Abstract
The rapid advances of high-throughput sequencing technologies dramatically prompted metagenomic studies of microbial communities that exist at various environments. Fundamental questions in metagenomics include the identities, composition and dynamics of microbial populations and their functions and interactions. However, the massive quantity and the comprehensive complexity of these sequence data pose tremendous challenges in data analysis. These challenges include but are not limited to ever-increasing computational demand, biased sequence sampling, sequence errors, sequence artifacts and novel sequences. Sequence clustering methods can directly answer many of the fundamental questions by grouping similar sequences into families. In addition, clustering analysis also addresses the challenges in metagenomics. Thus, a large redundant data set can be represented with a small non-redundant set, where each cluster can be represented by a single entry or a consensus. Artifacts can be rapidly detected through clustering. Errors can be identified, filtered or corrected by using consensus from sequences within clusters.
Keywords: clustering, metagenomics, next-generation sequencing, protein families, artificial duplicates, OTU
INTRODUCTION
Metagenomics [1, 2] is a genomic approach that uses culture-independent sequencing to study the microorganism populations under different environments. It offers unprecedented vision of the identities, composition, dynamics, functions and interactions of the diverse microbial world and has become an important tool in many fields such as ecology, energy, agriculture and medicine.
Earlier metagenomics projects, such as Sargasso Sea [3], human gut [4] and soil [5], relied on traditional Sanger sequencing technology, so most of these projects have limited throughput. In recent years, the rapid advances of next-generation sequencing (NGS) technologies [6], such as 454, Illumina, SOLiD, PacBio and Ion Torrent, dramatically propelled the expansion of metagenomics research, and large ‘waves’ of metagenomics sequencing projects were launched to study a diverse range of microbial communities in their environments, such as the virome [7], farm animals [8] and the human microbiome [9, 10]. It is widely expected that many more environmental and microbiome samples will be studied by NGS technologies. However, the intrinsic complexity and massive quantity of metagenomics data create tremendous challenges for data analysis.
First, because of the sheer number of sequences, all kinds of sequence analyses, including database search, multiple alignment, sequence mapping, assembly and phylogenetic analysis, are getting more time consuming and memory demanding and require more manual efforts for parsing the output results. Second, the growth of sequence data in the public databases has been very uneven due to highly biased efforts toward model organisms and those populations or environments of special interest. Metagenomic analyses that rely on comparison with these biased and redundant reference databases may lead to incorrect conclusions. Third, different NGS techniques and protocols show quite different bias and artifacts. For example, single-cell multiple displacement amplification produces very non-uniform coverage by orders of magnitude [11]. Many sequencers generate tens or hundreds of copies of artificially duplicated reads for same templates [12]. Fourth, NGS platforms have higher error rates than traditional Sanger sequencers and also have platform-specific error patterns, such as homopolymer indels for 454 and Ion Torrent reads and degraded quality at 3′-ends for Illumina reads. Finally, sequence errors and artifacts are propagated from reads to protein sequences, which can be false genes, fragmented or with frame-shift errors.
Clustering analysis, a method that identifies and groups similar objects, is a powerful tool to explore and study large-scale complex data. It can effectively resolve many of the challenges stated earlier. By sequence clustering, a large redundant data set can be represented with a small non-redundant (NR) set, which requires less computation. Errors can be identified, filtered or corrected by using consensus from sequences within clusters. In addition, many fundamental questions in metagenomics can be readily addressed by clustering, such as the identification of gene families and the classification of species in a population. So, since the infancy of metagenomics, clustering analysis has been an essential part of this field for applications, such as identification of artificial duplicates [12, 13], classification of operational taxonomic units (OTUs) [14], protein family analysis [15, 16] and transcriptomics analysis [17].
In this article, we will discuss several common clustering applications in metagenomics and the methodologies for different types of analysis.
CLUSTERING METHODS AND RESOURCES
Sequence clustering is not a new topic; it existed long before the emerging of metagenomics and NGS technologies. In the past, many available clustering programs were used for clustering protein sequences such as ProtoMap [18], ProtoNet [19], RSDB [20], GeneRAGE [21], TribeMCL [22], ProClust [23], UniqueProt [24], OrthMCL [25], MC-UPGMA [26], Blastclust [27] and CD-HIT [28–31]. Many methods were also used for clustering expressed sequence tags (ESTs), such as Unigene [32], TIGR Gene Indices [33], d2_cluster [34] and several others [35–37].
Many of the above clustering methods require all against all comparisons of sequences for optimal results, so they are very computational intensive for large data sets. A method for reducing the intensive requirement arose with CD-HIT. Thus, with the rapid growth of sequence data, the fast program CD-HIT become a very popular clustering tool; it has been widely used in many areas such as preparing NR reference databases [38]. CD-HIT uses a greedy incremental algorithm. Basically, sequences are first ordered by decreasing length, and the longest one becomes the seed of the first cluster. Then, each remaining sequence is compared with existing seeds. If the similarity with any seed meets a pre-defined cutoff, it is grouped into that cluster; otherwise, it becomes the seed of a new cluster. More recently, several new fast programs, including Uclust [39], DNACLUST [40] and SEED [41], have been developed using greedy incremental approaches similar to that introduced by CD-HIT. These methods use various heuristics and achieved high speed in clustering NGS sequences. Herein, we briefly introduce the features and functions of these programs.
