Background: How botulinum neurotoxin serotype C (BoNT/C) enters neurons is unclear.
Results: BoNT/C utilizes dual gangliosides as host cell receptors.
Conclusion: BoNT/C accesses gangliosides on the plasma membrane.
Significance: Plasma membrane accessibility of the dual ganglioside receptors suggests synaptic vesicle exocytosis may not be necessary to expose BoNT/C receptors.
Keywords: Bacterial Toxins, Botulinum Toxin, Ganglioside, Neurons, Receptors
Abstract
Botulinum neurotoxins (BoNTs) cleave SNARE proteins in motor neurons that inhibits synaptic vesicle (SV) exocytosis, resulting in flaccid paralysis. There are seven BoNT serotypes (A–G). In current models, BoNTs initially bind gangliosides on resting neurons and upon SV exocytosis associate with the luminal domains of SV-associated proteins as a second receptor. The entry of BoNT/C is less clear. Characterizing the heavy chain receptor binding domain (HCR), BoNT/C was shown to utilize gangliosides as dual host receptors. Crystallographic and biochemical studies showed that the two ganglioside binding sites, termed GBP2 and Sia-1, were independent and utilized unique mechanisms to bind complex gangliosides. The GBP2 binding site recognized gangliosides that contained a sia5 sialic acid, whereas the Sia-1 binding site recognized gangliosides that contained a sia7 sialic acid and sugars within the backbone of the ganglioside. Utilizing gangliosides that uniquely recognized the GBP2 and Sia-1 binding sites, HCR/C entry into Neuro-2A cells required both functional ganglioside binding sites. HCR/C entered cells differently than the HCR of tetanus toxin, which also utilizes dual gangliosides as host receptors. A point-mutated HCR/C that lacked GBP2 binding potential retained the ability to bind and enter Neuro-2A cells. This showed that ganglioside binding at the Sia-1 site was accessible on the plasma membrane, suggesting that SV exocytosis may not be required to expose BoNT/C receptors. These studies highlight the utility of BoNT HCRs as probes to study the role of gangliosides in neurotransmission.
Introduction
Botulinum neurotoxins (BoNTs)4 intoxicate α-motor neurons at the neuromuscular junction to elicit flaccid paralysis (1). There are seven BoNT serotypes (A–G); BoNT/A, B, E, and F are associated with human botulism, whereas BoNT/C and D are associated with animal botulism. A phenomenon observed within the BoNT/C and D family is the presence of naturally arising chimeric and mosaic toxins in addition to the prototypical serotypes (2). One mosaic toxin, BoNT/D-C is a combination of BoNT/C and BoNT/D with functional and antigenic properties that are similar but also unique relative to the parental neurotoxins (3–6). BoNTs are the most toxic proteins for humans and are category A select agents. Although historically limited to animal botulism, members of the BoNT/C and D family are toxic to non-human primates and may have potential therapeutic applications, justifying continued research efforts (7).
BoNTs, initially produced and secreted as a single polypeptide, are then processed proteolytically into di-chain proteins with AB structure-function organization. The N-terminal light chain (LC) (the A domain) is linked by a disulfide bond to the C-terminal heavy chain (HC) (the B domain). LC comprises a zinc protease that cleaves SNARE proteins, whereas the HC comprises two independent functional domains: the translocation domain (HCT) and the receptor binding domain (HCR) (8). BoNT neurotropism is attributed to the preferred affinity for neurons and the cleavage of neuron-specific SNARE proteins.
BoNTs enter neurons by a dual host receptor mechanism (9). For BoNT serotypes that have defined receptors, the dual receptors are a ganglioside and a synaptic vesicle (SV)-associated protein (10). The current model for BoNT entry involves the initial binding to gangliosides on resting neurons and then, upon SV exocytosis, association with a luminal domain of a SV-associated protein. Upon internalization, the lumen of the SV acidifies to trigger translocation of the LC into the cytosol followed by cleaving SNARE proteins (11). SNARE proteins are transmembrane proteins located on the plasma membrane and SVs (12). LC/A, C, and E cleave SNAP25 at unique sites; LC/C also cleaves syntaxin. LC/B, D, F, and G and tetanus toxin (TeNT) cleave synaptobrevin 2 (Syb2, also called VAMP2) at unique sites. LC/B and LC/TeNT cleave the same site on VAMP2. This shows the importance of intracellular trafficking in eliciting the unique pathologies of botulism and tetanus (13, 14). SNARE protein cleavage in peripheral motor neurons disrupts SV fusion to the plasma membrane and subsequent neurotransmitter release, inhibiting muscle contraction at the neuromuscular junction (14).
The BoNT HCR contains the binding sites for ganglioside and SV protein receptor (15, 16). Gangliosides are glycosphingolipids with the ceramide component imbedded in the outer leaflet of the plasma membrane, whereas the carbohydrate moiety extends into the extracellular space. The composition of the carbohydrate moiety of complex gangliosides includes a sugar backbone consisting of glucose, galactose, and N-acetylgalactosamine, with attached sialic acids to varying degrees and positions. Complex gangliosides are enriched in neuronal tissue (17, 18). Ganglioside binding was initially characterized in TeNT, which was shown to bind to a shallow pocket within the HCR that contains a conserved His … Ser-X-Trp-Tyr motif (19, 20). The indole ring of Trp stacks against the hydrophobic face of the terminal galactose moiety of the ganglioside backbone, whereas His, located opposite Trp, contacts hydroxyl groups present on the sugars of the ganglioside backbone. Tyr forms the back face of the ganglioside binding pocket, whereas Ser forms hydrogen bonds with sugar hydroxyl groups of the ganglioside backbone. This ganglioside binding site is structurally conserved among TeNT and BoNT/A, B, and F and has been designated the ganglioside binding pocket (GBP). BoNT/E and G possess the Ser, Trp, and Tyr characteristic of the GBP, but substitutions for the His residue set these serotypes apart. It is yet to be determined how the unique contacts in the GBP of BoNT/E and G influence ganglioside binding.
Resident SV proteins were subsequently identified as the second receptor; BoNT/A, E, and F binds synaptic vesicle protein 2 (SV2) with differential preference for SV2 isoforms (21–23), and BoNT/B and G bind synaptotagmin I and II (15, 16, 24). The crystal structure of HCR/B in complex with the luminal peptide of synaptotagmin showed that the C-terminal β-trefoil domain of HCR/B was lined by hydrophobic residues that contacted the luminal peptide of synaptotagmin, which will be designated the second receptor binding site (SRS). Less is known about how BoNT/C associates with host receptors.
An early study found that BoNT/C, but not BoNT/D, bound GD1b and GT1b and that BoNT/C toxicity decreased in ganglioside-deficient mice (25). This study concludes that BoNT/C uses gangliosides as functional receptors, whereas BoNT/D intoxication is ganglioside-independent (25). Subsequent studies found that BoNT/D toxicity is dependent on complex ganglioside expression and that both BoNT/C and/D utilize a synaptic vesicle-dependent pathway to enter neurons (26–29).
Recent studies implicate a unique mechanism of ganglioside binding by BoNT/C and BoNT/D relative to the other BoNT serotypes. Crystal structures of HCR/C, HCR/D, and a D-C mosaic HCR (HCR/D-C South Africa) show that although the overall architecture of the GBP is similar to other BoNT serotypes, three of the four conserved residues of the GBP (His, Ser, and Trp) are absent (3). Further analysis has identified a β-hairpin loop that contained a Trp residue with HCR/C, HCR/D, and HCR/D-C South Africa. This β-hairpin loop, which aligns near the SRS and is required for ganglioside and neuron binding, is termed the ganglioside binding loop (GBL) (3, 30). Additional studies have expanded our understanding of how BoNT/C and BoNT/D bind cells. Strotmeier et al. (28) found that the Trp within the GBL of BoNT/D of is necessary for the expression of toxicity and that another residue within the GBP (Asp-1233) also contributes to toxicity, allowing the conclusion that BoNT/D entry into neurons is dependent on multiple carbohydrate interactions. Other studies performed with BoNT/C have arrived at similar conclusions and also found an additional sialic acid binding site (Sia-1) that overlaps with the synaptotagmin binding site in BoNT/B and BoNT/G that contributes to toxicity (15, 16, 26, 31). These studies highlight the contribution of the GBL to BoNT/C and BoNT/D entry and identify two potential ganglioside binding sites in HCR/C and HCR/D: the established GBP and the Sia-1 site.
These studies provide a foundation for defining the mechanism of BoNT/C entry into neurons. Using a gain-of-function cell-based assay we confirmed that BoNT/C utilized gangliosides to enter cells and established that the ganglioside binding pocket bound sia5-containing gangliosides via a Trp-independent mechanism. This site was designated GBP2 to reflect the unique contacts mediating ganglioside binding. The previously identified Sia-1 site was found to bind sia7-containing gangliosides (GD1b > GT1b). Disruption of either ganglioside binding site ablated HCR/C entry into primary neurons. Unexpectedly, one HCR/C derivative deficient in GBP2 binding, but with an intact Sia-1 site, bound and entered Neuro-2A (N2A) cells enriched with exogenous ganglioside, albeit less efficient than wild-type HCR/C. These findings provided direct evidence for the presence of two independent ganglioside binding sites in HCR/C. Binding two gangliosides as host receptors was unique to BoNT/C, yet HCR/C entry into neurons was enhanced with synaptic activity. This suggests the dual receptors for HCR/C are present on the plasma membrane and that SV exocytosis may not be necessary to expose BoNT/C receptors but rather may enhance entry because of the compensatory increase in SV endocytosis following depolarization (32).
EXPERIMENTAL PROCEDURES
Materials
Escherichia coli codon-optimized DNA encoding the HCR domain of BoNT/C-Stockholm (residues 864–1290) and BoNT/D-C-5995 (residues 867–1285) were synthesized by EZ Biolab (Westfield, IN). Chemicals and reagents were obtained from Sigma-Aldrich, and restriction enzymes were purchased from New England Biolabs (Ipswitch, MA) or Invitrogen (Carlsbad, CA). Neuro-2A cells (CCL-131) were purchased from ATCC (Manassas, VA) and cultured as recommended. Cell culture reagents were purchased from Invitrogen.
HCR Expression and Purification
DNA encoding HCRs were subcloned into pET-28a (EMD Millipore, Billerica, MA), which introduced a 3-FLAG epitope on the N terminus of the HCR. HCRs were purified from E. coli BL-21(DE3) cells using affinity chromatography. For HCRs used for biochemical characterization, cells were lysed in a detergent-based buffer (B-Per II) (Thermo Scientific, Rockford, IL) supplemented with bacterial protease inhibitors and DNase/RNase followed by a single-step purification utilizing Ni2+-nitrilotriacetic acid spin columns (Qiagen, Valencia, CA). For crystallization, E. coli cells were lysed by French press followed by three sequential chromatography steps: Ni2+-nitrilotriacetic acid resin, S200-HR gel filtration, and DEAE-Sephacryl ion exchange resin. Typical purifications from a 1-liter culture yielded between 5 and 10 mg of HCR.
