Background: Strategies on the basis of doxycycline-inducible lentiviruses in mouse cells allowed the examination of mechanisms governing somatic cell reprogramming.
Results: Using a doxycycline-inducible human reprogramming system, we identified unreported miRs enhancing reprogramming efficiency.
Conclusion: We generated a drug-inducible human reprogramming reporter system as an invaluable tool for genetic or chemical screenings.
Significance: These cellular systems provide a tool to enable the advancement of reprogramming technologies in human cells.
Keywords: Embryonic Stem Cell, Induced Pluripotent Stem (iPS) Cell, MicroRNA, Reprogramming, Stem Cells, Doxycycline, Reporter System
Abstract
Reprogramming of somatic cells into induced pluripotent stem cells is achieved by the expression of defined transcription factors. In the last few years, reprogramming strategies on the basis of doxycycline-inducible lentiviruses in mouse cells became highly powerful for screening purposes when the expression of a GFP gene, driven by the reactivation of endogenous stem cell specific promoters, was used as a reprogramming reporter signal. However, similar reporter systems in human cells have not been generated. Here, we describe the derivation of drug-inducible human fibroblast-like cell lines that express different subsets of reprogramming factors containing a GFP gene under the expression of the endogenous OCT4 promoter. These cell lines can be used to screen functional substitutes for reprogramming factors or modifiers of reprogramming efficiency. As a proof of principle of this system, we performed a screening of a library of pluripotent-enriched microRNAs and identified hsa-miR-519a as a novel inducer of reprogramming efficiency.
Introduction
Induction of pluripotency in somatic cells by the transcription factors OCT4, SOX2, KLF4, and cMYC represented a breakthrough in regenerative medicine (1). Initially, generation of mouse-induced pluripotent stem cells (miPSCs)3 relied on the retroviral transduction and integration of the transcription factors Oct4, Sox2, Klf4, and cMyc (1). miPSC colonies were selected on the basis of the neomycin resistance provided by the reactivation of the Fbx15 promoter (1), a gene specifically expressed in mouse embryonic stem cells and in the early embryo. However, although these miPSCs were able to contribute to all three germ layers after injection into blastocysts, no live chimeric mice were obtained, most likely because of the incomplete reprogramming of the miPSCs (1). Later reports showed that selection based on the promoter reactivation of alternative stem cell markers, such as Oct4 or Nanog, rendered miPSCs capable of generating germ line-competent live chimeric mice (2–4) and even all-miPSC mice (5–7), demonstrating the full pluripotency of these cells. Together, these reports highlighted the importance in the use of selective stem cell markers to identify fully reprogrammed iPSC colonies.
In the last few years the development of reprogramming systems on the basis of doxycycline-inducible lentiviruses expressing the reprogramming factors enabled strategies for examining the mechanisms governing cell reprogramming (8, 9). This system was used to generate chimeric mice by injecting into blastocysts doxycycline-inducible miPSCs or mouse embryonic stem cells containing an inducible reprogramming cassette expressing the four factors (10–11). Cells derived from these mice allowed for the generation of “secondary” miPSCs from a wide variety of somatic tissues (10–11). Importantly, somatic cells from these mice can be reprogrammed only by the addition of doxycycline and are easily traceable by their re-expression of a GFP gene driven by the Nanog or Oct4 promoters. Interestingly, these somatic cells reprogram with 25- to 50-fold greater efficiencies than those observed using direct infection and drug selection for pluripotent markers (11). Furthermore, the generation of transgenic mice with defined doxycycline-inducible subsets of the four reprogramming factors has been reported (12). Mouse embryonic fibroblasts (MEFs) isolated from these transgenic mice could generate secondary GFP-positive miPSC only when the missing factor was reintroduced (12). Altogether, these systems have greatly facilitated the characterization of the reprogramming process and will serve as an invaluable tool for genetic or chemical screenings to identify functional substitutes of the reprogramming factors with easy fluorescent traceable markers.
Importantly, although these mouse reporter tools to date have provided methods of examining reprogramming that cannot be performed in a human system, the fact that there are key molecular mechanistic differences between mouse and human somatic cell reprogramming warrants the development of a similar reporter system using human cells. Previous studies have reported the generation of drug-inducible reprogramming systems in human cells with higher efficiencies compared with retroviral-based protocols (13–14). However, although these cellular systems can be used to dissect the underlying molecular and epigenetic events occurring during the reprogramming of human cells, the absence of a pluripotent reporter in these systems, which could allow for the identification of bona fide hiPSC colonies on the basis of the reactivation of endogenous stem cell promoters, have precluded their use for screening purposes. In this work, we report the generation of a drug-inducible human reprogramming system that incorporates a reporter gene driven by the OCT4 promoter, as it has been shown that its reactivation is a very reliable marker to identify fully reprogrammed cells (15–16).
EXPERIMENTAL PROCEDURES
hES Cell Culture and Differentiation
The H1 (WA01), H7 (WA07), H9 (WA09), and H1-OCT4GFP embryonic stem (17) cell lines were obtained from the WiCell Research Institute and maintained on MEFs or Matrigel (BD Biosciences) using mTeSR1 medium (Stem Cell Technologies). hESC colonies were split using a solution of dispase (2 mg/ml) or collagenase (1 mg/ml) and scraping the colonies with a glass pipette. Derived hiPSCs were cultured similarly as described above for hESCs. 293T cells, dFib-OCT4GFP fibroblast-like cells (18), and BJ human fibroblasts (ATCC, CRL-2522) were cultured in DMEM (Invitrogen) supplemented with 10% FBS and 0.1 mm non-essential amino acids. Commercial primary cells obtained from the ATCC, Lonza, and Promocell (supplemental Table S1) were cultured according to the recommendations of the supplier.
Human hiPSC Generation
For the generation of human primary hiPSCs derived from dFib-OCT4GFP cells, a mix of retroviruses plus lentiviruses was used to infect the fibroblast-like cells by spinfection at 800 × g for 1 h at room temperature in the presence of polybrene (4 μg/ml). As an example, for the generation of hiPSC-OCT4GFP-indSKC, the ratio of viruses used was 0.5:0.05:0.05:0.05:0.15 (pMX-OCT4:pLVFUtetO-SOX2:pLVFUtetO-KLF4:pLVFUtetO-cMYC:FUdeltaGW-rtTA). Similarly, the rest of hiPSC lines were obtained by using different combinations of retroviruses and lentiviruses. After infections at day 0 and day 1, cells were plated on day 2 onto fresh MEFs with DMEM (Invitrogen), 10% FBS, and 0.1 mm non-essential amino acids supplemented with 100 ng/ml (unless other specified) of doxycycline. The day after, cells were switched to hESC medium: DMEM/F12 (Invitrogen) supplemented with 20% knockout serum replacement (Invitrogen), 1 mm l-glutamine, 0.1 mm non-essential amino acids, 55 μm β-mercaptoethanol, 10 ng/ml bFGF (basic fibroblast growth factor, Joint Protein Central) and 100 ng/ml doxycycline. For the derivation of the hiPSC lines, GFP-positive colonies were manually picked and maintained on fresh MEF feeder layers for five passages before growth in Matrigel/mTesR1 conditions. For the generation of secondary hiPSC lines, the corresponding dFib-OCT4GFP–ind fibroblast-like cells were serially infected twice with retroviruses encoding the missing reprogramming factor or with miR-encoding lentiviruses. Immediately after the second infection, cells were incubated with DMEM (Invitrogen), 10% FBS, and 0.1 mm non-essential amino acids supplemented with 100 ng/ml (unless other specified) of doxycycline and, 2 days later, plated onto fresh MEFs or Matrigel. The day after, cells were switched to hESC medium supplemented with doxycycline until colonies appeared in the well. In all cases, hiPSC colonies were stained for either alkaline phosphatase or Nanog expression or used to establish independent cell lines.
