Parkin after mitochondrial damage (8–13, 15, 16). Together, these results validate our biochemical approach to identify proteins specifically associated with Parkin and indicate that VDACs are major binding partners of Parkin on defective mitochondria.
VDACs Recruit Parkin to Defective Mitochondria
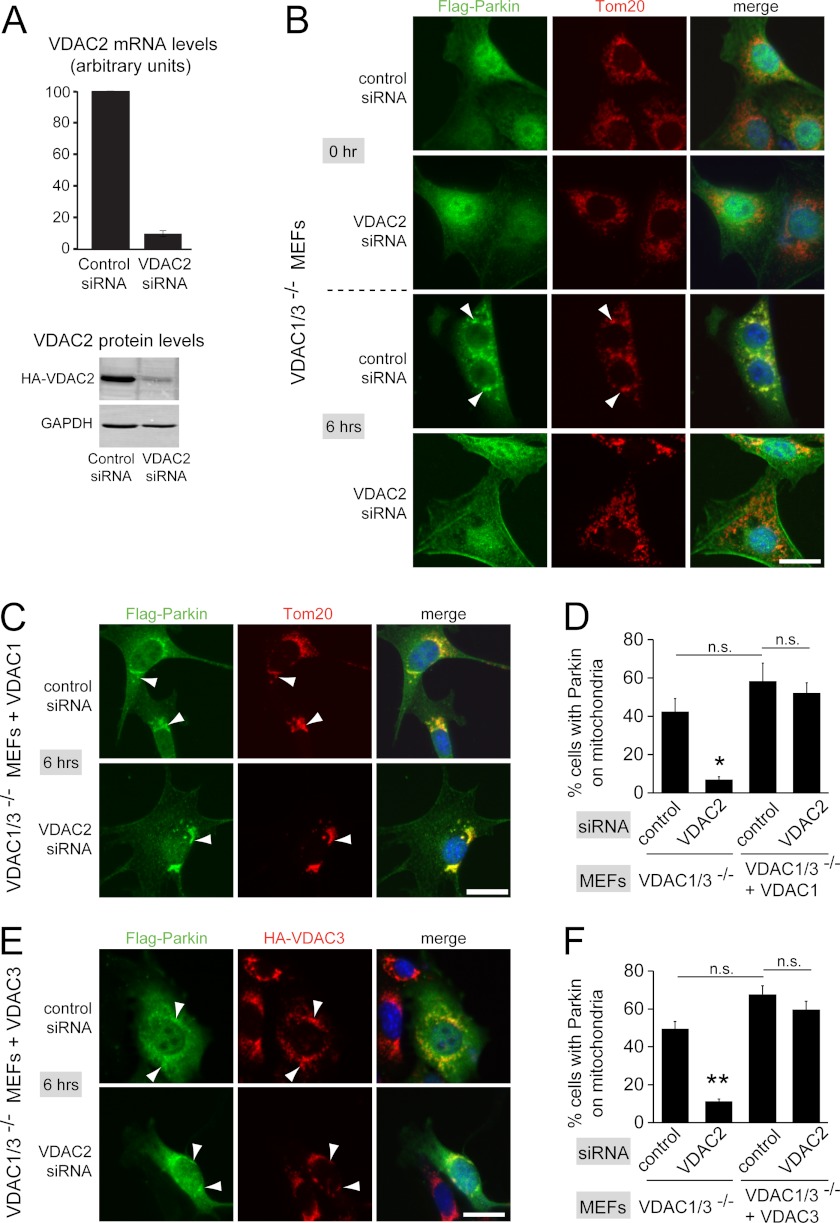
Based on our biochemical interaction data, we speculated that VDACs function to recruit Parkin to defective mitochondria. To test this idea, we took advantage of VDAC1/3−/− MEFs that express VDAC2 as the sole VDAC gene. These cells were generated from VDAC1/3−/− knock-out mice, which are viable and generally similar to wild-type control animals (28). VDAC1/3−/− MEFs have been shown to recruit Parkin to defective mitochondria after CCCP treatment and eliminate defective mitochondria as efficiently as control wild-type MEFs (18). We retrovirally expressed FLAG-tagged Parkin (FLAG-Parkin) in VDAC1/3−/− MEFs and confirmed efficient targeting of FLAG-Parkin to mitochondria and mitophagy after CCCP treatment (data not shown, and Fig. 2B (control siRNA/6 h CCCP); see also Fig. 4A (control siRNA). To determine whether VDAC2 in VDAC1/3−/− MEFs is necessary for Parkin targeting, we depleted VDAC2 by RNAi knockdown. VDAC2 siRNA reduced VDAC2 mRNA levels to approximately 10% of control levels, and protein levels of HA-tagged VDAC2 were strongly reduced (Fig. 2A). Depletion of VDAC2 in VDAC1/3−/− MEFs did not affect the diffuse localization of FLAG-Parkin throughout the cell, cell morphology appeared normal, and CCCP efficiently depolarized mitochondria (Fig. 2B and supplemental Fig. S2). After incubation with CCCP for 6 h, VDAC1/3−/− MEFs treated with control siRNA efficiently recruited FLAG-Parkin to mitochondria as determined by co-localization with the mitochondrial marker protein Tom20 (Fig. 2B). In contrast, when VDAC2 was depleted in VDAC1/3−/− MEFs, FLAG-Parkin remained diffuse throughout the cell, suggesting that recruitment of Parkin to mitochondria was impaired (Fig. 2B). We quantified recruitment of FLAG-Parkin to mitochondria by determining the fraction of cells that had FLAG-Parkin on mitochondria with two lines of VDAC1/3−/− MEFs expressing Parkin. In one line, 42 ± 7% of cells had Parkin localized to mitochondria when treated with control siRNA, and VDAC2 siRNA significantly reduced this to 6.9 ± 1.5% (Fig. 2D, 49 ± 7% and 11.3 ± 1.3%, respectively, for the second line, Fig. 2F). VDAC2 knockdown did not completely abolish targeting of Parkin to mitochondria, possibly due to incomplete depletion. These findings suggest that VDAC2 in VDAC1/3−/− MEFs is necessary for efficient targeting of Parkin to defective mitochondria.
