Background: Succinate dehydrogenase (SDH) requires a covalent addition of FAD for catalytic function.
Results: Mutational analyses of Sdh1 implicate C-terminal region Arg residues involvement in covalent flavinylation and SDH assembly.
Conclusion: SDH assembly is dependent on FAD binding to Sdh1 but not covalent binding.
Significance: These results document the basis for the SDH deficiency and pathology seen with mutations in human Sdh1.
Keywords: Bioenergetics/Electron Transfer Complex, Electron Transport System (ETS), Flavoproteins, Membrane Biogenesis, Metalloenzymes, Mitochondria, Mitochondrial Diseases
Abstract
The enzymatic function of succinate dehydrogenase (SDH) is dependent on covalent attachment of FAD on the ∼70-kDa flavoprotein subunit Sdh1. We show presently that flavinylation of the Sdh1 subunit of succinate dehydrogenase is dependent on a set of two spatially close C-terminal arginine residues that are distant from the FAD binding site. Mutation of Arg582 in yeast Sdh1 precludes flavinylation as well as assembly of the tetrameric enzyme complex. Mutation of Arg638 compromises SDH function only when present in combination with a Cys630 substitution. Mutations of either Arg582 or Arg638/Cys630 do not markedly destabilize the Sdh1 polypeptide; however, the steady-state level of Sdh5 is markedly attenuated in the Sdh1 mutant cells. With each mutant Sdh1, second-site Sdh1 suppressor mutations were recovered in Sdh1 permitting flavinylation, stabilization of Sdh5 and SDH tetramer assembly. SDH assembly appears to require FAD binding but not necessarily covalent FAD attachment. The Arg residues may be important not only for Sdh5 association but also in the recruitment and/or guidance of FAD and or succinate to the substrate site for the flavinylation reaction. The impaired assembly of SDH with the C-terminal Sdh1 mutants suggests that FAD binding is important to stabilize the Sdh1 conformation enabling association with Sdh2 and the membrane anchor subunits.
Introduction
Succinate dehydrogenase (SDH),4 also known as succinate:quinone oxidoreductase, is a citric acid cycle enzyme that links directly to the aerobic respiratory chain. The enzyme catalyzes the FAD-dependent oxidation of succinate to fumarate coupled with the reduction of ubiquinone to ubiquinol. SDH is a heterotetrameric integral membrane protein complex. The eukaryotic enzyme is embedded within the mitochondrial inner membrane by a hydrophobic module associated with the catalytic module protruding into the mitochondrial matrix (1). The hydrophilic catalytic module consists of Sdh1 and Sdh2 subunits and the electron transfer cofactors. Succinate oxidation occurs in the FAD-containing Sdh1 with the abstracted electrons from the reaction shuttled via three iron-sulfur centers in Sdh2 to the ubiquinone reduction site (QP -proximal) at the interface of Sdh2 and the membrane anchor (1–4). The Sdh3/Sdh4 membrane domain contains a bound heme b moiety at the subunit interface of Sdh3 and Sdh4 with each providing one of the two axial His ligands, although the role of the heme in eukaryotic SDH is unresolved (5).
The FAD of Sdh1 is covalently attached at an active site His residue (2). This covalent bond increases the FAD redox potential by ∼60 mV to permit succinate oxidation (6). SDH is the major mitochondrial protein containing a covalent bound flavin (7). Sdh1 containing a H90S substitution is enzymatically inactive in succinate oxidation but assembles into a tetrameric complex that exhibits fumarate reductase activity (8). Fumarate reductase activity in SDH does not require covalent flavinylation. SDH is related to the bacterial fumarate reductase and both enzymes can catalyze succinate oxidation and fumarate reduction with different efficiencies (9). Flavinylation of Sdh1 was found to occur after import into the matrix and to be influenced by the presence of the iron/sulfur cluster subunit Sdh2 but largely independent of the membrane anchor (10). The presence of citric acid intermediates stimulated the flavinylation process (10).
The covalent addition of FAD was proposed to be autocatalytic (7), but recently, a dedicated assembly factor Sdh5 was identified that is required for covalent flavinylation (11). The role of Sdh5 in Sdh1 flavinylation was discovered by the interaction of the two proteins and the demonstration that sdh5Δ yeasts were devoid of SDH activity and lacked flavinylated Sdh1. The direct role of Sdh5 in Sdh1 flavinylation was shown by the enhanced covalent addition of FAD to recombinant Sdh1 expressed in bacteria in the presence of Sdh5. However, the mechanism by which the 22-kDa Sdh5 protein mediates flavinylation of Sdh1 remains unresolved.
The human Sdh5 ortholog, SDHAF2, was shown to have a corresponding role in covalent flavinylation in human cells (11). Mutations in SDHAF2 are associated with a neuroendrocrine tumor designated as paraganglioma. Mutations in the SDH structural genes are also detected in a range of tumors, including paraganglioma, pheochromocytomas, gastrointestinal stromal tumors, and renal cell carcinoma (12). Recently, the bacterial ortholog of Sdh5 was identified in the bacterium Serratia. The factor designated SdhE was shown to interact with the flavoprotein (SdhA) and independently associate with FAD (13). This is the first suggestion that the assembly factor may mediate Sdh1 flavinylation by presenting bound FAD. Whereas sdh5Δ cells exhibited a specific defect in SDH, the bacterial sdhE mutant showed a more pleiotropic defect, suggesting SdhE may flavinylate other flavoproteins (13).
SDH is one of many flavoproteins with a covalently bound cofactor. SDH has an 8α-N3-histidyl-FAD linkage, but other flavoproteins use cysteinyl-FAD or tyrosyl-FAD linkages (14). Two large families of covalent flavoproteins are the glucose oxidase/methanol oxidase family and the vanillyl-alcohol oxidase family. Within the vanillyl-alcohol oxidase family, FAD linkages to histidine, cysteine, and tyrosine are known, although no ancillary enzyme is known to catalyze the covalent addition. Covalent flavinylation with FAD is known to proceed nonenzymatically, albeit slowly.
In the present study, we identify essential arginines in the C-terminal tail in yeast Sdh1 that are required for flavinylation and SDH assembly. The impaired assembly of the tetrameric enzyme with these Sdh1 mutants correlates with the lack of FAD binding.
MATERIALS AND METHODS
Yeast Strains and Vectors
All Saccharomyces cerevisiae strains used in this study were derivatives of Trp303 (Mata ura3-1 ade2-1 trp1-1 his3-11,15 leu2-3 112). Deletion strains sdh1Δ, sdh2Δ, sdh3Δ, and sdh4Δ, in addition to plasmid pRS414 Sdh2-His6Myc2, were kindly provided by Dr. Jared Rutter at the University of Utah. Additional deletion strains (flxΔ, sdh5Δ, sdh3Δsdh4Δ double) were constructed by homologous recombination using either the KanMX4 or the HIS3MX6 disruption cassettes (15). The C-terminal mutants of SDH1 along with SDH1 WT under the control of its own promoter and CYC1 terminator were expressed in sdh1Δ strain or subcloned into integrating pRS405 vectors for chromosomal expression. The integrating constructs were further digested with AflII and integrated at the leu2-3,112 locus of sdh1Δ to be able to express wild type and mutants SDH1 chromosomally. All integrated strains were confirmed by PCR analysis of the locus. The C-terminal point mutations were introduced by QuikChange mutagenesis PCR system (Agilent Technology). All mutations were confirmed by DNA sequencing. Yeast strains were transformed using lithium acetate. Strains were grown in synthetic complete medium lacking the amino acid(s) to maintain plasmid selection with either 2% galactose or 2% glycerol/lactate as the carbon source.
