Abstract
Metagenomic sequencing provides a unique opportunity to explore earth’s limitless environments harboring scores of yet unknown and mostly unculturable microbes and other organisms. Functional analysis of the metagenomic data plays a central role in projects aiming to explore the most essential questions in microbiology, namely ‘In a given environment, among the microbes present, what are they doing, and how are they doing it?’ Toward this goal, several large-scale metagenomic projects have recently been conducted or are currently underway. Functional analysis of metagenomic data mainly suffers from the vast amount of data generated in these projects. The shear amount of data requires much computational time and storage space. These problems are compounded by other factors potentially affecting the functional analysis, including, sample preparation, sequencing method and average genome size of the metagenomic samples. In addition, the read-lengths generated during sequencing influence sequence assembly, gene prediction and subsequently the functional analysis. The level of confidence for functional predictions increases with increasing read-length. Usually, the most reliable functional annotations for metagenomic sequences are achieved using homology-based approaches against publicly available reference sequence databases. Here, we present an overview of the current state of functional analysis of metagenomic sequence data, bottlenecks frequently encountered and possible solutions in light of currently available resources and tools. Finally, we provide some examples of applications from recent metagenomic studies which have been successfully conducted in spite of the known difficulties.
Keywords: functional annotation, metagenomics, bioinformatics, next-generation sequencing, pathway-mapping, comparative analysis
INTRODUCTION
The microbial world shows vast diversity, and microbes inhabit almost every niche on the planet. Many of them have been shown to be important members of their given ecosystems and to play crucial roles in various environmental and host-associated biological processes. However, due to their general unculturability (it is believed that only a small percentage of bacteria in nature can be cultured [1]), up until just a few years ago it was practically impossible to sequence and analyze them in greater detail. As a result, a large fraction of microbes still remain poorly characterized and unstudied; and the means by which they exert beneficial or other effects in different environments remain largely unknown.
The recent culture independent technology to study microbes inhabiting different environments, termed metagenomics [2], has opened new avenues for answering questions commonly asked in microbiology, such as ‘Which species inhabit a given environment?’ and ‘What are these microbes doing and how are they doing it?’ The basic steps involved in a typical metagenomic project to estimate the number of species and the functional repertoire of an environment include DNA or RNA sequencing using next-generation sequencers (such as Illumina and Roche 454), sequence assembly, gene prediction, functional and metabolic analysis, taxonomic binning and comparative analysis of the sequence data using specialized bioinformatics methods and tools (Figure 1, Tables 1 and 2). However, each stage of the analysis suffers heavily due to inherent problems of the metagenomic data generated, including incomplete coverage, massive volumes of raw sequence data produced by the next-generation sequencers, generally short read-lengths, species abundance and diversity and so on [3, 4].
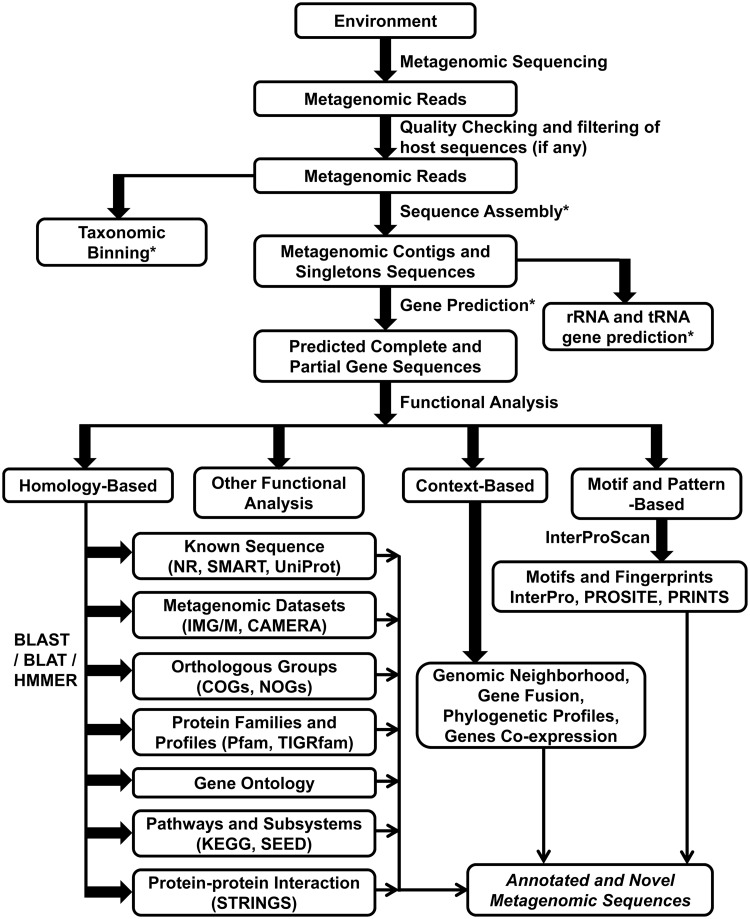
Figure 1:
Flow chart for the analysis of a metagenome from sequencing to functional annotation. Only the basic flow of data is shown up to the gene prediction step. For the context-based annotation approach, only the gene neighborhood method has been implemented thus far on metagenomic data sets; although in principal, other approaches which have been used for whole genome analysis can also be implemented and tested. *: A list of tools commonly used for these processes is provided in Table 1. Table 3 provides a list of some of the additional functional analyses that can be performed on the metagenomic sequences.