FIGURE 2.
VDACs are necessary for efficient recruitment of Parkin to defective mitochondria. A, depletion of VDAC2 by siRNA knockdown was confirmed by quantitative PCR (top graph). Measured transcript levels were normalized to β-actin and compared with the nontargeting control siRNA. Bars show the mean of three experiments ± S.E (error bars). To demonstrate VDAC2 depletion at the protein level, VDAC1/3−/− MEFs expressing HA-tagged VDAC2 were treated with siRNAs and analyzed by Western blotting with anti-HA antibodies and anti-GAPDH antibodies as a control. B, VDAC1/3−/− MEFs expressing FLAG-Parkin were transfected with the indicated siRNAs. 48 h after transfection, cells were incubated with 20 μm CCCP for 0 h or 6 h and immunostained for Parkin and the mitochondrial marker Tom20. Punctate FLAG-Parkin structures in the 6 h/control siRNA sample co-localize with mitochondria (arrowheads point to examples). C, as in B, except that VDAC1/3−/− MEFs expressing FLAG-tagged Parkin also expressed HA-VDAC1. Cells were treated with CCCP for 6 h. HA-VDAC1 expression rescues the defective mitochondrial targeting of Parkin caused by depletion of VDAC2. D, the fraction of cells with punctate, mitochondria-localized Parkin after 6 h of CCCP treatment was determined from randomized images of the experiments in B and C. E, as in B, except that VDAC1/3−/− MEFs expressing FLAG-tagged Parkin also expressed HA-VDAC3, and mitochondria were immunostained for HA-VDAC3 instead of Tom20. HA-VDAC3 expression rescues the defective mitochondrial targeting of Parkin caused by depletion of VDAC2. F, quantification as in D, except that images were from experiments shown in E for VDAC1/3−/− MEFs expressing FLAG-tagged Parkin and HA-VDAC3 and from experiments not shown for VDAC1/3−/− MEFs expressing FLAG-tagged Parkin (note that this cell line is different from the one in B and was used to generate VDAC1/3−/− MEFs expressing FLAG-tagged Parkin and HA-VDAC3). Scale bars, 20 μm. Data are means ± S.E. of three independent experiments (>100 cells for each experiment). *, p < 0.02; **, p < 0.001, denoting significant difference from the three other groups. Other comparisons are not significantly different (p > 0.05).
