Abstract
There is extensive evidence that post-transcriptional mechanisms of gene regulation, such as mRNA turnover, critically affect the patterns of expressed mRNAs. Conventional microarray analysis measures steady-state messenger RNA (mRNA) levels, which represents the dynamic balance between new transcription and mRNA degradation. Accordingly, only de novo transcription can accurately reflect the temporal and spatial events of transcriptional regulation. In this chapter, we describe a recently reported method to study transcription systematically. It involves the genome-wide labeling of nascent transcripts using non-radioactive modified nucleotides, their isolation for amplification, and their hybridization and analysis using commercial microarrays.
Keywords: nascent RNA, nuclear run-on (NRO), biotin, microarray, post-transcriptional regulation
1. Introduction
Biological processes are critically controlled by changes in gene expression. Many such changes are initiated through the altered binding of transcription factors (TFs) to specific DNA cis-elements that control the initiation of transcription and by factors that affect the elongation and termination of transcription. Together, the temporal and spatial regulation of transcription is the primary determinant of both the fate and function of the cell. Virtually all microarray studies to-date have measured changes in the levels of steady-state mRNA by harvesting total cellular RNA and using it to generate mRNA-specific probes through a variety of strategies. However, an increasing appreciation for the scope and importance of post-transcriptional regulatory mechanisms such as changes in mRNA stability [1–4] has highlighted the need for a more precise and direct measurement of TF-controlled transcription. Conventional microarray analysis cannot be used towards this end, as it employs steady-state/total RNA as the starting material to study a process that represents a changing balance between the rates of mRNA synthesis and decay. This approach almost certainly masks true transcriptional events, since mRNA turnover contributes measurably (sometimes exclusively) to changes in the abundance of expressed mRNAs.
In recent years, powerful array-based methods such as ChIP on Chip (chromatin immunoprecipitation using specific TFs crosslinked to genomic DNA binding sites followed by array interrogation of the captured nucleic acid sequences) have emerged to catalog and quantify the binding of TFs to DNA [5–7]. These strategies are capable of identifying a large majority of TF-binding sites at the genomic level, including previously unknown regulatory regions. However, studies using these high-throughput ChIP approaches reveal that TFs often bind to gene loci whose transcription is not altered [8,9]. In addition, their application is limited by (i) the requirement of prior knowledge about the involvement of a particular TF in a specific biological event, (ii) the inherent difficulty in identifying functionally coordinated TFs by this method, and (iii) insufficient information about the functional consequences of the TF upon gene transcription (i.e., whether the TF-DNA binding activates, represses, or leaves unchanged the rate of transcription) [7,10]. Attempts to link the ChIP-Chip studies to actual changes in mRNA expression profiles are limited by the confounding contributions of transcription and decay rates, as described above. Thus, a reliable genome-wide method for detecting nascent mRNA transcripts is useful for the interpretation ChIP-Chip data by properly identifying changes in gene expression which appear to be directly influenced by TF binding.
Classical nuclear run-on (NRO) techniques have been used extensively for many years on a single-gene basis to directly measure nascent gene transcription rates. When used in conjunction with RNA polymerase inhibitors, these techniques have also been used to calculate mRNA decay rates. Because single-gene NRO methods both are quite laborious and require considerable skill, we first developed a simplified medium-throughput NRO assay. Based on the hybridization of metabolically radiolabeled nascent RNAs with early-generation nylon membrane microarrays, this assay permitted the simultaneous measurement of nascent transcription for thousands of genes per sample per experiment [3]. We extended the use of this technique to map nascent gene transcription profiles in human cervical cancer cells following stress stimulation [10]. Using bioinformatic approaches to locate the possible regulatory cis-elements (promoters) of genes that were coordinately transcribed, we identified possible TFs involved; by subsequent RNAi and ChIP-PCR analyses, we validated the interactions between the predicted TFs and corresponding cis-elements [10]. This strategy is advantageous because: (i) it can potentially identify coordinately acting TFs, (ii) it does not require prior knowledge of the involvement of TFs, and (iii) the results directly reflect changes in nascent transcription as the functional consequences of the TF-DNA interactions.