Table 1:
List of commonly used tools for sequence assembly, protein coding gene prediction, RNA gene prediction and phylogenetic classification steps of metagenomic data analysis
| Process | Tools | URL/ References |
|---|---|---|
| Sequence assembly | Phrap | http://www.phrap.org/ |
| Forge | http://combiol.org/forge/ | |
| Arachne | [5] | |
| JAZZ | [6] | |
| Celera | [7] | |
| Velvet | [8] | |
| Newbler | 454 Life Sciences | |
| SOAPdenovo | [9] | |
| EULER | [10] | |
| ORFome assembly | [11] | |
| IDBA-UD | [12] | |
| Gene prediction | Metagene | [13] |
| GeneMark | [14] | |
| ORF-Finder | http://www.ncbi.nlm.nih.gov/ projects/gorf/ | |
| FragGeneScan | [15] | |
| fgenesB | http://www.softberry.com | |
| GLIMMER | [16] | |
| BLAST | [17] | |
| RNA gene prediction | tRNAscan-SE | [18] |
| Similarity-based searches for rRNA in reference databases | – | |
| Taxonomic binning | MetaBin | [19] |
| MEGAN | [20] | |
| WebCARMA | [21] | |
| PhyloPythia | [22] | |
| TETRA | [23] | |
| NBC | [24] | |
| TACOA | [25] |
Table 2:
Current list of commonly used publicly available pipelines for the functional annotation of metagenomic data sets
| Pipeline/tools | IMG/M | METAREP | CAMERA | RAMMCAP | MG-RAST | Smash community | MEGAN4 | CoMet | WebMGA |
|---|---|---|---|---|---|---|---|---|---|
| Functional analysis | |||||||||
| Homology-based | |||||||||
| Known sequence | NCBI (NR), SMART, UniProt | NCBI (NR), UniProt | NCBI (NR) | – | NCBI (NR), SMART, UniProt | SMART, UniProt | NCBI (NR) | – | NCBI (NR) |
| Metagenomic data sets | IMG/M | – | – | – | IMG/M | – | – | – | – |
| Orthologous groups | COGs | – | COGs | COGs | COGs, eggNOGs | COGs, eggNOGs | – | – | COGs |
| Protein families | Pfam, TIGRfam | Pfam, TIGRfam | Pfam, TIGRfam | Pfam, TIGRfam | FIGfams | Pfam | – | Pfam | Pfam, TIGRfam |
| Ontology | GO | GO | GO | GO | GO | – | – | GO | GO |
| Enzymes, pathways and subsystems | KEGG, SEED | PRIAM | KEGG, SEED | – | KEGG, SEED | KEGG | KEGG, SEED | – | KEGG |
| Protein interactions | – | – | – | – | STRING | STRING | – | – | – |
| Motif- and pattern-based | |||||||||
| Database | InterPro | – | – | – | – | – | – | – | – |
| Context-based | |||||||||
| Approach | Gene neighborhood | – | – | – | – | Gene Neighborhood | – | – | – |
| Other functional analysis | |||||||||
| Types of predictions | CRISPRs, enzymes, transporter classes | Enzymes, transmembrane helices, lipoprotein motifs | – | – | – | Protein networks | – | – | – |
| URL | http://img.jgi.doe.gov/m/doc/uiMap.html | http://www.jcvi.org/metarep/ | http://camera.calit2.net/ | – | http://metagenomics.nmpdr.org/ | http://www.bork.embl.de/software/smash/ | http://ab.inf.uni-tuebingen.de/software/megan/ | http://comet .gobics.de/ | http://weizhong-lab.ucsd.edu/metagenomic-analysis/ |
| References | [26] | [27] | [28] | [29] | [30] | [31] | [32] | [33] | [34] |
These problems also adversely affect the downstream functional analysis process. For example, due to shorter read-length the overall functional composition is comparatively poor for shorter pyrosequencing- or Illumina-sequencing derived reads than for longer Sanger reads [35]. Additionally, for very complex communities, partial or poor assemblies are obtained due to incomplete coverage, resulting in many short contigs and unassembled sequences. This leads to the prediction of a large number of small, fragmented genes which may not exhibit any matches in the reference sequence databases, or match with very low significance [36]. Although sequence assembly and gene prediction tools specifically developed for metagenomic data sets offer some advantages over similar tools developed for more complete genome sequences, surprisingly, no such ‘metagenome specific’ tools have yet been developed for functional analysis. Thus, appropriate tools, from the current repertoire, and parameters must be used to achieve comprehensive and biologically meaningful functional analysis of metagenomic data sets. The steps for sequence assembly and gene prediction of metagenomic data sets are compared in several recent comprehensive reviews [3, 4, 37, 38].
The scope of this review is to comprehensively discuss the prime objectives, methods and problems for functional and metabolic analysis of metagenomic sequence data, and to propose some solutions for the latter. Toward this, we first try to familiarize the reader with the aims of functional metagenomic analysis and the most commonly adopted publicly available tools and resources to achieve them. Next, we discuss how the problems arising from metagenomic sequencing affect this process, and we suggest various strategies for addressing some of these issues under the present scenario. Lastly, we demonstrate that, despite these issues, metagenomic functional analysis can still be reliably used to address globally important environmental and biological questions.
OBJECTIVES OF FUNCTIONAL METAGENOMIC ANALYSIS STUDIES
Interestingly, the same microbial communities sampled at different times or from different hosts can vary significantly. For example, the gut microbiomes of 13 healthy Japanese individuals were quite different, yet they still shared many microbes [39]. Also, the community members for any given environment commonly play different roles. For example, in the human gut microbiome, segmented filamentous bacteria are known to play important roles in maintaining intestinal immunity [40, 41], whereas bifidobacteria are known to utilize complex carbohydrates and thereby exert beneficial effects on human health [42]. Thus, there are mainly two broad objectives of the functional analysis for metagenomic studies: the first is to determine what are the functional and metabolic repertoires of the different community members that enable them to exert different effects, and the second is to identify the variations, if any, within the functional compositions of the different communities, e.g. those found between healthy and diseased individuals that may be related to the cause of the disease. To determine the functional content of the member species of a microbiome, the coding and functional capacity for all (or at least the dominant) members should be comprehensively analyzed. Alternatively, if the goal of the study is to analyze and contrast the functional and metabolic capacities of different communities, then the functional and metabolic pathway profiles for the communities need to be generated and compared.
PUBLICLY AVAILABLE RESOURCES AND TOOLS FOR FUNCTIONAL ANNOTATION OF METAGENOMIC DATA
Dedicated tools for functional annotation and analysis of metagenomic data sets lag far behind the rate at which the data is being generated. Recently, some web-based, as well as local-use based, pipelines have been developed for the analysis of metagenomic data sets. Table 2 provides a list of a few well-known representative pipelines and compares the functional analysis capacity of each. Almost all of these pipelines provide integrated platforms for the functional prediction of metagenomic sequences using multiple tools and databases, which are also commonly used for the analysis of whole genome sequences. Most of the pipelines offer sufficient resources for the functional analysis of user data. However, to account for the inherent problems associated with the metagenomic data sets, it is highly recommended to evaluate the computational workflow and parameters for any given project. This can be achieved by using simulated sequencing reads generated by MetaSim [43], to assess and compare different tools before actually using them on full data sets. The analysis time of any pipeline typically depends on the size of the data sets and, in the case of web-based servers, the load of requests that are already in progress submitted by other users. Web-based servers such as CAMERA [28], MG-RAST [30] and IMG/M [26] host pre-computed results for most published metagenomes that enable users to perform comparative analysis with their own data sets. In most cases, the computed data can be visualized in the form of simple plots. However, KEGG [44] pathway maps and abundance profiles can also be obtained using the IMG/M and MG-RAST servers.
STRATEGIES COMMONLY ADOPTED BY THE PIPELINES FOR THE FUNCTIONAL ANALYSIS OF METAGENOMIC DATA
Protein function is a very broad term, as function can be predicted at several different levels. For example, the Gene Ontology database [45] adopts three broad domains for classifying gene products viz., the cellular location of the protein, the overall biological process it takes part in and the molecular function of the protein. On the other hand, the subsystem-based classification approach adopted by the SEED database [46] relies mainly on the grouping of functional roles into subsystems by curation experts. The defined subsystems may be thought of as a generalization of the term ‘pathway’. Similarly, the KEGG database [44] is a resource of pathway maps built from both genomic and chemical information of the biological systems. However, such specific functional assignment may be lacking for completely novel proteins or for those which share very weak homology with known proteins both of which are ample in metagenomic data sets. For such proteins, even minimal information that can be extracted related to their function can be useful, and may be the only available clues to their function.
As shown in Figure 1 and Table 2, the basic tools that are implemented in almost all of the available pipelines for functional analysis of metagenomic data are the same as those which are commonly used for whole genome studies and are well known. However, their performance in the metagenomic context have yet to be evaluated and reviewed. Thus, in the current review, we have divided these tools into four categories based on their inherent approach. In the following sections, we review each approach in context to its application to metagenomic data analysis, keeping in mind the associated problems of the data itself.