FIGURE 4.
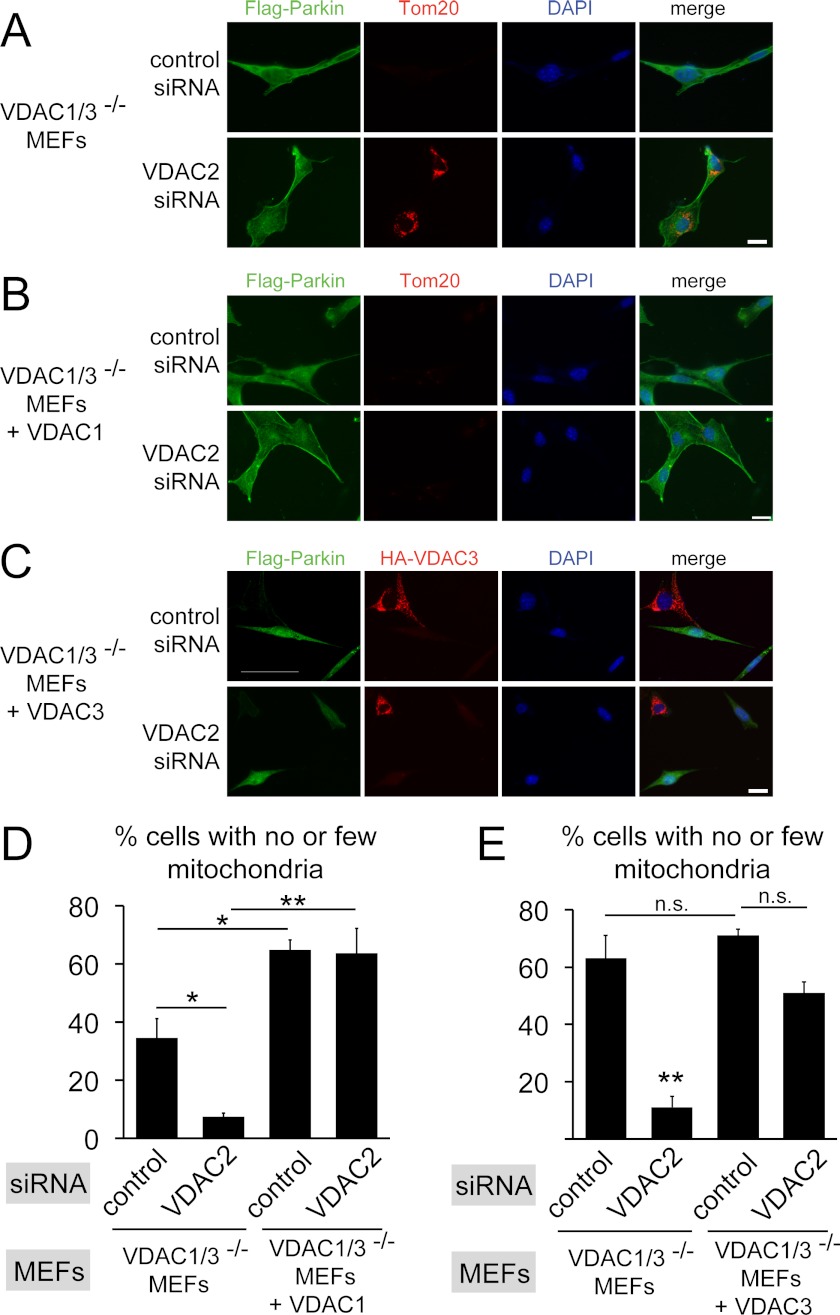
VDACs are necessary for efficient elimination of mitochondria. A, VDAC1/3−/− MEFs expressing FLAG-Parkin were transfected with the indicated siRNAs. 48 h after transfection, cells were incubated with 20 μm CCCP for 24 h and immunostained for Parkin and the mitochondrial marker Tom20. Absence of the Tom20 signal indicates that mitochondria are efficiently eliminated in cells transfected with control siRNA, but not when VDAC2 is depleted by siRNA. B, as in A except that HA-VDAC1 was expressed in VDAC1/3−/− MEFs expressing FLAG-tagged Parkin. Expression of HA-VDAC1 rescues the mitochondrial elimination defect caused by VDAC2 depletion. C, as in A except that HA-VDAC3 was expressed in VDAC1/3−/− MEFs expressing FLAG-tagged Parkin and cells were immunostained for HA-VDAC3 instead of Tom20. Expression of HA-VDAC3 rescues the mitochondrial elimination defect caused by VDAC2 depletion. Note that HA-VDAC3 is not expressed in all cells. Scale bars, 20 μm. D, the fraction of cells having no or few detectable mitochondria was determined from randomized images of the experiments in A and B by Tom20 immunofluorescence. Data are means ± S.E. (error bars) of three independent experiments (>100 cells each). *, p < 0.05; **, p < 0.01. E, the fraction of cells having no or few detectable mitochondria was determined from randomized images of the experiments in C and from data not shown for VDAC1/3−/− MEFs expressing FLAG-tagged Parkin. Note that this cell line is different from the one in A and was used to generate VDAC1/3−/− MEFs expressing FLAG-tagged Parkin and HA-VDAC3. This may explain the