For carbon swap cultures overnight, 50-ml glucose-grown cultures were used to inoculate 1 liter of medium containing 2% galactose as the carbon source. Cells were grown to an A600 nm ∼ 0.4 to 0.5 and harvested in a sterile manner. The cell pellet was resuspended in fresh medium containing 2% glycerol/lactate as the carbon source and grown for ∼10 h (∼3–4 doublings) before harvesting.
Mitochondria Isolation
Intact mitochondria were isolated using previously described methods of Glick and Pon (16) and Diekert et al. (17). For HPLC experiments, isolated mitochondria were further purified using ultracentrifugation through a Histodenz (Sigma Aldrich) step gradient (14 and 22%). Total mitochondrial protein was quantified using either the Bradford (18) or the bicinchoninic acid assays (19).
Immunoblotting and Blue-native PAGE
Steady-state levels of mitochondrial proteins were analyzed using the NuPAGE Bis-Tris gel system (Invitrogen) using MES as the buffer system. Proteins were subsequently transferred to nitrocellulose membrane and probed using the indicated primary antibodies and visualized using enhanced chemiluminescence (ECL) reagents with horseradish peroxidase-conjugated secondary antibodies. Primary antibodies were obtained from the following: anti-Sdh1, Sdh2, Sdh3, and Sdh5 were generated in this study (21st Century Biochemicals). Anti-HA, anti-Myc, and anti-porin were purchased from Rockland, Roche Applied Science, and Molecular Probes, respectively. Anti-F1 ATP synthase was a generous gift from Alex Tzagoloff.
Analysis of yeast mitochondrial native membrane complexes was performed using the native PAGE gel system (Invitrogen) that is based on the blue-native polyacrylamide gel electrophoresis (BN-PAGE) technique developed by Schägger and von Jagow (20). Solubilized mitochondria (1% digitonin for 20–40 μg of mitochondrial protein) were separated on either 3–12% or 4–16% Bis-Tris native gels, transferred to PVDF membrane, and analyzed by immunoblotting.
Hemin-agarose Pulldown Assays and Immunoprecipitation
Purified mitochondria (0.75 to 1 mg of total protein) were solubilized using either 0.1% DDM or 1% digitonin in PBS buffer and clarified at 20,000 × g for 5 min. For hemin-agarose pulldown assay, the clarified lysates were incubated with 20 to 40 μl of hemin-agarose beads (Sigma), which had been prereduced using DTT. Binding was performed for 2 h or overnight at 4 °C. Beads were subsequently washed (15 min × 4) with PBS buffer, 0.1% DDM or 1% digitonin, ± DTT, and eluted with 2× SDS-PAGE loading buffer. For immunoprecipitation, the clarified lysates were incubated with anti-HA or anti-Myc beads for 2 h or overnight at 4 °C, washed four times with PBS buffer + 0.1% DDM or 1% digitonin, and eluted with 2× SDS-PAGE loading buffer. The clarified lysate, final wash, and eluate fraction were analyzed by SDS-PAGE and immunoblotting. Bands from Coomassie-stained SDS-PAGE gel from hemin-agarose pulldown assay were also excised for identification using MS analysis of the tryptic digests.
SDH Activity Assay
SDH activities in isolated yeast mitochondria were measured spectrophotometrically at 22 °C following the succinate-dependent, phenazine methosulfate-mediated reduction of either 2,6-dichlorophenolindophenol or cytochrome c as the terminal electron acceptor described for intact mitochondria (21).
Analysis of Sdh1-bound FAD and Total Mitochondrial FAD Levels
Levels of covalently attached FAD to Sdh1 were analyzed as described previously (11, 22). Mitochondrial proteins were resolved on SDS-PAGE, and the gel was placed in a 10% acetic acid solution for 20 min to oxidize flavin. The FAD band was visualized upon exposure to UV light using a Bio-Rad Gel Doc transilluminator.
Freely soluble mitochondrial FAD levels were analyzed as described previously (23). Purified mitochondria (500 μg) were pelleted at 20,000 × g for 10 min and resuspended in 500 μl of deionized water and boiled at 105 °C for 10–15 min to precipitate proteins. The resulting yellowish supernatant was clarified at 20,000 × g for 5 min and analyzed on a Waters Sunfire C18 HPLC column (5 μm 4.6 × 150 mm) equilibrated with 15% MeOH in 10 mm ammonium acetate, pH 6.4. Flavin was eluted using a linear MeOH gradient to 75% in 10 mm ammonium acetate, pH 6.4, for 25 min and monitored at 450 nm. Elution times were compared with FAD and FMN flavin mononucleotide standards (Sigma). The concentration of FAD for the peak area was determined using an extinction coefficient of 11,300 m−1 cm−1 at 450 nm (9).
RESULTS
Adsorption of Sdh1 on Heme Agarose
The focus on SDH flavinylation initiated from a goal to isolate heme-binding proteins within the mitochondria using affinity purification with a heme-agarose matrix. Detergent-solubilized mitochondrial lysates were chromatographed on heme-agarose affinity beads either in the oxidized (Fe3+; hemin) or reduced (Fe2+; heme) state (Fig. 1A). Proteins adsorbed on the heme matrix were eluted by SDS treatment and analyzed by SDS-PAGE. With reduced heme-agarose, a single major band was apparent with Coomassie staining that was identified as Sdh1 by mass spectrometry. The 67-kDa band was further verified to be Sdh1 by immunoblotting using Sdh1 antisera. Subunits Sdh2 and Sdh3 were also detected with their respective antiseras but in less abundance compared with Sdh1 (data not shown). The adsorption of Sdh1 on heme-agarose was competed by preincubation of the lysate with free heme, and no Sdh1 was retained on activated agarose beads lacking the heme moiety.
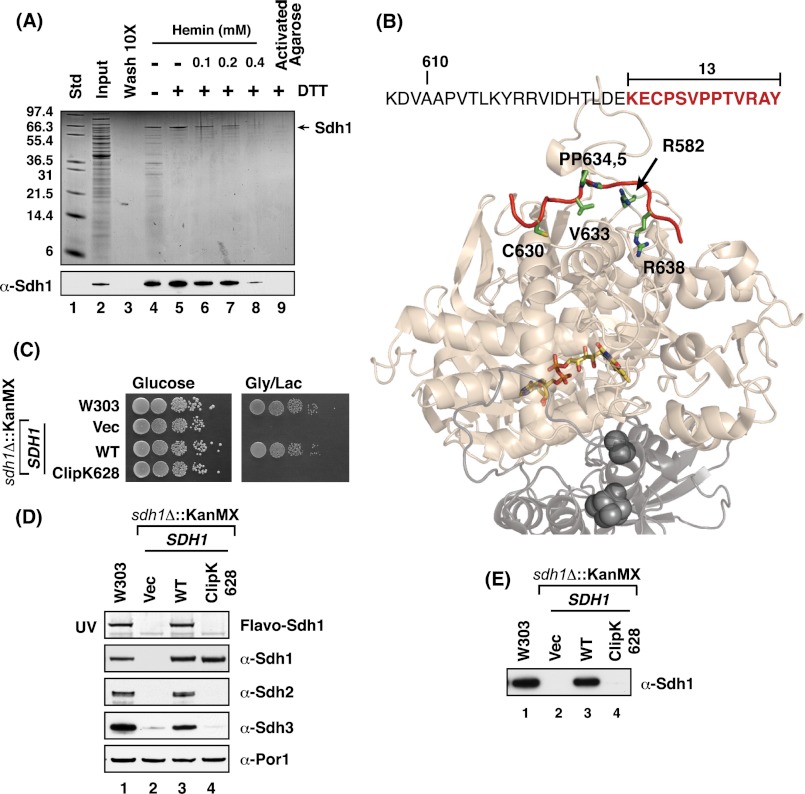
FIGURE 1.