Homology-based approach
As shown in Table 2, the ‘simplest’ and most common approach adopted by all of the available pipelines for functional prediction is by comparison of the predicted query proteins to existing resources of reference protein sequences, including NCBI NR [47], SMART [48] and UniProt/UniRef [49]. The IMG/M [26] and MG-RAST [30] servers also search the publicly available metagenomic data sets for homologs of the query sequences. The databases of clusters of orthologous groups (COGs) [50], non-supervised orthologous groups (NOGs) [51], protein families and domains including Pfam [52] and TIGRFAM [53], etc. are used by several pipelines to infer functional categories or to identify families and domains embedded in the query proteins. In some cases, similarities to genes found in the GO database are further explored to infer hierarchical annotations. Pathway and subsystem information for the query proteins is inferred by searching for homologs in the KEGG and SEED databases, respectively, by almost all of the pipelines.
For these searches, different variants of BLAST [17] are the most preferred algorithms, including BLASTX, BLASTP, RPS-BLAST, etc. For less sensitive, but faster, searches BLAT [54] may also be used, as in the case of MG-RAST server. Additionally, more sensitive profile- and pattern-based search methods are used by almost all of the pipelines in which sequence profiles generated from alignments of protein families in Pfam or TIGRfam databases are searched using the hidden Markov model-based algorithm, HMMER [55]. For all these methods, best hits are identified based on statistical calculations and annotation information is directly applied to the query proteins.
Homology-based approaches mainly suffer from the long computation time required to search for homologs for each of the sequences within the typically massive metagenomic data sets. Additionally, BLAST-based functional predictions have been estimated to include 13–15% database propagation errors [56]. Moreover, to detect a true match, the reference database being searched needs to contain at least one homolog of the query sequence. And, the fragmentary nature of the shotgun-generated metagenomic data leading to partial proteins negatively impacts homology-based function prediction. This is discussed in more detail below.
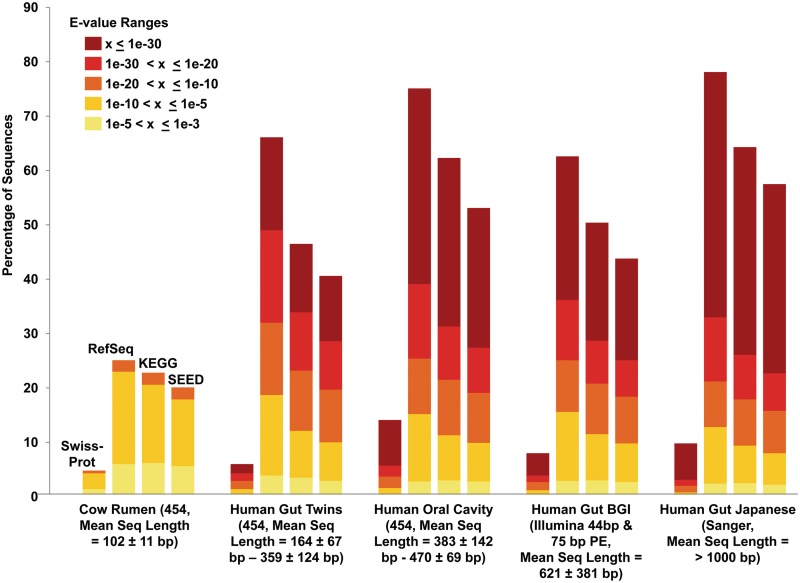
The extent to which metagenomic functional annotation has been achieved using different databases is demonstrated in Figures 2 and 3. The highest fraction of metagenomic sequences were annotated using the NCBI RefSeq database, which is a comprehensive collection of non-redundant well-annotated protein sequences. On the other hand, only a small fraction of sequences could be annotated using the Swiss-Prot database, which harbors manually annotated and reviewed protein sequences. The number of proteins annotated using the COGs database was slightly less than RefSeq. Among the protein family and profile databases, more predictions were made using Pfam as compared to the TIGRFAM database. This could mainly be due to the great number of protein families that are included in the Pfam database (13 672 in Pfam 26.0 release) than in the TIGRFAM database (4209 in TIGRFAM 12.0 release). The annotation using KEGG metabolic pathways is relatively low mainly due to the inherent problems of the metagenomic data sets, as discussed below. The SEED system of classification performs similar to that of KEGG, although the number of predictions is slightly lower.
Figure 2:
Distribution of metagenomic sequence matches in the SwissProt, RefSeq, KEGG and SEED databases at various E-value cut-offs. Smaller sequences match at lower confidence (higher E-values; lighter colors) or do not match at all in the databases. More sequences match with higher confidence (lower E-values; darker colors) as the sequence length used for the analysis increases. Pre-computed data for the metagenomes shown was derived from the MG-RAST server.
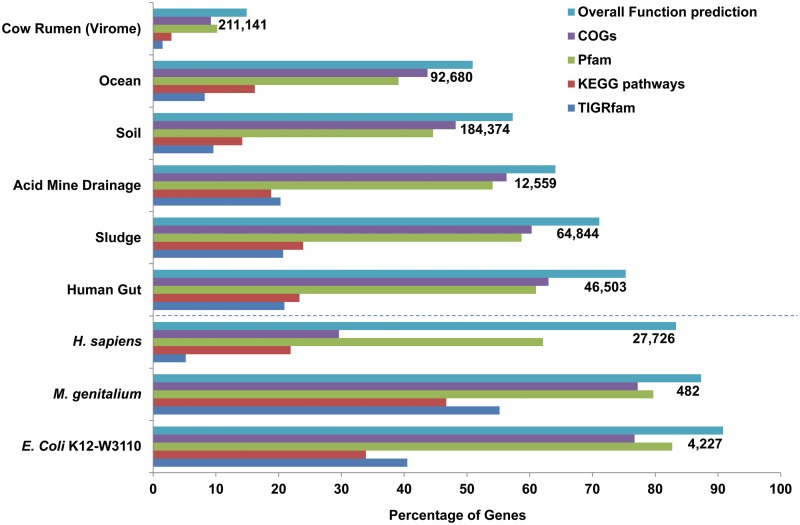
Figure 3:
Status of functional prediction of protein-coding genes from different metagenomic data sets and representatives of completely sequenced genomes. The overall functional prediction bars represent the fraction of protein-coding genes that map to at least any one of the four databases including cluster of orthologous groups (COGs), Pfam, TIGRFAM and KEGG pathways. For comparative purposes, the functional annotation status for the well-studied model microbial genome, E. coli K12-W3310, the smallest microbial genome, M. genitalium, and the human genome are also shown. The data for this graph was derived from the IMG/M database. It should be noted that for uniform comparison, the prokaryotic COGs version was also used for Homo sapiens. The number of matches to eukaryotic COGs (KOG database [57]) may be higher for H. sapiens. The numbers next to the bars represent the total number of predicted protein-coding genes in each data set using the IMG/M annotation pipeline. For the Sludge [58] community, data from only the Phrap assembly, a widely used program for DNA sequence assembly, was used. Except for the Cow Rumen Viral community [59], which was sequenced using the 454 platform (average read-length > 300 bp), all other metagenomes were sequenced using the Sanger method (average read-length > 1000 bp). The following additional data sets were used: Ocean [60], Soil [61], Acid Mine Drainage [62], Human Gut [63].