Molecular Biology of HCR Domain
DNA encoding HCR/C was mutagenized using a QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Plasmid DNA was sequenced to verify the specificity of the mutation. A chimeric HCR (HCR/D-C(GBL/C)) was generated by overlap PCR (33). The PCR fragment was digested with KpnI and PstI and subcloned into pET28a. The plasmid was sequenced to verify DNA manipulation. HCR domains with a 3-HA epitope were generated for cell-based trafficking studies. The HCR DNA was digested with KpnI and NotI and subcloned into a 3-HA expression vector (34).
HCR-Ganglioside Binding Assay
Gangliosides (Matreya, Pleasant Gap, PA) were diluted to a stock concentration of 20 mg/ml in DMSO for storage at −20 °C. A final concentration at 20 μg/ml of the indicated ganglioside was prepared in 100 μl of methanol, added to a nonbinding 96-well plate (Corning Inc.), and allowed to evaporate overnight. The plate was washed three times with PBS and blocked with carbonate buffer (50 mm Na2CO3, pH 9.6) plus 2% bovine serum albumin (BSA) for 1 h at 4 °C. Following three washes with PBS, wells were incubated with the indicated HCR in PBS plus 2% BSA for 1 h at 4 °C. Bound HCR was detected with a complex of α-FLAG (Sigma) primary and HRP (Thermo Scientific) secondary antibody for 30 min at 4 °C. The plate was washed three times with PBS and developed with room temperature 3,3′,5,5′-tetramethylbenzidene (TMB) substrate for 20 min. The reaction was stopped with 1 m H2SO4. The plate was read at 450 nm on a plate reader. The signal detected in a well containing HCR minus ganglioside was used for nonspecific binding.
HCR Entry into Neuro-2A Cells
Gangliosides were diluted to the final desired concentration in N2A medium with 0.5% fetal bovine serum and sonicated for 20 min. Neuro-2A cells were cultured to confluence on coverslips coated with collagen. Cells were washed and incubated in untreated medium or with sonicated ganglioside in N2A medium for 4 h to allow ganglioside enrichment of membranes. Cells were washed and incubated with the indicated 3-HA-HCR (40 nm) in PBS for 30 min at 37 °C. Cells were washed and processed for microscopy. Briefly, cells were fixed in 4% paraformaldehyde in PBS at room temperature for 20 min, permeabilized with 4% formaldehyde plus 0.1% Triton X-100, quenched with glycine in PBS, and blocked in 10% FBS, 2.5% fish skin gelatin, 0.1% Triton X-100, and 0.05% Tween 20. Bound HCR and endogenous synaptophysin were identified by immunofluorescence using α-HA (Sigma) and α-synaptophysin antibody (Synaptic Systems, Goettingen, German), respectively. Cells were washed and incubated with fluorescent secondary antibodies (Invitrogen). Following washes and a final fixation in 4% paraformaldehyde, the coverslips were mounted with Prolong Gold (Invitrogen). Images were captured on an Eclipse TE2000 inverted microscope (Nikon Instruments Inc., Melville, NY) equipped with a CFI Plan Apo VC60 oil, NA1.4-type objective, and Cool Snap HQ2 camera (Photometrics, Tucson, AZ). MetaMorph image acquisition software (Molecular Devices, Sunnyvale, CA) was used. Representative images were pseudo-colored and set to equivalent brightness and contrast levels using ImageJ software (National Institutes of Health, Bethesda, MD).
Microscopy Image Analysis
For quantification of fluorescence intensity, exposure times and instrument settings were consistent between treatments. Three images of similar cell density and morphology were captured for each treatment. Images were threshold equivalently, and the arbitrary HCR intensity was measured using ImageJ. The average HCR intensity measured in cells without exogenous ganglioside was subtracted from the average HCR intensity in cells enriched with ganglioside. For cell enumeration, the fluorescence intensity of GT1b-enriched cells established image threshold conditions that were applied directly to untreated cells or cells enriched with other gangliosides; this was done using ImageJ. Data were analyzed using Excel (Microsoft, Seattle, WA) and GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). Statistical significance was determined using a two-tailed t test.
HCR Entry into Primary Cortical Neurons
Rat E18 cortical neurons were cultured on poly-d-lysine-coated glass coverslips in neurobasal medium with 2 mm glutamine and B27 supplement for 10–14 days prior to use. Entry and trafficking of HCRs in neurons were examined after incubation of 40 nm HCR/A1 or HCR/C in low potassium (15 mm HEPES, 145 mm NaCl, 5.6 mm KCl, 2.2 mm CaCl, and 0.5 mm MgCl, pH 7.4) or high potassium (15 mm HEPES, 95 mm NaCl, 56 mm KCl, 2.2 mm CaCl, and 0.5 mm MgCl, pH 7.4) buffer for 5 min at 37 °C. Cells were washed with Dulbecco's phosphate-buffered saline (DPBS), fixed with 4% (w/v) paraformaldehyde in DPBS, and permeabilized with 4% formaldehyde plus 0.1% Triton X-100 in DPBS. Cells were incubated with 150 mm glycine in DPBS, washed with DPBS, and blocked with 10% (v/v) normal goat serum, 2.5% (w/v) cold fish skin gelatin (Sigma), 0.1% Triton-X-100, and 0.05% Tween-20 in DPBS (blocking solution) for 1 h at room temperature followed by incubation with rat α-HA antibody and guinea pig anti-synaptophysin antibody in 5% (v/v) normal goat serum, 1% (w/v) cold fish skin gelatin, 0.1% Triton X-100, and 0.05% Tween-20 in DPBS (antibody solution) overnight at 4 °C. Cells were washed with DPBS and incubated with goat α-rat IgG Alexa 488 and goat α-guinea pig IgG Alexa 568 for 1 h at room temperature. Follow the addition of a secondary antibody, cells were washed, fixed, washed, and mounted using Citifluor AF-3 (Electron Microscopy Sciences, Hatfield, PA). Images were captured with a Nikon TE2000 TIRF (total internal reflection fluorescence) microscope equipped with a CFI Plan Apo VC 100× oil, NA 1.4-type objective. Image acquisition and analyses were performed using Nikon Elements, version 4.0 (Nikon Instruments). Briefly, the resulting images from the various HCRs tested were set to equivalent brightness and contrast levels. The arbitrary fluorescence was then measured. A minimum of five random fields was captured for each HCR, quantified, and averaged. The data presented are the average of four independent experiments.
Crystallization and Data Collection
Purified HCR/D-C(GBL/C) was concentrated to 5 mg/ml in 10 mm Tris-HCl, pH 7.6, and 200 m NaCl. Crystals produced by the vapor diffusion hanging drop method contained 2 μl of the concentrated protein solution mixed with 2 μl of 0.1 m HEPES, pH 8.5, 16% PEG 5000 MME, and 50 mm MgSO4. The complex of HCR/D-C(GBL/C) bound to sialic acid (referred to as HCR/D-C(GBL/C)-Sia) was obtained by soaking the crystals of HCR/D-C(GBL/C) in mother liquid containing 50 mm sialic acid overnight. Diffraction data for HCR/D-C(GBL/C)-Sia were collected using an R-AXIS IV2+ with a MicroMax 007 generator at 100 K. HKL2000 was used for data processing (35). Data collection and processing statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
Numbers in parentheses are values for the highest resolution shell. r.m.s.d., root mean square deviation.
| Crystal (PDB: 4FVV) | HCR/D-C-GBL/C1-Sia |
|---|---|
| Diffraction data | |
| Resolution range (Å) | 50–2.70 (2.80–2.70) |
| No. total reflections | 95,326 |
| No. unique reflections | 31,121 (3,024) |
| Completeness (%) | 96.4 (95.1) |
| Redundancy | 3.1 (3.1) |
| I/σ(I) | 9.8 (2.0) |
| Unit cell a, b, c (Å) | 82.6, 154.3, 181.0 |
| Space group | I222 |
| Rsymm | 0.113 (0.505) |
| Vm (Å3/Da)/solvent content (%) | 2.9/56 |
| Monomers in an asymmetric unit | 2 |
| Refinement | |
| Rcrystal/Rfree | 0.206 (0.263) |
| r.m.s.d. bond length/bond angle | 0.007/1.4 |
| No. protein atoms | 6,741 |
| No. water molecules | 190 |
| No. glycerol atoms | 18 |
| No. SO4 atoms | 15 |
| No. sialic acid atoms | 21 |
| Average B-factors | |
| Protein atoms (Å2) | 34.8 |
| Water molecules (Å2) | 24.2 |
| Glycerol (Å2) | 47.0 |
| SO4 (Å2) | 56.7 |
| Sialic acid (Å2) | 58.3 |
| Ramachandran statistics | |
| Most favored regions (%) | 82.0 |
| Additional allowed regions (%) | 17.6 |
| Generously allowed regions (%) | 0.4 |
| Disallowed regions (%) | 0 |
Structure Determination and Refinement
The structure of HCR/D-C(GBL/C) was solved by the molecular replacement method using the PHASER (36) program and the structure of HCR/D-C (residues 863–1284; PDB code, 3N7L) (3). The initial structure obtained from molecular replacement trials was refined using the program CNS (37). The refinement procedure consisted of rigid body and positional refinements followed by a simulated annealing protocol. Iterative rounds of positional and temperature factor refinement were followed by manual fitting and rebuilding using the graphics program COOT (38) with [2Fo − Fc] and [Fo − Fc] difference Fourier maps (38). The final model was completed with Rcrystal/Rfree values of 0.206/0.263.