CD-HIT [28–31] is a comprehensive clustering package. The current version (v 4.5) has seven programs. CD-HIT and CD-HIT-EST cluster protein and deoxyribonucleic acid (DNA) data sets, respectively. CD-HIT-454 identifies duplicates from 454 reads. PSI-CD-HIT clusters proteins at low-identity cutoff (20–50%). CD-HIT-DUP identifies duplicates from single or paired Illumina reads. CD-HIT-LAP identifies overlapping reads. CD-HIT-OTU is a multi-step pipeline to generate OTU clusters for ribosomal ribonucleic acid (rRNA) tags from 454 and Illumina platforms. CD-HIT uses a heuristics based on statistical k-mer filtering to speed up clustering calculations. It also has a multi-threading function, so it can run in parallel on multi-core computers. CD-HIT is open source software available from http://cd-hit.org. It is also available from the cd-hit web server [28], the CAMERA web portal [42] and the WebMGA server [43] for metagenomic data analysis.
Uclust [39] follows CD-HIT’s greedy incremental approaches, but it uses a heuristics called Usearch for fast sequence comparison. It also gains speed by comparing a few top sequences instead of the full database. Uclust can run on DNA, protein and rRNA sequences. Currently, its 32-bit pre-compiled binaries are freely available from http://www.drive5.com/usearch/. DNACLUST [40] also follows greedy incremental approach; it uses a suffix array to index the input data set. Unlike CD-HIT and Uclust, which can process both proteins and DNAs, DNACLUST only works on DNA sequences, and it is suitable for clustering highly similar DNAs, especially for rRNA tags. It is available as open source program at http://dnaclust.sourceforge.net/. SEED [41] only works with Illumina reads and only identifies up to three mismatches and three overhanging bases. It uses an open hashing technique and a special class of spaced seeds, called block spaced seed. SEED is also an open source software available at http://manuals.bioinformatics.ucr.edu/home/seed.
Although some programs are claimed to be faster than other programs, those claims are usually based on a certain type of sequences and clustering parameters (e.g. an identity cutoff). Herein, we do not intent to make a side-by-side performance comparison but simply list some examples that we ran to give some hints on the speed and results for some common clustering analyses by these programs (Table 1). CD-HIT and Uclust often produce comparable results in both protein and DNA clustering tests. SEED is faster than other programs in clustering Illumina reads, but it yields many more clusters. Except for SEED, the other three programs all work on rRNA sequences, where Uclust is fastest and CD-HIT gives the fewest clusters.
Table 1:
Clustering speed and results for common data sets
| Data seta | Program and parametersb | Timec (minutes) | Clusters |
|---|---|---|---|
| NCBI NR, proteins, 4.3 GB: 12 054 819 sequencesd | cd-hit v4.5.7 ‘-n 5 -M 0 -c 0.9’ | 1405/181 | 7 036 029 |
| cd-hit v4.5.7 ‘-n 5 -M 0 -c 0.7’ | 962/152 | 4 933 074 | |
| Swissprot, proteins, 222 MB: 437 168 sequences | cd-hit 4.5.7 ‘-n 5 -M 0 -c 0.9’ | 3.7/0.8 | 298 617 |
| Uclust v5 ‘-id 0.9’ | 17.3 | 301 076 | |
| cd-hit 4.5.7 ‘-n 5 -M 0 -c 0.7’ | 4.6/0.8 | 190 695 | |
| Uclust v5 ‘-id 0.7’ | 7.6 | 192 847 | |
| Illumina (SRR061270), 380 MB, 5 million reads | cd-hit v4.5.7 ‘-n 10 -M 0 -c 0.95’ | 56.8/9.2 | 956 734 |
| Uclust v5 ‘-id 0.95’ | 164.6 | 958 887 | |
| cd-hit v4.5.7 ‘-n 10 -M 0 -c 0.9’ | 347.5/46.0 | 751 581 | |
| Uclust v5 ‘-id 0.9’ | 227.5 | 734 981 | |
| cd-hit v5.0 beta ‘-c 0.9’ | 23.5/4.0 | 750 276 | |
| SEED (default parameters) | 7.9 | 1 056 109 | |
| 1.1 million 16s rRNAs: 454 reads Ref. [44] | cd-hit v4.5.7 ‘-n 10 -M 0 -c 0.97’ | 47.9/7.5 | 24 842 |
| Uclust v5 ‘-id 0.97’ | 4.3 | 29 586 | |
| DNACLUST ‘-s 0.97’ | 15.3 | 31 151 |
aNR and Swissprot were downloaded from NCBI at ftp://ftp.ncbi.nih.gov/blast/db/FASTA/. Illumina reads from SRR061270 was downloaded from NCBI at http://www.ncbi.nlm.nih.gov/sra. The 16s rRNAs was kindly provided by the authors from Ref. [44]. b‘-c 0.9’, ‘-id 0.9’ and ‘-s 0.9’ mean 90% identity. However, DNACLUST’s definition is slightly different from CD-HIT and Uclust (Ref. [40]). cThe second number is the time for eight cores; currently, only CD-HIT has a multiple threading function. dThe free 32-bit version of Uclust cannot process NR, so only CD-HIT is used.
IDENTIFICATION OF ARTIFICIAL DUPLICATES
NGS platforms, such as 454 and Illumina, commonly produce artificially duplicated reads, which can lead to an overestimated abundance of species, genes or functions. The duplicates originate from the same template but are separately sequenced, so they can be exactly identical or can be nearly identical with variable read lengths (454 reads) and mismatches due to sequence errors.