However, the radioactive/nylon NRO arrays were largely limited by: (i) the requirement of large amounts of radioactive material (typically 0.5 mCi [α33P]-UTP per reaction) and large numbers of cells (typically >50 million per sample) and (ii) technically demanding procedures and potential artifacts from hybridization of labeled antisense transcripts to double-stranded spotted cDNAs. To circumvent these issues, we have established an efficient genome-wide, non-radioactive labeled, NRO array which is compatible with most commercially available oligomer arrays (11).
2. Materials
Precautions should be taken to minimize RNase contamination throughout all of the steps (see Notes 1 and 2).
2.1. Equipment
Low-speed table-top centrifuge.
Microcentrifuge.
Dynal MPC®-S Magnet Separator.
Dynal® RKDynal Sample Mixer.
Eppendorf Thermomixer® R Dry Block Heating and Cooling Shaker.
Hybridization Oven.
Nanodrop ND1000 Spectrophotometer.
Illumina Hybridization and Scanner System.
Affymetrix Hybridization and Wash Station and GeneChip® Scanner.
2.2. Materials and Reagents for Preparation of Nuclei from Culture Cells
Phosphate Buffered Saline (PBS), which can be prepared from 10× stock and pre-cooled at 4°C.
DEPC-treated water, molecular biology grade.
Cell Lysis Buffer: 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 20 mM NaCl, 100 mM Sucrose (Sigma cat. # S0389), 0.25% NP-40 (Sigma cat. # I8896), prepared in nuclease-free water (see Note 3).
2× Nuclei Resuspension Buffer: 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 0.1 mM EDTA, 45% Glycerol (Fluka cat. # 49767), prepared in nuclease-free water (see Note 4).
Qiagen RNeasy Mini Kit (Qiagen, cat. # 74106).
2.3. Materials and Reagents for Nascent RNA Labeling and RNA Purification
2× NRO Reaction Buffer: 300 mM KCl, 10 mM MgCl2, 2 mM each of rATP, rCTP, rGTP (Promega cat. # E6000), 1 mM Bio-16-UTP (Enzo Life Sciences, cat. # ENZ-42814), 800 Units/ml RNaseOUT (Invitrogen cat. # 10777-019).
rUTP (Promega cat. # 6000).
DNase I, RNase-free (Roche Applied Sciences cat. # 04716728001).
Proteinase K, RNA Grade (Invitrogen cat. #25530-049).
Qiagen RNeasy® Mini Kit (Qiagen cat. # 74106).
Ethanol, absolute (Sigma cat. # 7023).
TURBO DNase™ (Ambion cat. # 2239).
(Optional) RNeasy MinElute Cleanup Kit (Qiagen cat. # 74024).
2.4. Materials and Reagents for Selection of Biotin-labeled RNA, cDNA Synthesis and cRNA Amplification
1× Phosphate Buffered Saline (PBS) (Invitrogen cat. # 10010).
BeadWash Solution A: 100 mM NaOH (Fluka cat. # 72068), 50 mM NaCl, prepared with nuclease-free water.
Bead Wash Solution B: 100 mM NaCl, prepared with nuclease-free water.
Dynabeads® kilobaseBINDER™ Kit (Invitrogen/Dynal cat. # 601-01).
RNaseOUT (Invitrogen cat. # 10777-019).
DEPC-treated water, molecular biology grade.
T7N6 Primer: 5'- GTA ATA CGA CTC ACT ATA GGG CTN NNN NN -3' (Integrated DNA Technologies)
Illumina® TotalPrep RNA Amplification Kit (Ambion cat. # AMIL 1791).
2.5. Materials and Reagents for Affymetrix Exon Array
GeneChip® Whole Transcript (WT) cDNA Synthesis and Amplification Kit (Affymetrix part # 900673).
GeneChip® WT Terminal Labeling Kit (Affymetrix part # 900671).
GeneChip® Hybridization, Wash, and Stain Kit (HWS kit) (Affymetrix parts # 900720, 900721, 900722).