more efficient mitochondrial elimination compared with D. Data are means ± S.E. of three independent experiments (>100 cells each). **, p < 0.01, significantly different from the three other groups. Other comparisons are not significantly different (p > 0.05).
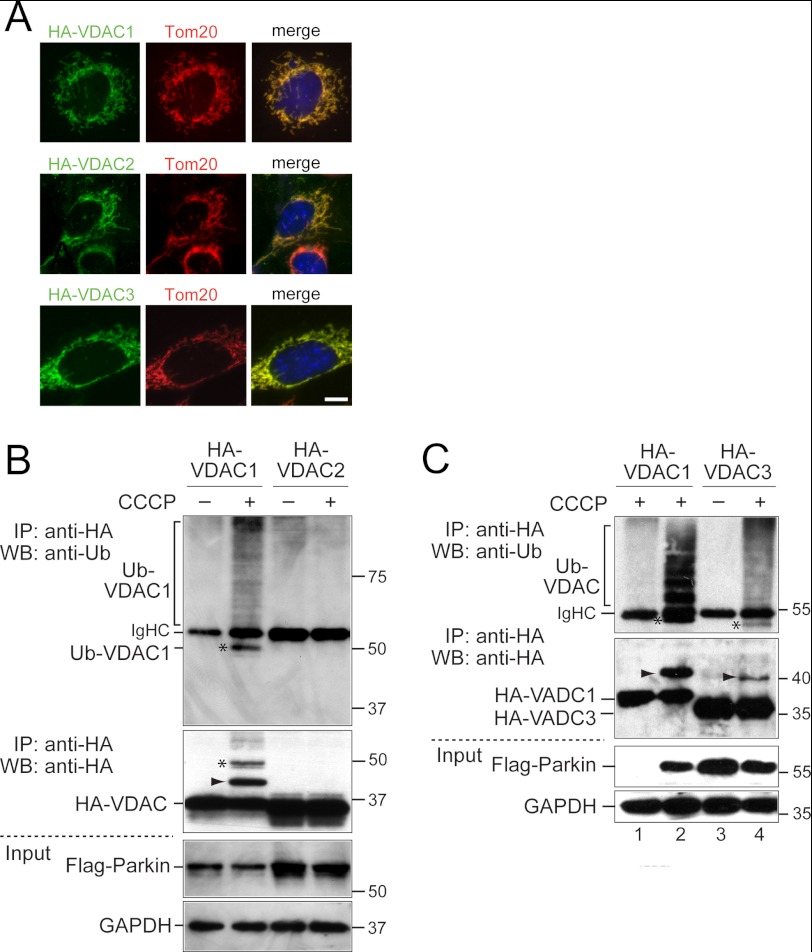
Because we detected the three VDACs as the top hits by mass spectrometry, we wanted to test whether VDACs may function redundantly to recruit Parkin. To this end, we expressed HA-tagged human VDAC1 (HA-VDAC1) in VDAC1/3−/− MEFs. Based on sequence comparison, HA-VDAC1 should be resistant to VDAC2 siRNAs. As expected, HA-VDAC1 localizes to mitochondria in VDAC1/3−/− MEFs as determined by co-localization with Tom20 and did not affect Parkin localization (Fig. 3A and supplemental Fig. S3A). We found that in VDAC1/3−/− MEFs expressing HA-VDAC1, Parkin recruitment to mitochondria was resistant to VDAC2 knockdown (Fig. 2, C and D), suggesting that VDAC1 as the sole VDAC can recruit Parkin. This experiment also confirmed that the effect of VDAC2 siRNA is specific. Expression of HA-VDAC1 in VDAC1/3−/− MEFs slightly increased the efficiency of Parkin targeting after CCCP treatment, but this effect was not statistically significant. Next, we determined that expression of HA-VDAC3 in VDAC1/3−/− MEFs also restored Parkin recruitment to mitochondria after VDAC2 knockdown (Fig. 2, E and F, and supplemental Figs. S3B and S4A). Together, these results suggest that the three VDACs can function redundantly to target Parkin to defective mitochondria.
FIGURE 3.