Motif- or pattern-based approach
The partial proteins generated from short contigs and unassembled sequences which arise due to short read-lengths or complex environments generally exhibit very poor similarities using homology-based approaches (Figure 2). Additionally, some proteins, despite sharing a common function, are more diverse at the sequence level. The overall sequence similarity of such proteins is usually lower than the thresholds used for homology-based functional prediction; however, they still share one or more common sequence or structural patterns or motifs necessary to maintain their structure and function. Currently, databases like PROSITE [64] and PRINTS [65] present a reliable repository of such patterns or motifs against which the query metagenomic sequences may be searched either independently or through the integrated InterPro database [66]. Currently, only the IMG/M server incorporates the InterPro database. However, a general problem with motif-based annotation is that short sequence matches typically show low statistical significance and false-positive rates can be high [67]. Nevertheless, given the amount of novelty inherent in metagenomic data sets, it is recommended to run motif-based analysis in parallel with other functional prediction approaches.
Context-based annotation
Metagenomic data sets contain a large number of novel sequences which share no homology with known sequences and thus remain unannotated by the previous two approaches. To overcome these limitations, gene context-based approaches may also be used. A few examples from single genome annotation projects include genomic neighborhood [68, 69], gene fusion [70, 71], phylogenetic profiling [72] and gene co-expression analysis [73]. Among these, only the genomic neighborhood approach has been implemented in the case of metagenomics. In 2007, Harrington et al. [74] applied a combination of homology-based searches and customized gene neighborhood methods to four metagenomic data sets derived from a variety of complex environments. Whereas BLAST-based methods alone annotated 70% of the sequences, their combined method inferred specific functions for 76% and non-specific functions for 83% of the sequences. However, due to the paucity of complete genomes in metagenomic data sets and the lack of knowledge about the true species origin of the sequences, this approach has its limitations. These problems may be ameliorated by increasing the sequencing depth and by improving the taxonomic assignment of the sequences. Additionally, better assemblies resulting in longer contigs will also improve the efficiency of context-based annotation methods. Currently, only IMG/M and SmashCommunity [31] can be used to view predicted genes in the genomic neighborhood context.
Other types of functional prediction
Lastly, the putative roles of the metagenomic sequences can also be inferred by running more specific analyses using dedicated tools that target prediction of carbohydrate active enzymes, glycosyl hydrolases, protein localizations, lipoproteins, adhesins, secretory proteins, transporters, CRISPRs (Clustered Regulatory Interspaced Short Palindromic Repeats), insertion sequences, virulence factors, etc. A list of a few representative tools for such analysis is given in Table 3. It should be noted that the list is not comprehensive, and that a discussion about all the tools for the above-mentioned purpose is beyond the scope of this review.
Table 3:
List of commonly used available resources for functional analysis (other than homology-, motif- and context-based) that can be performed on metagenomic data sets
GENE-CENTRIC ANALYSIS OF METAGENOMIC DATA SETS
To explore the effect of environment on the functional and metabolic contents of different communities, comparative functional analysis may be performed on the total gene-content of the communities, i.e. gene-centric analysis. For this purpose, functional profiles can be compared and contrasted across different metagenomic data sets to look for functional characteristics responsible for community differences. Normally two levels of comparison are performed, viz., comparison of abundance of functional families and pathways, and estimation of statistical parameters to ensure that the observed differences in abundance are not merely chance occurrences. Different types of abundance profiles may be generated and compared using, for example, COGs functional categories, Pfam functional families, KEGG metabolic pathways, or SEEDs subsystems. However, before comparing the metagenomes, proper normalizations of the data sets should be performed to account for the data-associated problems, such as partial genes and effective genome sizes (discussed later). Heat-maps are commonly used to visualize the differences in communities with respect to the above-mentioned functional or metabolic profiles (for example [60, 61, 76–78]). In addition, statistical methods, such as principal component analysis (PCA) and multidimensional scaling (MDS), may be used to reveal which factors most affect the observed data (for example [79, 80]). The common approaches and limitations of the gene-centric analysis are discussed and reviewed by Kunin et al. [3].
PROBLEMS ASSOCIATED WITH FUNCTIONAL ANALYSIS OF METAGENOMIC DATA
The analysis and annotation of metagenomic data sets differ from that of whole genome studies mainly because the former is a complex mixture of sequences from multiple species. Even draft quality bacterial whole genome sequences represent most of the chromosomes, except for a few of the more complex regions that include repeats, insertion sequences, tRNAs, rRNAs, etc. When sequence coverage is sufficient, the assemblies obtained usually result in very long contigs with few gaps. The efficiency of gene prediction algorithms on such long contigs is quite high and most of the full-length coding DNA sequences (CDSs) can be predicted with high confidence. Functional prediction analysis can next be applied to obtain the functional repertoire of the genome. The functionally annotated CDSs can then be viewed in the context of metabolic pathways to predict the metabolic capabilities of the species under study.
A metagenome can be viewed as a collection of several whole genomes. To fully understand an environment, in principal, draft quality whole genome sequences for every member should be achieved by complete DNA sequencing. However, in spite of the availability of high throughput second-generation sequencers, this is still a very expensive and daunting task. What can be best captured from a metagenomic sample is a mixture of fragmented sequences from the community members, and mostly from dominant members of the environment. When the sequencing depth is sufficient, and by the use of sequence assemblers developed specifically for metagenomic data (Table 1), draft quality assemblies for some of the member species may be achieved; e.g. a draft methanogen genome was recently assembled from a permafrost microbial community [78]. However, this still did not suffice for completely understanding the environment, as the assemblies for many other members remained poor due to the inherent complexity of the environments and lower sequencing coverage for these genomes. Thus, for most metagenomic studies, we are left with only enormous volumes of fragmented sequences (comprised of a mixture of short contigs and singletons) from multiple species to perform analysis on. In the case of contigs, gene predictions will be more accurate, whereas the predicted genes from singletons will almost always be partial in spite of using gene prediction tools specifically developed for metagenomic data (Table 1), unless very long read-lengths were obtained during sequencing. This is mainly because the typical average read-lengths generated by next-generation sequencers providing deeper coverage, including Illumina, are still smaller (up to 300 bp for paired-end reads) than the average size of the typical prokaryotic protein coding gene (∼1000 bp [81]). The 454 pyrosequencing platform can be an alternative technology due to the longer average read-lengths it can generate (up to 700 bp for 454 GS FLX+ pyrosequencer, http://454.com/downloads/GSFLXApplicationFlyer_FINALv2.pdf), but it is not the preferred choice mainly due to its lower coverage and higher cost as compared to Illumina sequencing.
To obtain the most complete information of the functional repertoire for any metagenome it is recommended to use the genes predicted from both the contigs and the singletons, even though many of the predicted CDSs are partial. In general, short query lengths negatively impact homology-based functional prediction as they may decrease the significance of pairwise similarities due to added noise. This is clearly evident from Figure 2, which shows that there are no matches for sequences of length ∼100 bp for the ‘Cow Rumen’ metagenome [79] in the lower and more significant E-value bins (E-value < 1e − 10). On the other hand, as sequence length increases, the E-value bins with lower values become more populated, as in the case of the ‘Human Gut Japanese’ [39] data set. Additionally, for short sequence lengths, homology-based approaches have limited sensitivity. For example, only ∼25% of the ‘Cow Rumen’ sequences could be annotated using GenBank, whereas >75% of the ‘Human Gut Japanese’ sequences could be annotated using the same database with the same parameters (Figure 2). These problems may be ameliorated to some extent by increasing sequencing depth or read-length so that better assemblies and gene predictions can be obtained.