GeneChip® Human Exon 1.0 ST Array (Affymetrix part # 900650)
3. Methods
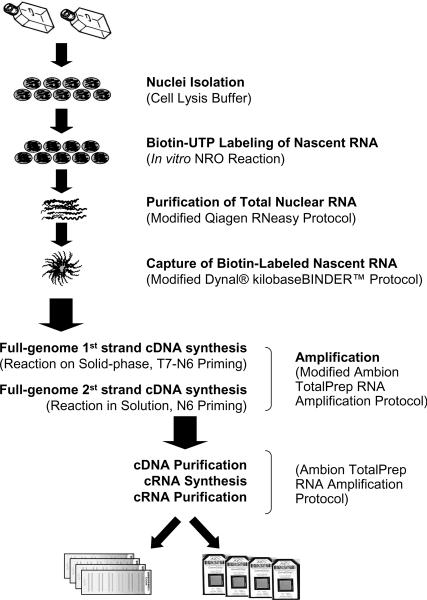
The method described here has been successfully applied to both human and mouse cell lines including human Jurkat T cells, human A549 lung adenocarcinoma epithelial cells, human P493-6 B lymphocytes, human H9 stem cells, and mouse embryonic fibroblasts (see Note 5). The major steps of this method are outlined in Figure 1. Biotin UTP labeling of all of the nascent transcription allows for the potential labeling of nascent non-protein-coding transcripts (antisense RNA, primary microRNA, etc), as well as nascent pre-mRNA (see Note 6).
Figure 1.
Schematic of the protocol
Fan et al.,
3.1. Preparation of Nuclei from Culture Cells
For adherent cultured cells, collect all cells of each condition into a 50-ml conical tube after trypsinization; for suspension cultured cells, directly collect cells into a 50-ml conical tube.
Pellet cells by spinning for 5 min at 216×g (~1,000 rpm, table-top centrifuge).
Carefully remove and discard supernatant; wash cells with 40 ml of precooled 1× PBS and spin as in previous step.
Carefully remove and discard supernatant; add 10 ml of precooled 1× PBS, resuspend cells by pipetting up and down several times or by inverting the tube several times.
Aliquot ~1 ml (~10%) of the cell suspension into microcentrifuge 1.5-ml tubes and spin 3–5 min at 2,000×g in a microcentrifuge. These cells will be used for total RNA isolation and conventional microarrays (see Note 7).
For the remaining 90% of cells, spin again for 5 min at 216×g in a table-top centrifuge; decant supernatant carefully, and proceed to isolate nuclei as follows.
Add 10 ml of cold Cell Lysis Buffer to the above cell pellet; mix by pipetting up and down several times or by inverting the tube several times; let tubes sit on ice for 6 min and invert tubes several times.
Spin 5 min. at 216×g (~1,000 rpm, table top centrifuge).
Aspirate carefully (so as not to disturb the nuclei pellet) and discard supernatant. Invert the 50-ml tube (with nuclei pellet at bottom) upside down on a piece of clean wipe paper on the laboratory bench to drain out excess solution inside the tube (see Note 8).
Place all tubes on ice horizontally to avoid collecting excess solution; resuspend each nuclei pellet with 100 μl Nuclei Resuspension Buffer; transfer nuclei suspensions into appropriately-labeled 1.5-ml microcentrifuge tubes and keep on ice until all of samples have been processed.
3.2. NRO Reaction for Nascent RNA Labeling and Purification of Total Nuclear RNA
Add equal volume of 2× NRO Reaction Buffer (usually the nuclei suspension reaches ~120 μl), mix well by inverting the tubes several times.
Incubate the NRO labeling reactions at 30°C for 30 min with constant mixing in an oven (see Note 9).
(Optional) Add cold rUTP to 1 mM (2 μl of 100 mM of stock per 200 μl NRO Reaction); continue the incubation for an additional 5–10 min.
Remove NRO Reaction tubes from 30°C incubation and reset temperature at 37°C. Add 200 Units of DNase I (10 Units/μl, Roche Applied Sciences) to each reaction and incubate for 20 min at 37°C.