In 454 data sets, duplicated reads can make up 11–35% of the raw reads [12]. As finding identical sequences is very easy, only exact duplicates were identified and removed in some early studies [7]. Nearly identical duplicates were considered ever since the study by Gomez-Alvarez et al. [12]. Gomez-Alvarez’s method applies CD-HIT-EST to cluster the reads at 90% identity and then parses the clustering results. Later, CD-HIT-454 [13] was introduced by reengineering CD-HIT-EST. CD-HIT-454 is faster and more accurate than Gomez-Alvarez’s method. It identifies duplicates that are either exactly identical or meet the following criteria: (i) reads must be aligned at 5′-ends; (ii) for sequences of different length, a shorter read must be fully aligned with the longer one (the seed) and (iii) they have less than user-defined percentage of indels and substitutions (default 4%). The default cut-off value, which is trained according to the pyrosequencing’s error model, maximizes the sensitivity and specificity of identification of duplicates from 454 reads. Another common, easy way for finding duplicates is to compare prefixes and consider that the reads are duplicates if they share a common prefix of a certain length. Both MG-RAST [45] and IMG/M [46] use prefix checking for identification of duplicates. Prefix checking is faster but less accurate than CD-HIT-454. CD-HIT-454 only needs a few minutes to run a typical 454 data set with less than a million reads, so it is still very efficient, similar to the original CD-HIT.
For Illumina data sets, prefix checking has more advantages, because it is relatively faster than CD-HIT-454, and it fits features of Illumina reads, which have fewer indels and exhibit worse quality at the 3′-ends. For pair-ended Illumina reads, a reasonable way for finding duplicates is to check prefixes at both ends. This function is available in CD-HIT-DUP.
When removing duplicates, a question that needs to be considered is ‘are these duplicates all artificial’? The experimentally observed duplicated sequences also include natural duplicates, i.e. those that happen to be duplicates by chance. So, simply removing all duplicates may also cause an underestimation of abundance associated with natural duplicates. The CD-HIT-454 article investigated the occurrence of natural duplicates for different types of metagenomic samples and found that (i) the rate of natural duplicates highly correlates with the read density (the number of reads divided by genome size); (ii) for high-complexity metagenomic samples, natural duplicates make up a few percent of all duplicates and (iii) for viral metagenomic samples or metatranscriptomics, natural duplicates can be more abundant than artificial duplicates. These guidelines help to decide whether to remove or to keep duplicated reads in a metagenomic sample [13].
DIVERSITY
Metagenomic projects (e.g. [9, 47, 48]) often survey both genomic DNAs and 16S rRNAs. The later are used to estimate the microbial diversity, which is often quantitatively described in OTUs. Because of read length limitation, it is not practical to sequence the full length of 16s rRNA (∼1.5 kb), so 16s rRNA studies often use individual variable regions (V1–V9) or sections that cover a few variable regions (e.g. V1–V3 and V3–V5). Pyrosequencing of 16S rRNA amplicons has been the dominant approach in rRNA studies. Finding OTUs from 16S rRNA tags can be readily addressed by clustering. Conventionally, tags with ≥97% identity are placed in the same OTUs at the species level. CD-HIT [29] and DOTUR [49] were often used for OTU clustering during early studies.
However, a big problem in OTU analysis is that directly clustering the raw rRNA reads or even the high-quality reads often greatly over-estimates the diversity. A recent review [50] analyzed a list of methods and discussed solving this problem at the clustering algorithm level. This article suggested using average linkage-based hierarchical clustering methods such as ESPRIT [51], instead of greedy incremental methods such as CD-HIT [29] and Uclust [39] for OTU clustering.
In the meantime, many other studies [52–56] found that the single biggest cause of the over-estimation problem is the sequence errors or noise, so new methods such as SLP [52], PyroNoise [54], Denoiser [55] and Ampliconnoise [56] focus at identifying and removing sequence noise. All these methods find sequence errors by clustering analysis and are based on a principle that a high-abundance cluster can recruit small clusters and singletons, which have more sequence errors. SLP clusters the actual rRNA tags, and the rest of the methods cluster the original flowgram data. Currently, the best performing method among them is AmpliconNoise [56], which has been benchmarked by several commonly used Mock data sets; these data sets are artificial mixtures of 16S rRNA clones at different abundance levels from a number of known species.
Although the speed of AmpliconNoise is considerably improved over its predecessor version (PyroNoise), it is still quite computational intensive. Recently, CD-HIT-OTU was introduced to the CD-HIT package. CD-HIT-OTU also uses a multi-step clustering method to remove reads with sequence errors and achieves results comparable with AmpliconNoise. However, as CD-HIT-OTU clusters sequences instead of flowgram data and inherits unique heuristics from CD-HIT, it is orders of magnitude faster than AmpliconNoise and other methods such as Denoiser. Table 2 lists the performance of CD-HIT-OTU, AmpliconNoise and Denoiser (implemented in QIIME [57]) on clustering the Mock benchmark data sets [56] at 97% identity level.
Table 2:
Accuracy and speed for OTUs identificationa
| Datab | True OTUsc | Number of predicted OTUs, sensitivity (%), specificity (%), CPU time (h, min, s) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD-HIT-OTU | AmpliconNoise | Denoiser | |||||||||||
| Divergent | 23 | 26 | 100 | 88 | 11 s | 28 | 100 | 82 | 32 h | 35 | 100 | 65 | 15 m |
| Artificial | 33 | 32 | 100 | 100 | 13 s | 34 | 96 | 91 | 22 h | 38 | 96 | 81 | 13 m |
| Even1 | 53 | 71 | 100 | 74 | 8 s | 85 | 100 | 62 | 68 h | NAd | |||
| Even2 | 53 | 57 | 96 | 89 | 7 s | 83 | 100 | 63 | 49 h | NAd | |||
| Even3 | 52 | 60 | 100 | 86 | 7 s | 90 | 100 | 57 | 65 h | NAd | |||
| Uneven1 | 49 | 56 | 91 | 80 | 5 s | 76 | 97 | 63 | 39 h | NAd | |||
| Uneven2 | 41 | 45 | 85 | 77 | 7 s | 67 | 95 | 58 | 35 h | NAd | |||
| Uneven3 | 38 | 42 | 100 | 90 | 7 s | 73 | 97 | 50 | 44 h | NAd | |||
| Titanium | 69 | 69 | 98 | 98 | 7 s | 90 | 100 | 76 | 388 h | 146 | 100 | 47 | 6 h |
aAll data sets were downloaded from http://people.civil.gla.ac.uk/∼quince/Data/AmpliconNoise.html according to an article [56]. bParameters are based on each programs default setting. cTrue OTUs were calculated by clustering the reference sequences that are covered by the raw reads. dFlowgram data are only available in AmpliconNoise-specific format, so we can run AmpliconNoise but not Denoiser. However, Denoiser’s performance for these data sets can be referenced from an article [56].