RESULTS
Ganglioside Enrichment Enhances HCR/C Entry into Neuro-2A Cells
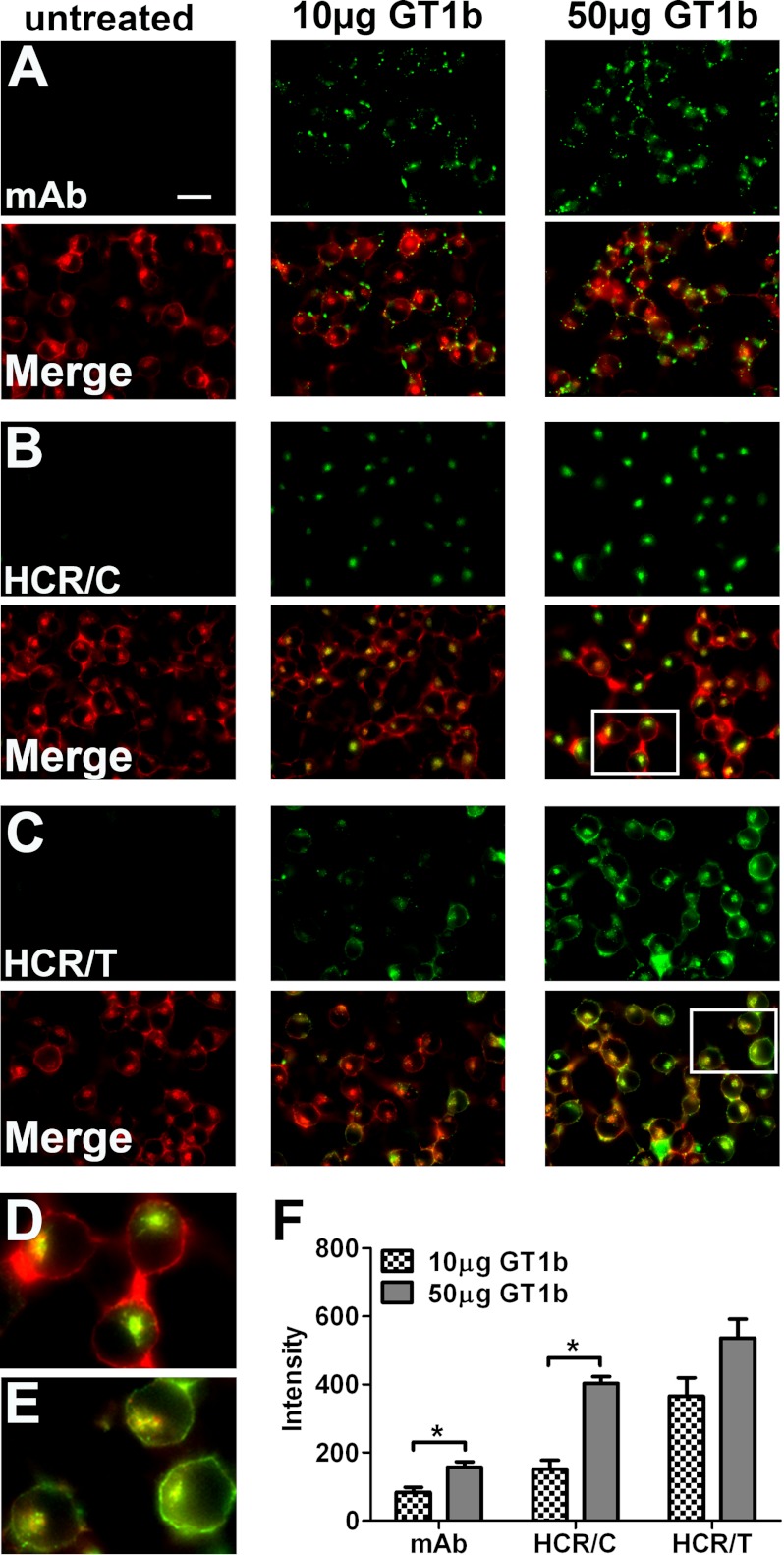
A cell-based assay was used to evaluate the role of gangliosides in HCR/C entry into N2A cells, a mouse neuroblastoma cell line lacking complex gangliosides in the plasma membrane. The plasma membrane of Neuro-2A cells was enriched with GT1b, a complex ganglioside, to establish the assay parameters for studying HCR trafficking. N2A cell morphology was detected by actin staining, which localized on the cell periphery and in an intracellular compartment (Fig. 1). Endogenous GT1b was below detection level in untreated N2A cells, whereas GT1b-enriched cells possessed detectable GT1b on the cell periphery that was proportional to the amount of GT1b enrichment (Fig. 1, A and F). HCR/C did not enter untreated N2A cells, but binding was detected within an intracellular compartment upon GT1b enrichment (Fig. 1, B and D). The amount of cell-associated HCR/C was proportional to the amount of GT1b enrichment (Fig. 1F). Reducing the assay temperature allowed detection of cell surface HCR/C, which was not observed at 37 °C, indicating a rapid transition from the cell surface to intracellular compartments (data not shown). Control experiments showed that HCR/TeNT, which utilizes dual gangliosides as host receptors (39), binding to N2A cells was dependent on and proportional to the amount of GT1b enrichment. Interestingly, the HCR/TeNT signal was detected on the cell periphery and in an intracellular compartment (Fig. 1, C, E, and F).
FIGURE 1.
GT1b-enriched Neuro-2A cells support HCR/C entry. N2A cells were untreated or enriched with 10 or 50 μg of GT1b/well for 4 h. Cells were washed and incubated in PBS alone or with 40 nm of the indicted HCR in PBS for 30 min at 37 °C. Unbound HCR was washed away, and cells were processed for microscopy. Exogenous GT1b was detected with an anti-GT1b antibody (mAb). HCR was identified with an anti-HA antibody. Cellular actin was identified by staining with phalloidin. Image exposure settings were identical between treatments. Representative ×60 micrographs are shown. HCR images were set to equivalent brightness and contrast levels using ImageJ. The brightness of the mAb images was increased relative to the HCR images for publication. A–C, the upper rows show the signals detected for the anti-GT1b antibody (A), HCR/C (B), and HCR/T (C). The lower rows show the merged images created from the antibody or HCR signal overlaid with the actin signal. Scale bar = 20 μm. D, the boxed area from B is expanded and shown here. Notice how the HCR/C signal is exclusively intracellular, as the actin positive compartment appears green/yellow and the cell periphery is red. E, the boxed area from C is expanded and shown here. Notice how the HCR/T signal is intracellular and bound to the plasma membrane as the intracellular compartment and the cell periphery are green/yellow in the merged image. F, quantification of microscopic data. All images (mAb and HCRs) were set to equivalent brightness and contrast for quantification using ImageJ. Three fields were captured per treatment. The average GT1b antibody (mAb) or HCR signal was determined. The average intensity measured in N2A cells without exogenous ganglioside was determined and subtracted from the average intensity in ganglioside-treated wells, and the ganglioside-dependent mAb or HCR intensity is reported. The average of two independent experiments is shown. Asterisk denotes statistical significance (*, p < 0.05).
Identification of a Novel Ganglioside Binding Pocket within a HCR/C Chimera
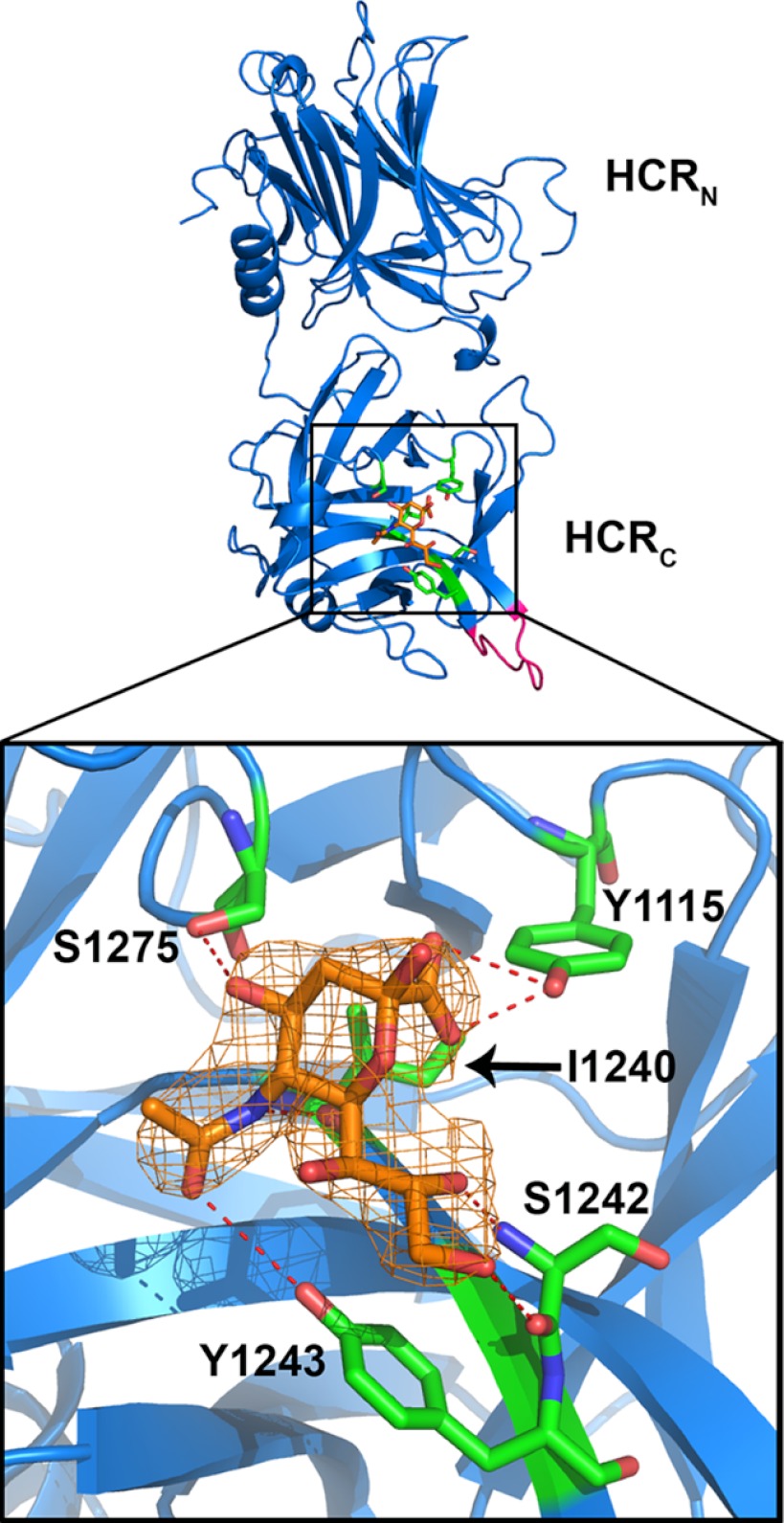
The ganglioside-HCR interaction was tested by creating a chimeric protein between HCR/C and a closely related HCR/D-C. When the structures of HCR/C and HCR/D-C are superimposed, the average root mean square deviation for all α-carbons in the backbones is 0.46Å (3). Thus, HCR/C and HCR/D-C have essentially the same structure. This similarity was utilized to characterize the mechanism of ganglioside binding by BoNT/C. A nine-amino acid segment of HCR/C was swapped with the analogous region of HCR/D-C (BoNT/C-(1252–1260) into BoNT/D-C-(1245–1254)). The resulting chimeric protein, HCR/D-C(GBL/C) was readily crystallized and was used to generate a crystal in complex with sialic acid (PDB code, 4FVV). Bound sialic acid formed hydrogen bonds with Tyr-1115, Ile-1240, Ser-1242, Tyr-1243, and Ser-1275 of the HCR/C chimera (Fig. 2). The side chain hydroxyl of Tyr-1115 formed two H-bonds with O1A and O1B of the carboxylic acid of sialic acid. An H-bond was detected between the backbone carbonyl of Ile-1240 with the N5 of sialic acid. The backbone amide and carbonyl groups of Ser-1242 contacted O8 and O9 in sialic acid, respectively, and the hydroxyl Tyr-1243 H-bonded with O10 of sialic acid. The hydroxyl group of Ser-1275 (OG) contacted the O4 of sialic acid (Table 2). The bound sialic acid, which overlapped physically with the GBP of other BoNT serotypes, was defined as ganglioside binding pocket 2 (GBP2) to reflect the similar location of but different contacts between the ganglioside and HCR than those observed for other BoNT serotypes (3).