Add 400 Units of Proteinase K (20 mg/ml, Ambion, premixed with 10% SDS at 3:1) and incubate 15 min at 37°C.
Adjust the NRO Reaction volume to 600 μl by adding RLT Buffer (Qiagen RNeasy Mini Kit); add equal volume (600 μl) absolute ethanol and mix well.
Load onto RNeasy column and follow RNeasy Mini Kit instructions thereafter.
Elute total nuclear RNA with 100 μl of nuclease-free water.
Measure RNA concentration using a Nanodrop ND1000 Spectrophotometer.
(Optional) Aliquot 10 μg of total nuclear RNA, treat with Ambion TURBO DNase™ and clean it using Qiagen RNeasy MinElute Cleanup Kit.
3.3. Preparation of Dynabeads and Immobilization of Biotin-labeled Nascent RNA
Aliquot total amount of kilobaseBINDER Binding Solution (60 μl per sample, Dynabeads® kilobaseBINDER Kit) in a 1.5-ml microcentrifuge tube; add RNaseOUT at 5 μl per 100 μl Binding Solution; put on ice for later use.
Aliquot total amount of Dynabeads needed (30 μl per sample, Dynabeads® kilobaseBINDER™ Kit) in a 1.5-ml microcentrifuge tube, remove solution on the Magnetic Separator Stand.
Wash beads sequentially with two volumes of the total bead pellet volume using the following buffer: 1× PBS (cell culture grade), Bead Wash Solution A, and Bead Wash Solution B. For each wash, let beads settle completely on the Magnetic Separator Stand before discarding the supernatant.
Wash beads once with one volume of the above-described kilobaseBINDER Binding Solution (see Note 10).
Resuspend beads in equal volume (total bead amount) of above prepared kilobaseBINDER Binding Solution; put on ice for later use.
Denature 10 μg (in 30 μl nuclease-free water) purified biotinylated nuclear run-on RNA at 68°C for 3 min; pulse-spin to collect samples and immediately place on ice (see Note 11).
Dispense 30 μl prepared beads into each RNA tube, pipette up and down to mix; mount onto Dynal® RKDynal Sample Mixer and set rotation speed at 8–10 rpm; incubate for 3 hrs at room temperature (see Note 12).
3.4. Bead Wash and T7N6 Primer Annealing for cDNA Synthesis
Prepare T7N6 Primer at 3 mM in nuclease-free water; put on ice for later use.
Carefully add 600 μl of nuclease-free water to each bead-RNA binding tube (section 3.3.); close tubes and invert several times to mix, spin 5–10 sec at 2,000×g in a microcentrifuge; place tubes on the Magnetic Separator Stand and remove supernatant.
Thoroughly wash beads with nuclease-free water as follows: once with 600 μl, twice with 400 μl, and twice with 200 μl; each time allow the beads to settle down completely on the Magnetic Separator Stand (see Note 13).
After removing water from last wash, immediately resuspend beads in 24 μl of 3 mM T7N6 primer and place on ice (see Note 14).
Denature the T7N6 primer and beads-NRO RNA mix at 68°C for 3 min in a Thermomixer with constant shaking (1,000 rpm); immediately afterwards, transfer tubes onto Sample Mixer and incubate at room temperature for 5–10 min with constant rotation (8–10 rpm).
3.5. Solid-phase 1st-strand and solution-based 2nd-strand cDNA Synthesis
Prepare Master Mix for 1st-strand cDNA Synthesis at 16 μl for each sample (4 μl of 10× 1st-strand Buffer, 8 μl of dNTP solution, 2 μl of RNase Inhibitor, and 2 μl of ArrayScript, Illumina® TotalPrep RNA Amplification Kit).
Remove bead tubes from Sample Mixer, spin 5–10 sec at 2,000×g in a microcentrifuge to collect all beads and solutions at the bottom of the tubes.