VDAC1 and VDAC3 but not VDAC2 are ubiquitinated in VDAC1/3−/− MEFs. A, VDAC1/3−/− MEFs expressing HA-VDAC1, HA-VDAC2, or HA-VDAC3 were immunostained with anti-HA and anti-Tom20 antibodies. HA-VDACs co-localize with Tom20, indicating correct mitochondrial localization of the HA-tagged VDACs. Scale bar is 10 μm. B, VDAC1/3−/− MEFs expressing FLAG-Parkin and either HA-VDAC1 or HA-VDAC2 were treated for 6 h with 20 μm CCCP or DMSO as a control. Cell lysates were immunoprecipitated (IP) with anti-HA antibodies and analyzed by Western blotting (WB) with anti-ubiquitin antibodies (1st panel) or anti-HA antibodies (2nd panel). Higher molecular mass bands corresponding to ubiquitinated VDACs are detected for HA-VDAC1 when cells are treated with CCCP in the anti-ubiquitin and anti-HA Western blots (Ub-VDAC1). The band marked with an arrowhead in the anti-HA blot is not consistently detected in anti-ubiquitin blots, and the band labeled with an asterisk in the anti-ubiquitin blot is not consistently detected in anti-HA blots. Note that the cell line expressing HA-VDAC2 has somewhat higher expression levels of VDAC and Parkin than the HA-VDAC1 cell line (2nd and 3rd panels). GAPDH was used as a loading control (4th panel). C, VDAC1/3−/− MEFs expressing HA-VDAC1 with or without FLAG-Parkin were treated as in B (lanes 1 and 2). No ubiquitinated, higher molecular mass bands of VDAC1 are detected in the absence of Parkin (lane 1). VDAC1/3−/− MEFs expressing FLAG-Parkin and HA-VDAC3 were treated as in B (lanes 3 and 4). Bands are labeled with an asterisk and arrowhead as in B. Note that HA-VDAC3 migrates faster than HA-VDAC1. IgHC indicates the immunoglobulin heavy chain of the anti-HA antibodies. Molecular masses are indicated in kDa.