Another problem in metagenomic functional analysis stems from the lack of knowledge of the species of origin of the sequences. Although phylogenetic classification and binning methods specific to metagenomic sequences may be able to classify 40–93% of the reads [19] at the genus level, depending on the novelty of the data set, at the species level this percentage is expected to decrease. This indicates that at least 7–60% of the sequences still remain unclassified due to the limitations of the available tools and the paucity of reference genomes in the public databases. Thus, in spite of gaining some functional information, due to the absence of specific species information, it is extremely difficult to put together many functionally annotated metagenomic sequences in context of their actual metabolic pathways. Additionally, because most of the metagenomic sequences will be derived from the dominant species, the complete functional and metabolic repertoire of the less abundant members cannot be obtained. Other techniques complimentary to metagenomics, such as single cell genomics [82], may help in overcoming this problem by providing access to the genomic DNA from unculturable microbes. However, even single cell genomics has many challenges remaining [82]. Nevertheless, if the objective of the metagenomic study is to only analyze the overall metabolic capacity of the entire community, then putting the sequences in context of their individual genomes of origin may not pose a serious problem.
Given that metagenomic studies are aimed at exploring complex environments harboring many yet uncultured and unknown microbes, the data sets are expected to possess a large number of novel sequences. As shown in Figure 3, the overall functional annotation achieved in the case of some example bacterial metagenomes is 50–75%, with the remaining sequences being unannotated. Even for ‘complete’ genomes, functional annotation is not complete. In the most studied model organism, Escherichia coli K12-W3110, and the smallest studied genome, Mycoplasma genitalium, both of which are considered ‘simpler’ systems, the overall functional annotation remains ∼90%. And, in a more complex system viz., the human genome, only ∼82% of the predicted proteins are currently annotated. For the even more complex human gut metagenome, this number decreases to ∼75%. Interestingly, while ocean and soil are also considered as ‘complex metagenomes’ on the scale of the human gut microbiome, only ∼50–55% of the sequences in these communities can be annotated. This difference in level of annotation could be due to a bias in the number of human-associated microbial genomes that have thus far been sequenced and are included in the reference sequence databases. To deal with the novelty of metagenomic data, reference genome sequencing efforts should be initiated for other environments as has been done under the Human Microbiome Project [83], which plans to sequence a large number of reference genomes from different body sites for the human microbiome.
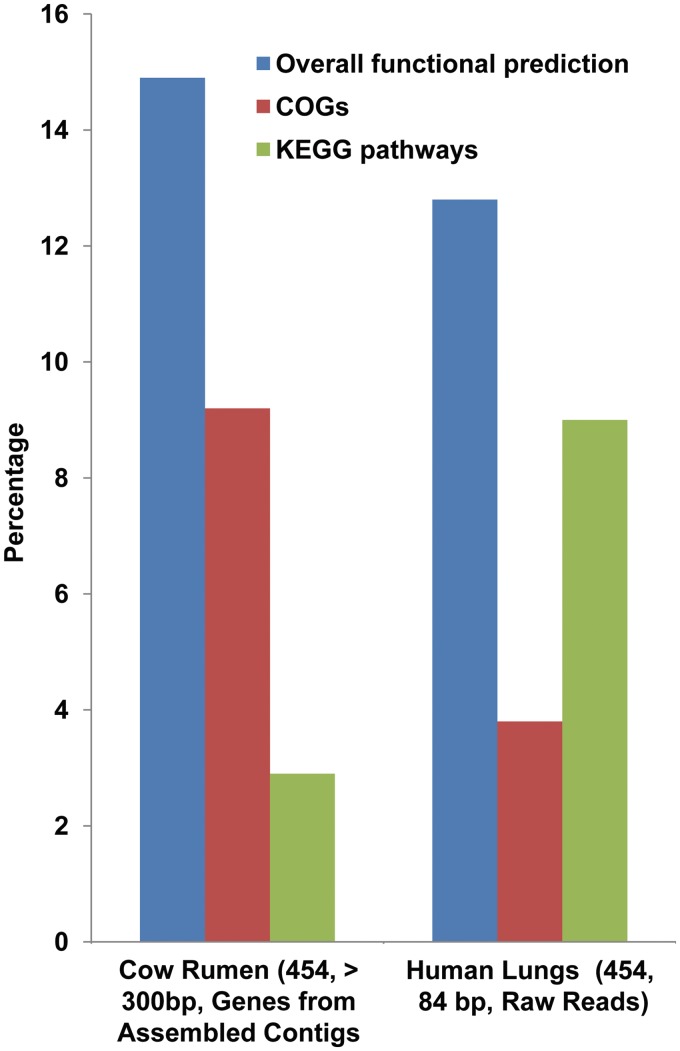
While the functional annotation of bacterial metagenomes is at a reasonable level and is gradually improving, the situation for viral metagenomes, or viromes, lags far behind. The extent of virome annotation for cow rumen [59] and human lung [80] drops to as low as 13–15% (Figure 4) in comparison to bacterial annotation (cow rumen: 32%) for similar environments. The average metagenomic read-length used for the human lung virome was only 84 bp. One might argue that this reduction in the percentage of functional annotation may be due to the short read-length, which is known to affect the extent and confidence level of the functional prediction process, as discussed earlier. But, surprisingly, the percentage of functional annotation for the cow rumen virome is also low (15%), despite using a longer read-length (>300 bp). Thus, this reduction in the extent of functional prediction for viromes could be mainly due to the limited number of completely sequenced viral species in the reference databases.
Figure 4:
Status of functional prediction for viral metagenomes. The bars for the Cow Rumen viral metagenome data set represent the percentage of genes predicted from assembled contigs, while those for the Human Lung viral metagenome data set [80] represent the percentage of raw reads.
The genome sizes of the individual microbial members of a community can vary greatly. It is known that larger genomes harbor a smaller relative fraction of universal and housekeeping genes, and thus contain a large number of novel genes [84, 85]. Indeed, a weakly significant positive correlation was found between the effective genome size and the potential for carrying novel genes [86]. Therefore, the average genome size in an environmental sample could also affect the comparative functional analysis of the metagenome. Recently, Beszteri et al. [87] demonstrated how, among metagenomic samples, the differences in relative gene abundance, which are often used to interpret habitat-specific adaptations, are biased by the average genome size of the communities sampled. Thus, before arriving at biological conclusions from functional analysis of metagenomic data sets, the latter should be normalized to account for their different average genome sizes.