Add 16 μl of the above prepared Master Mix for 1st-strand Synthesis into each beads-NRO RNA tube and pipette up and down to mix well (now the total volume is 40 μl per reaction). Incubate the solid-phase 1st-strand cDNA Synthesis reaction for 2 hrs at the preset 42°C oven with constant rotation (8–10 rpm) (see Note 15).
Prepare Master Mix for RNase H digestion at 20 μl per reaction: 10 μl of nuclease-free water, 6 μl of 10× 2nd Strand Buffer, 2 μl of Invitrogen Random Primer N6 (Invitrogen), and 2 μl of RNase H, Illumina® TotalPrep RNA Amplification Kit.
Remove all samples from the 42°C oven, spin 5–10 sec at 2,000×g in a microcentrifuge to collect all beads and solutions; at the same time, reset oven temperature at 37°C.
Add 20 μl of the Master Mix for RNase H digestion prepared above into each tube and pipette up and down to mix well (now the total volume reaches 60 μl per reaction). Incubate 30 min at 37°C oven with constant rotation (8–10 rpm).
Prepare Master Mix for 2nd-strand cDNA Synthesis at 40 μl for each sample (30 μl of nuclease-free water, 4 μl of 10× 2nd-strand cDNA Buffer, 4 μl of dNTP solution, and 2 μl of DNA Polymerase, Illumina® TotalPrep RNA Amplification Kit).
Remove all samples from 37°C oven, spin 5–10 sec at 2,000×g in a microcentrifuge to collect all beads and solutions.
Incubate tubes in 68°C for 3 min in a Thermomixer with vigorous mixing (above 1,000 rpm); immediately spin 5–10 sec at 2,000×g in a microcentrifuge to collect all beads and solutions and place tubes in the Magnet Separator Stand; remove solution fraction (will contain the 1st-strand cDNA) into appropriately labeled 1.5-ml microcentrifuge tubes and place on ice until all of the samples are collected (see Note 16).
Dispense 40 μl of above prepared Master Mix for 2nd-strand cDNA Synthesis to each 1st-strand cDNA tube, pipette up and down to mix well (now the total volume is 100 μl per reaction); incubate 2 hrs in a Thermomixer at 16°C with constant shaking (650–850 rpm).
3.6. cDNA Purification, cRNA Synthesis, cRNA Purification and Illumina Array Hybridization, Wash, Scan and Data Analysis
For cDNA purification and cRNA synthesis, follow instructions of Illumina® TotalPrep RNA Amplification Kit (see Note 17).
For cRNA purification, directly add 350 μl of cRNA Binding Buffer and 250 μl of absolute ethanol into the cRNA synthesis reaction tube, and follow instructions of Illumina® TotalPrep RNA Amplification Kit.
We recommend loading 5–10 μg of total nuclear run-on cRNA on Illumina BeadsChip for hybridization at 58°C for 14–16 hrs. For array wash, scan and data analysis, follow instructions as for conventional Illumina arrays.
3.7. Application to other Microarray Platforms
The biotin-labeled NRO cRNA prepared here can be applied to other microarray platforms with minor protocol modifications. Here we describe our application to Affymetrix Exon Arrays (see Note 18).
1st-strand cDNA Synthesis: Aliquot 10 μg NRO cRNA and follow instructions of GeneChip Whole Transcript (WT) cDNA Synthesis and Amplification Kit.
Cleanup Single-strand DNA (ssDNA): follow instructions of GeneChip WT cDNA Synthesis Kit.
Fragmentation of ssDNA: follow instructions of GeneChip® WT Terminal Labeling Kit.
Labeling of fragmented ssDNA: follow instructions of GeneChip® WT Terminal Labeling Kit.
Microarray hybridization and wash: follow instructions of GeneChip Hybridization, Wash and Stain Kit (HWS Kit).
4. Notes
For general precautions when working with RNA, all instruments that touch sample tubes should be kept nuclease-free. We typically wipe laboratory bench and centrifuge (including rotor and tube slots) and any other equipment in direct contact with tubes with RNaseZap® (Ambion, cat. # 9780).
We suggest purchasing solutions which are certified DNase-free and RNase-free from the manufacturers. In our experience, RNase-free solutions from Quality Biologicals perform well.