FIGURE 2.
A novel sialic acid binding pocket within HCR/D-C(GBL/C). The crystal structure of HCR/D-C(GBL/C) was solved in complex with sialic acid and is shown here (PDB code, 4FVV). Sialic acid is shown in orange. Residues of HCR/D-C(GBL/C) in contact with sialic acid are shown in green and define the GBP2. The N (HCRN)- and C (HCRC)-terminal subdomains of the HCR are labeled. GBL/C is shown in pink. The boxed area is expanded and shown (bottom). The sialic acid electron density is shown. [2Fo − Fc] contoured at 1σ. The residues TYR-1115, Ile-1240, Ser-1242, Tyr-1243, and Ser-1275 in HCR/D-C form direct H-bonds with sialic acid (red dashed lines). Nitrogen atoms are shown in blue and oxygen atoms in red.
TABLE 2.
The distance (Å) of the hydrogen bonds observed between sialic acid and residues within the GBP2 of HCR/D-C
| Sialic acid | Distance | HCR/D-C |
|---|---|---|
| Å | ||
| O1A | 2.8 | Tyr-1115 (OH) |
| O1B | 3.2 | Tyr-1115 (OH) |
| O4 | 2.9 | Ser-1275 (OG) |
| O8 | 2.8 | Ser-1242 (N) |
| O9 | 3.0 | Ser-1242 (O) |
| O10 | 3.5 | Tyr-1243 (OH) |
| N5 | 2.8 | Ile-1240 (O) |
HCR/C Has Two Independent Ganglioside Binding Sites
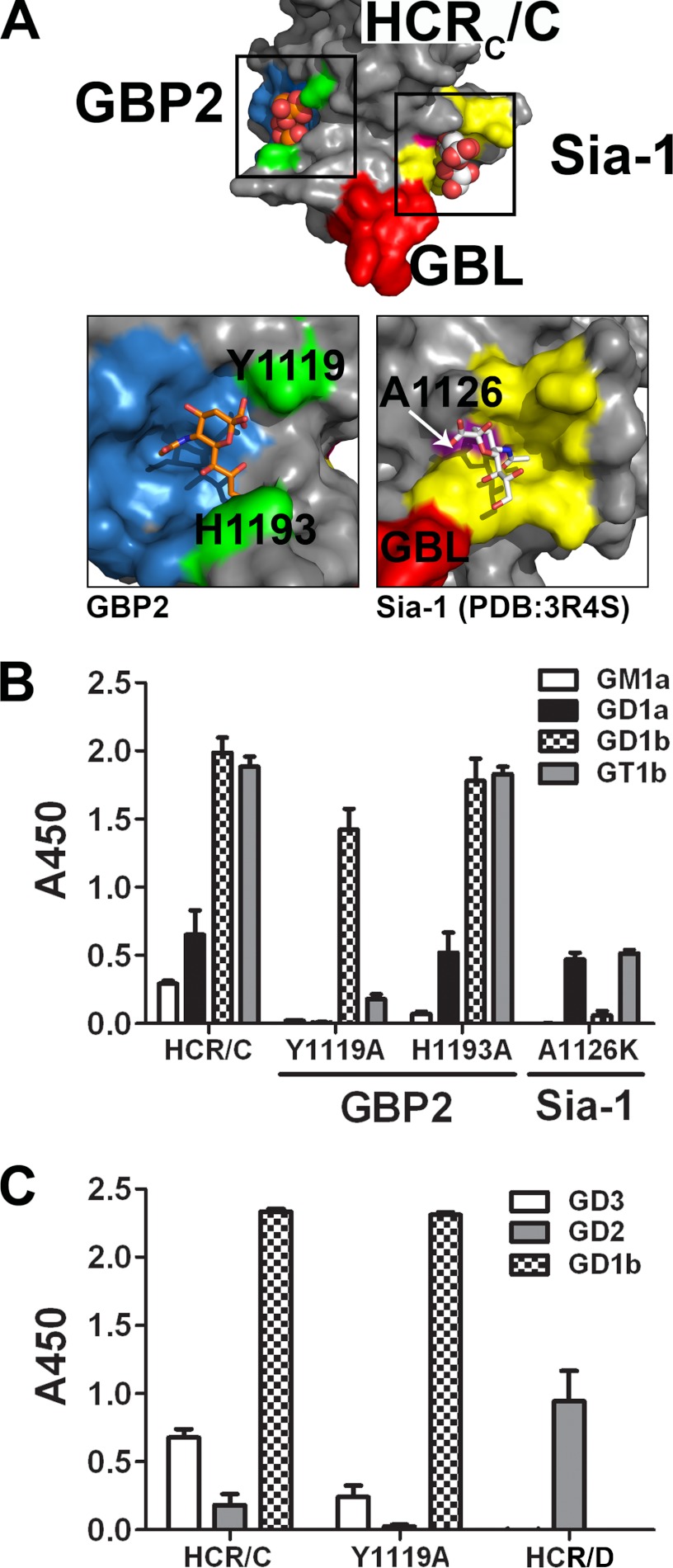
These studies suggest that HCR/C possesses two potential ganglioside binding sites: the GBP2 identified above and a previously identified sialic acid binding site, the Sia-1 site (26). The sialic acid bound to HCR/D-C(GBL/C) was modeled onto the HCR/C structure; from this, the residues predicted to contact sialic acid in the GBP2 of HCR/C include Tyr-1119, Ile-1247, Thr-1249, Tyr-1250, and Ser-1281, as the side chains of these residues align in space with the ganglioside-contacting residues of the HCR/D-C GBP2. The Sia-1 site is formed by several discontiguous peptides (Fig. 3A), Ala-1126 maps to a β-hairpin loop that forms the center of the Sia-1 site and directly contacts the O4 of sialic acid. There are also multiple residues on the central β-hairpin loop that contacted sialic acid either directly or through H2O with residues Tyr-1179 and Leu-1203 (26) on one face and residues Leu-1254, Gly-1255, and Gly-1256 that were part of a previously identified region of the HCR implicated in ganglioside binding, termed the GBL (3).
FIGURE 3.
HCR/C has two independent ganglioside binding sites. A, HCR/C has multiple sialic acid binding sites. The crystal structure of HCR/C (PDB code, 3N7K) (3) is shown with GBP2 highlighted in blue. The GBL is shown in red. A sialic acid site termed the Sia-1 site is shown in yellow (PDB code, 3R4S) (26). The boxed areas were expanded and shown below. Analysis of the HCR/C GBP2 indicates that multiple signature residues of the GBP2 are conserved in space and in identity. In HCR/C, the GBP2 is defined by Tyr-1119, Ile-1246, Thr-1259, Tyr-1250, and Ser-1281. Unlike HCR/D-C, a distal loop segment partially shields Tyr-1250 and contributes His-1193 to the pocket that is structurally analogous to the His of the GBP. The Sia-1 site is shown on the right. Residues in contact with the bound sialic acid (white) or that were identified through mutagenesis are shown in yellow. Ala-1126 is highlighted in pink. GBL residues adjacent to the sialic acid molecule are shown in red. B, point mutations were introduced into the sialic acid binding sites and evaluated for binding to a panel of common gangliosides. Purified gangliosides were immobilized in wells of a 96-well plate and probed with the indicated HCR/C derivatives. Disruption of the GBP2 with the mutation Y1119A eliminated high affinity GT1b binding and low affinity GD1a and GM1a binding. GD1b binding is retained. His-1193 does not contribute to ganglioside binding by the GBP2. Disruption of the Sia-1 site with the mutation A1126K eliminated high affinity GD1b binding. High affinity GT1b binding was reduced to GD1a levels. GM1a binding was eliminated. These results indicate the GBP2 binds the sia5 moiety of GD1a = GT1b and the Sia-1 site binds the sia7 of GD1b > GT1b. C, the indicated HCR (5 nm) was incubated in wells coated with GD3, GD2, or GD1b or uncoated. The plate was processed as described in B. HCR/D (10 nm) was included as a positive control for GD2 binding (27). These results indicate that ganglioside binding by the Sia-1 site requires a terminal galactose ring on the sugar backbone to bind efficiently.
To evaluate the contribution of the GBP2 and the Sia-1 sites in ganglioside binding by HCR/C, directed mutagenesis generated two independent point mutations that were predicted to physically disrupt sialic acid-GBP2 interactions (Y1119A) and sialic acid-Sia-1 site interactions (A1126K) (Fig. 3A). Tyr-1119 was chosen because the analogous GBP2 residue of HCR/D-C (TYR-1115) contacted sialic acid through two hydrogen bonds as opposed to one hydrogen bond for all other GBP2 residues. Ala-1126 was changed to lysine as the bulky, charged R-group is predicted to occlude the Sia-1 site. Mutated proteins were evaluated for ganglioside binding using a solid-phase assay (Fig. 3B). Wild-type HCR/C bound gangliosides with the following efficiency: GT1b = GD1b > GD1a > GM1a. HCR/C(Y1119A) had reduced binding to GT1b, did not bind GD1a or GM1a, but retained GD1b binding. Analysis of the biosynthetic precursors of GD1b indicated that neither GD2 nor GM3 (Fig. 3C) supported HCR/C(Y1119A) binding, showing that the backbone sugars contributed to high affinity binding of GD1b to the Sia-1 binding site of HCR/C and that the Sia-1 site preferentially bound complex gangliosides with sia7-sialic acid (GD1b > GT1b). HCR/C(A1126K) did not bind GD1b or GM1a but retained GD1a binding and had low binding to GT1b (Fig. 3B), showing that the GBP2 site preferentially bound complex gangliosides with sia5-sialic acids (GD1a = GT1b). Overall, GBP2 preferentially bound sia5-gangliosides (GT1b and GD1a), whereas the Sia-1 site preferentially bound sia7-gangliosides (GD1b > GT1b). The residual ganglioside binding observed for the mutated proteins suggests that the point mutations did not disrupt the structural integrity of the proteins.