Apart from the aforementioned problems, the analysis of metagenomic data sets can also be influenced by the sequencing technology used. For example, 454 pyrosequencing technology produces between 11–35% artificial replicates, both identical reads (duplicates) and reads that begin at the same position but vary in length or contain sequencing discrepancies, which lead to biased functional annotations [88]. Replicates were also observed in an Illumina sequenced permafrost microbial community analysis [78]. Thus, the metagenomic reads should be de-replicated before in-depth functional analysis is performed. Both 454 pyrosequencing and the more recent Ion Torrent sequencing technologies are known to introduce frameshift errors in the reads, mostly due to homopolymer runs. Almost none of the available bioinformatics tools for functional annotation of metagenomic sequences are capable of handling such errors; although several specialized tools for frameshift detection are currently available [89–93] in the public domain and should be used for more in-depth functional analysis. In some cases, the protocols used for sample preparation, particularly the use of filters or other sample selection methods, can also lead to inappropriate biological interpretations. For example, in the first Sargasso Sea data set [94], some nitrogen-fixing genes were found to be lacking [95]. However, the lack of these genes was later attributed to the absence of their main contributors, cyanobacteria, which were likely removed during the filtration step [96].
APPLICATIONS OF METAGENOMIC FUNCTIONAL ANALYSIS
Despite the challenges for metagenomic functional analysis, many studies exploring different environments are being conducted with varying degrees of success. The applications of metagenomic functional analysis is an extremely important and versatile subject; and, given the scope of the current review, it is impossible to comprehensively discuss it here. Therefore, to exemplify the successful implementation of metagenomic functional analysis to answer some biologically and environmentally important issues, a few recent example studies are presented in the following sections. For a discussion of other studies of major interest, we recommend the comprehensive review by Wooley et al. [4].
Comparative metagenomic-based studies
Recently, in a large-scale metagenomic analysis of 124 European individuals, a catalogue of over 3.3 million human gut microbial genes was created [97]. This led to the identification of bacterial functions that are necessary for a bacterium to thrive in the gut context, and to those functions involved in homeostasis of the entire ecosystem. This catalogue not only provides a good resource for annotating new human gut-related metagenomes and for comparative analysis, it also enables future studies to discover associations between the microbial genes and human phenotypes. In another study, the gut metagenomes of four healthy individuals were compared to those of individuals with autoimmune disorders, including type I diabetes [98]. This analysis suggested that increased adhesion and flagella synthesis in diseased individuals may be involved in triggering type I diabetes associated autoimmune response. Recently, a comparison between the human gut environment and the oral cavity was made by comparing the two metagenomes, and clear distinctions in the functional capacities of the two niches were observed [99]. In the same study, another comparison between oral metagenomes from supragingival dental plaque and cavities of healthy and diseased individuals, respectively, suggested that the dental plaque of healthy individuals (those who have never suffered from caries) may be a genetic reservoir for novel anticaries compounds and probiotics, which are live microorganisms thought to be beneficial to the host organism.
Metagenomics studies to date have not only aimed at exploring human health-related issues, but have also attempted to address various environmental issues. Global warming resulting from the emission of greenhouse gases is a major concern worldwide. Rising global temperatures cause permafrost, a vast reservoir of natural carbon, to thaw, resulting in microbial degradation of organic matter and emission of more greenhouse gases. Comparative metagenomics of permafrost was recently applied to both the frozen and thawed states to analyze the shifts in microbial and functional composition [78]. Multiple genes involved in carbon and nitrogen cycling were found to shift rapidly during thaw. From this study, important insights about the microbial species and functional components involved in greenhouse gas emissions may be obtained.
Metagenomic data-mining-based studies
The natural diversity and affluence of metagenomic data is enormous. Over 300 independent metagenomic projects have already been completed or are underway. These facts provide a great opportunity for in-depth mining of metagenomic data and exploration of novel gene candidates useful under a variety of different scenarios. For example, the metagenomic data sets from 10 diverse sources were used to identify several novel candidates for commercially useful enzymes (CUEs) [100]. A catalogue of 510 CUEs was prepared using literature search followed by manual curation, and then the catalogue was used to find homologues in the metagenomic data sets. High-throughput functional metagenomic screening may be used to look for the presence of CUEs and other specific enzymes of interest in the metagenomes [101]. In another study, the recruitment of genomes from pathogens against the metagenomes of healthy individuals containing commensal strains of the same species was used to identify the genomic regions of individual bacterial isolates missing in the metagenomes [102]. These regions are referred to as metagenomic islands and are found to harbor several virulence-related genes specific to the pathogenic strain.
CONCLUSIONS
Metagenomic sequencing provides a unique opportunity to explore yet unknown environments in great detail. Functional analysis of the metagenomic data plays a central role in such studies by providing important clues about functional and metabolic diversity, as well as variation. While metagenomic studies continue to suffer from certain caveats that make the downstream data analysis a challenging task for bioinformaticians, the gradual improvement in metagenomic technologies and development of tools and resources that account for the known problems will relieve some of the burdens. For example, the use of next-generation sequencers producing longer read-lengths (>300 bp) will usually lead to better sequence coverage. This can then be followed by the use of sequence assembly and gene prediction tools and parameters specifically developed for metagenomic sequences which will further help in improving assembly and gene prediction efficiency, respectively, and will result in a greater number of complete predicted proteins. Better functional assignments for metagenomic data sets can be obtained by using more complete proteins. However, while comparing the abundance profiles of functions between communities, the frequencies of the functions should not be masked by the assembly, and the read depths of the contigs should be accounted for. Another common problem that is usually encountered in metagenomic data functional analysis is the long computational time that is required for BLAST-based homology searches for orthologs. The use of alternative search algorithms, such as BLAT, can provide analysis results in shorter times; however, the loss of sensitivity by BLAT-based searches should be taken into account when analyzing the results. Alternatively, profile-based search methods using the HMMER algorithm may also be used whenever pre-computed sequence profiles are available. Certain issues, including large volumes of metagenomic sequence data, large storage requirements for the analyzed data, and the typically large number of unknown sequences in the metagenomic data still pose serious challenges for its analysis. Therefore, there is great need for the development of new, faster, more sensitive tools and more thorough resources dedicated to the functional analysis of metagenomic data sets. Also, it is strongly advised that when analyzing the data, one must be aware of any additional factors that can influence the functional analysis, including sample preparation, sequencing method, diversity of the environments, etc. Proper calibrations, normalizations and statistical tests for significance should always be performed in order to arrive at the most reliable conclusions.
DNA sequence-based metagenomic functional analysis is limited in that it only provides information about the functional content of an environment. Thus, it may be complemented by other independent approaches that help to gain further insights about the more dynamic aspects of a given community. For example, a few metatranscriptomic projects have been undertaken to address which genes are actually being expressed in different environments and to what extent [103, 104]. Given that proteins are much more stable than mRNAs [105], a proteome-based analysis is expected to provide a more accurate view of the functionality of a given environment. Toward this, a few metaproteomic studies have been conducted to explore which protein products are formed and how are they involved in the cross-talk within the environment under different conditions [106–109]. The metabolome, which represents the complete set of small molecules in an organism, can influence gene expression and protein function. Therefore, metabolomics also plays a key role in understanding cellular systems and decoding the functions of genes [110, 111]. A few metabolomic analyses have been conducted to determine which metabolites are produced as a result of the underlying metabolic pathways that are being exerted in a given community and to study host-microbe interactions [112–117]. Another alternative to the DNA-based studies used for determining microbial community composition, metalipidomics, is being implemented mainly to identify the living microbial cells in an environment [118]. Intact polar lipids (IPLs), which are the basic building blocks of biomembranes, are ubiquitous in nature and have several characteristics that make them useful as proxies for living microbial cells. To date, metabolomic studies have not been directly used for the functional analysis of environments. However, studies seeking to identify microbes of specific functional interest may be conducted, as has been done for ammonia-oxidizing microbes from marine and estuarine sediments [119]. The functional component of the environment may then be extensively analyzed using different approaches to gain more insights about the cross-talk taking place in that environment. Thus, the application of metalipidomics to study host-associated microbial composition and functional analysis, while not yet explored, appears promising.