The hiPSC43A2, hiPSC43B2, hiPSC43D6, hiPSC57A5, hiPSC57A7, and hiPSC57B7 lines were generated from the human fibroblasts lines CRL-2429 and CRL-2522 respectively, using the commercial polycistronic lentivirus STEMCCA encoding the four reprogramming factors (Millipore, SCR510). The F1hiPSC4F line was generated from F1 fibroblasts using the polycistronic lentivirus STEMCCA. The CBhiPSC2F3, CBhiPSC3F12, and CBhiPSC4F3 lines were generated from human cord blood samples by retroviral infection. These hiPSC lines were maintained on MEFs or human feeder fibroblasts using KO DMEM medium (Invitrogen) in the presence of knockout serum replacement (Invitrogen) supplemented with 0.1 mm non-essential amino acids, 0.1 mm mercaptoethanol, 1× glutamax, and 10 ng/ml bFGF (Invitrogen).
Evaluation of Reprogramming Efficiency
To calculate the efficiency of reprogramming, we plated the same number of dFib-OCT4GFP fibroblast-like cells on MEFs after infection with either retroviruses (to supply the missing factor) or lentiviruses (to supply the indicated miR). Eighteen days (in experiments where OSKC were expressed) or 25 days (in experiments where OSK were expressed) after the initial infection, the cell cultures were fixed and stained to detect AP activity or NANOG expression. Reprogramming efficiency was assessed by the number of AP+ or NANOG + hiPSC colonies per number of initial seed cells or as a relative percentage of AP+ or NANOG + hiPSC colonies. This percentage was calculated as fold change relative to the value of the number of colonies generated with dFib-OCT4GFP fibroblast-like cells infected with pmiR-000 lentiviruses (which encoded for a control miRNA). At least three independent experiments were performed in triplicate in each case.
Reprogramming with miR Mimics
For the evaluation of reprogramming efficiency after transfection with miR mimics, BJ fibroblasts were infected only once by spinfection with a mix of retroviruses encoding OCT4, SOX2, and KLF4 (day 0). At days 0 and 5 after initial infection, cells were transfected with miR mimics (obtained from Qiagen) at 30 nm final concentration using Lipofectamine (Invitrogen) following the recommendations of the manufacturer. At day 6, cells were transferred onto fresh MEFs with DMEM (Invitrogen), 10% FBS, and 0.1 mm non-essential amino acids. The day after, cells were switched to hESC medium (see above) until hiPSC colonies developed.
Derivation and Validation of dFib-OCT4GFP-ind Cells
An embryoid body-mediated protocol was used to differentiate pluripotent cells into fibroblast-like cells. Pluripotent cell colonies growing on Matrigel were loosely detached by dispase treatment, resuspended in DMEM/F12 supplemented with 10% FBS (Atlanta Biologicals), 0.5 mm l-glutamine, 0.1 mm non-essential amino acids, and 55 μm β-mercaptoethanol, and maintained on low attachment plates with daily media changes. After 4 days on suspension, embryoid bodies were plated onto gelatin-coated tissue culture plates and maintained in embryoid body medium for two additional days, followed by their maintenance in DMEM, 10% FBS, and 1% nonessential amino acids (NEAA) until cells showed fibroblast morphology. Derived fibroblasts-like cells were serially passaged by using Tryple (Invitrogen) and tested for loss of pluripotent markers as well as GFP expression and gain of expression of fibroblast markers.
Statistical Analyses
Results are reported as mean ± S.D. (see the figure legends for specific details regarding the number of biological replicates, independent experiments, and technical replicates). Statistics were performed using two-tailed Student's t tests. Values with p < 0.05 were considered statistically significant.
RESULTS
Generation of a Drug-inducible Human Reporter System for Reprogramming
We have previously developed a human reporter system where the expression of a GFP gene is driven by the endogenous OCT4 promoter (18). This system was obtained by differentiating a knockin OCT4GFP human H1 embryonic stem cell line (17) into a fibroblast-like population of cells (dFib-OCT4GFP) (18). We verified that the population of dFib-OCT4GFP cells displayed the expected morphology and expressed fibroblast markers at a similar level to what is observed in human fibroblasts (18). Moreover, we determined that these cells no longer expressed GFP because of the silencing of the OCT4 promoter, or several pluripotent markers, detected in the H1-OCT4GFP cell line. After reprogramming dFib-OCT4GFP cells by transduction with a combination of retroviruses encoding for OCT4, SOX2, KLF4, and/or cMYC (OSKC), we were able to observe the reactivation of the endogenous OCT4 locus, which correlated with the appearance of GFP (18). However, we were not able to reprogram dFib-OCT4GFP cells by transduction of only OCT4 or OCT4/SOX2 (supplemental Fig. S1).
To first create a similar human reprogramming reporter system that was drug-inducible (Fig. 1A), we transduced dFib-OCT4GFP cells with inducible lentiviruses expressing the four factors and selected two primary hiPSC-OCT4GFP–ind-OSKC cell lines for further analysis (Fig. 1B). We first verified that these hiPSC lines had regained the expression of GFP, expressed endogenous pluripotent markers, down-regulated fibroblast markers, and contributed in vivo to the three embryonic germ layers (Fig. 1, B–D). We then differentiated the hiPSC-OCT4GFP cell lines toward fibroblast-like cells (dFib-OCT4GFP-ind-OSKC) (Fig. 1E). We determined that these cells regained the expression of fibroblast markers, down-regulated the expression of pluripotent markers, and lost the expression of GFP (Fig. 1C). Furthermore, we verified that treatment of dFib-OCT4GFP-ind-OSKC cells with doxycycline induced the expression of the lentiviral-delivered reprogramming factors (Fig. 1F) and, when grown under hESC culture conditions, generated secondary hiPSCs that re-express GFP, pluripotent markers, and down-regulated fibroblasts markers (Fig. 1, C and G). These data demonstrated the generation of a human reprogramming reporter system that can be used to investigate the process of reprogramming.