In a control experiment to address the relationship between GBP2 of BoNT/C and the GBP of other BoNT serotypes, His-1193 of GBP2, which is structurally analogous to the conserved His of GBP, was mutated and tested for ganglioside binding. HCR/C(H1193A) had the same complex ganglioside binding profile as HCR/C, showing that His-1193 did not participate in ganglioside binding (Fig. 3B) (26). This supported the model that GBP and GBP2 utilize different mechanisms to bind gangliosides.
HCR/C Binds Dual Gangliosides to Enter N2A Cells
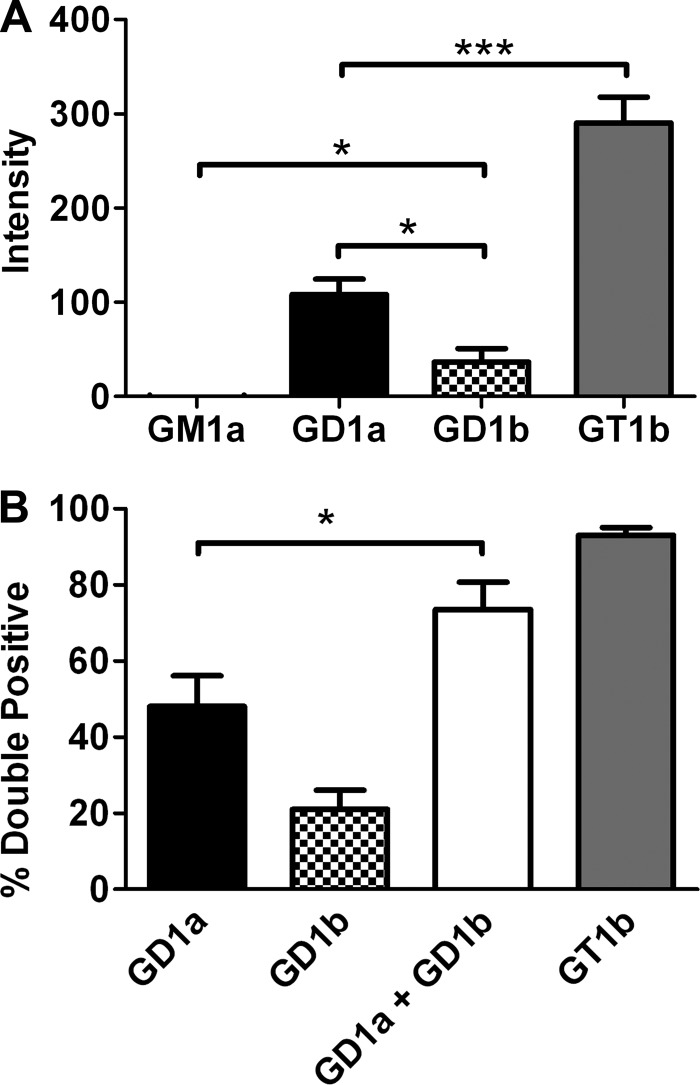
The capacity of specific complex gangliosides to support HCR/C entry into N2A cells was also tested. Although minimal HCR/C entry was detected in untreated N2A cells, enrichment of membranes with gangliosides increased HCR/C entry with the following affinity: GT1b > GD1a > GD1b. GM1a failed to support activity above background (Fig. 4A). HCR/C fluorescence intensity was greater in N2A cells enriched with GD1a relative to GD1b. The greater capacity of GT1b to support HCR/C entry may reflect the ability of GT1b to bind into both the GBP2 and Sia-1 pockets (Fig. 3B). Control experiments showed that HCR/TeNT entry into GD1b- and GT1b-enriched cells was similar (data not shown), indicating comparable GT1b and GD1b enrichment within the cells.
FIGURE 4.
HCR/C entry into N2A cells enriched with complex gangliosides. A, quantification of microscopic data. N2A cells were cultured with or without the indicated ganglioside (25 μg/well) for 4 h. 40 nm HCR/C was incubated with N2A cells with or without ganglioside for 30 min at 37 °C. Unbound HCR was washed away, and the cells were processed for microscopy. HCR/C was detected with an anti-HA antibody. Cells were stained for synaptophysin. Exposure times were constant between treatments. The average HCR/C fluorescence intensity was determined from three independent fields of consistent cellular morphology and density. The HCR/C intensity detected in N2A cells not supplemented with exogenous ganglioside was determined and subtracted from the ganglioside-dependent HCR intensity, and that value is presented here. Data presented are the average of three independent experiments. B, N2A cells were cultured with or without 5 μg of the indicated ganglioside for 4 h. 40 nm HCR/C was incubated with N2A cells with or without ganglioside for 30 min at 37 °C. Unbound HCR was washed away, and the cells were processed for microscopy. The percentage of cells in a given field that were double positive for HCR/C and synaptophysin was determined using ImageJ. Data were prepared using Excel and GraphPad Prism. The average of three independent experiments is shown. Asterisks denote statistical significance (*, p < 0.05; ***, p < 0.0001). A combination of GD1a and GD1b resulted in a statistically significant increase in the number of double positive cells relative to GD1a alone as determined by a two-tailed t test.
These studies provided a basis for assessing the role of the two ganglioside binding sites, GBP2 and Sia-1, in HCR/C entry into N2A cells. To measure the contribution of each site, HCR/C entry was measured in N2A cells enriched with GD1a, which mediates binding at the GBP2 site, and GD1b, which mediates binding at the Sia-1 site, or cells enriched with a combination of GD1a and GD1b. Parameters were established so that most cells were double positive for HCR/C and synaptophysin upon GT1b enrichment. Under these conditions, the efficiency of ganglioside-dependent HCR/C entry was GT1b > GD1a >GD1b (Fig. 4A). HCR/C entered ∼50% of the GD1a-enriched cells and ∼20% of the GD1b-enriched cells, whereas HCR/C entered ∼75% of the cells enriched with both GD1a and GD1b (Fig. 4B). The enhanced entry of HCR/C upon enrichment with GD1a and GD1b, which support binding to GBP2 and Sia-1, respectively, was consistent with HCR/C utilizing both ganglioside binding sites to enter cells.
Binding of Dual Gangliosides Is Required for Efficient HCR/C Entry into Cells
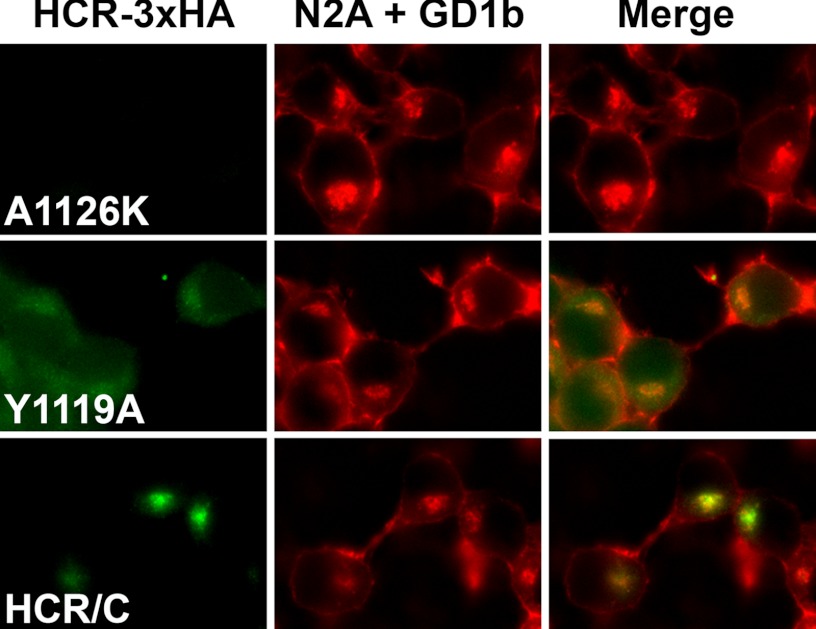
The contributions of the GBP2 site and the Sia-1 site to cell entry were assessed by evaluating the entry of HCR/C(Y1119A) and HCR/C(A1126K) relative to HCR/C in ganglioside-enriched N2A cells. Ganglioside enrichment was increased to allow detection of cell binding by the point-mutated HCR/C derivatives. Following GD1b enrichment, however, a Y1119A signal was detectable above background (Fig. 5). The preferred binding of HCR/C(Y1119A) to GD1b over GT1b is in line with the results of the solid-phase binding assay (Fig. 3B). Unexpectedly HCR/C(Y1119A) did not exclusively localize within the synaptophysin positive compartment like wild-type HCR/C but appeared diffuse throughout the cytoplasm, indicating that trafficking to the synaptophysin positive compartment was impaired (Fig. 5). This indicates that whereas HCR/C bound to the plasma membrane through a GD1b-Sia-1site interaction, this interaction was not sufficient to efficiently traffic HCR/C into cells. Cell enriched with GT1b supported robust HCR/C entry but failed to support the entry of either Y1119A or A1126K (data not shown).
FIGURE 5.
Binding of two gangliosides enhances HCR/C entry into N2A cells. Assessment of HCR/C, HCR/C(Y1119A), and HCR/C(A1126K) trafficking in N2A cells enriched with exogenous ganglioside. N2A cells were enriched with 50 μg of GD1b/well for 4 h. The indicated HCR (40 nm) was incubated in N2A cells with or without exogenous ganglioside for 30 min. Unbound HCR was washed away, and cells were processed for microscopy. Instrument settings and exposure times were consistent between treatments. Images were prepared for publication and set to equivalent brightness and contrast settings using NIS-Elements (Nikon Instruments). Representative micrographs are shown. Magnification is similar to that described in the legend for Fig. 1, D and E. A Sia-1 site-competent derivative of HCR/C (HCR/C(Y1119A)) is able to enter N2A cells with ganglioside. These results indicate that two intact ganglioside binding sites are required for efficient trafficking of HCR/C into cells. However, binding GD1b through the Sia-1 site appears to support entry.