KEY POINTS.
Read-lengths generated during metagenomic sequencing influence assembly, gene prediction and eventually functional analysis. The enormous volume of sequence data, which leads to long computational times and massive storage requirements, also impedes metagenomic functional prediction.
Factors that potentially influence functional analysis of metagenomic data, including sample preparation, sequencing method, average genome size, etc. should be considered prior to analysis.
A higher fraction of metagenomic sequences are annotated using BLAST against data-rich reference sequence databases such as NCBI NR as compared to SwissProt, COGs, KEGG, etc.
Integrated methods using more than one approach can improve the efficiency and reliability of functional predictions.
DNA-sequence-based metagenomic functional analysis should be complemented with other types of approaches, such as metatranscriptomics, metaproteomics, metabolomics and metalipidomics, to gain better insights of the dynamics of a community.
FUNDING
This work was supported by the operational expenditure fund of RIKEN.
Biographies
Tulika Prakash is a Research Scientist in the Laboratory for MetaSystems Research at RIKEN, Japan. With specialization in functional and metabolic analysis of prokaryotic genomes, her current research involves bioinformatic analysis of genomes and metagenomes.
Todd Taylor is Team Leader of the Laboratory for MetaSystems Research at RIKEN, Japan. A former member of the International Human Genome Sequencing Consortium, his current research involves bioinformatic approaches to metagenomics.
References
- 1.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–40. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 2.Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet. 2005;6:805–14. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 3.Kunin V, Copeland A, Lapidus A, et al. A bioinformatician’s guide to metagenomics. Microbiol Mol Biol Rev. 2008;72:557–78. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667–79. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batzoglou S, Jaffe DB, Stanley K, et al. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–89. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aparicio S, Chapman J, Stupka E, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–10. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 7.Myers EW, Sutton GG, Delcher AL, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 8.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li R, Zhu H, Ruan J, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–72. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci USA. 2001;98:9748–53. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Y, Tang H. An ORFome assembly approach to metagenomics sequences analysis. J Bioinform Comput Biol. 2009;7:455–71. doi: 10.1142/s0219720009004151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng Y, Leung HC, Yiu SM, et al. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 13.Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–30. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132–46. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38:e191–202. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delcher AL, Harmon D, Kasif S, et al. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–41. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma VK, Kumar N, Prakash T, et al. Fast and accurate taxonomic assignments of metagenomic sequences using MetaBin. PLoS One. 2012;7:e34030–8. doi: 10.1371/journal.pone.0034030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huson DH, Auch AF, Qi J, et al. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–86. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlach W, Junemann S, Tille F, et al. WebCARMA: a web application for the functional and taxonomic classification of unassembled metagenomic reads. BMC Bioinformatics. 2009;10:430–9. doi: 10.1186/1471-2105-10-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McHardy AC, Martin HG, Tsirigos A, et al. Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 23.Teeling H, Waldmann J, Lombardot T, et al. TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics. 2004;5:163–9. doi: 10.1186/1471-2105-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosen GL, Reichenberger ER, Rosenfeld AM. NBC: the Naive Bayes Classification tool webserver for taxonomic classification of metagenomic reads. Bioinformatics. 2011;27:127–9. doi: 10.1093/bioinformatics/btq619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz NN, Krause L, Goesmann A, et al. TACOA: taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach. BMC Bioinformatics. 2009;10:56–71. doi: 10.1186/1471-2105-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markowitz VM, Chen IM, Chu K, et al. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res. 2012;40:D123–9. doi: 10.1093/nar/gkr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goll J, Rusch DB, Tanenbaum DM, et al. METAREP: JCVI metagenomics reports–an open source tool for high-performance comparative metagenomics. Bioinformatics. 2010;26:2631–2. doi: 10.1093/bioinformatics/btq455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun S, Chen J, Li W, et al. Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res. 2011;39:D546–51. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W. Analysis and comparison of very large metagenomes with fast clustering and functional annotation. BMC Bioinformatics. 2009;10:359–67. doi: 10.1186/1471-2105-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass EM, Wilkening J, Wilke A, et al. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- 31.Arumugam M, Harrington ED, Foerstner KU, et al. SmashCommunity: a metagenomic annotation and analysis tool. Bioinformatics. 2010;26:2977–8. doi: 10.1093/bioinformatics/btq536. [DOI] [PubMed] [Google Scholar]
- 32.Huson DH, Mitra S, Ruscheweyh HJ, et al. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–60. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lingner T, Asshauer KP, Schreiber F, et al. CoMet–a web server for comparative functional profiling of metagenomes. Nucleic Acids Res. 2011;39:W518–23. doi: 10.1093/nar/gkr388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S, Zhu Z, Fu L, et al. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444–52. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mende DR, Waller AS, Sunagawa S, et al. Assessment of metagenomic assembly using simulated next generation sequencing data. PLoS One. 2012;7:e31386–96. doi: 10.1371/journal.pone.0031386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raes J, Foerstner KU, Bork P. Get the most out of your metagenome: computational analysis of environmental sequence data. Curr Opin Microbiol. 2007;10:490–98. doi: 10.1016/j.mib.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Pignatelli M, Moya A. Evaluating the fidelity of de novo short read metagenomic assembly using simulated data. PLoS One. 2011;6:e19984–92. doi: 10.1371/journal.pone.0019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yok NG, Rosen GL. Combining gene prediction methods to improve metagenomic gene annotation. BMC Bioinformatics. 2011;12:20–31. doi: 10.1186/1471-2105-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurokawa K, Itoh T, Kuwahara T, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–81. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prakash T, Oshima K, Morita H, et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe. 2011;10:273–84. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Ventura M, O’Connell-Motherway M, Leahy S, et al. From bacterial genome to functionality; case bifidobacteria. Int J Food Microbiol. 2007;120:2–12. doi: 10.1016/j.ijfoodmicro.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Richter DC, Ott F, Auch AF, et al. MetaSim: a sequencing simulator for genomics and metagenomics. PLoS One. 2008;3:e3373–84. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overbeek R, Begley T, Butler RM, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–5. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–5. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–7. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 51.Powell S, Szklarczyk D, Trachana K, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:D284–9. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selengut JD, Haft DH, Davidsen T, et al. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–4. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–3. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- 57.Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41–54. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia MH, Ivanova N, Kunin V, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol. 2006;24:1263–9. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- 59.Berg Miller ME, Yeoman CJ, Chia N, et al. Phage-bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ Microbiol. 2012;14:207–27. doi: 10.1111/j.1462-2920.2011.02593.x. [DOI] [PubMed] [Google Scholar]