Subunit 1 of SDH (Sdh1) binds hemin in vitro, and truncation of its last 13 residues abrogates this heme binding and renders the cells respiratory defective. A, Coomassie-stained SDS-PAGE gel of hemin-agarose purified Sdh1 from DDM-solubilized mitochondria ± DTT (lane 4 versus 5) and in the presence of increasing concentrations of free hemin as a competitor to the hemin-agarose beads (lanes 6–8). Activated agarose beads conjugated to a six-carbon linker (minus hemin) failed to capture Sdh1 (lane 9) std, protein standards; wash, final wash of the resin concentrated 10-fold. B, the last 13 residues of Sdh1 is a featureless strand (highlighted in red) located on the surface of the protein and away from the buried FAD cofactor (colored sticks). Sdh1 is shown in light beige, Sdh2 is shown in light gray, and its iron-sulfur clusters are shown as dark gray spheres. PyMOL was used to generate the figure from Protein Data Bank code 2H88 (avian). C, WT and sdh1Δ strains transformed with vectors (Vec) encoding full-length and truncated Sdh1 were spotted on fermentable (glucose) and non-fermentable (glycerol/lactate) medium with serial dilution and incubated at 30 °C. Cells with truncation of the last 13 residues in Sdh1 (Clip K628) failed to grow on respiratory medium. D, SDS-PAGE and immunoblot analysis of mitochondria isolated from strains in B. Covalent flavinylation of Sdh1 is lost in cells with truncation of the last 13 residues in Sdh1 (Clip K629) as analyzed by UV illumination of SDS-PAGE gel. Immunoblots show that steady-state levels of Sdh2 and Sdh3 are also affected by C-terminal truncation. E, immunoblot analysis of hemin-agarose purification of Sdh1 using mitochondria isolated from strains indicated in B shows that heme binding is lost in the Sdh1 C-terminal truncate (Clip K629). Flavo, flavoprotein. KanMX is the deletion cassette.
The retention of Sdh1 on heme-agarose suggested Sdh1 may possess a heme-binding motif. Inspection of the yeast Sdh1 protein sequence showed a single CP motif in the C-terminal tail found in a subset of heme-binding proteins (Fig. 1B). This C-terminal tail strand lies on the surface of Sdh1 and is located far from the buried FAD cofactor. Deletion of a C-terminal 13-residue segment in Sdh1 generated a truncate (designated ClipK628) that was stably expressed, although cells harboring this truncate failed to propagate on non-fermentable carbon sources (Fig. 1C). The presence of the Sdh1 truncate abrogated assembly of the tetrameric SDH enzyme and steady-state levels of Sdh2 and Sdh3 were markedly attenuated (Fig. 1D). The truncated Sdh1 was not flavinylated as shown by the lack of a FAD fluorescence band after SDS-PAGE (10). Chromatography of detergent lysates on the mutant cells on heme-agarose did not result in any retention of the Sdh1 truncate on the column (Fig. 1E).
C-terminal Segment of Sdh1 Is Critical for Flavinylation
Site-directed mutagenesis was carried out to map the residues responsible for flavinylation and heme-agarose binding. C630A, V633S, or a double P634A,P635A mutation in Sdh1 had no effect of glycerol/lactate growth (Fig. 2A). Furthermore, the C630A mutant in the CP motif did not attenuate SDH activity or compromise steady-state protein levels (Fig. 2B). Heme-agarose binding was also unaffected (Fig. 2C). However, cells harboring a mutant allele of Sdh1 with a R638A substitution were compromised in glycerol/lactate growth (Fig. 2A) and SDH activity (Fig. 2D). Flavinylation of the mutant Sdh1 was attenuated by ∼60% relative to the WT subunit. Assembly of the mutant Sdh1 into the tetrameric complex was not impaired as seen by BN-PAGE (Fig. 2E). The R638A mutant Sdh1 retained the ability to associate with the heme-agarose matrix (Fig. 2F).
FIGURE 2.
Double mutation of C630A and R638A residues leads to loss of respiratory growth and assembly of the SDH complex and loss of heme binding to Sdh1. A, growth test of strains with chromosomally integrated SDH1 C-terminal point mutants. Cells possessing R638A substitutions showed growth impairment on fermentable medium with the double C630A,R638A mutant showing the greatest growth defect. B, upper panel: SDH activities of isolated mitochondria from sdh1Δ strains transformed with vectors (vec) encoding WT and C630A Sdh1 mutant. Activities expressed as a percentage of WT (n ≥ 3 ± S.D.). Lower panels, corresponding immunoblot analysis of mitochondria isolated from above strains showing steady-state levels of covalent flavinylation (UV illumination) and SDH subunits. C, immunoblot analysis of hemin-agarose purification of Sdh1 using mitochondria isolated from strains indicated in B. D, upper panel: SDH activities of isolated mitochondria from strains expressing chromosomally integrated SDH1 C-terminal point mutants expressed as a percentage of WT (n ≥ 3 ± S.D.). Lower panel: steady-state of flavinylated-Sdh1 and SDH complex subunits in corresponding mitochondria of upper panel. The flavoprotein (Flavo) % WT is a relative comparison to WT based on band density as quantified by ImageJ software. E, BN-PAGE immunoblots of isolated mitochondria from B. 20 μg of isolated mitochondria were solubilized in 1% digitonin and separated on 3–12% native PAGE. Native protein complexes were transferred onto PVDF membrane and probed with antibodies to Sdh1 and F1 (loading control). F, immunoblot analysis of hemin-agarose purification of Sdh1. DDM-solubilized mitochondria lysates in B shows that heme binding is lost in the double C630A,R638A mutant.
A double C630A,R638A mutant revealed a more impaired phenotype than the single R638A mutant. The double mutant in Sdh1 failed to support SDH assembly of the tetrameric complex (Fig. 2E), resulting in a marked drop in steady-state levels of Sdh2 and Sdh3 (Fig. 2D). The mutant Sdh1 exhibited no flavinylation, although the mutant Sdh1 was stably expressed. Furthermore, the double mutant failed to associate with the heme-agarose matrix (Fig. 2F).
Sdh1-Sdh5 Interaction in the Sdh1 C-terminal Mutants
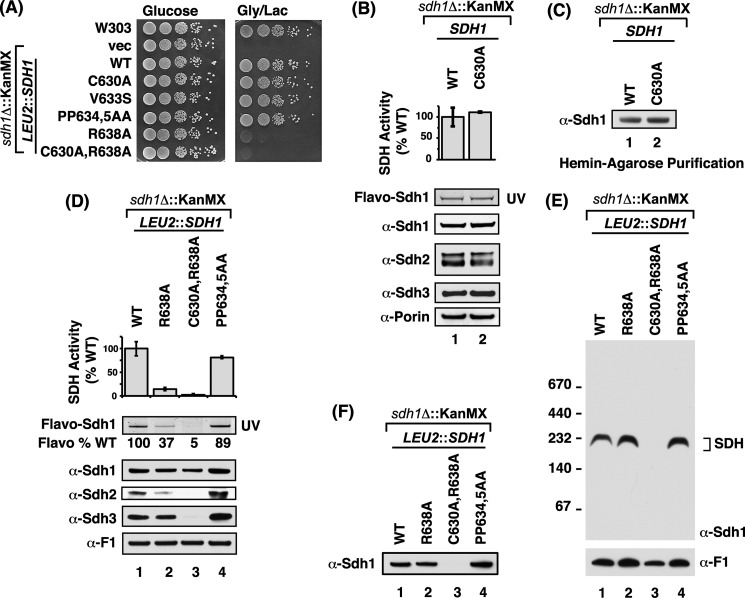
The diminished flavinylation in the C-terminal Sdh1 mutants raised the possibility that the mutations attenuated the known interaction of Sdh1 and Sdh5 that is important in Sdh1 flavinylation. The cellular level of Sdh5 has been observed to be dependent on Sdh1 (11) (i.e. Sdh5 is destabilized in cells lacking Sdh1). Steady-state levels of Sdh5 were tested in sdh1Δ cells expressing Sdh1 mutant alleles (Fig. 3A). A significant diminution in Sdh5 levels was apparent in cells with the double C630A,R638A Sdh1 mutant.