Binding of Dual Gangliosides Is Required for HCR/C Entry into Neurons
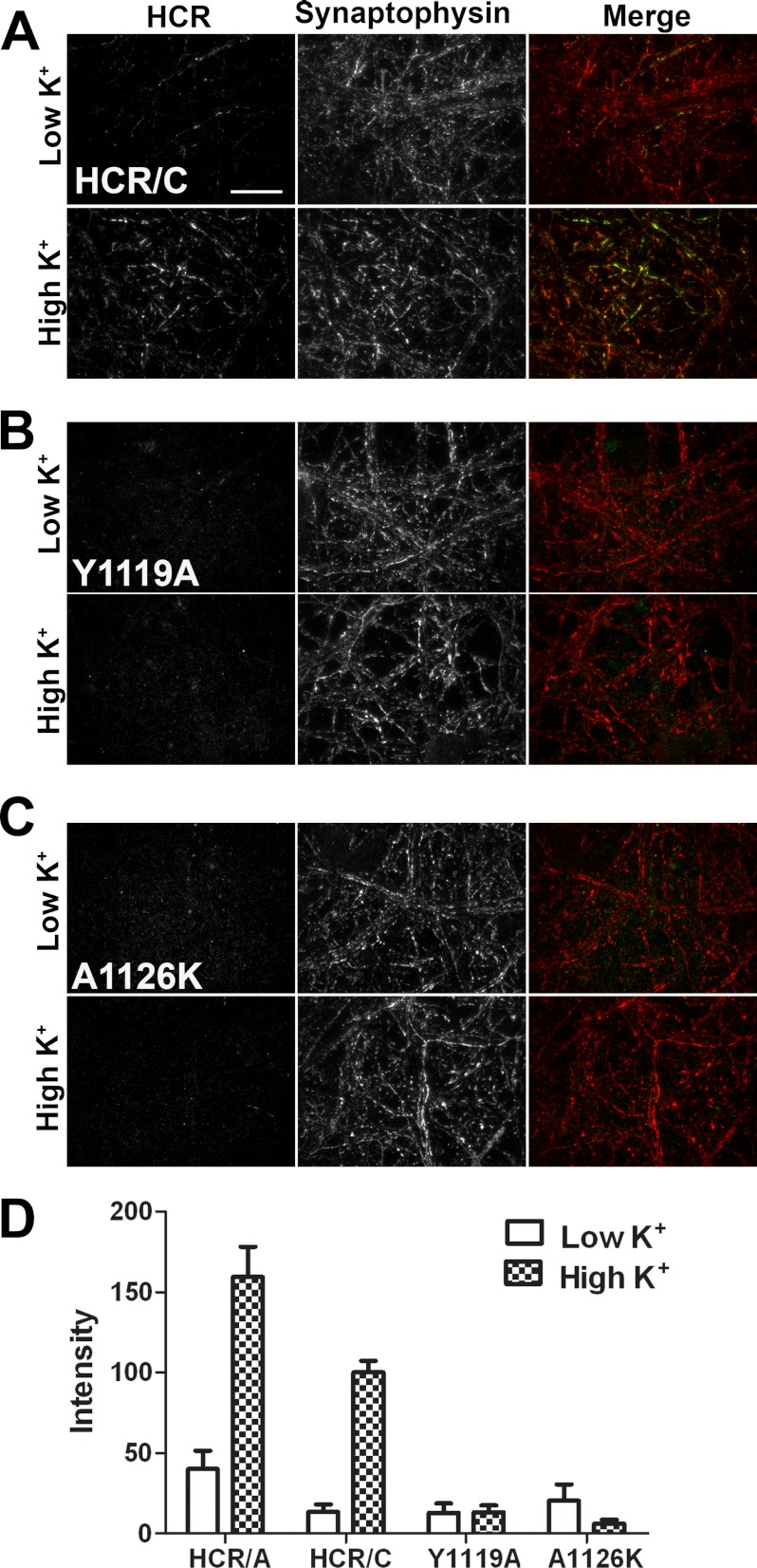
BoNTs enter neurons via synaptic vesicles during synaptic vesicle cycling. Using HCR/A as a control for synaptic vesicle-dependent entry, the entry of HCR/C was measured in unstimulated primary cortical neurons and in cortical neurons that were stimulated for SV cycling. SV cycling was stimulated by the incubation of cells with potassium-containing buffer (high K+). HCR/A entry was increased following membrane depolarization (Fig. 6D). Like HCR/A, HCR/C entry was enhanced with depolarization (Fig. 6, A and D). Colocalization between HCR/C and an SV marker, synaptophysin, is evident from the merged image, as the neuronal projections appear yellow/green, indicative of an HCR/C entering neurons through the cycling SVs (Fig. 6A).
FIGURE 6.
Synaptic activity enhances the uptake of HCR/C. A, primary hippocampal neurons were incubated in the presence of HCR/A, HCR/C, or HCR/C derivatives (40 nm) for 5 min under resting (low K+) or depolarizing (high K+) conditions. The cells were processed for microscopy. Bound/internalized HCR was detected with an anti-HA antibody followed by a fluorescent secondary antibody. Synaptic vesicles were identified by staining for synaptophysin. Micrographs were captured using TIRF microscopy with identical exposure settings between treatments. Representative ×100 micrographs are shown. Scale bar = 20 μm. Images were adjusted to the same brightness and contrast using NIS Elements. Representative merged micrographs are shown for HCR/C (A), HCR/C(Y1119A) (B), and HCR/C(A1126K) (C). Under resting conditions, the cells appear red in the merged images, as only the synaptophysin signal is detected. Following depolarization, some of the neurons projections in the field appear yellow or green, indicating uptake of the HCR and overlap with synaptophysin. D, quantification of microscopy data. Eight random, synaptophysin-positive fields per treatment were captured with identical exposure settings between treatments. The HCR fluorescence, in arbitrary units, was measured using NIS Elements. The graph was prepared for presentation using GraphPad Prism. An average of four independent experiments is shown.
To determine whether binding dual gangliosides is necessary for HCR/C entry into neurons, HCR/C(Y1119A) and HCR/C(A1126K) were tested for neuron entry capacity. Under the same conditions that support robust HCR/C entry following synaptic stimulation, neither Y1119A nor A1126K entered neurons (Fig. 6, B–D). The low level of signal detected above background was not cell-associated and was likely nonspecific binding of the HCRs to the tissue culture plate (Fig. 6, B and C). Based on these findings, we concluded that binding of dual gangliosides is required for the entry of HCR/C into neurons.
Organization of the Two Ganglioside Binding Sites within HCR/C
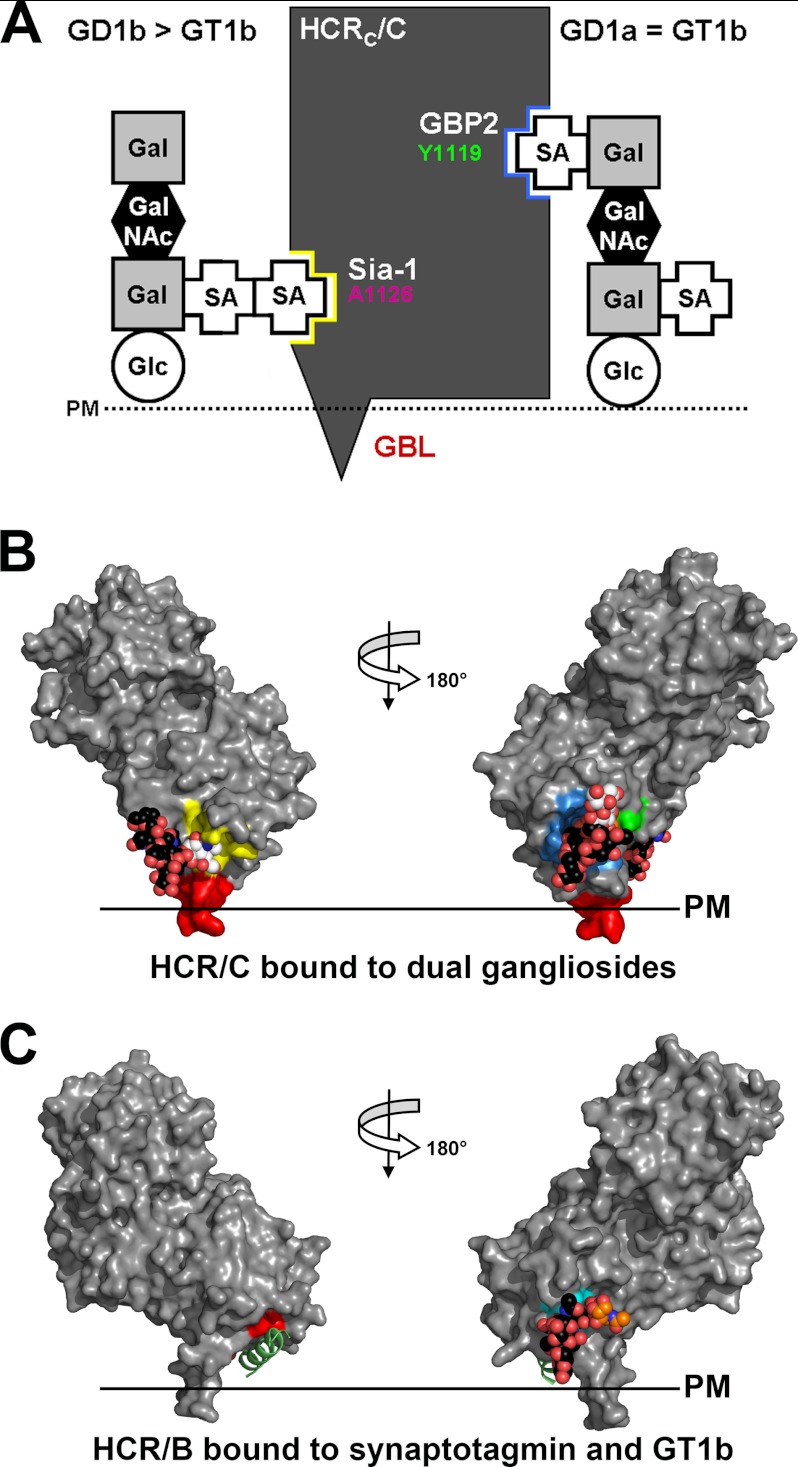
A schematic of the ganglioside interaction with the Sia-1 site and the GBP2 of HCR/C is shown in Fig. 7A. A manually generated model of HCR/C bound to dual gangliosides was also constructed to orient HCR/C on the plasma membrane. Sia5 of GD1a was modeled into the GBP2 site using the sialic acid bound to HCR/D-C(GBL/C) as a reference, and sia7 of GD1b was docked into the Sia-1 site using sialic acid bound to HCR/C as a reference. The geometry of HCR/C with the plasma membrane is similar to the orientation of HCR/B modeled onto a plasma membrane when bound to GT1b and synaptotagmin (15, 16). In both models a respective region of the HCR penetrates the lipid bilayer (Fig. 7, B and C). The possibility that a single molecule of GT1b could bind to the GBP2 and Sia-1 sites was addressed by measuring the distance between the two sites, which is ∼30 Å, by following the surface contour of the HCR. This is too long to allow a single molecule of GT1b to span the distance between GBP2 and the Sia-1 site (data not shown).
FIGURE 7.