Next, we wanted to exclude the possibility that VDAC2 in VDAC1/3−/− MEFs functioned in the targeting Parkin to defective mitochondria as a ubiquitination substrate of Parkin. Previous experiments with HeLa cells had established that human VDAC2, in contrast to VDAC1, is not ubiquitinated by Parkin (18). We confirmed these results for mouse VDAC2 in MEFs by expressing HA-tagged VDAC1 or 2 in VDAC1/3−/− MEFs also expressing FLAG-Parkin (Fig. 3). After CCCP treatment for 6 h, ubiquitinated HA-VDAC1 was detected as high molecular weight bands by immunoprecipitation with anti-HA antibodies and Western blotting with anti-ubiquitin antibodies. High molecular weight bands were also detected by Western blotting with anti-HA antibodies (Fig. 3B). HA-VDAC3 was also ubiquitinated after CCCP treatment (Fig. 3C). VDAC1 ubiquitination was dependent on Parkin because it was not detected in VDAC1/3−/− MEFs not expressing Parkin (Fig. 3C). In contrast, no ubiquitinated HA-VDAC2 was detected (Fig. 3B). Although VDAC2 is not a ubiquitination target of Parkin, we detected it as a Parkin interactor with similar efficiency as VDAC1 and 3 in our mass spectrometry analysis (Table 1). Together, these findings suggest that VDACs function redundantly to recruit Parkin to defective mitochondria and that targeting of Parkin to mitochondria and the interaction with VDACs do not require ubiquitination of VDACs.
VDAC-dependent Targeting of Parkin Is Required for Efficient Degradation of Mitochondria
Next, we asked whether VDAC-dependent recruitment of Parkin to mitochondria is required for CCCP-induced degradation of mitochondria. Recruitment of Parkin to mitochondria promotes their degradation by an autophagy mechanism, and typically, a significant fraction of cells expressing Parkin and treated with CCCP for 24 h have no or few detectable mitochondria (5). Prior to CCCP treatment of VDAC1/3−/− MEFs expressing FLAG-Parkin, all cells have detectable mitochondria by immunofluorescence with the mitochondrial marker protein Tom20. After treatment with CCCP for 24 h, more than one third of the cells had no or few detectable mitochondria, similar to published results (Fig. 4, A, D, and E). When VDAC2 was depleted in these cells prior to CCCP treatment, the fraction of cells that had lost their mitochondria was significantly reduced (Fig. 4, A, D, and E). Defective mitochondrial degradation due to depletion of VDAC2 by siRNA could be rescued by expressing HA-VDAC1 (Fig. 4, B and D) or HA-VDAC3 (Fig. 4, C and E). Expression of VDAC1 significantly increased the fraction of cells with no or few detectable mitochondria compared with cells not expressing VDAC1, perhaps due to overexpression of VDAC1. Together, these results support the idea that VDACs function redundantly to promote Parkin-dependent mitochondrial degradation, exclude the possibility of off-target effects of the VDAC2 siRNAs in these experiments, and suggest that the recruitment of Parkin to defective mitochondria by VDACs is necessary to efficiently degrade mitochondria in CCCP-treated cells.
DISCUSSION
Recent work in a variety of systems showed that a PINK1/Parkin pathway eliminates defective mitochondria by an autophagy mechanism (1, 2, 14). Our data support a model in which VDACs are part of this mechanism by recruiting Parkin to defective mitochondria. First, the three VDACs are the major mitochondrial interaction partners of Parkin after treatment of cells with CCCP, detectable by affinity purification of FLAG-tagged Parkin and mass spectrometry. Importantly, VDACs only co-immunoprecipitated with Parkin when cells were treated with CCCP, demonstrating that Parkin interacts with VDACs in a specific manner. Second, in the absence of all three VDACs after depleting VDAC2 by RNAi knockdown in VDAC1/3−/− MEFs, targeting of Parkin to defective mitochondria is impaired, and this could be rescued by expressing VDAC1 or VDAC3 in these cells. Third, depletion of VDAC2 in VDAC1/3−/− MEFs impaired elimination of defective mitochondria, and this defect could be rescued by expression of VDAC1 or VDAC3. Taken together, our results are consistent with the idea that VDACs can function redundantly to target Parkin to defective mitochondria to promote mitophagy.