FIGURE 3.
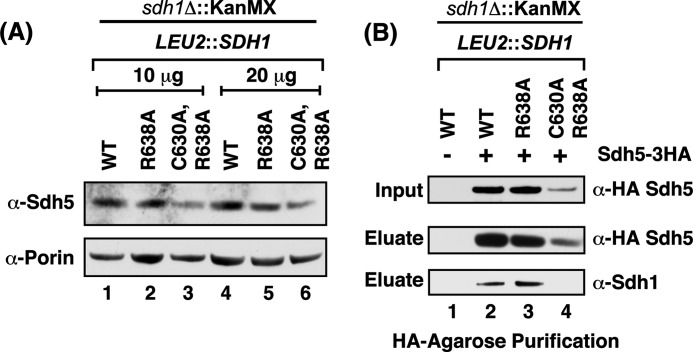
The steady-state level of Sdh5 is diminished in the C630A,R638A double mutant. A, steady-state immunoblot analysis of endogenous Sdh5 from isolated mitochondria from strains expressing chromosomally integrated SDH1 C-terminal mutants. B, immunoprecipitation of HA-tagged Sdh5. Isolated mitochondria from cells expressing exogenous HA-tagged Sdh5 on top of the strains in A were solubilized in digitonin and subject to IP. The Sdh1-Sdh5 interaction was analyzed by SDS-PAGE immunoblot analysis of the HA-agarose purification eluate and probed with antibodies to HA (Sdh5) and Sdh1.
The interaction of Sdh1 and Sdh5 is apparent by co-immunoprecipitation. Mitochondria from cells containing HA-tagged Sdh5 and wild-type Sdh1 were solubilized with digitonin and incubated with anti-HA beads. The immunoprecipitation of Sdh5 led to the co-purification of Sdh1 in the WT as expected (Fig. 3B). This was also true with Sdh1 containing the single R638A mutation. Attempts to test binding of Sdh5 to the double C630A,R638A Sdh1 mutant were compromised by the attenuated steady-state levels of Sdh5 in the Sdh1 mutant cells. Although the IP of Sdh5-HA failed to show any Sdh1, the reduction in input Sdh5 makes the absence of Sdh1 in the co-immunoprecipitate difficult to interpret.
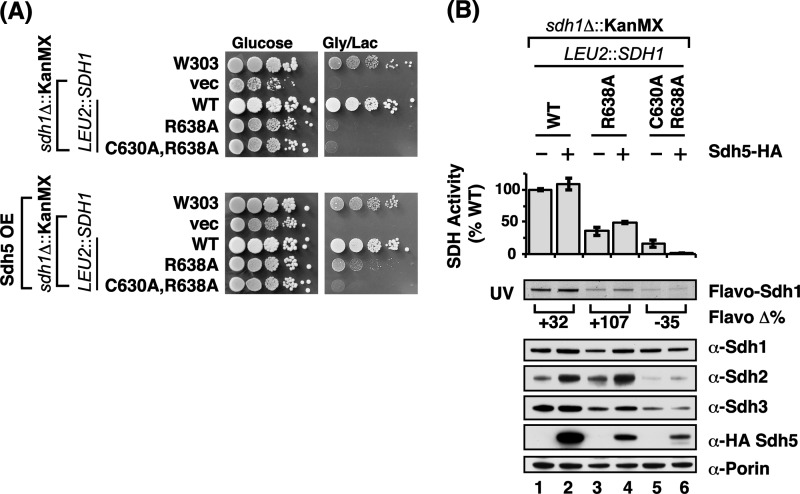
We evaluated whether the impaired SDH function observed in sdh1Δ cells expressing Sdh1 mutant alleles was suppressed by overexpressing SDH5 (Fig. 4A). Expression of SDH5 from a low-copy vector in the Sdh1 mutant cells did not significantly increase total Sdh5 levels as measured by immunoblotting with antisera to Sdh5 (data not shown). However, the expression of SDH5 gave a modest improvement in respiratory growth (Fig. 4A) as well as SDH activity (Fig. 4B) in cells harboring the R638A mutant Sdh1. Sdh1 flavinylation was also partially elevated by overexpression of Sdh5. The respiratory growth enhancement was not further increased by overexpression of SDH5 using a high-copy vector (data not shown). In contrast, no rescue was observed in growth or SDH activity of cells containing the double C630A,R638A Sdh1 mutant. The effect of overexpression of Sdh5 in cells with the Sdh1 double mutant actually led to a diminution in SDH activity rather than any restoration.
FIGURE 4.
Overexpression of Sdh5 partially restores growth of the single R638A mutant; the double C630A,R638A is unaffected. Similar effects are observed for steady-state covalent flavinylation and protein levels. A, growth test of strains with chromosomally integrated SDH1 C-terminal point mutants minus (top panel) or plus (bottom panel) low-copy vector (vec) expressing Sdh5. Cells were grown on glucose (fermentable) and glycerol/lactate (non-fermentable) media at 30 °C. B, upper panel: SDH activities of isolated mitochondria from strains in A expressed as a percentage of WT (n ≥ 3 ± S.D.). Lower panels: corresponding immunoblot analysis of mitochondria isolated from above strains showing steady-state levels of covalent flavinylation levels (UV illumination) and SDH subunit. The flavoprotein (Flavo) Δ% is a relative band density (quantified using ImageJ software) comparison of the Sdh5 overexpressing strain to that of the non-overexpressing strain.
Isolation of Intragenic SDH1 Suppressors
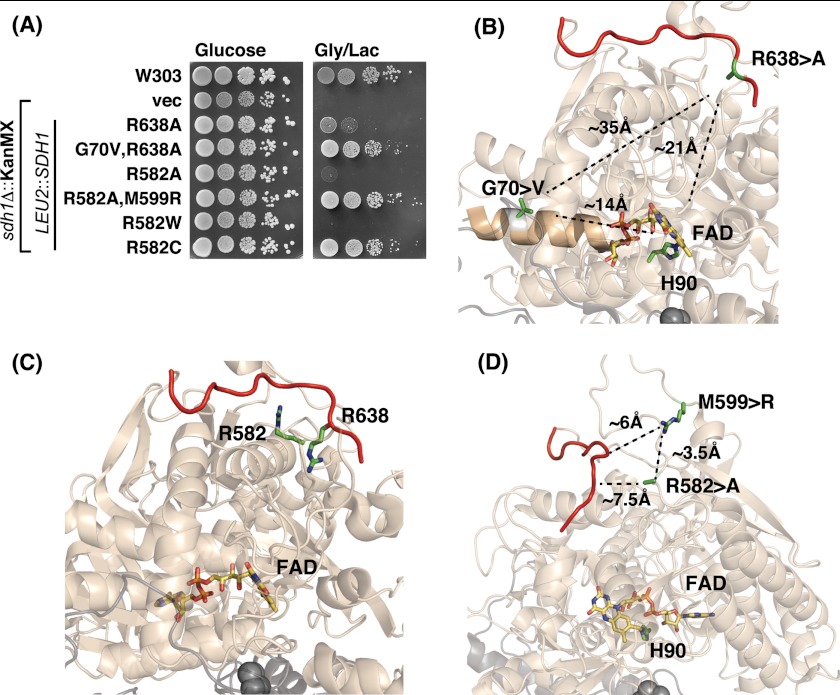
In the absence of a more marked rescue of the respiratory defect of R638A Sdh1 mutant cells by overexpression of SDH5, we isolated genetic suppressors in the mutant cells to gain possible insight into the mechanism underlying the point mutants. In addition to focusing on the R638A mutant, we analyzed a Sdh1 mutant in a second conserved arginine residue (Arg582) that is spatially adjacent to Arg638. The corresponding residue in human SDHA is Arg589 and a R589W mutation was reported in a patient afflicted with paraganglioma (24). We generated R582W and R582A mutations in yeast Sdh1 and observed that both substitutions resulted in lack respiratory growth (Fig. 5A).