CD-HIT-OTU has following steps: (i) the raw reads with ambiguous base calls are removed. Reads are also removed if their 5′-ends do not match user-provided primer sequence or a consensus, which is built from the 5′ of all reads of k bases (k = 6 by default, adjustable by users). For long reads, it also trims off the tails portion at 3′-ends that are beyond median read length. (ii) Processed reads are clustered at 100% identity using CD-HIT-DUP. At this step, the reads from a unique rRNA template will form one large primary cluster (it contains error-free reads) and some small clusters, which contain reads with sequence errors. (iii) The representative sequences from step 2 are sorted by abundance and then clustered by CD-HIT-EST at a threshold that allows up to two mismatches. For example, 200-bp reads are clustered at 99.0% identity, so that small clusters are recruited into their primary clusters. (iv) Let x to be the median size of small clusters recruited into the most abundant primary cluster with two mismatches. Clusters smaller than x are dominated by reads with more than two errors from the most abundant template; so these clusters are removed. Herein, x is often very small (2 or 3), so that rare species will still be kept in the analysis. (5) The remaining representative sequences from step 2 are clustered into OTUs using CD-HIT-EST (parameters: -c 0.97 -n 10 -l 11 -p 1 -d 0 -g 1). Herein, option ‘-c 0.97’ means 97% identity. (6) The non-representative tags are recruited into the OTUs using CD-HIT-EST-2D (parameters: -c 0.97 -n 10 -l 11 -p 1 -d 0 -g 1).
The ultra-high speed of CD-HIT-OTU allows clustering multi-million rRNA tags pooled from a series of related samples. Such clustering can significantly increase the accuracy of OTU identification, because tags shared by different samples validate each other. Clustering pooled samples may identify very rare OTUs, which may be missed if individual samples are processed independently. We applied CD-HIT-OTU on two pooled data sets, Human_gut_V6 [48] and Human_body_V2 [44]; these include 33 gut samples from obese and lean twin families and 815 samples from different body sites, respectively (Table 3). CD-HIT-OTU only used a few minutes for these two data sets.
Table 3:
OTU analysis for pooled human gut and human samples
| Data seta | Reads | Region | Platform | OTUs | CPU (s) |
|---|---|---|---|---|---|
| Human_gut | 817942 | V6 | GS 20 | 317 | 37 |
| Human_body | 1071335 | V2 | GS FLX | 238 | 295 |
aThe Human_gut data set was downloaded from http://gordonlab.wustl.edu/NatureTwins_2008/TurnbaughNature_11_30_08.html. The Human_body data set was kindly provided by the authors from Ref. [44].
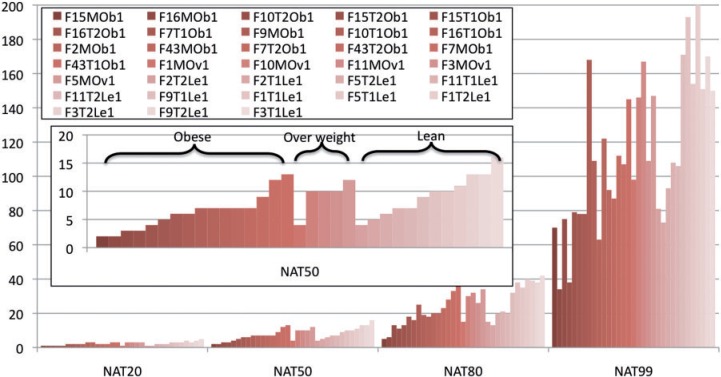
In this analysis, we found that clustering the pooled samples identified 19–80 more rare OTUs than clustering individual samples for the 33 human gut data sets. For the 815 human body data sets, clustering pooled samples found up to 50 more rare OTUs. Clustering pooled samples also provides a very straightforward way to define a ‘core microbiome’ and to compare the diversity and composition of samples. For example, we calculated NAT50 for each sample. Herein, NAT50 is a diversity indicator we defined, which stands for the number of most abundant taxonomic groups covering 50% populations. Figure 1 shows that obese samples have less diversity than lean samples. Please note the abundance of OTUs is the abundance of rRNA genes and may not be the abundance of species, because the rRNA copy numbers are unknown. However, rRNA genes abundance largely correlates with species abundance. The full results for human gut and human body are also available as examples with the CD-HIT-OTU software, which is available from http://weizhongli-lab.org/cd-hit-otu. CD-HIT-OTU is also available as a web server within WebMGA [43], a collection of web servers for metagenomic data analysis.
Figure 1:
Distribution of microbial diversity measured by NATs (NAT20, NAT50, NAT80 and NAT99) for 33 human gut samples. The x-axis is NAT category. The y-axis is NAT value. Samples are colored by sample type (obese, over weight or lean). The results show that obese samples have less average NAT50 than the lean samples.
FILTERING SEQUENCE ERRORS
As shown in the previous section, clustering-based approaches very well address sequencing errors in rRNA tags. Similar clustering analyses can also filter out errors in genomic and metagenomic reads and, therefore, improve sequence assembly, gene prediction and other analyses. However, finding errors from genomic reads is more difficult than from rRNA tags, which can be aligned at their 5′-ends, because they all start with the same universal primers. For genomic reads, there are several existing methods in detecting sequence errors by various clustering approaches. For example, FreClu [58] and EDAR [59] use k-mer frequency; Hammer [60] uses a Hamming graph and ECHO [61] clusters overlapping reads through k-mer hashing. These methods avoid very time-consuming full-length sequence alignment in clustering the reads. However, full-length sequence alignment is feasible using ultra-fast sequence clustering algorithms. For example, the analysis in the SEED article [41] shows that genome assembly can be notably improved by only assembling cluster representatives.