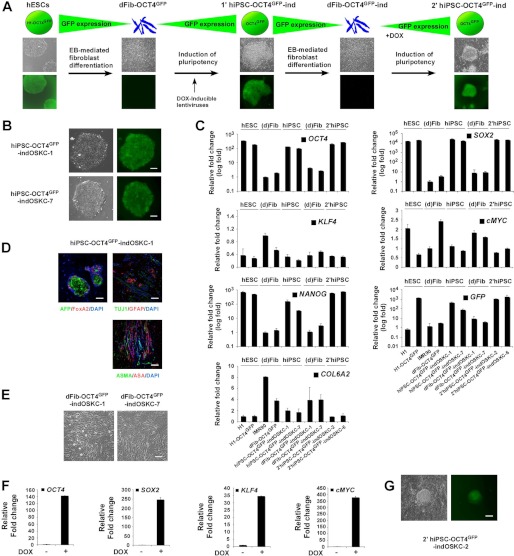
FIGURE 1.
Generation of the dFib-OCT4GFP–ind-OSKC reporter system. A, schematic representation of the methodology used to obtain dFib-OCT4GFP–ind fibroblast-like cells expressing the four reprogramming factors. EB, embryoid body; DOX, doxycycline. B, primary hiPSC colonies obtained from dFib-OCT4GFP after infection with inducible lentiviruses expressing the four reprogramming factors. Note that hiPSC colonies have regained the expression of GFP. Scale bar = 100 μm. C, real-time PCR analysis was performed for the pluripotent markers OCT4, SOX2, NANOG, the reprogramming factors KLF4 and cMYC, the fibroblast marker COL6A2, and for GFP. Data are shown as relative means ± S.D. of two biological replicates analyzed in triplicate. D, teratoma formation was assessed by injection of the hiPS-OCT4GFP–ind-OSKC-1 cell line into the testes or kidney of SCID mice. Immunofluorescence analysis demonstrates the existence of the three main embryonic germ layers as defined by the expression of specific endodermal (α-fetoprotein (AFP) and FoxA2), ectodermal (TUJ1 and glial fibrillary acidic protein (GFAP), and mesodermal (α-smooth muscle actin (ASMA) and α sarcomeric actin (ASA)) markers. All images were obtained from the same tumor. DAPI was used to visualize the nuclei. Scale bar = 200 μm. E, morphology of the dFib-OCT4GFP–ind-OSKC fibroblasts cell lines after embryoid body-mediated differentiation. Scale bar = 50 μm. F, dFib-OCT4GFP–indOSKC-7 cells were treated with 100 ng/ml doxycycline for 48 h. Real-time PCR analysis was performed for OCT4, SOX2, KLF4, and cMYC. Data are shown as relative means ± S.D. of two biological replicates analyzed in triplicate. G, Secondary hiPSC colonies isolated from dFib-OCT4GFP-ind-OSKC cells after doxycycline treatment. Note that hiPSC colonies are GFP-positive. Scale bar = 100 μm.
We were next interested in expanding this approach to generate additional cellular systems that expressed different subsets of reprogramming factors to use them as a tool to screen for functional substitutes of the reprogramming factors or to identify new mediators in the reprogramming process. To do this, we took advantage of the fact that retroviral-mediated expression of exogenous reprogramming factors is epigenetically silenced after the reactivation of the pluripotent endogenous network, whereas the expression of reprogramming factors delivered by lentiviruses can still be modulated by doxycycline independent of the somatic or pluripotent cell state. Thus, infecting dFib-OCT4GFP cells with a combination of retrovirus expressing one factor and a mix of inducible lentiviruses expressing the remaining three factors would allow us to generate primary hiPSCs (hiPSC-OCT4GFP) (see Fig. 2, A and B). Then, these cells can be differentiated toward fibroblast-like cells that will retain the ability to re-express the three lentiviral-delivered reprogramming factors after the addition of doxycycline, whereas the remaining retroviral-delivered factor would no longer be expressed. For example, by infecting dFib-OCT4GFP cells with retroviruses expressing OCT4 (pMX-OCT4) and lentiviruses expressing SOX2, KLF4, and cMYC (pLVFUtetO-SOX2, -KLF4, and -cMYC), we would generate hiPSCs-OCT4GFP–indSKC that could be further differentiated toward fibroblast-like cells (dFib-OCT4GFP–indSKC) (Fig. 2, A and B). dFib-OCT4GFP–indSKC cells will express SOX2, KLF4, and cMYC after the addition of doxycycline but not OCT4. Under the proper culture conditions, these cells would not reprogram unless exogenous OCT4 or a functional OCT4 substitute is supplied to the cells where detection of GFP can be used to identify fully reprogrammed cells (Fig. 2B). Thus, this cellular system represents an ideal tool to screen for functional substitutes of the reprogramming factors or new modifiers of the reprogramming process.
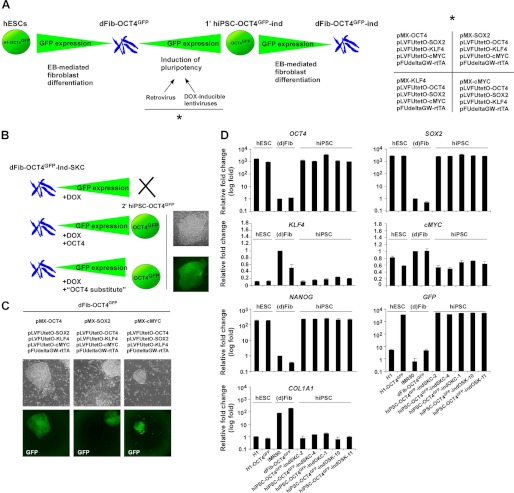
FIGURE 2.
Generation of hiPSC lines with doxycycline-inducible expression of different subsets of reprogramming factors. A, schematic representation of the rationale followed to obtain dFib-OCT4GFP–ind fibroblast-like cells expressing different subsets of reprogramming factors. The right panel shows the different combination of retroviruses and lentiviruses used to express the reprogramming factors. B, schematic representation showing the example of dFib-OCT4GFP–ind-SKC that would not reprogram by only the addition of doxycycline (DOX) unless either OCT4 or a functional substitute for OCT4 was provided to the cells. C, primary hiPSC colonies obtained from dFib-OCT4GFP after the infection with the combination of viruses depicted at the top of the images. Note that hiPSC colonies are GFP-positive. D, real-time PCR analysis was performed for the pluripotent markers OCT4, SOX2, NANOG, the reprogramming factors KLF4 and cMYC, the fibroblast marker COL1A1, and for GFP. Data are shown as relative means ± S.D. of two biological replicates analyzed in triplicate.
We infected dFib-OCT4GFP cells with different combinations of retroviruses and drug-inducible lentiviruses and successfully generated primary hiPSC-OCT4GFP colonies (Fig. 2, A and C). Unfortunately, we were unable to obtain hiPSC-OCT4GFP lines when KLF4 expression was driven by retroviruses for unknown reasons. We isolated and expanded, in the absence of doxycycline, four hiPSC-OCT4GFP–SKC, four hiPSC-OCT4GFP–OKC, and nine hiPSC-OCT4GFP–OSK lines. On the basis of a similar level of expression between the reprogramming factors after the addition of doxycycline in these hiPSC-OCT4GFP lines (data not shown), we selected the hiPSC-OCT4GFP–SKC-2 and -4, hiPSC-OCT4GFP–OKC-2, and hiPSC-OCT4GFP–OSK-10 and -11 lines for further analysis. We first verified that these reprogrammed cells had regained the expression of GFP, expressed endogenous pluripotent markers, and down-regulated fibroblast markers (Fig. 2D). Moreover, we confirmed the pluripotency of these hiPSC lines as they differentiated in vitro (Fig. 3A) and contributed in vivo (B) to the three embryonic germ layers.