FIGURE 5.
Spontaneous intragenic second-site suppressors of the Sdh1 flavinylation defect shows restoration of growth. A, growth test of intragenic second-site spontaneous suppressors to Sdh1 Arg mutants. The plasmid-borne SDH1 second site mutations were integrated chromosomally into sdh1Δ strains at LEU2 locus along with the original Arg mutation and tested for growth on fermentable (glucose) and non-fermentable (glycerol/lactate) medium at 30 °C. B, PyMOL-generated representation of avian SDH (Protein Data Bank code 2H88) showing the two critical Arg residues that are located on (Arg638) or near (Arg582) the C terminus (red strand) that when mutated causes a covalent flavinylation defect. C, the second-site mutation G70V that restores the growth defect of the R638A mutation lies on a helix that makes contact with the phosphate group of FAD but lies 35 Å away from the Arg638 and 14 Å away from FAD. PyMOL generated representation of avian SDH (Protein Data Bank code 2H88). D, the second-site mutation M599R is spatially close to the C-terminal tail (∼6 Å) of Sdh1 and to the original R582A (∼3.5 Å) mutation. In eukaryotes, the residue corresponding to Met599 in yeast has already been replaced to Arg. PyMOL generated figure using avian SDH (Protein Data Bank code 2H88). vec, vector.
Plating sdh1Δ cells expressing either R638A, R589A, or R589W Sdh1 from high-copy plasmids at high density on glycerol/lactate medium resulted in the appearance of papillae that contained mutants proficient in respiratory growth. The suppressors failed to respire in each case when the plasmid expressing the Sdh1 point mutation was shed, suggesting intragenic suppression. Rescue of the plasmid in Escherichia coli and retransformation in parent sdh1Δ cells resulted in glycerol/lactate growth.
DNA sequencing of the R638A mutant SDH1 gene revealed a second-site mutation resulting in a G70V substitution. The respiratory competency of the G70V,R638A Sdh1 mutant is shown in Fig. 5A. Gly70 is located 35 Å from Arg638 but is close to the FAD. It is also on the opposite face from the His90 that forms the covalent bond (Fig. 5B). SDH activity was restored in these second-site mutant cells (Fig. 6A).
FIGURE 6.
Spontaneous intragenic second-site suppressors of the Sdh1 flavinylation defect restores SDH activity, SDH assembly, and heme binding to Sdh1. A, upper panel: SDH activities of isolated mitochondria from strains in Fig. 5A expressed as a percentage of WT (n ≥ 3 ± S.D.) when cells were grown initially on 2% glucose and swapped to 2% glycerol/lactate (carbon swap; see “Material and Methods”). Lower panels: corresponding immunoblot analysis of mitochondria isolated from above strains showing steady-state levels of covalent flavinylation levels (UV illumination) and SDH subunit. B, corresponding blue native immunoblot analysis of mitochondria isolated from strains in A. The streaking in lane 3 (R638A) is likely a result of fractional amounts of soluble aggregates of Sdh2 formed in this mutant background. C, BN-PAGE immunoblot analysis of mitochondria isolated from selected strains in A but cultured in 2% glycerol/lactate showing the full restoration of complex assembly under respiratory condition for the two spontaneous suppressors (R582C and the double R582A,M599R). D, hemin-agarose purification of Sdh1 from isolated mitochondria from selected strains in C showing the restored heme-binding to Sdh1 in the spontaneous suppressors (R582C and the double R582A,M599R) but not in the original point mutant (R582A). Flavo, flavoprotein; vec, vector.
The cultures used for the studies in Fig. 6 were propagated using a carbon swap protocol in which cultures initially grown in glucose to an A600 nm ∼ 0.5 were switched to glycerol/lactate for the last 10 h of the experiment. Under these conditions, even the sdh1Δ cells maintained viability during the last phase of growth. Propagation of sdh1Δ cells with the R638A Sdh1 mutant on galactose medium rather than the carbon swap protocol resulted in markedly reduced SDH activity as seen in Figs. 2D and 4B; however, the presence of the G70V,R638A Sdh1 suppressor mutant did not exhibit enhanced SDH activity. The abundance of wild-type and mutant SDH complexes is markedly enhanced in glycerol/lactate medium relative to galactose medium (data not shown).
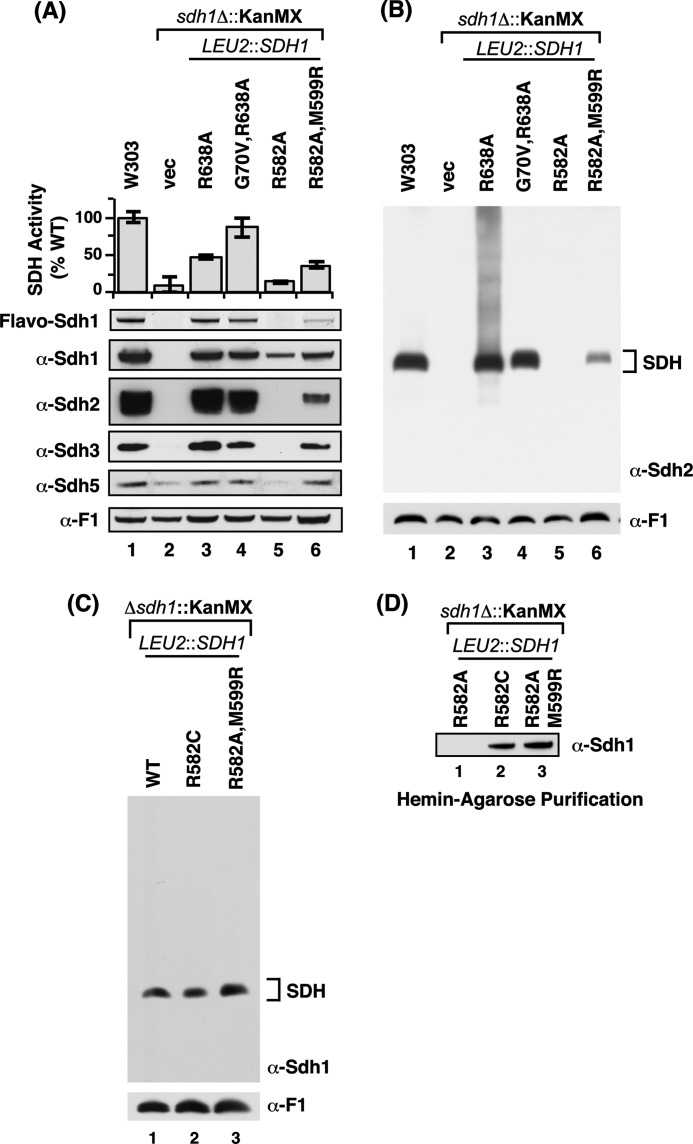
The genetic suppressor screen carried out with the R582A Sdh1 mutant cells resulted in the recovery of a second site suppressor with a M599R Sdh1 substitution in addition to the original R582A mutation (Fig. 5A). Met599 is spatially close to Arg582 and Arg638 (Fig. 5, C and D). The double R582A,M599R Sdh1 mutant was catalytically active unlike the R582A single mutant (Fig. 6A). Steady-state levels of Sdh1, Sdh2, and Sdh3 were restored. The suppressor was able to partially assemble, unlike the R582A single mutant, into a tetrameric SDH complex that could be visualized upon extended exposure of the blue-native immunoblot (Fig. 6B). In addition, the double mutant was competent to bind the heme-agarose matrix (Fig. 6D). The suppressor mutation resulted in a stabilized Sdh5 polypeptide.