Herein, we show an example using clustering-based filtering to improve metagenome assembly. Metagenomic samples often contain a small number of dominant organisms along with hundreds or more less abundant species. Because of sequencing errors, major problems in metagenome assembly often occur for the high-abundance species. Clustering methods, including the k-mer frequency-based approaches, benefit from high sequence redundancy, from which better consensus can be derived. So, the assembly difficult for the dominant species in metagenome can be effectively corrected.
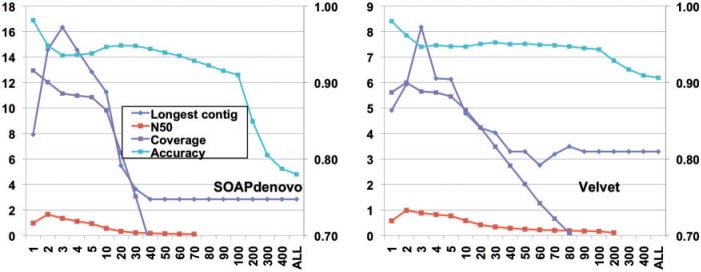
Herein, as a demonstration, we use an Illumina data set representing the high-abundance species of a human gut sample (MH0006) from MetaHIT project [9] at http://gutmeta.genomics.org.cn. The MetaHIT study also provided Sanger reads for sample MH0006 as reference and assembled them into 995 contigs. We mapped the Illumina reads to these reference contigs using SOAP2 [62] (option: -M 4); 144 contigs have a coverage of at least 200. The reads mapped to these contigs are selected as a high-abundance subset, which contains 36 175 286 of 75 bp pair-ended reads. The clustering-based approach has the following steps: (i) reads are clustered with CD-HIT-EST (options: ‘-c 0.96 -n 10 -r 1 –aS 0.5 -b 2 -G 0’); (ii) for each cluster, we only kept at most N reads that have the best average quality score per base and filtered out the extra sequences, where N is a redundancy cutoff parameter and (iii) the remaining reads were assembled at different N levels and optimal assembles achieved. The comparison of contigs between original reads and filtered reads using Velvet [63] and SOAPdenovo [64] is shown in Figure 2. The filtered data sets largely improve the N50 and longest contig. Actually, for the unfiltered data set, because the coverage is so low, there is no valid N50. The accuracy and coverage are also much higher with the filtered reads.
Figure 2:
Assembly performance of the filtered reads for metagenomic sample MH0006. x-axis is the redundancy cutoff N. The length of the longest contig (kb) and N50 (kb) are plotted against the left y-axis. The accuracy and genome coverage are against the right y-axis. The assembly results for original reads are at far right side marked as ‘ALL’ on x-axis. The accuracy of contigs is the total length of correct contigs divided by the total length of all contigs. The genome coverage is the fraction of reference genome covered by the correct contigs.
There are two reasons that we used the high-abundance subset instead of the full MH0006 data. First, sequence errors deteriorate sequence assembly for high-abundant reads. So, our filtering method only improves the high-abundance species. Second, we evaluated the contigs assembled from Illumina reads by comparing them with high-quality references (contigs from Sander reads). Most contigs assembled from the low-abundance Illumina reads cannot be mapped to any reference sequences. So, we cannot evaluate these contigs.
DATABASE SEARCH
In metagenomic projects, an important annotation step is to query the reads or Open Reading Frames (ORF) against reference databases of known genomes, DNAs or proteins with an alignment program such as basic local alignment search tool (BLAST) [27], BWA [65], BLAT [66], FR-HIT [67] or Rapsearch [68]. Because of the huge size of both reference databases and the query, such database searches can be very time consuming. However, both reference databases and the query sample can be very redundant, so simply using NR data sets may save a great deal of computation time and, in some cases, also improve the accuracy of database search [69].
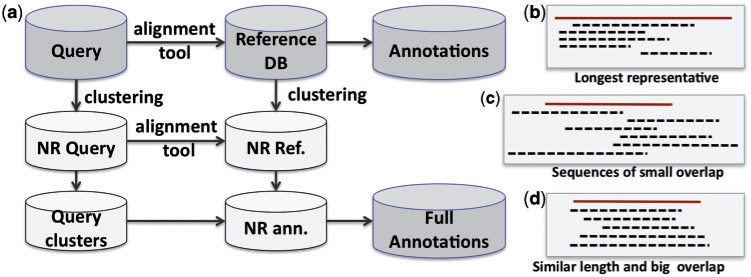
As illustrated in Figure 3, before database searching, both the reference database and the query are clustered at certain similarity thresholds. Then, the NR sequences (i.e. representatives) from the query are aligned to the NR sequences in the reference database. Finally, the annotation results are copied from the representatives to other sequences in the same clusters.
Figure 3:
Using NR query and NR reference database for metagenome annotation.
A big concern of this approach is to ascertain how much difference there is between annotations calculated from the NR data sets and from the original full data sets. The key of this approach is to use appropriate, conservative clustering parameters, such that the clusters are homogeneous, as required by annotation goals. For example, we can use 97% as the identity cutoff for 16S rRNAs at species level, to obtain a taxonomy annotation for 16S rRNA reads at a species level by aligning them to a reference rRNA database such as Silva [70], RDP [71] and Greengene [72]. Then, the query and references should be clustered at a similarity cutoff greater than 97%. If the goal is to annotate ORFs using the KEGG database [73], then clustering both KEGG reference sequences and the ORFs at 90% will be harmless, because sequences sharing 90% identity will rarely belong to different KEGG orthology groups.