- 60.DeLong EF, Preston CM, Mincer T, et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 61.Tringe SG, von MC, Kobayashi A, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–7. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 62.Tyson GW, Chapman J, Hugenholtz P, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 63.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sigrist CJ, Cerutti L, de CE, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–6. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Attwood TK, Bradley P, Flower DR, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–2. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hunter S, Jones P, Mitchell A, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–12. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol. 2007;8:995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- 68.Dandekar T, Snel B, Huynen M, et al. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–8. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 69.Overbeek R, Fonstein M, D’Souza M, et al. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96:2896–901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Enright AJ, Iliopoulos I, Kyrpides NC, et al. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 71.Marcotte EM, Pellegrini M, Ng HL, et al. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–3. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 72.Pellegrini M, Marcotte EM, Thompson MJ, et al. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–8. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marcotte EM, Pellegrini M, Thompson MJ, et al. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–6. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 74.Harrington ED, Singh AH, Doerks T, et al. Quantitative assessment of protein function prediction from metagenomics shotgun sequences. Proc Natl Acad Sci USA. 2007;104:13913–8. doi: 10.1073/pnas.0702636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sachdeva G, Kumar K, Jain P, et al. SPAAN: a software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics. 2005;21:483–91. doi: 10.1093/bioinformatics/bti028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 77.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mackelprang R, Waldrop MP, DeAngelis KM, et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011;480:368–71. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- 79.Brulc JM, Antonopoulos DA, Miller ME, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA. 2009;106:1948–53. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Willner D, Furlan M, Haynes M, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One. 2009;4:e7370–81. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu L, Chen H, Hu X, et al. Average gene length is highly conserved in prokaryotes and eukaryotes and diverges only between the two kingdoms. Mol Biol Evol. 2006;23:1107–8. doi: 10.1093/molbev/msk019. [DOI] [PubMed] [Google Scholar]
- 82.Yilmaz S, Singh AK. Single cell genome sequencing. Curr Opin Biotechnol. 2011;23:1–7. doi: 10.1016/j.copbio.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raes J, Korbel JO, Lercher MJ, et al. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8:R10–20. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van NE. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–84. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 86.Raes J, Harrington ED, Singh AH, et al. Protein function space: viewing the limits or limited by our view? Curr Opin Struct Biol. 2007;17:362–9. doi: 10.1016/j.sbi.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 87.Beszteri B, Temperton B, Frickenhaus S, et al. Average genome size: a potential source of bias in comparative metagenomics. ISME J. 2010;4:1075–7. doi: 10.1038/ismej.2010.29. [DOI] [PubMed] [Google Scholar]
- 88.Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–7. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 89.Peltola H, Soderlund H, Ukkonen E. Algorithms for the search of amino acid patterns in nucleic acid sequences. Nucleic Acids Res. 1986;14:99–107. doi: 10.1093/nar/14.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guan X, Uberbacher EC. Alignments of DNA and protein sequences containing frameshift errors. Comput Appl Biosci. 1996;12:31–40. doi: 10.1093/bioinformatics/12.1.31. [DOI] [PubMed] [Google Scholar]
- 91.Brown NP, Sander C, Bork P. Frame: detection of genomic sequencing errors. Bioinformatics. 1998;14:367–71. doi: 10.1093/bioinformatics/14.4.367. [DOI] [PubMed] [Google Scholar]
- 92.Halperin E, Faigler S, Gill-More R. FramePlus: aligning DNA to protein sequences. Bioinformatics. 1999;15:867–73. doi: 10.1093/bioinformatics/15.11.867. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Y, Sun Y. HMM-FRAME: accurate protein domain classification for metagenomic sequences containing frameshift errors. BMC Bioinformatics. 2011;12:198–207. doi: 10.1186/1471-2105-12-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Venter JC, Remington K, Heidelberg JF, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 95.Johnston AW, Li Y, Ogilvie L. Metagenomic marine nitrogen fixation–feast or famine? Trends Microbiol. 2005;13:416–20. doi: 10.1016/j.tim.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 96.Remington KA, Heidelberg K, Venter JC. Taking metagenomic studies in context. Trends Microbiol. 2005;13:404. doi: 10.1016/j.tim.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 97.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brown CT, vis-Richardson AG, Giongo A, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792–800. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, et al. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma VK, Kumar N, Prakash T, et al. MetaBioME: a database to explore commercially useful enzymes in metagenomic datasets. Nucleic Acids Res. 2010;38:D468–72. doi: 10.1093/nar/gkp1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tasse L, Bercovici J, Pizzut-Serin S, et al. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 2010;20:1605–12. doi: 10.1101/gr.108332.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Belda-Ferre P, Cabrera-Rubio R, Moya A, et al. Mining virulence genes using metagenomics. PLoS One. 2011;6:e24975–80. doi: 10.1371/journal.pone.0024975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Turnbaugh PJ, Quince C, Faith JJ, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA. 2010;107:7503–8. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gosalbes MJ, Durban A, Pignatelli M, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6:e17447–55. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Taverna DM, Goldstein RA. Why are proteins marginally stable? Proteins. 2002;46:105–9. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- 106.Klaassens ES, de Vos WM, Vaughan EE. Metaproteomics approach to study the functionality of the microbiota in the human infant gastrointestinal tract. Appl Environ Microbiol. 2007;73:1388–92. doi: 10.1128/AEM.01921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Verberkmoes NC, Russell AL, Shah M, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–89. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 108.Li X, LeBlanc J, Truong A, et al. A metaproteomic approach to study human-microbial ecosystems at the mucosal luminal interface. PLoS One. 2011;6:e26542–55. doi: 10.1371/journal.pone.0026542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kolmeder CA, de BM, Nikkila J, et al. Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS One. 2012;7:e29913–26. doi: 10.1371/journal.pone.0029913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–83. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 111.Saito K, Matsuda F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol. 2010;61:463–89. doi: 10.1146/annurev.arplant.043008.092035. [DOI] [PubMed] [Google Scholar]
- 112.Claus SP, Tsang TM, Wang Y, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fukuda S, Nakanishi Y, Chikayama E, et al. Evaluation and characterization of bacterial metabolic dynamics with a novel profiling technique, real-time metabolotyping. PLoS One. 2009;4:e4893–902. doi: 10.1371/journal.pone.0004893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han J, Antunes LC, Finlay BB, et al. Metabolomics: towards understanding host-microbe interactions. Future Microbiol. 2010;5:153–61. doi: 10.2217/fmb.09.132. [DOI] [PubMed] [Google Scholar]
- 115.Claus SP, Ellero SL, Berger B, et al. Colonization-induced host-gut microbial metabolic interaction. MBIO. 2011;2:e00271–10. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 117.Nakanishi Y, Fukuda S, Chikayama E, et al. Dynamic omics approach identifies nutrition-mediated microbial interactions. J Proteome Res. 2011;10:824–36. doi: 10.1021/pr100989c. [DOI] [PubMed] [Google Scholar]
- 118.Schubotz F, Wakeham SG, Lipp JS, et al. Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environ Microbiol. 2009;11:2720–34. doi: 10.1111/j.1462-2920.2009.01999.x. [DOI] [PubMed] [Google Scholar]
- 119.Pitcher A, Hopmans EC, Mosier AC, et al. Core and intact polar glycerol dibiphytanyl glycerol tetraether lipids of ammonia-oxidizing archaea enriched from marine and estuarine sediments. Appl Environ Microbiol. 2011;77:3468–77. doi: 10.1128/AEM.02758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]