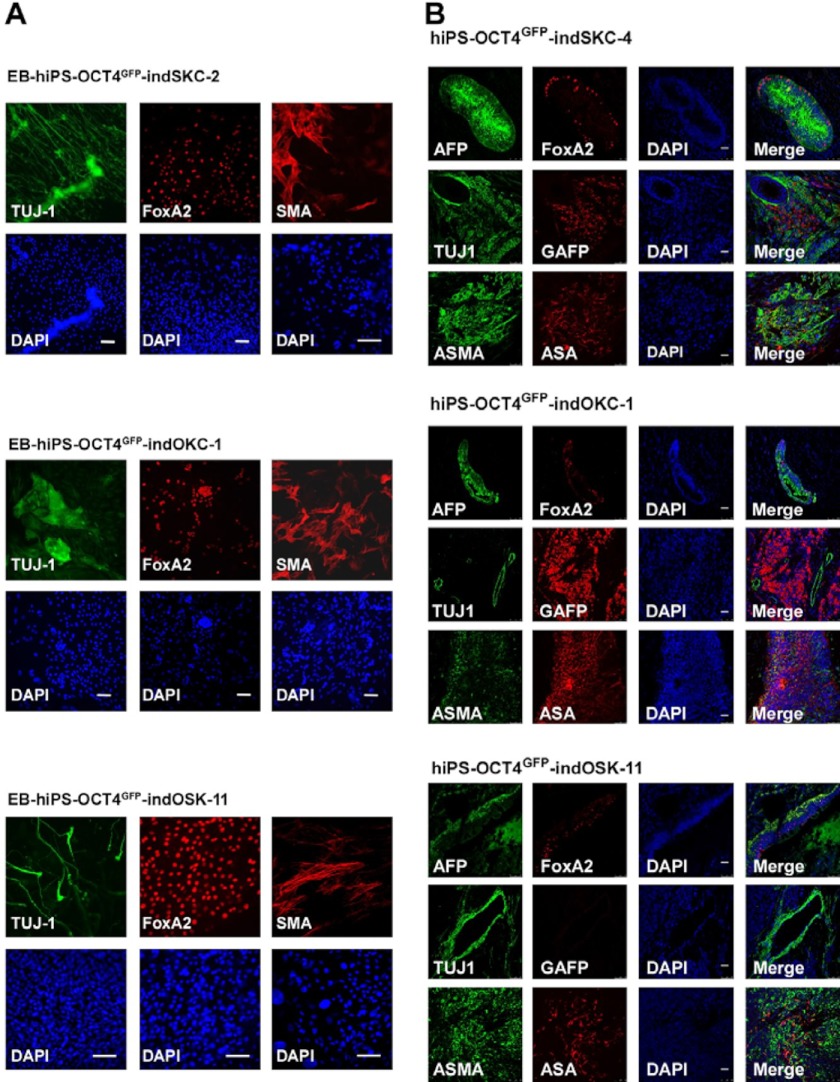
FIGURE 3.
hiPS-OCT4GFP–ind cells are pluripotent. A, embryoid bodies derived from the indicated hiPS-OCT4GFP–ind cells lines were differentiated for 15 days. Immunofluorescence was performed for specific differentiation markers from the three embryonic germ layers as defined by the expression of mesodermal (α-smooth muscle actin (SMA)), ectodermal (TUJ1), and endodermal (FoxA2) markers. Scale bar = 50 μm. B, teratoma formation was assessed by injection of the hiPS-OCT4GFP–ind cells lines into the testes or kidney of SCID mice. Immunofluorescence analysis demonstrate the existence of the three main embryonic germ layers as defined by the expression of specific endodermal (α-fetoprotein (AFP) and FoxA2), ectodermal (TUJ1 and glial fibrillary acidic protein (GFAP), and mesodermal (ASMA and α sarcomeric actin (ASA)) markers. All images were obtained from the same tumor. DAPI was used to visualize the nuclei. Scale bar = 50 μm.
We then differentiated the hiPSC-OCT4GFP lines using an embryoid body-mediated protocol to obtain morphologically fibroblast-like cells (dFib-OCT4GFP-ind) (Fig. 4A and supplemental Fig. S2A) (18). We next determined that the different dFib-OCT4GFP-ind fibroblast-like cells regained the expression of fibroblasts markers, down-regulated the expression of pluripotent markers, and lost the expression of GFP (Fig. 4B). Furthermore, we verified that treatment of dFib-OCT4GFP-ind cells with doxycycline induced the expression of the lentiviral-delivered reprogramming factors but not the expression of the factor initially supplied by retroviral infection (Fig. 4C and supplemental Fig. S2B).
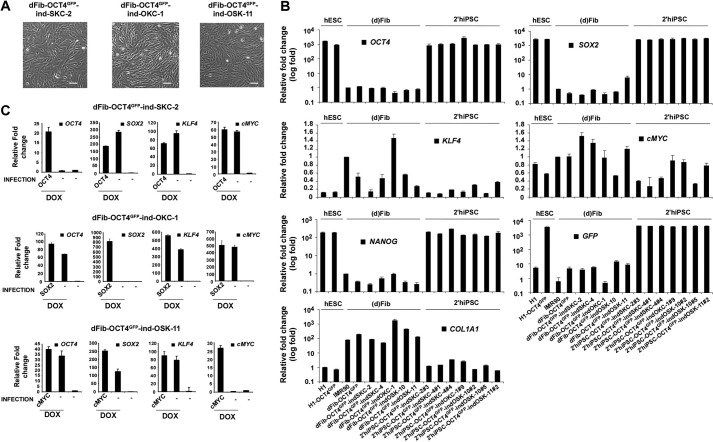
FIGURE 4.
Generation of dFib-OCT4GFP–ind lines with doxycycline-inducible expression of different subsets of reprogramming factors. A, morphology of the dFib-OCT4GFP–ind fibroblasts-like cells (at passage 7) after embryoid body-mediated differentiation. Scale bar = 50 μm. B, real-time PCR analysis was performed for the pluripotent markers OCT4, SOX2, NANOG, the reprogramming factors KLF4 and cMYC, the fibroblast marker COL1A1, and for GFP. Data are shown as relative means ± S.D. of two biological replicates analyzed in triplicate. C, dFib-OCT4GFP–indSKC, dFib-OCT4GFP–ind-OKC, and dFib-OCT4GFP–ind-OSK cells were infected with retroviruses encoding OCT4, SOX2, and cMYC, respectively, and either treated or not treated with 100 ng/ml of doxycycline for 48 h. Real-time PCR analyses were performed to detect the transcripts corresponding to the four reprogramming factors. Note that there is no residual expression from any of the factors delivered by retroviruses to generate the original primary hiPSC lines. Data are shown as relative means ± S.D. of two biological replicates analyzed in triplicate.