To further reduce the annotation difference between NR data sets and full data sets, this approach should only be used to cluster sequences of similar length and with enough overlapping regions (Figure 3D), instead of other clustering settings (Figure 3B, C). The CD-HIT program has many parameters such as sequence length, alignment length and alignment coverage for users to finely tune the clustering process to form more homogeneous clusters.
Table 4 lists the clustering results for commonly used reference databases in metagenomic studies at conservative thresholds. Herein, the sizes of the NR data sets are 28–58% of the original ones. After clustering, the size of a NR query data set, which highly depends on the sequencing depth, can often be 50% to many times smaller than the original data set. So overall, the annotation using NR data sets can be easily accelerated by 10-fold.
Table 4:
Clustering results of reference databases by CD-HIT package////
| Data seta | Number sequences | Total | Cutoff b (%) | Clusters | Reduced to (%) | Time (minutes)c |
|---|---|---|---|---|---|---|
| NCBI NR | 12 054 819 | 4.3 GB | 90 | 7 036 029 | 58 | 181 |
| 16S (Silva + Greengene) | 555 530 | 799 MB | 98 | 154 170 | 28 | 90 |
| NCBI microbial genomes | 3 355 | 6.4 GB | 90 | 1 279 | 38 | 389 |
| NCBI virus sequences | 1 042 347 | 1.3 GB | 95 | 288 701 | 28 | 480 |
aNCBI NR was downloaded from NCBI at ftp://ftp.ncbi.nih.gov/blast/db/FASTA/. 16S sequences from Silva and Greengene were downloaded from http://www.arb-silva.de/download/archive/ and http://greengenes.lbl.gov/Download/Sequence_Data/, respectively. NCBI microbial genomes were downloaded from ftp://ftp.ncbi.nih.gov/genomes/Bacteria/ (file: all.fna.tar.gz). NCBI virus sequences were kindly provided by the CAMERA project (Ref. 42). bParameters for NR and 16S rRNA are ‘-c 0.9 -n 5 -g 1 -M 0 -T 0’ and ‘-c 0.98 -n 11 -b 5 -M 0 -T 0 -G 1’, respectively. NCBI microbial genomes and virus sequences are clustered by a beta version of CD-HIT that can process very long sequences with parameter ‘-c 0.9’ and ‘-c 0.95’. cTime on computer with eight cores.
PROTEIN FAMILY IDENTIFICATION
Reference-based metagenome annotation by comparison with known sequences is essential but has drawbacks, with the biggest limitation being the inability to annotate novel sequences. Large metagenomes and those from under-explored environments contain a large number of novel genes, which might be specific to the environment. These novel proteins can well be overlooked by reference-based annotation.
Clustering analysis is the most effective way to discover novel gene families from large data sets. This has been demonstrated by the global ocean sampling (GOS) study, which identified 3995 novel protein clusters from 17.4 million ORFs [16]. Other large-scale studies, such as MetaHIT [9], also found novel gene families through sequence clustering.
Clustering metagenomic proteins into families is more complicated than creating a NR data set. In both the GOS and MetaHIT projects, the analyses started by removing highly similar sequences (95–98% identity), followed by several steps of protein clustering. In GOS, these steps include (i) calculation of all-against-all similarities using BLAST; (ii) construction of core sequence clusters, which are dense sub-graphs in the whole graph where the vertices are sequences and the edges are defined by a set of very strong similarities cutoffs; (iii) calculation of sequence profiles for large core clusters using FFAS [74] and PSI-BLAST [27], (iv) creation of protein families by merging core clusters using FFAS profiles and (5) recruitment of small clusters and singletons into large core clusters using PSI-BLAST profiles. In MetaHIT, families were clustered from all-against-all BLAST results with an algorithm called MCL, whose details were not described in the MetaHIT article.
The above clustering pipelines require very time-consuming BLAST calculation (e.g. GOS used 1 million CPU hours), so they are not very feasible for small labs, which now also generate large-scale metagenomic data by using NGS technologies. Earlier, the speed of the GOS clustering pipeline was improved [75] by adopting CD-HIT as a fast clustering and recruiting tool. An independent study using only CD-HIT to build protein families from GOS proteins was also introduced later [15]. This CD-HIT-based clustering produced comparable results to the original GOS study but only used ∼10 000 CPU hours. Thus, this approach is more suitable for those projects with large data sets that lack the computation resources for exploring protein families.