BoNT/C binds dual gangliosides. A, schematic showing how BoNT/C binds ganglioside was generated based on the results of the mutagenesis study from Fig. 4. B, a manually generated model for HCR/C bound to dual ganglioside molecules (bound sialic acid moiety is shown in white, and the remainder of the ganglioside in black) was built by docking the sia7 from GD1b into the Sia-1 site using the Sia-1 molecule as a reference (PDB code, 3R4S) (26). The sia5 from GD1a was modeled into the GBP2 using the sialic acid bound by the HCR/D-C(GBL/C) as a reference. Ganglioside binding sites are colored as described in the legend for Fig. 3. Binding of dual gangliosides positioned HCR/C to interact with the plasma membrane as shown with the GBL (red), penetrating the plasma membrane (PM) lipid bilayer. C, HCR/B bound to dual receptors: synaptotagmin and GT1b. Alignment of HCR/B with the synaptotagmin peptide (green; PDB code, 2NM1) (15, 16) and GT1b bound to HCR/A (PDB code, 2VU9) (45) are shown. Residues that contact the synaptotagmin peptide and GT1b are shown in red and cyan, respectively.
DISCUSSION
We report here that BoNT/C binds dual gangliosides to enter cells through two independent ganglioside binding sites (termed GBP2 and Sia-1) where each binding site has unique ganglioside specificity. Although the GBP2 of BoNT/C is physically analogous to the GBP of BoNT/A, the ganglioside-HCR contacts are unique. Likewise, although the Sia-1 site of BoNT/C is physically analogous to the synaptotagmin binding site of BoNT/B, the respective ganglioside-HCR/C and peptide-HCR/B contacts are unique. Conversely, the functions of the two binding site pairs appear analogous, which allowed extension of the model for how BoNTs utilize dual receptors to enter neurons. The observed accessibility of HCR/C(Y1119A) to GD1b-enriched N2A cells shows the direct binding of the HCR to the plasma membrane via the Sia-1 site. Extrapolation suggests that either gangliosides or SV proteins support BoNT interactions at the Sia-1 site or the SRS, respectively, and that SV exocytosis need not precede BoNT entry to present SV protein receptors on the plasma membrane. The enhanced BoNT entry observed with the stimulation of SV exocytosis may present additional SV protein receptor on the plasma membrane. Earlier, Neale et al. (40) reported that BoNT/A-intoxicated neurons retain endocytosis. The acceptance that SV exocytosis and SV endocytosis are uncoupled and that there are pools of SV components resident on the plasma membrane supports the initial interactions between BoNTs and host receptors on the surface of neurons (32, 41–43).
The crystal structure of HCR/D-C(GBL/C) bound to sialic acid, in conjunction with structural modeling, provides a mechanism for ganglioside recognition by GBP2 of BoNT/C that can be analyzed relative to ganglioside recognition by GBP of BoNT/A. Similar properties between GBP2 and GBP include interactions with the sia5 component of gangliosides, where HCR/A Tyr-1117 and Ser-1275 coordinate the binding to sia5 of GD1a and GT1b, whereas HCR/C Tyr-1119 and Ser-1281 coordinate sia5 binding by gangliosides (44, 45). In contrast, GBP2 and GBP differed in the coordination of the backbone components of gangliosides. GBP2 of HCR/C lacks the conserved Trp within the GBP of HCR/A that coordinates binding to the backbone components of gangliosides, and His-1193 within the GBP2 of HCR/C does not contribute to ganglioside binding, whereas His-1253 within the GBP of HCR/A does contribute to ganglioside binding. The lack of contacts between HCR/C and the ganglioside backbone may be compensated for by the additional contacts between the residues within the GBP2 and the sialic acid. Earlier studies observed an electron density of a sialic acid near the GBP in HCR/D-C (46) but did not identify contacts between the HCR and sialic acid; these studies determined that Ile-1240 and Tyr-1243 contribute to ganglioside binding, which is shown in our current study to contact sialic acid.
The Sia-1 binding site of BoNT/C was initially identified as a sialic acid binding site. The current study has characterized the ganglioside specificity of the Sia-1 site. Unexpectedly, the Sia-1 site preferentially bound GD1b (Fig. 3B). GD1b binding was eliminated with the A1126K mutation, confirming that the Sia-1 site is a ganglioside binding site that binds sia7-containing gangliosides (GD1b > GT1b). Analysis of Sia-1 site binding specificity showed that the terminal galactose (gal4) of the ganglioside backbone was necessary for high affinity binding, as HCR/C bound GD1b with greater affinity than GD2 (Fig. 3C). The lower affinity of HCR/C for GT1b relative to GD1b may reflect the modification of gal4 with sialic acid, which may interfere with the contacts between the Sia-1 site and gal4. Protein modeling showed that the replacement of Ala-1126 with Lys occludes sialic acid interactions within the Sia-1 site. Ala-1126 maps to a β-hairpin that is conserved in multiple BoNT serotypes and overlaps with the second receptor site of BoNT/B. Val-1118 of the synaptotagmin binding site in HCR/B is on the same conserved β-hairpin as Ala-1126 in HCR/C (15, 16). Based on their structural similarity, the Sia-1 site and the second receptor site may have a conserved function between BoNT/C and BoNT/B but via interactions with unique receptor molecules.
The multiple roles that gangliosides perform in neuronal homeostasis can complicate the resolution of the direct versus indirect effects of ganglioside ablation on BoNT function (47). Using a gain-of-function, cell-based assay allowed evaluation of the capacity of gangliosides to support HCR/C entry. Protein modeling predicted two independent ganglioside binding sites within BoNT/C that have different preferences for complex gangliosides: GBP2 binds sia5-containing gangliosides, whereas the Sia-1 site binds sia7-containing gangliosides. This explains why GT1b is a better overall receptor for mediating HCR/C entry, because GT1b contains a sia5 and sia7. The observation that HCR/C bound GD1b > GD1a in the solid-phase assay (Fig. 3B) but entry into N2A cells was GD1a > GD1b indicates that the arrangement of the sialic acids on gangliosides is relevant to the entry process. Enrichment with a combination of GD1a and GD1b enhanced HCR/C entry relative to enrichment with either GD1a or GD1b alone, supporting a role for dual gangliosides as receptors for BoNT/C.
Using primary neurons in culture, the influence of disrupting either ganglioside binding site on the uptake and trafficking of HCR/C was evaluated. HCR/C entry was enhanced following synaptic stimulation, similar to HCR/A (Fig. 6). Disruption of either ganglioside binding site ablated the ability of HCR/C to enter neurons. These results agree with previous reports that characterize the dual ganglioside binding sites of HCR/TeNT (48). This indicates that eliminating either ganglioside binding site influences ganglioside binding affinity as well as binding specificity. This is consistent with the finding that BoNT/C with the Sia-1 site disrupted by the mutation L1203F or with the GBP2 disrupted with the mutation S1281Y decreases toxicity to 0.5 and 15% of wild-type BoNT/C, respectively (26). Thus, one functional ganglioside binding site can allow entry into neurons but with reduced activity.
Binding multiple ganglioside molecules as functional receptors is not a property unique to the clostridia neurotoxins, as Shiga toxin and cholera toxin both utilize multiple ganglioside molecules to enter cells. The mechanism of Shiga toxin entry is dependent on the unique geometry of the ganglioside binding sites in the binding domain to drive membrane ganglioside clustering. Shiga toxin-mediated ganglioside clustering results in a localized membrane perturbation characterized by negative curvature in the plasma membrane, which leads to invagination formation and toxin entry in the absence of a preexisting cellular pathway (49–51). This cargo-induced entry provides one possible mechanism for the ganglioside-mediated entry of BoNTs into neurons. In this case, the tethering of the HCR domain by the two binding sites may provide sufficient contortion (curvature) of the membrane to initiate uptake. Shiga toxin molecules are predicted to cluster in the plasma membrane of target cells, which also contributes to entry, whereas the extreme potency of BoNT indicates that a minute amount of toxin very efficiently reaches target cells, suggesting that cooperativity between toxin molecules is not required. Multimeric forms of TeNT and BoNT HCRs have been observed (52). Whether HCR/C has the ability to contort the plasma membrane like Shiga toxin will be the subject of future experiments.
Neuron-specific tropism of BoNT suggests that simply binding to plasma membrane gangliosides is not sufficient for entry but rather HCR/C utilizes a neuron-specific component or endocytic pathway for entry. Along these lines, numerous studies have attempted to define a protein-based receptor for BoNT/C and have failed (25, 26, 29, 53), but it is still unknown whether gangliosides are sufficient for BoNT/C intoxication. Continued studies may resolve this process and provide insight into developing neurotoxins as targeted therapeutic agents.
Acknowledgment
We thank Amanda Przedpelski for assistance with HCR production.
This work was supported, in whole or in part, by National Institutes of Health grants from the NIAID and the Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research Program (to J. J. K. and J. T. B).
The atomic coordinates and structure factors (code 4FVV) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- BoNT
- botulinum neurotoxin
- LC
- light chain
- HC
- heavy chain
- HCR
- HC receptor binding domain
- SV
- synaptic vesicle
- TeNT
- tetanus toxin
- GBP
- ganglioside binding pocket
- SRS
- second receptor binding site
- GBL
- ganglioside binding loop
- N2A
- Neuro-2A
- PDB
- Protein Data Bank.