Functional Validation of the Human Reprogramming System
We next validated that each population of dFib-OCT4GFP-ind cells had the ability to reprogram back into pluripotency by the addition of doxycycline and exogenous expression of the missing respective reprogramming factor. To test this, we infected dFib-OCT4GFP-indSKC, dFib-OCT4GFP-indOKC, and dFib-OCT4GFP-indOSK cells with retroviruses encoding OCT4, SOX2, and cMYC, respectively, and grew the cells in the presence or absence of doxycycline and compared them with uninfected cells to evaluate their reprogramming abilities. We first determined the optimal concentration of doxycycline to reprogram the dFib-OCT4GFP-ind cells, as it has been reported that the level of expression of the reprogramming factors can influence the efficiency of the reprogramming process (supplemental Fig. S2C) (19). Next, we observed that dFib-OCT4GFP-indSKC and dFib-OCT4GFP-indOKC cells reprogrammed only when doxycycline and the missing factor (OCT4 or SOX2, respectively) was supplied to the cells (Fig. 5, A–D). However, dFib-OCT4GFP-indOSK could reprogram, although with much lower reprogramming efficiency, by the addition of doxycycline only (Fig. 5, E and F). This is in agreement with reports demonstrating that cMYC is not required for reprogramming but contributes to enhance the efficiency of the process (20). The appearance of reprogrammed colonies from dFib-OCT4GFP-ind cells correlated with the expression of GFP (Fig. 5). Moreover, GFP-positive hiPSCs also expressed pluripotent markers such as NANOG or TRA-1–60 (Fig. 5). These hiPSC colonies could be isolated and expanded as independent hiPSC cell lines that retained GFP expression, showed the expression of endogenous pluripotent markers, and down-regulated fibroblast markers (Fig. 4B). Furthermore, we also determined that dFib-OCT4GFP-ind can be reprogrammed in feeder-free conditions (data not shown). Together, these data demonstrate a simple and reliable human reprogramming reporter system that utilizes a GFP signal to visualize pluripotency. This system can be used to screen functional substitutes for the reprogramming factors or identify new mediators in the reprogramming process in a high-throughput manner by the addition of doxycycline only without any requirement of further viral manipulation.
FIGURE 5.
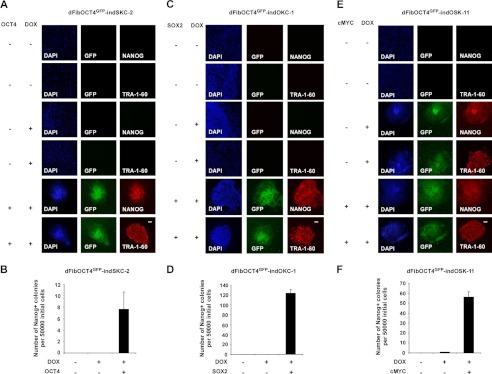
Validation of the dFib-OCT4GFP–ind reporter system. A, C, and E, GFP detection and NANOG/TRA-1-60 immunofluorescence analysis in dFib-OCT4GFP–indSKC-2 (A), dFib-OCT4GFP–indOKC-1 (C), and dFib-OCT4GFP–indOSK-11 (E) cells infected with retroviruses encoding the missing factor and/or treated with doxycycline (DOX) for 18 days. Scale bar = 150 μm. B, D, and F, evaluation of reprogramming efficiency in dFib-OCT4GFP–indSKC-2 (B), dFib-OCT4GFP–indOKC-1 (D), and dFib-OCT4GFP–indOSK-11 (F) cells infected with retroviruses encoding the missing factor and/or treated with doxycycline for 18 days. Reprogramming efficiency was evaluated as the number of Nanog+ colonies per 50,000 dFib-OCT4GFP–ind plated cells on MEFs after retroviral infection.
Expression of hsa-miR-519a Increases Reprogramming Efficiency
MicroRNAs are 22-nucleotide-long non-coding RNAs that regulate the expression of downstream targets by mRNA destabilization and translational inhibition (21). Most of the mRNA-miR targeting occurs through incomplete nucleotide complementation between a short sequence located in the 5′ region of the miR, the so-called seed sequence, and its mRNA target. A single miR can target hundreds of different mRNAs and multiple pathways, which makes them powerful regulators of cell function. In the last few years, miRs have emerged as critical factors in regulating cell fate as well as in the maintenance and acquisition of the pluripotency during cell reprogramming (22). In fact, introduction of mouse embryonic stem cell-specific miRs, such as members of the miR-290 cluster (mmu-miR-291, mmu-miR-294, or mmu-miR295), the miR-106b∼25 cluster (mmu-miR93 and mmu-miR106b), miR-302b∼367, or depletion of fibroblast-specific miRs such as mmu-miR-21 or mmu-miR-29a promoted the formation of miPSCs in the absence of cMYC (23–26). Furthermore, the expression of specific subsets of different miRs is sufficient to reprogram somatic cells (27–28). Although the role of miRs in self-renewal and pluripotency or during the generation of iPSCs from mouse cells has been reported (23–26), little is known about their role in a human context. Therefore, we reasoned that a thorough analysis of the effect on the reprogramming efficiency of hESC-specific miRs would probe the utility of the screening system described above and provide new insights in miR regulation of cell reprogramming.
We first profiled the miR population of 18 samples comprising different subsets of pluripotent and somatic cells (supplemental Table S1). Statistical analysis on the basis of similarity with the hsa-miR-302a expression profile and principal component analysis combined with expression level comparisons led us to the identification of 26 miRs specifically expressed in pluripotent cells or strongly down-regulated in somatic cells (supplemental Fig. S3, A and B and supplemental Table S2). We identified different miRs belonging to clusters that have already been described to have a role in stem cell maintenance or cell reprogramming, such as hsa-miR-302a-d, hsa-miR-372, and hsa-miR-373 (human orthologous to mouse mmu-miR-290); hsa-miR-200; or hsa-miR-520 (supplemental Fig. S3B) (23–26, 29, 30).