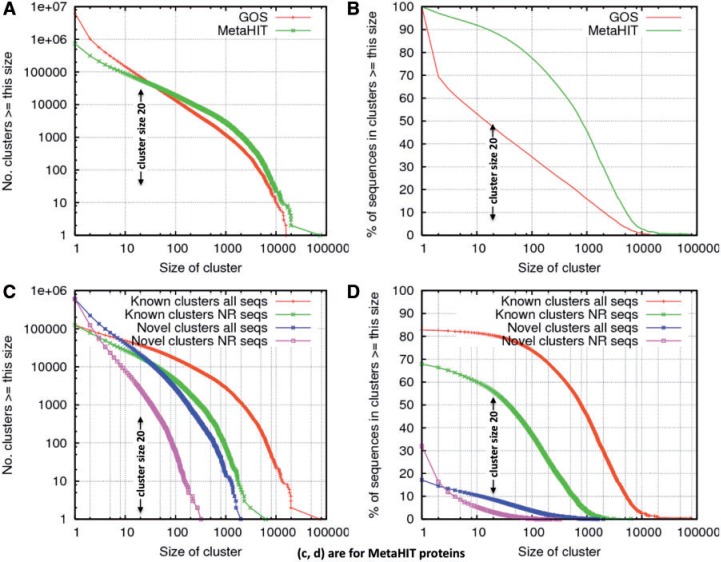
The above CD-HIT-based protein family finding process has three clustering steps where each subsequent clustering uses the representative sequences generated in the previous step. The similarity thresholds for these three clustering steps are 90%, 60% and 30%, respectively. The first two steps perform regular CD-HIT, and the last step uses PSI-CD-HIT, which also allows an alterative e value threshold. The details of this method are described in Refs. [15] and [76], where this method was further improved. The GOS data set is available from CAMERA project at http://camera.calit2.net. Herein, we demonstrate this easy approach for protein family identification using MetaHIT data. The original 14 792 886 proteins were downloaded from MetaHIT project at from http://gutmeta.genomics.org.cn/. These proteins were hierarchically clustered at 90%, 80%, 60% and 30% identity or an e value of 1e-6. These four steps cost 10, 2, 102 and 720 CPU hours and got 3 076 514; 2 471 148; 1 554 866 and 732 063 clusters, respectively. We used a 4-step clustering for MetaHIT data set, instead of the 3-step clustering we used earlier on the GOS data set, because the 4-step clustering provides better classification accuracy. The cluster distributions of MetaHIT are illustrated along with GOS clusters produced in Ref. [15] (Figure 4A, B). Compared with GOS ORFs, which contain more than half spurious ORFs due to the six reading frame translation, the MetaHIT genes predicted using Metagene [77] have far fewer false ORFs. So, using similar clustering parameters, MetaHIT data set has far fewer small clusters and singletons than GOS (Figure 4A). Cluster distributions for MetaHIT clusters grouped by known and novel are shown in Figure 4C and D. Herein, novel clusters are those clusters with no detectable similarity to Pfam families [78] using HMMER3 [79]. The 732 063 MetaHIT clusters contain 20 328 large clusters with at least 20 NR proteins, and 2580 of them are novel (Figure 4C), which covers ∼9% all MetaHIT sequences (Figure 4D). These novel clusters may represent human gut-specific gene families and should be further investigated.
Figure 4:
Distribution of GOS and MetaHIT protein clusters. The x-axis is the cluster size X. The y-axis in left figures is the number of clusters of size at least X; the y-axis in right figures is the percentage of total sequences included in the clusters of size at least X. Graphs in (A) and (B) are for all GOS and MetaHIT sequences. Graphs in (C) and (D) are only for MetaHIT sequences, grouped by Known and Novel clusters. In addition, two separate lines are made for NR sequences (i.e. the 3 076 514 representative sequences clustered at 90% identity).
LIST OF TOOLS AND THEIR ALGORITHM CHARACTERISTICS
As a summary, the tools tested in this article are listed in Table 5.
Table 5:
A list of clustering tools for metagenomic sequence analysis used in this study
| Tool and reference | Description | Key parameters |
|---|---|---|
| CD-HIT [28–31] | Cluster protein sequences | -c identity cutoff |
| -n word size | ||
| CD-HIT-EST [28–31] | Cluster nucleotide sequences | -c identity cutoff |
| -n word size | ||
| Uclust [39] | Cluster protein or nucleotide sequences | -id identity cutoff |
| –w word size | ||
| SEED [41] | Cluster highly similar Illumina reads (up to 3 mismatches and overhanging bases) | –mismatch allowed mismatches |
| DNACLUST [40] | Cluster highly similar DNA sequences (e.g. 16S rRNAs) | -s similarity cutoff |
| -k word size | ||
| CD-HIT-454 [13] | Identify duplicates for 454 reads | -c identity cutoff |
| CD-HIT-DUP | Identify duplicates for single or pair-ended Illumina reads | -e allowed mismatches |
| CD-HIT-LAP | Identify overlapping Illumina reads | -m overlapping length |
| -p overlapping coverage | ||
| PSI-CD-HIT [28–31] | Cluster proteins at low identity cutoff (20–50%) | -c identity cutoff |
| -ce expect value cutoff | ||
| CD-HIT-OTU | Identify operational taxonomic units (OTUs) from rRNAs | Identity cutoffa |
| AmpliconNoise [56] | Cluster flowgram data to remove noises from reads for OTU clustering | Identity cutoffa |
| Denoiser [55] | Cluster flowgram data to remove noises from reads for OTU clustering | Identity cutoffa |
| Cluster-based filtering | Filter sequence errors for improved sequence assembly | See CD-HIT-EST |
| Protein family clustering [15] | Identify protein families from metagenomic sequences | See CD-HIT and PSI-CD-HIT |
aCD-HIT-OTU, AmpliconNoise and Denoiser have multiple steps involves many parameters, which usually do not need to be modified.
The key parameters of these programs include the clustering similarity cutoff and some algorithmic parameters. For clustering similarity cutoff, CD-HIT, CD-HIT-EST, Uclust, CD-HIT-454, PSI-CD-HIT and OTU clustering packages use sequence identity; DNACLUST uses a similarity cutoff based on edit distance, which is very similar to sequence identity; SEED and CD-HIT-DUP allow certain number of mismatches. Word size is the most important algorithmic parameter for many programs (Table 5). The choice of word size depends on the clustering similarity cutoff and the type of sequences (protein or DNA). A higher similarity cutoff works with a longer word, which yields higher clustering speed. An important property of clustering methods is whether the results will change when the order of inputted sequences is different. Most methods introduced herein, including programs in CD-HIT package, Uclust and DNACLUST sort sequences by length and process them from long to short. The OTU clustering packages (e.g. CD-HIT-OTU) sort sequences by abundance and process them from high to low. So the order of inputted sequences does not change the output clusters except when the inputted sequences of the same length (or abundance) are in different order. Reads in most Illumina data sets have identical length, so the clustering results of Illumina reads depend on the order of inputted sequences.