We typically prepare 10% NP-40 as regular stock and add the needed amount when preparing solutions. Cell Lysis Buffer can be stored at 4°C for long term use (more than 6 months).
The 2× Nuclei Resuspension Buffer can be dispensed into 1 ml aliquots and stored at −80°C for extended periods of time (more than 6 months).
To get consistent adequate labeling, we typically use 20–30×106 of either adherent or suspension cells for isolation of nuclei for each condition (untreated and treated cells); we use trypsinization to collect adherent cells. For comparison between nascent transcription and steady-state RNA expression profiles, we recommend aliquoting a small portion of cells (such as 1/10) for total RNA isolation and conventional microarray analysis.
We have successfully detected nascent primary microRNA transcripts using real-time PCR with the double-stranded cDNAs (before cRNA synthesis) as the template. Modified procedures of the cRNA synthesis step (no addition of biotin-modified ribonucleotides) followed by labeling of 1st-strand cDNA (using the NRO cRNA as prepared above as template) with biotin-modified dCTP can be applied to detect antisense transcripts.
Cell pellet can be stored at −80°C for total RNA isolation at later times. We typically use Qiagen RNeasy Mini Kit with DNase I treatment to isolate total RNA for conventional microarray.
Watch out for the nuclei pellet sliding down the wall of the conical tube!
We recommend carrying out the reaction in a hybridization oven. First, set the oven temperature at 30°C; then fix the microcentrifuge 1.5-ml tubes on the rotor with rubber bands; remount the rotor and keep rotation at 8–10 rpm.
The kilobaseBINDER Binding Solution is viscous and it takes longer for the beads to settle onto the wall of the tube on the Magnet Separator Stand. After place tubes on the Magnetic Separator Stand and solution, we recommend pipetting up and down several times while the solution turns clear to prevent the loss of beads retained in the tips.
We typically apply 10 μg of the total NRO RNA for bead capture although it can go down to 5 μg depending on the labeling efficiency.
As mentioned above, the kilobaseBINDER Binding Solution is viscous, thus be careful to not make too many bubbles when pipette mixing the beads with RNA samples. On the other hand, a few unavoidable bubbles are actually helpful in maintaining a homogenous mixing state in the Dynal® RKDynal Sample Mixer. We typically use two ways of binding depending on the time frame: one is incubating 3 hrs at room temperature and then forward to the next step immediately; the other is to incubating 2 hrs at room temperature, and then transfer the Sample Mixer to the 4°C cold room to incubate over night and begin subsequent steps the following day.
We recommend resuspending the beads at each wash by flipping or inverting the tubes. For the last 200 μl wash, we typically remove 150 μl and flip the remainder to mix beads, spin 2,000×g in a microcentrifuge, and finally remove the remaining 50 μl water on the Magnetic Separator Stand. In addition, to prevent the beads from over drying during handling, we suggest handling no more than four samples for each wash, and no more than two samples when removing 150 μl from the last 200 μl wash.
We recommend adding 24 μl of 3 mM T7N6 primer solution right above the beads on the wall of each tube, tightening the tube, gently flipping to resuspend all beads in solution, then putting on ice until all samples are finished.
We recommend placing the tubes immediately on the rotor in the 42°C oven to keep constant rotation after mixing each sample tube. To further prevent beads from settling in the tube, after mounting all tubes on the rotor, one measure that can be taken is to set the rotor a little unevenly for a slightly asymmetric rotation.
We recommend handling no more than two samples at the 68°C 3-min step. After putting two tubes on the Magnetic Separator Stand, load the next two samples for the 68°C 3 min step while waiting for the beads of the first two samples to settle on the Magnetic Separator Stands for separating the 1st-strand cDNA in solution.
For consistent results, we recommend using Zymo DNA Clean & Concentrator™-5 Kit (Zymo Research cat. #D4014) for cDNA purification.