A second site suppressor was also found for the non-functional R582W Sdh1 mutant (Fig. 5A). The suppressor mutant consisted of a conversion of the Trp to a Cys residue (Fig. 5A). The R582C Sdh1 mutant is active and assembles into the tetrameric complex (Fig. 6C) and is capable of adsorption on heme-agarose beads (Fig. 6D). Thus, Sdh1 with either an Arg or Cys at sequence position 582 is functional.
Role of FAD in SDH Complex Assembly
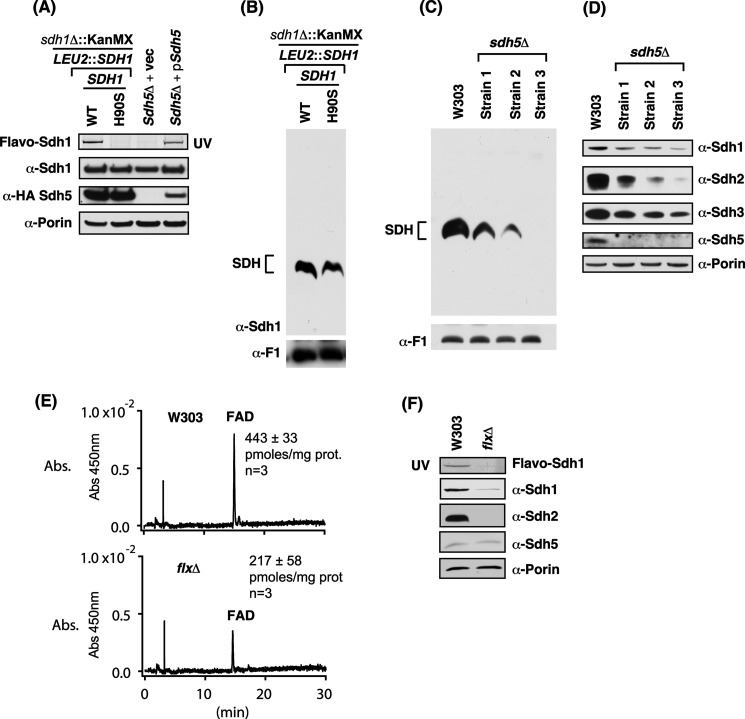
The lack of assembly in the R582A and the double C630A,R638A mutant Sdh1 and the lack of Sdh1 flavinylation in those mutants raised the question of whether SDH assembly is dependent on flavinylation. Yeast lacking Sdh5 were reported to be impaired in SDH assembly (11), but cells containing a mutant H90S Sdh1 mutant with a replacement of the histidyl residue forming the covalent FAD adduct was able to assemble into the tetrameric complex (8). We confirmed the latter result; the H90S Sdh1 yeast lacked any observable covalent FAD (Fig. 7A), yet it exhibited a tetrameric complex on BN-PAGE (Fig. 7B). Although no covalent FAD was seen, Robinson et al. (8) reported that FAD was still associated non-covalently with the enzyme. Sdh5 levels were normal in cells containing the H90S mutant Sdh1 (Fig. 7A).
FIGURE 7.
Covalent flavinylation of Sdh1 is not required for assembly of SDH as indicated by sdh5Δ and H90S Sdh1 mutants. However, Sdh1-FAD binding is likely required for stability of Sdh1 and SDH assembly. A, steady-state immunoblot analysis of isolated mitochondria from sdh1Δ strains expressing either wild type Sdh1 or H90S substitution (lanes 1 and 2) and sdh5Δ cells transformed with the indicated vectors (vec; lanes 3 and 4). B, corresponding BN-PAGE immunoblot analysis of mitochondria from A showing the assembly of SDH even in the absence of covalent flavinylation. C, BN-PAGE immunoblot analysis of mitochondria from WT and three different sdh5Δ cells showing the assembly of SDH even in the absence of covalent flavinylation. D, steady-state immunoblot analysis of isolated mitochondria from C. E, HPLC analysis of FAD levels in purified mitochondria from WT and flxΔ strains showing the ∼50% decrease in FAD levels in the flxΔ strain. F, steady-state SDS-PAGE UV and immunoblot analysis of isolated mitochondria from strains in E. Note the dramatic decrease in Sdh1 and the absence of Sdh2 in the flxΔ strain likely resulting from the decrease in mitochondrial FAD levels. Abs., absorbance. prot, total mitochondrial protein.
In the case of sdh5Δ cells, despite no covalent flavinylation (Fig. 7A), we observed varying degrees of SDH assembly in multiple independent experiments with three different sdh5Δ strains (Fig. 7C). This variability occurs within a given deletion strain. The variable assembly states seen by BN-PAGE may imply that the SDH complex is unstable and dissociates under the Coomassie gel conditions. In these mitochondrial lysates, we quantified steady-state Sdh2 and Sdh3 levels to assess the in vivo stability of the SDH complex (Fig. 7D). The level of Sdh2 correlated with the abundance of the assembled SDH complex. Thus, it appears likely that the complex is destabilized in vivo and that an equilibrium exists between assembly and disassembly in which some mutants are more prone to the latter step.
A correlation between SDH assembly and Sdh1 flavinylation is also seen in flx1Δ cells. Flx1 is a mitochondrial carrier protein implicated in FAD utilization (23, 25, 26). Sdh1 flavinylation is impaired in yeast lacking the mitochondrial carrier Flx1 (23, 25, 26). Although some uncertainty exists on whether Flx1 functions to import or export FAD from mitochondria, a careful quantitation of matrix FAD levels in flx1Δ cells showed a marked diminution (Fig. 7E) as was reported previously (23). Steady-state levels of Sdh1 and Sdh2 were markedly attenuated in the flx1Δ cells, yet Sdh5 levels were normal (Fig. 7F). As expected from the reduced Sdh1 and Sdh2 levels, no assembled SDH was observed on BN-PAGE (data not shown).