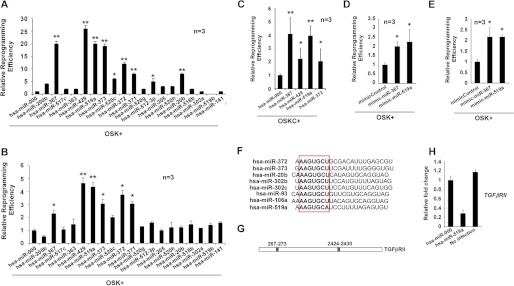
To screen for novel miRs that can enhance reprogramming efficiency, we investigated whether the expression of these miRs influence reprogramming efficiency in the absence of cMYC in human fibroblasts. To this end, we infected dFib-OCT4GFP-indOSK-11 cells with lentiviruses encoding 19 miRs selected for analysis, added doxycycline to the medium, and followed the formation of hiPSC colonies. We observed that although the expression of hsa-miR-200b, -520c, -371, -512–3p, or -20b promoted the formation of hiPSC colonies to some extent, the expression of hsa-miR-367, -372, -373, -429, and -519a greatly improved reprogramming efficiency in the absence of cMYC (Fig. 6A). Moreover, to validate these effects, we performed a similar reprogramming experiment using BJ fibroblasts and observed similar results (Fig. 6B). Overall, these data demonstrate that expression of stem cell-specific miRs, as reported for mouse cells, have a profound impact on reprogramming efficiency. We next focused on investigating hsa-miR-519a, not only because of its previously unreported dramatic enhancement of reprogramming efficiency but also because of the fact that this miR is only found in primates. We first analyzed the effect of hsa-miR-519a in the presence of cMYC. It has been shown that expression of some miRs, such as mmu-miR-429 or members of the mmu-miR-290 cluster, is directly regulated by the binding of cMYC to their promoters in ES cells (31). Thus, although the expression of these miRs enhances reprogramming efficiency when cMYC is absent, this positive effect is reduced when cMYC is coexpressed with them (23–24). Similarly, expression of hsa-miR-429 and -373 showed little increase in the formation of hiPSC colonies when exogenous cMYC is included in the reprogramming experiments, whereas the effect of hsa-miR-519a was further increased in a similar experimental setting (Fig. 6C). Furthermore, we also determined that reprogramming efficiency increased after transfection of hsa-miR-519a mimics in BJ fibroblasts or dFib-OCT4GFP cells (Fig. 6, D and E). We next sought to identify the putative targets of hsa-miR-519a. On the basis of bioinformatic predictions, we chose a subset of genes to test whether their expression was modulated by hsa-miR-519a (see supplemental Experimental Procedures). Thus, we performed real-time PCR analysis on RNA obtained from BJ fibroblasts infected with hsa-miR-519a and its corresponding control. We determined that about half of the genes assayed were down-regulated after the expression of hsa-miR-519a (supplemental Fig. S4A). Importantly, hsa-miR-519a shares almost the identical seed sequence recently described for many mouse embryonic stem cell-specific cell cycle-regulating microRNAs and their corresponding human orthologs (Fig. 6F) (23). Several of these miRs (i.e. mmu-miR-302b, -372, -106a, or -93 and has-miR-302 and -372) target TGFβRII which, when down-regulated, has been shown to enhance reprogramming efficiency (24, 29). Interestingly, the 3′ UTR region of the TGFβRII mRNA contains two well conserved recognition sequences for hsa-miR-519a (Fig. 6G). We observed that expression of hsa-miR-519a down-regulates the level of TGFβRII mRNA in fibroblasts and, therefore, could explain, at least partly, its effect on reprogramming efficiency (Fig. 6H).
FIGURE 6.
Expression of hsa-miR-519a enhances reprogramming efficiency. A, cell cultures of dFib-OCT4GFP–indOSK-11 were infected with lentiviruses encoding the indicated miRs. Relative reprogramming efficiency normalized to the efficiency observed in pmiR-000-infected cells is shown, with the fold changes indicated. Uninfected cells were used as a negative control for all experiments. n = number of independent experiments. The number of alkaline-positive (AP) colonies was used to calculate the relative reprogramming efficiency. Error bars depict the S.D. pmiR-000, control miR. *, p < 0.05; **, p < 0.01. B and C, similar experiment as described in A, but BJ fibroblasts infected with retroviruses encoding the three factors (B, OSK) or the four factors (C, OSKC) plus lentiviruses encoding the indicated miRs, were used instead. *, p < 0.05; **, p < 0.01. The number of Nanog-positive colonies was used to calculate the relative reprogramming efficiency. D and E, cultures of BJ fibroblasts (D) or dFib-OCT4GFP-ind cells (E) were infected with retroviruses encoding the three factors (OSK) and transfected at days 0 and 5 with 30 nm of the indicated miR mimics. Relative reprogramming efficiency normalized to the efficiency observed in mimic control cells is shown with the fold changes indicated. n = number of independent experiments. Error bars depict the S.D. *, p < 0.05; **, p < 0.01. The number of NANOG-positive colonies was used to calculate the relative reprogramming efficiency. F, alignment of different embryonic stem cell-enriched miRs. The seed sequence is highlighted in the box. G, schematic representation of the putative target sites of miR-519a for the TGFβRII mRNA. H, real-time PCR analysis to detect the transcripts of TGFβRII in BJ fibroblasts that were either uninfected or infected with the indicated lentiviruses. Data are shown as relative means ± S.D. of two biological replicates analyzed in triplicate.
Taken together, these data describe the existence of human pluripotent-specific miRs able to increase reprogramming efficiency. Of those, hsa-miR-519a emerged as a new promoter of pluripotency.
Expression of hsa-miR-519a Promotes an Increase in Proliferation
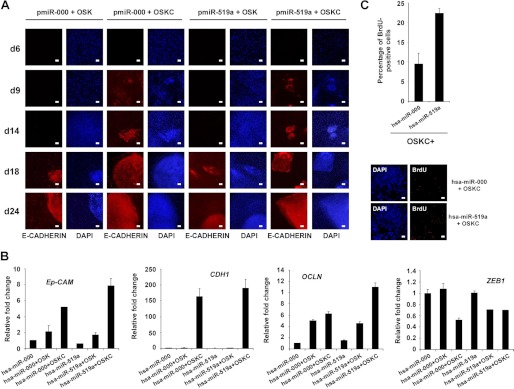
We next investigated the possible molecular mechanisms by which hsa-miR-519a could mediate the observed increase in reprogramming efficiency. Recent reports have shown that fibroblasts undergo a process of mesenchymal-to-epithelial transition (MET) during the reprogramming process (26, 32–33). Interestingly, the expression of certain ESCC (hsa-miR-302b and hsa-miR-372) accelerated the kinetics of MET, increasing reprogramming efficiency (26, 29). This effect is mediated by down-regulation of specific targets, such as TGFβRII or RHOC, that regulate the choice between epithelial and mesenchymal fates (29). Thus, we hypothesized that hsa-miR-519a could facilitate MET through a similar mechanism, as we have shown that this miR can also down-regulate the expression of TGFβRII. To test this possibility, we infected BJ fibroblasts with three (OSK) or four (OSKC) reprogramming factors, together with either hsa-miR-519a or its corresponding control, and followed the expression of the epithelial marker E-cadherin through the reprogramming process at different time points. E-cadherin has been shown to be crucial for embryonic stem cell pluripotency and is as a very reliable marker to identify bona fide hiPSCs derived from mesenchymal cells (34, 35). We observed that modulating hsa-miR-519a levels did not affect the kinetics of E-cadherin induction independently of the expression of cMYC (Fig. 7A). Furthermore, we also analyzed the endogenous expression of several pluripotent markers (OCT4, NANOG, ZPF42, and DPPA4) in BJ fibroblasts infected with three (OSK) or four (OSKC) reprogramming factors, together with either hsa-miR-519a or its corresponding control. Real-time PCR analysis 6 days after the initial infection determined that expression of hsa-miR-519a does not influence the induction of pluripotent markers during the first steps of reprogramming (supplemental Fig. S4B). Additionally, we analyzed the expression of different epithelial (Ep-CAM, E-cadherin and OCL (Occludin)) and mesenchymal (ZEB1) markers in RNA obtained form BJ fibroblasts expressing OSK or OSKC and either hsa-miR-519a or its corresponding control. We observed a small increase in the expression of Ep-CAM and OCLN when hsa-miR-519a was coexpressed with OSKC compared with its respective control (Fig. 7B). However, we did not detect changes in the expression of E-cadherin in similar experimental conditions (Fig. 7B). Moreover, we also observed a decrease in the expression of ZEB1 when hsa-miR-519a was coexpressed with OSK compared with its respective control (Fig. 7B). Importantly, these small changes in the expression of epithelial and mesenchymal markers could reflect not a change in the kinetics of MET but an increase in the number of cells being reprogrammed. Thus, we monitored the proliferation level of BJ fibroblasts infected with OSKC and either hsa-miR-519a or its corresponding control 6 days after viral infection. We observed a consistent increase in the number of BrdU+ cells when hsa-miR-519a was expressed compared with cells expressing the control miR (Fig. 7C). Overall, these results suggest that hsa-miR-519a increases reprogramming efficiency by modulating the proliferation levels of the cells subjected to reprogramming.