In contrast to VDAC1 and VDAC3, VDAC2 is not a ubiquitination target of Parkin in HeLa cells and VDAC1/3−/− MEFs, yet VDAC2 alone is sufficient to support Parkin targeting and mitophagy (Figs. 2, 3, and 4 in this study) (17, 18). Ubiquitination of VDACs by Parkin should, therefore, not be necessary for mitophagy. Instead, we hypothesize that VDACs function by providing docking sites for Parkin on mitochondria by directly interacting with Parkin. Binding to VDACs could then trigger ubiquitination of mitochondrial target proteins, including mitofusin and Miro, as a step toward mitochondrial degradation. Our experiments do not distinguish whether the observed interaction between Parkin and VDACs is direct or indirect. Answering that question will require reconstitution of Parkin-VDAC binding with purified proteins in vitro. We consider an indirect interaction mediated by additional proteins less likely because the three VDACs were by far the most abundant Parkin-interacting proteins detected by mass spectrometry.
PINK1 is required to recruit Parkin to mitochondria by an unknown mechanism. Point mutations that disrupt the kinase activity of PINK1 do not recruit Parkin to mitochondria, suggesting that the kinase activity of PINK1 is required (17, 20, 23). It is possible that phosphorylation of Parkin by PINK1, observed in some cases but not others, is responsible for targeting Parkin to mitochondria (23, 29, 30, 33). Alternatively, PINK1 could phosphorylate a mitochondrial protein to promote the interaction of this protein with Parkin (1), and VDACs would be attractive phosphorylation targets.
Direct interaction between Parkin and PINK1 has been reported in some cases and could, in principle, be sufficient to target Parkin to mitochondria (2). However, whereas mutations in PINK1 that disrupt its kinase activity also disrupt PINK1 ability to recruit Parkin to mitochondria, the same mutations did not affect the reported interaction with Parkin (30). This is more consistent with the idea that the PINK1-Parkin interaction is not relevant for recruitment of Parkin to mitochondria (1). In support of this, our mass spectrometry analysis did not detect PINK1 in Parkin immunoprecipitations. Furthermore, a recent study found that targeting PINK1 to mitochondria, peroxisomes, or lysosomes was sufficient to recruit Parkin to these organelles, but in agreement with our data, no interaction between Parkin and PINK1 was detected in co-immunoprecipitation experiments or by blue native polyacrylamide gel electrophoresis (31). Whereas it is possible that the interaction between PINK1 and Parkin is weak and difficult to detect by various biochemical approaches, we favor a model in which Parkin interacts with some other molecule on mitochondria for efficient targeting, and VDACs are attractive candidates for this function. They are abundant proteins in the outer mitochondrial membrane and interact robustly and specifically with Parkin when mitochondria are defective. VDACs have also been found to provide functionally important binding sites for other proteins that are targeted from the cytosol to mitochondria under certain cellular conditions, for example hexokinase and proteins of the Bcl-2 family (27). We propose a similar function for VDACs to recruit Parkin to defective mitochondria to promote mitophagy.
Supplementary Material
Acknowledgments
We thank Dr. William J. Craigen (Baylor College of Medicine) for the VDAC1/3−/− MEFs; X. William Yang (UCLA) for a human Parkin cDNA clone; Alex van der Bliek for insightful discussions; Kathrin Plath, laboratory members, and Bernadett Papp for assistance with tissue culture approaches and FACS analysis.
This work was supported in part by National Institutes of Health Grant R01GM089778. This work was also supported by funds from the Jonsson Comprehensive Cancer Center at UCLA (to J. A. W.).

This article contains supplemental Figs. S1–S3.