Role of Other SDH Subunits in Sdh1 Flavinylation
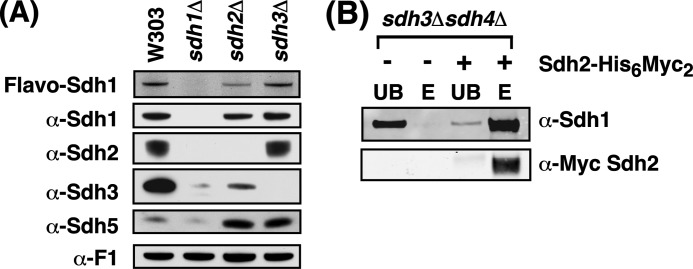
In the pioneering work of Bernard Lemire on SDH assembly and flavinylation, Sdh1 flavinylation was shown to occur upon mitochondrial import and proteolytic processing of Sdh1. In a pulse-chase study, the flavinylation of Sdh1 was assessed (10) in strains lacking one of the other SDH subunits to assess the dependence of Sdh1 flavinylation on the presence of other subunits (10). Robinson and Lemire (10) reported that Sdh1 flavinylation is independent of the membrane anchor subunits Sdh3 and Sdh4 and partially attenuated in cells lacking Sdh2. We extended their observations and found that steady-state Sdh1 levels were normal in cells lacking Sdh2 and the membrane anchor Sdh3 (Fig. 8A). Cells lacking Sdh4, or Sdh3 and Sdh4 double, possess a similar phenotype as cells lacking Sdh3 only (data not shown). Sdh2 steady-state levels were near normal in cells lacking Sdh3, but Sdh2 levels were absent in cells lacking Sdh1. Sdh1 flavinylation was partially depressed in sdh2Δ cells. However, flavinylation of Sdh1 occurred normally in sdh3Δ (Fig. 8A). The Sdh1 and Sdh2 subunits persisting in double sdh3Δ,sdh4Δ mutant formed a complex as indicated by affinity purification (Fig. 8B). Thus, flavinylation of Sdh1 occurs to a limited extent in the absence of other SDH subunits, but Sdh2 is important for efficient covalent FAD binding. Interestingly, the Sdh5 steady-state levels were elevated in sdh2Δ or sdh3Δ cells (Fig. 8A).
FIGURE 8.
Sdh1 is flavinylated in the absence of the membrane anchor subunits and forms a stable dimeric complex with Sdh2. A, steady-state immunoblot and Sdh1 flavinylation analysis (UV illumination). B, nickel-nitrilotriacetic acid purification of Sdh2-His6Myc2 in mitochondria isolated from sdh3Δsdh4Δ cells. UB, unbound; E, eluate; Flavo, flavoprotein.
DISCUSSION
We show presently that flavinylation of the Sdh1 subunit of succinate dehydrogenase is dependent on a set of two spatially close Arg residues near the C terminus, which are distant (>20 Å) from the FAD binding site but are critical in flavinylation. These residues are also important for the assembly of Sdh1 into the tetrameric enzyme complex. Mutant Sdh1 proteins with either a R582A or double C630A,R638A substitution are neither flavinylated nor assembled into the SDH complex (Table 1).
TABLE 1.
Molecular properties of strains with mutations or deletions that result in the loss of covalent flavinylation in Sdh1
The double C630A,R638A and the single R582W mutations at or near the C terminus lead to loss of SDH assembly as well as binding to heme-agarose beads. However, mutations or deletions that maintain heme-agarose binding also maintain SDH assembly. ND, not determined.
| Strain | Heme binding | SDH assembly | Sdh2 steady-state | Sdh5 steady-state | Sdh1-Sdh5 interaction |
|---|---|---|---|---|---|
| H90S | Yes | Yes | Down | Stable | Attenuated |
| sdh5Δ | Yes | Yes | Down | ||
| ClipK | No | No | Absent | ND | ND |
| C630A,R638A | No | No | Absent | Unstable | Attenuated |
| R582A | No | No | Absent | Unstable | Attenuated |
With each mutant Sdh1, second-site Sdh1 suppressor mutations were recovered in Sdh1 permitting both flavinylation and SDH assembly. In the case of the single R638A mutation, the second site suppressor was a G70V substitution, whereas the R582A second site suppressor was a M599R substitution. The presence of the Arg at residue 599 restores a positively charged residue in proximity to residue position 582. It is of interest that the corresponding residue to Met599 in humans and metazoans is Arg.
In the human SDHA (Sdh1 equivalent), the Arg residue corresponding to yeast Arg582 is Arg589. Substitution of this Arg589 to a Trp (R589W) has been reported in a patient afflicted with paraganglioma (24). Yeast harboring a corresponding R582W mutant Sdh1 are compromised in SDH assembly and flavinylation, but a reversion mutant of R582C restores both Sdh1 flavinylation and SDH assembly.
One major question emerging from the present studies concerns the role of the C-terminal Arg residues in Sdh1 flavinylation. These mutations do not appear to destabilize the Sdh1 polypeptide; rather, they only impair Sdh1 maturation. Three candidate roles for the C-terminal Arg residues may be envisioned. One candidate link involves the binding of Sdh5 and its importance in Sdh1 flavinylation. Sdh5 levels are dependent on the presence of Sdh1. The reduced steady-state levels of Sdh5 in the Sdh1 mutants may reflect attenuated binding. However, cells harboring the mutant Sdh1 alleles are markedly impaired in SDH assembly, whereas sdh5Δ cells are only partially attenuated in SDH stability as seen by the variable levels of the assembled SDH complex in our series of isolates. Thus, the phenotypes observed with either the R582A or double C630A,R638A mutants appear distinct from that of sdh5Δ cells.
A second scenario is that the flavinylation may occur in a nascent conformation of Sdh1 that is somewhat distinct from the final mature conformation. In this scenario, the C-terminal Arg residues may be in juxtaposition for FAD flavinylation. The G70V second site suppressor mutation in the SDH-deficient R638A Sdh1 mutant is consistent with this postulate of a distinct nascent conformation in which these two residues are now in closer proximity. However, two observations argue against this model. The observation that citric acid cycle intermediates can stimulate the flavinylation process (10) suggests that a flavinylation-competent conformation may have a preformed native-like substrate binding site. The dependence of Sdh2 on efficient Sdh1 flavinylation suggests that the flavinylation-competent conformation of Sdh1 must be quite similar to the final mature fold enabling Sdh2 association. In the flavoprotein vanilly-alcohol oxidase containing a histidyl-linked FAD, structural similarity between the holo- and the apo-forms indicates that FAD and substrate bind to a folded, highly preorganized cofactor/active site cavity, followed by autocatalytic covalent flavinylation (27).
A third candidate role for the C-terminal Arg residues may relate to FAD binding. Although the C-terminal Arg residues are distantly removed from the FAD or substrate site in the mature Sdh1 structure, the Arg residues may be important in recruitment and/or guidance of FAD and or succinate to the substrate site. As mentioned, flavinylation of Sdh1 is dependent on succinate (10).
Another conserved Arg residue (human Arg408) stabilizes succinate or inhibitor binding through two hydrogen bonds. Mutations in the human gene yielding a R408C substitution was reported in a patient with a late onset neurodegenerative disease (28). Engineering the corresponding mutation in the E. coli enzyme resulted in impaired covalent flavinylation and the absence of membrane-associated enzyme (28).
The impaired assembly of SDH with the C-terminal Sdh1 mutants suggests that FAD binding is important to stabilize the Sdh1 conformation enabling association with Sdh2 and the membrane anchor subunits. To address the role of FAD binding in SDH assembly, we utilized a yeast flx1Δ deletion strain. This mutant was reported to have attenuated levels of matrix FAD levels (23), and we confirmed this observation. Cells lacking Flx1 are known to be deficient in two FAD-containing enzymes SDH and lipoamide dehydrogenase (22). We show the mutant cells are also impaired in SDH assembly and stability of Sdh2. The impaired SDH assembly in the FAD-deficient flx1Δ cells suggests that FAD binding is important for Sdh1 maturation enabling assembly of the tetrameric enzyme. Cells containing the H90S Sdh1 mutant that precludes covalent flavinylation assemble into the SDH complex consistent with a non-covalent association of FAD as was reported previously (8).
The covalent addition of FAD to Sdh1 likely occurs in a specific folded conformation of Sdh1 that brings a set of amino acids in juxtaposition for the autocatalytic addition. Sdh5 as well as succinate as a substrate are proposed to stabilize the flavinylation-competent conformation of Sdh1 for the reaction. The conserved C-terminal Arg residues (Arg582 and Arg638) could contribute to FAD recruitment and or its binding prior to formation of the covalent attachment. The G70V second site suppressor mutation in the SDH-deficient R638A Sdh1 mutant may merely partially deform the Sdh1 conformation allowing FAD binding in the absence of Arg638. The C-terminal Arg residues may also have a secondary role in the binding of Sdh5.