REFERENCES
- 1. Simpson L. L. (1981) The origin, structure, and pharmacological activity of botulinum toxin. Pharmacol. Rev. 33, 155–188 [PubMed] [Google Scholar]
- 2. Hill K. K., Smith T. J., Helma C. H., Ticknor L. O., Foley B. T., Svensson R. T., Brown J. L., Johnson E. A., Smith L. A., Okinaka R. T., Jackson P. J., Marks J. D. (2007) Genetic diversity among botulinum neurotoxin-producing clostridial strains. J. Bacteriol. 189, 818–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karalewitz A. P., Kroken A. R., Fu Z., Baldwin M. R., Kim J. J., Barbieri J. T. (2010) Identification of a unique ganglioside binding loop within botulinum neurotoxins C and D-SA . Biochemistry 49, 8117–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moriishi K., Koura M., Abe N., Fujii N., Fujinaga Y., Inoue K., Ogumad K. (1996) Mosaic structures of neurotoxins produced from Clostridium botulinum types C and D organisms. Biochim. Biophys. Acta 1307, 123–126 [DOI] [PubMed] [Google Scholar]
- 5. Moriishi K., Syuto B., Kubo S., Oguma K. (1989) Molecular diversity of neurotoxins from Clostridium botulinum type D strains. Infect. Immun. 57, 2886–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Webb R. P., Smith T. J., Wright P. M., Montgomery V. A., Meagher M. M., Smith L. A. (2007) Protection with recombinant Clostridium botulinum C1 and D binding domain subunit (Hc) vaccines against C and D neurotoxins. Vaccine 25, 4273–4282 [DOI] [PubMed] [Google Scholar]
- 7. Arnon S. S., Schechter R., Inglesby T. V., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Fine A. D., Hauer J., Layton M., Lillibridge S., Osterholm M. T., O'Toole T., Parker G., Perl T. M., Russell P. K., Swerdlow D. L., Tonat K., and Working Group on Civilian Biodefense (2001) Botulinum toxin as a biological weapon: medical and public health management. JAMA 285, 1059–1070 [DOI] [PubMed] [Google Scholar]
- 8. Montal M. (2010) Botulinum neurotoxin: a marvel of protein design. Annu. Rev. Biochem. 79, 591–617 [DOI] [PubMed] [Google Scholar]
- 9. Montecucco C. (1986) How do Tetanus and Botulinum toxins bind to neuronal membranes? Trends Biochem. Sci. 11, 314–317 [Google Scholar]
- 10. Baldwin M. R., Kim J. J., Barbieri J. T. (2007) Botulinum neurotoxin B-host receptor recognition: it takes two receptors to tango. Nat. Struct. Mol. Biol. 14, 9–10 [DOI] [PubMed] [Google Scholar]
- 11. Koriazova L. K., Montal M. (2003) Translocation of Botulinum neurotoxin light chain protease through the heavy chain channel. Nat. Struct. Mol. Biol. 10, 13–18 [DOI] [PubMed] [Google Scholar]
- 12. Jahn R., Scheller R. H. (2006) SNAREs: engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 13. Chen S., Hall C., Barbieri J. T. (2008) Substrate recognition of VAMP-2 by botulinum neurotoxin B and tetanus neurotoxin. J. Biol. Chem. 283, 21153–21159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schiavo G., Benfenati F., Poulain B., Rossetto O., Polverino de Laureto P., DasGupta B. R., Montecucco C. (1992) Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature 359, 832–835 [DOI] [PubMed] [Google Scholar]
- 15. Jin R., Rummel A., Binz T., Brunger A. T. (2006) Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature 444, 1092–1095 [DOI] [PubMed] [Google Scholar]
- 16. Chai Q., Arndt J. W., Dong M., Tepp W. H., Johnson E. A., Chapman E. R., Stevens R. C. (2006) Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature 444, 1096–1100 [DOI] [PubMed] [Google Scholar]
- 17. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds (2009) Essentials of Glycobiology, 2nd Ed., Chapter 9, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 18. Yowler B. C., Schengrund C. L. (2004) Glycosphingolipids: sweets for botulinum neurotoxin. Glycoconj. J. 21, 287–293 [DOI] [PubMed] [Google Scholar]
- 19. Shapiro R. E., Specht C. D., Collins B. E., Woods A. S., Cotter R. J., Schnaar R. L. (1997) Identification of a ganglioside recognition domain of tetanus toxin using a novel ganglioside photoaffinity ligand. J. Biol. Chem. 272, 30380–30386 [DOI] [PubMed] [Google Scholar]
- 20. Binz T., Rummel A. (2009) Cell entry strategy of clostridial neurotoxins. J. Neurochem. 109, 1584–1595 [DOI] [PubMed] [Google Scholar]
- 21. Dong M., Liu H., Tepp W. H., Johnson E. A., Janz R., Chapman E. R. (2008) Glycosylated SV2A and SV2B mediate the entry of botulinum neurotoxin E into neurons. Mol. Biol. Cell 19, 5226–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong M., Yeh F., Tepp W. H., Dean C., Johnson E. A., Janz R., Chapman E. R. (2006) SV2 is the protein receptor for botulinum neurotoxin A. Science 312, 592–596 [DOI] [PubMed] [Google Scholar]
- 23. Fu Z., Chen C., Barbieri J. T., Kim J. J., Baldwin M. R. (2009) Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F. Biochemistry 48, 5631–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dong M., Tepp W. H., Liu H., Johnson E. A., Chapman E. R. (2007) Mechanism of botulinum neurotoxin B and G entry into hippocampal neurons. J. Cell Biol. 179, 1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsukamoto K., Kohda T., Mukamoto M., Takeuchi K., Ihara H., Saito M., Kozaki S. (2005) Binding of Clostridium botulinum type C and D neurotoxins to ganglioside and phospholipid. Novel insights into the receptor for clostridial neurotoxins. J. Biol. Chem. 280, 35164–35171 [DOI] [PubMed] [Google Scholar]
- 26. Strotmeier J., Gu S., Jutzi S., Mahrhold S., Zhou J., Pich A., Eichner T., Bigalke H., Rummel A., Jin R., Binz T. (2011) The biological activity of botulinum neurotoxin type C is dependent upon novel types of ganglioside binding sites. Mol. Microbiol. 81, 143–156 [DOI] [PubMed] [Google Scholar]
- 27. Kroken A. R., Karalewitz A. P., Fu Z., Kim J. J., Barbieri J. T. (2011) Novel ganglioside-mediated entry of botulinum neurotoxin serotype D into neurons. J. Biol. Chem. 286, 26828–26837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strotmeier J., Lee K., Völker A. K., Mahrhold S., Zong Y., Zeiser J., Zhou J., Pich A., Bigalke H., Binz T., Rummel A., Jin R. (2010) Botulinum neurotoxin serotype D attacks neurons via two carbohydrate-binding sites in a ganglioside-dependent manner. Biochem. J. 431, 207–216 [DOI] [PubMed] [Google Scholar]
- 29. Rummel A., Häfner K., Mahrhold S., Darashchonak N., Holt M., Jahn R., Beermann S., Karnath T., Bigalke H., Binz T. (2009) Botulinum neurotoxins C, E, and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J. Neurochem. 110, 1942–1954 [DOI] [PubMed] [Google Scholar]
- 30. Tsukamoto K., Kozai Y., Ihara H., Kohda T., Mukamoto M., Tsuji T., Kozaki S. (2008) Identification of the receptor-binding sites in the carboxyl-terminal half of the heavy chain of botulinum neurotoxin types C and D. Microb. Pathog. 44, 484–493 [DOI] [PubMed] [Google Scholar]
- 31. Rummel A., Karnath T., Henke T., Bigalke H., Binz T. (2004) Synaptotagmins I and II act as nerve cell receptors for botulinum neurotoxin G. J. Biol. Chem. 279, 30865–30870 [DOI] [PubMed] [Google Scholar]
- 32. Dittman J., Ryan T. A. (2009) Molecular circuitry of endocytosis at nerve terminals. Annu. Rev. Cell Dev. Biol. 25, 133–160 [DOI] [PubMed] [Google Scholar]
- 33. Higuchi R., Krummel B., Saiki R. K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sato M. H., Wada Y. (1997) Universal template plasmid for introduction of the triple-HA epitope sequence into cloned genes. BioTechniques 23, 254–256 [DOI] [PubMed] [Google Scholar]
- 35. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode, in Methods in Enzymology (Carter C. W., Jr., ed) pp. 307–326, Academic Press, New York, NY: [DOI] [PubMed] [Google Scholar]
- 36. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brunger A. T. (2007) Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 38. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39. Chen C., Fu Z., Kim J. J., Barbieri J. T., Baldwin M. R. (2009) Gangliosides as high affinity receptors for tetanus neurotoxin. J. Biol. Chem. 284, 26569–26577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Neale E. A., Bowers L. M., Jia M., Bateman K. E., Williamson L. C. (1999) Botulinum neurotoxin A blocks synaptic vesicle exocytosis but not endocytosis at the nerve terminal. J. Cell Biol. 147, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fernández-Alfonso T., Kwan R., Ryan T. A. (2006) Synaptic vesicles interchange their membrane proteins with a large surface reservoir during recycling. Neuron 51, 179–186 [DOI] [PubMed] [Google Scholar]
- 42. Hua Y., Sinha R., Thiel C. S., Schmidt R., Hüve J., Martens H., Hell S. W., Egner A., Klingauf J. (2011) A readily retrievable pool of synaptic vesicles. Nat. Neurosci. 14, 833–839 [DOI] [PubMed] [Google Scholar]
- 43. Willig K. I., Rizzoli S. O., Westphal V., Jahn R., Hell S. W. (2006) STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature 440, 935–939 [DOI] [PubMed] [Google Scholar]
- 44. Benson M. A., Fu Z., Kim J. J., Baldwin M. R. (2011) Unique ganglioside recognition strategies for clostridial neurotoxins. J. Biol. Chem. 286, 34015–34022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stenmark P., Dupuy J., Imamura A., Kiso M., Stevens R. C. (2008) Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b: insight into the toxin-neuron interaction. PLoS Pathog. 4, e1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nuemket N., Tanaka Y., Tsukamoto K., Tsuji T., Nakamura K., Kozaki S., Yao M., Tanaka I. (2011) Structural and mutational analyses of the receptor binding domain of botulinum D/C mosaic neurotoxin: insight into the ganglioside binding mechanism. Biochem. Biophys. Res. Commun. 411, 433–439 [DOI] [PubMed] [Google Scholar]
- 47. Posse de Chaves E., Sipione S. (2010) Sphingolipids and gangliosides of the nervous system in membrane function and dysfunction. FEBS Lett. 584, 1748–1759 [DOI] [PubMed] [Google Scholar]
- 48. Chen C., Baldwin M. R., Barbieri J. T. (2008) Molecular basis for tetanus toxin coreceptor interactions. Biochemistry 47, 7179–7186 [DOI] [PubMed] [Google Scholar]
- 49. Römer W., Berland L., Chambon V., Gaus K., Windschiegl B., Tenza D., Aly M. R., Fraisier V., Florent J. C., Perrais D., Lamaze C., Raposo G., Steinem C., Sens P., Bassereau P., Johannes L. (2007) Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450, 670–675 [DOI] [PubMed] [Google Scholar]
- 50. Ewers H., Römer W., Smith A. E., Bacia K., Dmitrieff S., Chai W., Mancini R., Kartenbeck J., Chambon V., Berland L., Oppenheim A., Schwarzmann G., Feizi T., Schwille P., Sens P., Helenius A., Johannes L. (2010) GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 12, 11–18 [DOI] [PubMed] [Google Scholar]
- 51. Johannes L., Mayor S. (2010) Induced domain formation in endocytic invagination, lipid sorting, and scission. Cell 142, 507–510 [DOI] [PubMed] [Google Scholar]
- 52. Yeh F. L., Zhu Y., Tepp W. H., Johnson E. A., Bertics P. J., Chapman E. R. (2011) Retargeted clostridial neurotoxins as novel agents for treating chronic diseases. Biochemistry 50, 10419–10421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baldwin M. R., Barbieri J. T. (2007) Association of botulinum neurotoxin serotypes a and B with synaptic vesicle protein complexes. Biochemistry 46, 3200–3210 [DOI] [PubMed] [Google Scholar]