- VDAC
- voltage-dependent anion channel
- CCCP
- carbonyl cyanide p-chlorophenylhydrazone
- DMSO
- dimethyl sulfoxide
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Narendra D. P., Youle R. J. (2011) Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid. Redox Signal. 14, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Springer W., Kahle P. J. (2011) Regulation of PINK1-Parkin-mediated mitophagy. Autophagy 7, 266–278 [DOI] [PubMed] [Google Scholar]
- 3. Lin M. T., Beal M. F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795 [DOI] [PubMed] [Google Scholar]
- 4. Schon E. A., Przedborski S. (2011) Mitochondria: the next (neurode) generation. Neuron 70, 1033–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Youle R. J., Narendra D. P. (2011) Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ziviani E., Tao R. N., Whitworth A. J. (2010) Drosophila Parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. U.S.A. 107, 5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poole A. C., Thomas R. E., Yu S., Vincow E. S., Pallanck L. (2010) The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/Parkin pathway. PLoS One 5, e10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gegg M. E., Cooper J. M., Chau K. Y., Rojo M., Schapira A. H., Taanman J. W. (2010) Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/Parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 19, 4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanaka A., Cleland M. M., Xu S., Narendra D. P., Suen D. F., Karbowski M., Youle R. J. (2010) Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191, 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rakovic A., Grünewald A., Kottwitz J., Brüggemann N., Pramstaller P. P., Lohmann K., Klein C. (2011) Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS One 6, e16746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glauser L., Sonnay S., Stafa K., Moore D. J. (2011) Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 118, 636–645 [DOI] [PubMed] [Google Scholar]
- 14. Vives-Bauza C., Przedborski S. (2011) Mitophagy: the latest problem for Parkinson's disease. Trends Mol. Med. 17, 158–165 [DOI] [PubMed] [Google Scholar]
- 15. Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y. L., Selkoe D., Rice S., Steen J., LaVoie M. J., Schwarz T. L. (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu S., Sawada T., Lee S., Yu W., Silverio G., Alapatt P., Millan I., Shen A., Saxton W., Kanao T., Takahashi R., Hattori N., Imai Y., Lu B. (2012) Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 8, e1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 18. Narendra D., Kane L. A., Hauser D. N., Fearnley I. M., Youle R. J. (2010) p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6, 1090–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Itakura E., Kishi-Itakura C., Koyama-Honda I., Mizushima N. (2012) Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J. Cell Sci. 125, 1488–1499 [DOI] [PubMed] [Google Scholar]
- 20. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jin S. M., Lazarou M., Wang C., Kane L. A., Narendra D. P., Youle R. J. (2010) Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Greene A. W., Grenier K., Aguileta M. A., Muise S., Farazifard R., Haque M. E., McBride H. M., Park D. S., Fon E. A. (2012) Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13, 378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morita S., Kojima T., Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 25. Vashisht A. A., Zumbrennen K. B., Huang X., Powers D. N., Durazo A., Sun D., Bhaskaran N., Persson A., Uhlen M., Sangfelt O., Spruck C., Leibold E. A., Wohlschlegel J. A. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Florens L., Carozza M. J., Swanson S. K., Fournier M., Coleman M. K., Workman J. L., Washburn M. P. (2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shoshan-Barmatz V., Ben-Hail D. (2012) VDAC, a multi-functional mitochondrial protein as a pharmacological target. Mitochondrion 12, 23–34 [DOI] [PubMed] [Google Scholar]
- 28. Baines C. P., Kaiser R. A., Sheiko T., Craigen W. J., Molkentin J. D. (2007) Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat. Cell Biol. 9, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim Y., Park J., Kim S., Song S., Kwon S. K., Lee S. H., Kitada T., Kim J. M., Chung J. (2008) PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 377, 975–980 [DOI] [PubMed] [Google Scholar]
- 30. Sha D., Chin L. S., Li L. (2010) Phosphorylation of Parkin by Parkinson disease-linked kinase PINK1 activates Parkin E3 ligase function and NF-κB signaling. Hum. Mol. Genet. 19, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lazarou M., Jin S. M., Kane L. A., Youle R. J. (2012) Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell 22, 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chaugule V. K., Burchell L., Barber K. R., Sidhu A., Leslie S. J., Shaw G. S., Walden H. (2011) Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H. I., Campbell D. G., Gourlay R., Burchell L., Walden H., Macartney T. J., Deak M., Knebel A., Alessi D. R., Muqit M. M. (2012) PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2, 120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.