The propensity of Sdh1 to adsorb onto heme-agarose beads may relate to either a hydrophobic pocket that fortuitously accommodates heme with no physiological consequence of this binding. Alternatively, the presence of the positively charged Arg residues in the C-terminal segment could interact electrostatically with the dianionic propionate groups of heme, facilitating the association of Sdh1 with heme-agarose. This interaction may be analogous to the possible dianionic succinate or phosphates of FAD. Thus, in this scenario, heme is acting merely as a dianionic mimic of FAD or succinate. This notion is supported by the fact that protoporphyrin IX (heme lacking the iron center, but still possessing the dianionic propionates) can compete for heme binding to Sdh1 (results not shown). This observation argues that the iron center is not important for binding to Sdh1. Thus, the increased affinity of Sdh1 to heme in the presence of a reductant may be more related to the reduced state of Sdh1 than the heme iron.
A third less likely scenario is that heme has an effector role in the flavinylation reaction. We have no evidence that SDH biogenesis requires heme for Sdh1 maturation. Because heme is essential for cell survival and important in yeast for Hap1-mediated gene expression of mitochondrial proteins, the investigation of a role of heme in Sdh1 maturation is challenging and will be the topic of future studies.
This work was supported by Grant ES03817 from the NIEHS, National Institutes of Health (to D. R. W.), funding from the Huntsman Cancer Institute (P30 CA042014), and support from the National Institutes of Health (R24DK092784-01).
- SDH
- succinate dehydrogenase
- BN-PAGE
- blue native PAGE
- FAD
- flavin adenine dinucleotide
- DDm
- n-dodecyl-β-d-maltoside.
REFERENCES
- 1. Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z. (2005) Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121, 1043–1057 [DOI] [PubMed] [Google Scholar]
- 2. Hägerhäll C. (1997) Succinate: quinone oxidoreductases. Variations on a conserved theme. Biochim. Biophys. Acta 1320, 107–141 [DOI] [PubMed] [Google Scholar]
- 3. Oyedotun K. S., Lemire B. D. (2001) The quinone-binding sites of the Saccharomyces cerevisiae succinate-ubiquinone oxidoreductase. J. Biol. Chem. 276, 16936–16943 [DOI] [PubMed] [Google Scholar]
- 4. Silkin Y., Oyedotun K. S., Lemire B. D. (2007) The role of Sdh4 Tyr-89 in ubiquinone reduction by the Saccharomyces cerevisiae succinate dehydrogenase. Biochim. Biophys. Acta 1767, 143–150 [DOI] [PubMed] [Google Scholar]
- 5. Maklashina E., Rajagukguk S., McIntire W. S., Cecchini G. (2010) Mutation of the heme axial ligand of Escherichia coli succinate-quinone reductase: implications for heme ligation in mitochondrial complex II from yeast. Biochim. Biophys. Acta 1797, 747–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tomasiak T. M., Maklashina E., Cecchini G., Iverson T. M. (2008) A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II. J. Biol. Chem. 283, 15460–15468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lemire B. D., Oyedotun K. S. (2002) The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim. Biophys. Acta 1553, 102–116 [DOI] [PubMed] [Google Scholar]
- 8. Robinson K. M., Rothery R. A., Weiner J. H., Lemire B. D. (1994) The covalent attachment of FAD to the flavoprotein of Saccharomyces cerevisiae succinate dehydrogenase is not necessary for import and assembly into mitochondria. Eur. J. Biochem. 222, 983–990 [DOI] [PubMed] [Google Scholar]
- 9. Hägerhäll C., Sled V., Hederstedt L., Ohnishi T. (1995) The trinuclear iron-sulfur cluster S3 in Bacillus subtilis succinate:menaquinone reductase; effects of a mutation in the putative cluster ligation motif on enzyme activity and EPR properties. Biochim. Biophys. Acta 1229, 356–362 [DOI] [PubMed] [Google Scholar]
- 10. Robinson K. M., Lemire B. D. (1996) Covalent attachment of FAD to the yeast succinate dehydrogenase flavoprotein requires import into mitochondria, presequence removal, and folding. J. Biol. Chem. 271, 4055–4060 [DOI] [PubMed] [Google Scholar]
- 11. Hao H. X., Khalimonchuk O., Schraders M., Dephoure N., Bayley J. P., Kunst H., Devilee P., Cremers C. W., Schiffman J. D., Bentz B. G., Gygi S. P., Winge D. R., Kremer H., Rutter J. (2009) SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 325, 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rutter J., Winge D. R., Schiffman J. D. (2010) Succinate dehydrogenase - Assembly, regulation, and role in human disease. Mitochondrion 10, 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McNeil M. B., Clulow J. S., Wilf N. M., Salmond G. P., Fineran P. C. (2012) SdhE is a conserved protein required for the flavinylation of succinate dehydrogenase in bacteria. J. Biol. Chem. 287, 18418–18428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heuts D. P., Scrutton N. S., McIntire W. S., Fraaije M. W. (2009) What's in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J. 276, 3405–3427 [DOI] [PubMed] [Google Scholar]
- 15. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 16. Glick B. S., Pon L. A. (1995) Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260, 213–223 [DOI] [PubMed] [Google Scholar]
- 17. Diekert K., De Kroon A. I., Kispal G., Lill R. (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65, 37–51 [DOI] [PubMed] [Google Scholar]
- 18. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 19. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 20. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 21. Robinson K. M., Lemire B. D. (1995) Flavinylation of succinate: ubiquinone oxidoreductase from Saccharomyces cerevisiae. Methods Enzymol. 260, 34–51 [DOI] [PubMed] [Google Scholar]
- 22. Bafunno V., Giancaspero T. A., Brizio C., Bufano D., Passarella S., Boles E., Barile M. (2004) Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J. Biol. Chem. 279, 95–102 [DOI] [PubMed] [Google Scholar]
- 23. Tzagoloff A., Jang J., Glerum D. M., Wu M. (1996) FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J. Biol. Chem. 271, 7392–7397 [DOI] [PubMed] [Google Scholar]
- 24. Burnichon N., Brière J. J., Libé R., Vescovo L., Rivière J., Tissier F., Jouanno E., Jeunemaitre X., Bénit P., Tzagoloff A., Rustin P., Bertherat J., Favier J., Gimenez-Roqueplo A. P. (2010) SDHA is a tumor suppressor gene causing paraganglioma. Hum. Mol. Genet. 19, 3011–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu M., Repetto B., Glerum D. M., Tzagoloff A. (1995) Cloning and characterization of FAD1, the structural gene for flavin adenine dinucleotide synthetase of Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giancaspero T. A., Wait R., Boles E., Barile M. (2008) Succinate dehydrogenase flavoprotein subunit expression in Saccharomyces cerevisiae–involvement of the mitochondrial FAD transporter, Flx1. FEBS J. 275, 1103–1117 [DOI] [PubMed] [Google Scholar]
- 27. Fraaije M. W., van Den Heuvel R. H., van Berkel W. J., Mattevi A. (2000) Structural analysis of flavinylation in vanillyl-alcohol oxidase. J. Biol. Chem. 275, 38654–38658 [DOI] [PubMed] [Google Scholar]
- 28. Birch-Machin M. A., Taylor R. W., Cochran B., Ackrell B. A., Turnbull D. M. (2000) Late-onset optic atrophy, ataxia, and myopathy associated with a mutation of a complex II gene. Ann. Neurol. 48, 330–335 [PubMed] [Google Scholar]