FIGURE 7.
Expression hsa-miR-519a increases proliferation. A, BJ fibroblasts were infected with three (OSK) or four (OSKC) reprogramming factors together with either hsa-miR-519a or has-miR-000 (scrambled control). Expression of E-cadherin was monitored by immunofluorescence analysis at different time points during reprogramming. DAPI was used to visualize the nuclei. d, day. Scale bar = 200 μm. B, real-time PCR analysis to detect the endothelial (Ep-CAM, E-cadherin, and OCLN) and mesenchymal (ZEB1) markers were performed with RNA obtained from BJ fibroblasts infected with three (OSK) or four (OSKC) reprogramming factors together with either hsa-miR-519a or has-miR-000 (scrambled control). RNA extraction was performed 6 days after infection. Data are shown as mean ± S.D. of two biological replicates analyzed in triplicate. C, graph showing the percentage of BrdU+ cells in BJ fibroblast 6 days after infection with OSKC plus lentiviruses expressing the indicated miRs (upper panel). Three independent experiments were performed, and the results are represented as means ± S.D. of at least 200 cells in four different fields in each experiment. Immunofluorescence analysis showed a representative field (lower panel). DAPI was used to visualize nuclei. Scale bar = 200 μm.
DISCUSSION
Somatic cell reprogramming in induced pluripotent stem cells is a process not yet completely understood. Different parameters such as low reprogramming efficiency, heterogeneity among the cellular systems, or variable protocols used for cell reprogramming further complicate the analysis of this process. In the last few years, the development of reprogramming strategies on the basis of doxycycline-inducible lentiviruses, which allowed for temporal control of the expression of the reprogramming factors, facilitated the definition of the molecular cornerstones that occur during the transition toward the iPSC state (8, 9). Moreover, these systems have also been very valuable for screening purposes using mouse cells when drug-inducible somatic cell lines harbor reporter genes that allow for the identification of fully reprogrammed cells (15, 16). These screenings have led to the identification of numerous chemical compounds or transcription factors that shed light onto how cell reprogramming occurs. Importantly, although similar reprogramming systems have been developed using human cells (13, 14), the absence of reporter genes precluded their implementation in high-throughput screenings. Furthermore, the existence of molecular and mechanistic differences between the reprogramming process in mouse and human cells warrants the generation of human reporter reprogramming systems in human cells.
In this work, we report a drug-inducible human reprogramming reporter system that serves as a reliable and simple system that can be used for screening purposes in a high-throughput manner. The reprogramming of these cells can be readily tracked by the reactivation of the endogenous OCT4 promoter through the appearance of GFP and by the addition of doxycycline without the need of additional viral infections. We described the derivation of drug-inducible human fibroblast-like cell lines that express the four reprogramming factors or different subsets of them that can be used to screen functional substitutes of the factors as well as to gain insight into the reprogramming process. When performing a high-throughput screening (e.g. to identify chemical substitutes of reprogramming factors) using these types of cellular tools, proper screening optimization is critical. For example, we determined that different concentrations of doxycycline yield a variable number of hiPSC colonies. Moreover, the selection of specific endpoints in the screening must be defined carefully. The kinetics in the appearance of GFP-positive colonies can vary depending on the experimental setting (e.g. performing reprogramming on MEFs versus Matrigel) and the type of screening being performed (e.g. drug screenings involving chemicals in solvents such as dimethyl sulfoxide could alter reprogramming kinetics). Importantly, the development of automated devices able to scan multi-well plates, obtain live-cell images, and readily detect GFP-positive colonies can rapidly facilitate high-throughput screens. Positive “hits” obtained by this type of streamlined method should still be validated in different somatic cell types (e.g. primary fibroblasts). This process of validation is important to rule out any false positives that may exist (e.g. chemical molecules that activate the previously silenced retroviral constructions used to generate the fibroblast-like cell lines).
As a proof of principle of this system, we performed a screening of pluripotent-enriched miRs that allowed us to identify miRs that significantly enhance the reprogramming process. Of those, we found hsa-miR-519a as a novel inducer of reprogramming efficiency. Furthermore, we determined that results obtained from our human reprogramming reporter system were validated in independent human somatic cells.
To get insights into the molecular mechanisms by which hsa-miR-519a can increase reprogramming efficiency, we first sought to identify its putative targets. Among them, we observed that expression of hsa-miR-519a down-regulates TGFβRII. Importantly, a variety of different miRs with a similar seed sequence had been described to increase reprogramming efficiency by down-regulating TGFβRII (24, 29). In fact, inhibition of TGFβ signaling facilitates the induction of iPSC formation, whereas its activation blocks reprogramming (16, 36). This effect seems to be mediated by facilitating the process of MET during reprogramming (26, 29). Although we observed small increases in the expression of certain epithelial markers when hsa-miR-519a is coexpressed with OSKC, we did not detect accelerated kinetics in the appearance of E-cadherin+ hiPSC colonies. We determined that this might be due to the accelerated proliferation mediated by hsa-miR-519a, which increases the number of cells that are being reprogrammed. In fact, it has been reported that a high proliferation rate in reprogramming cells resulted in accelerated kinetics of iPSC formation (17, 37). Although down-regulation of TGFβRII could help to promote proliferation after hsa-miR-519a expression, we cannot exclude the possibility that modification in the levels of direct or indirect additional targets could also contribute to that effect. Overall, this cellular system provides a valuable tool to enable the advancement of reprogramming technologies in human cells.
Supplementary Material
Acknowledgments
We thank the members of the Stem Cell Core at the Salk Institute for continuous support, Marc Daniel and Nicole Méritet for technical assistance, Mercè Marti and Cristina Morera for excellent work at the Histology and Bioimaging Department in the Center of Regenerative Medicine in Barcelona, and the rest of the Belmonte laboratory.
This work was supported, in whole or in part, by grants from National Institutes of Health Grant HL107442, MINECO (Ministerio de Economía y Competitividad), Fundacion Cellex, Sanofi, The Leona M. and Harry B. Helmsley Charitable Trust, and the G. Harold and Leila Y. Mathers Charitable Foundation.