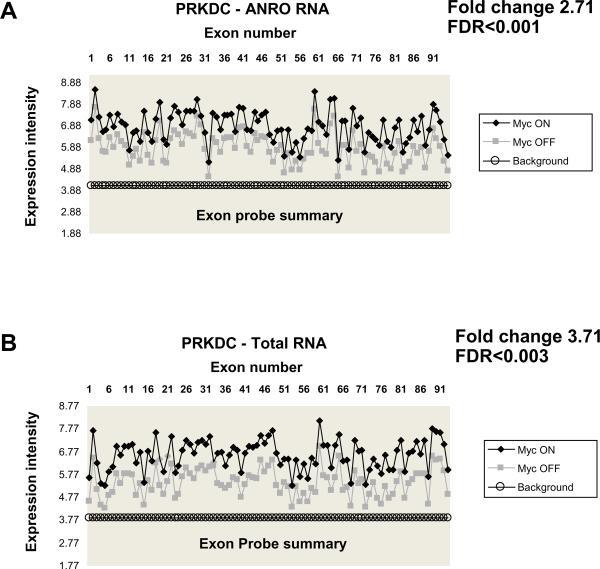
The protocols for application of NRO cRNA to other Affymetrix Arrays are not described in detail here. The Affy exon array data highlighted here demonstrates that the ANRO array method described above can effectively detect full-length, nascent transcripts as exemplified by the detection of the nascent transcript of PRKDC gene (Figure 2) which is composed of 91 exons and spans 190 KB of genomic DNA. In our experience, the Affymetrix Exon Arrays generated far fewer statistically significant regulated genes as compared to the Illumina Ref 8 Beadchips although there is a high correlation between the genes detected by both platforms.
Figure 2.
Detection of full-length nascent transcripts by the ANRO method. Affymetrix exon arrays were used to detect the 91 exons of the PRKDC gene spanning 190 KB of genomic DNA. As the PRKDC gene transcription is under control by the Myc promoter, PRKDC transcription rates and mRNA abundance were compared under conditions of high Myc expression (Myc ON) and no Myc expression (Myc OFF). Graphs represent the expression intensity of each exon using nascent pre-mRNA prepared by the ANRO method (A) and using total RNA prepared using conventional methods (B). As shown, all exons are represented in ANRO arrays, indicating that the nascent PRKDC pre-mRNA is labeled evenly throughout the entire transcript (A). Moreover, the label incorporation in the newly transcribed PRKDC pre-mRNA (A) mirrors the abundance of each exon in the mature PRKDC mRNA (B). When comparing Myc ON vs. Myc OFF, PRKDC pre-mRNA levels are induced by 2.71-fold (A), PRKDC mRNA levels by 3.71-fold. FDR, false discovery rate.
Fan et al.,
References
- 1.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acid Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and Functional Specificity in mRNA Decay. Proc Natl Acad Sci USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan J, Yang X, Wang W, Wood WH, III, Becker KG, Gorospe M. Global Analysis of Stress-regulated mRNA Turnover Using cDNA Arrays. Proc Natl Acad Sci USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheadle C, Fan J, Cho-Chung YS, et al. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei CL, Wu Q, Vega VB, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 6.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 7.Weinmann AS. Novel ChIP-based strategies to uncover transcription factor target genes in the immune system. Nature Reviews – Immunology. 2004;4:381–386. doi: 10.1038/nri1353. [DOI] [PubMed] [Google Scholar]
- 8.Zeller KI, Zhao X, Lee CW, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan J, Zhan M, Shen J, Martindale JL, Yang X, Kawai T, Gorospe M. En masse nascent transcription analysis to elucidate regulatory transcription factors. Nucleic Acids Res. 2006;34:1492–1500. doi: 10.1093/nar/gkj510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan J, Zeller K, Chen YC, Watkins T, Barnes KC, Becker KG, Dang CV, Cheadle C. Time-Dependent c-Myc Transactomes Mapped by Array-Based Nuclear Run-On Reveal Transcriptional Modules in Human B Cells. PLoS ONE. 2010;5:e9691. doi: 10.1371/journal.pone.0009691. [DOI] [PMC free article] [PubMed] [Google Scholar]