This article contains supplemental Figs. S1–S4, Tables S1 and S2, and Experimental Procedures.
- miPSC
- mouse-induced pluripotent stem cell
- MEF
- mouse embryonic fibroblast
- AP
- alkaline phosphatase
- miR
- microRNA
- MET
- mesenchymal-to-epithelial transition
- GFP
- green fluorescent protein
- hES cell
- human embryonic stem cell
- hiPSC
- human-induced pluripotent stem cell
- OSKC
- OCT4, SOX2, KLF4 and cMYC combination
- OSK
- OCT4, SOX2 and KLF4 combination
- SCID
- severe combined immunodeficiency
- bFGF
- basic fibroblast growth factor
- NEAA
- non essential amino acids.
REFERENCES
- 1. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 2. Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Plath K., Hochedlinger K. (2007) Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1, 55–70 [DOI] [PubMed] [Google Scholar]
- 3. Okita K., Ichisaka T., Yamanaka S. (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 4. Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 5. Zhao X. Y., Li W., Lv Z., Liu L., Tong M., Hai T., Hao J., Guo C. L., Ma Q. W., Wang L., Zeng F., Zhou Q. (2009) iPS cells produce viable mice through tetraploid complementation. Nature 461, 86–90 [DOI] [PubMed] [Google Scholar]
- 6. Boland M. J., Hazen J. L., Nazor K. L., Rodriguez A. R., Gifford W., Martin G., Kupriyanov S., Baldwin K. K. (2009) Adult mice generated from induced pluripotent stem cells. Nature 461, 91–94 [DOI] [PubMed] [Google Scholar]
- 7. Kang L., Wang J., Zhang Y., Kou Z., Gao S. (2009) iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell 5, 135–138 [DOI] [PubMed] [Google Scholar]
- 8. Brambrink T., Foreman R., Welstead G. G., Lengner C. J., Wernig M., Suh H., Jaenisch R. (2008) Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stadtfeld M., Maherali N., Breault D. T., Hochedlinger K. (2008) Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2, 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stadtfeld M., Maherali N., Borkent M., Hochedlinger K. (2010) A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat. Meth. 7, 53–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wernig M., Lengner C. J., Hanna J., Lodato M. A., Steine E., Foreman R., Staerk J., Markoulaki S., Jaenisch R. (2008) A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 26, 916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Markoulaki S., Hanna J., Beard C., Carey B. W., Cheng A. W., Lengner C. J., Dausman J. A., Fu D., Gao Q., Wu S., Cassady J. P., Jaenisch R. (2009) Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat. Biotechnol. 27, 169–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hockemeyer D., Soldner F., Cook E. G., Gao Q., Mitalipova M., Jaenisch R. (2008) A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3, 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maherali N., Ahfeldt T., Rigamonti A., Utikal J., Cowan C., Hochedlinger K. (2008) A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell, 3, 340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi Y., Desponts C., Do J. T., Hahm H. S., Schöler H. R., Ding S. (2008) Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell 3, 568–574 [DOI] [PubMed] [Google Scholar]
- 16. Ichida J. K., Blanchard J., Lam K., Son E. Y., Chung J. E., Egli D., Loh K. M., Carter A. C., Di Giorgio F. P., Koszka K., Huangfu D., Akutsu H., Liu D. R., Rubin L. L., Eggan K. (2009) A small-molecule inhibitor of tgf-β signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5, 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zwaka T. P., Thomson J. A. (2003) Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 21, 319–321 [DOI] [PubMed] [Google Scholar]
- 18. Ruiz S., Panopoulos A. D., Herrerías A., Bissig K. D., Lutz M., Berggren W. T., Verma I. M., Izpisua Belmonte J. C. (2011) A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 21, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carey B. W., Markoulaki S., Hanna J. H., Faddah D. A., Buganim Y., Kim J., Ganz K., Steine E. J., Cassady J. P., Creyghton M. P., Welstead G. G., Gao Q., Jaenisch R. (2011) Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 9, 588–598 [DOI] [PubMed] [Google Scholar]
- 20. Wernig M., Meissner A., Cassady J. P., Jaenisch R. (2008) c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2, 10–12 [DOI] [PubMed] [Google Scholar]
- 21. Bartel D. P. (2009) MicroRNAs. Target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mallanna S. K., Rizzino A. (2010) Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev. Biol. 344, 16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Judson R. L., Babiarz J. E., Venere M., Blelloch R. (2009) Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 27, 459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z., Yang C. S., Nakashima K., Rana T. M. (2011) Small RNA-mediated regulation of iPS cell generation. EMBO J. 30, 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang C. S., Li Z., Rana T. M. (2011) microRNAs modulate iPS cell generation. RNA 17, 1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liao B., Bao X., Liu L., Feng S., Zovoilis A., Liu W., Xue Y., Cai J., Guo X., Qin B., Zhang R., Wu J., Lai L., Teng M., Niu L., Zhang B., Esteban M. A., Pei D. (2011) MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem. 286, 17359–17364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anokye-Danso F., Trivedi C. M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P. J., Epstein J. A., Morrisey E. E. (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8, 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miyoshi N., Ishii H., Nagano H., Haraguchi N., Dewi D. L., Kano Y., Nishikawa S., Tanemura M., Mimori K., Tanaka F., Saito T., Nishimura J., Takemasa I., Mizushima T., Ikeda M., Yamamoto H., Sekimoto M., Doki Y., Mori M. (2011) Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8, 633–638 [DOI] [PubMed] [Google Scholar]
- 29. Subramanyam D., Lamouille S., Judson R. L., Liu J. Y., Bucay N., Derynck R., Blelloch R. (2011) Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 29, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y., Baskerville S., Shenoy A., Babiarz J. E., Baehner L., Blelloch R. (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 40, 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin C. H., Jackson A. L., Guo J., Linsley P. S., Eisenman R. N. (2009) Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J., 28, 3157–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., Qin D., Feng S., Song H., Yang D., Zhang B., Zeng L., Lai L., Esteban M. A., Pei D. (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 33. Samavarchi-Tehrani P., Golipour A., David L., Sung H. K., Beyer T. A., Datti A., Woltjen K., Nagy A., Wrana J. L. (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 [DOI] [PubMed] [Google Scholar]
- 34. Chen H. F., Chuang C. Y., Lee W. C., Huang H. P., Wu H. C., Ho H. N., Chen Y. J., Kuo H. C. (2011) Surface marker epithelial cell adhesion molecule and E-cadherin facilitate the identification and selection of induced pluripotent stem cells. Stem Cell Rev. 7, 722–735 [DOI] [PubMed] [Google Scholar]
- 35. Redmer T., Diecke S., Grigoryan T., Quiroga-Negreira A., Birchmeier W., Besser D. (2011) E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 12, 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maherali N., Hochedlinger K. (2009) Tgfβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 19, 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanna J., Saha K., Pando B., van Zon J., Lengner C. J., Creyghton M. P., van Oudenaarden A., Jaenisch R. (2009) Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.