Key Points.
Sequence clustering is an effective method to answer and address many fundamental questions and challenges in metagenomics. The applications include but are not limited to finding duplicates, diversity analyses, filtering sequence errors, database searches and finding protein families.
Ultra-fast clustering methods, such as CD-HIT, use less accurate algorithms than some sophisticated algorithms that rely on all-against-all similarities. However, when being used intelligently (e.g. multi-step clustering using parameters that fit a sequencing error model), the ultra-fast methods can produce comparable results to those sophisticated methods and can still be orders of magnitude faster.
Artificial duplicates should be removed for correct abundance calculation. However, attention should be paid to high-abundance viral and transcriptomic samples, where natural duplicates may be more abundant than artificial ones.
Using NR data sets saves significant database search time in metagenome annotation. However, conservative clustering parameters need to be used to ensure the clusters are homogeneous according to the annotation goal.
Clustering analysis is effective in finding novel gene families that might be overlooked using only reference-based annotation. Multi-step hierarchical clustering using ultra-fast methods can rapidly produce protein families from very large data sets.
FUNDING
National Institutes of Health [R01RR025030 and R01HG005978 to W.L.] and the Gordon and Betty Moore Foundation [CAMERA project].
Biographies
Weizhong Li is an Associate Research Scientist at the Center for Research in Biological Systems at University of California San Diego. Dr. Li has a background in computational biology. His research focuses on developing computational methods for sequence, genomic and metagenomic data analysis.
Limin Fu is a Postdoctoral Associate at the Center for Research in Biological Systems at University of California San Diego. Dr. Fu’s background is mathematics. His research focuses on bioinformatics algorithm development.
Beifang Niu is a Postdoctoral Associate at the Center for Research in Biological Systems at University of California San Diego. Dr. Niu was trained as a computer scientist. His research focuses on next-generation sequence analysis.
Sitao Wu is a Staff Scientist at the Center for Research in Biological Systems at University of California San Diego. Dr. Wu has a background in electric engineering. His research interests include protein structure prediction and metagenomics.
John Wooley is a Professor of Pharmacology and Associate Vice Chancellor, Research at the University of California San Diego, as well as a member of the Center for Research in Biological Systems and the California Institute of Telecommunications and Information Technology. Dr. Wooley’s background is biophysics and his current research interests include structural genomics and metagenomics.
References
- 1.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–85. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venter JC, Remington K, Heidelberg JF, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 4.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tringe SG, von Mering C, Kobayashi A, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–7. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 6.Mardis ER. A decade's perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 7.Dinsdale EA, Edwards RA, Hall D, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–32. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 8.Hess M, Sczyrba A, Egan R, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–7. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 9.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson J, Garges S, Giovanni M, et al. The NIH human microbiome project. Genome Res. 2009;19:2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chitsaz H, Yee-Greenbaum JL, Tesler G, et al. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat Biotechnol. 2011;29:915–21. doi: 10.1038/nbt.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–7. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 13.Niu B, Fu L, Sun S, et al. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics. 2010;11:187. doi: 10.1186/1471-2105-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Wooley JC, Godzik A. Probing metagenomics by rapid cluster analysis of very large datasets. PLoS ONE. 2008;3:e3375. doi: 10.1371/journal.pone.0003375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yooseph S, Sutton G, Rusch DB, et al. The sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert JA, Field D, Huang Y, et al. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS ONE. 2008;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yona G, Linial N, Linial M. ProtoMap: automatic classification of protein sequences and hierarchy of protein families. Nucleic Acids Res. 2000;28:49–55. doi: 10.1093/nar/28.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasson O, Vaaknin A, Fleischer H, et al. ProtoNet: hierarchical classification of the protein space. Nucleic Acids Res. 2003;31:348–52. doi: 10.1093/nar/gkg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J, Holm L, Heger A, et al. RSDB: representative protein sequence databases have high information content. Bioinformatics. 2000;16:458–64. doi: 10.1093/bioinformatics/16.5.458. [DOI] [PubMed] [Google Scholar]
- 21.Enright AJ, Ouzounis CA. GeneRAGE: a robust algorithm for sequence clustering and domain detection. Bioinformatics. 2000;16:451–7. doi: 10.1093/bioinformatics/16.5.451. [DOI] [PubMed] [Google Scholar]
- 22.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pipenbacher P, Schliep A, Schneckener S, et al. ProClust: improved clustering of protein sequences with an extended graph-based approach. Bioinformatics. 2002;18(Suppl. 2):S182–91. doi: 10.1093/bioinformatics/18.suppl_2.s182. [DOI] [PubMed] [Google Scholar]
- 24.Mika S, Rost B. UniqueProt: Creating representative protein sequence sets. Nucleic Acids Res. 2003;31:3789–91. doi: 10.1093/nar/gkg620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loewenstein Y, Portugaly E, Fromer M, et al. Efficient algorithms for accurate hierarchical clustering of huge datasets: tackling the entire protein space. Bioinformatics. 2008;24:i41–9. doi: 10.1093/bioinformatics/btn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Niu B, Gao Y, et al. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–2. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li WZ, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 30.Li WZ, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–3. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 31.Li WZ, Jaroszewski L, Godzik A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics. 2002;18:77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- 32.Boguski MS, Schuler GD. ESTablishing a human transcript map. Nature Genetics. 1995;10:369–71. doi: 10.1038/ng0895-369. [DOI] [PubMed] [Google Scholar]
- 33.Pertea G, Huang X, Liang F, et al. TIGR Gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–2. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 34.Burke J, Davison D, Hide W. d2_cluster: a validated method for clustering EST and full-length cDNAsequences. Genome Res. 1999;9:1135–42. doi: 10.1101/gr.9.11.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malde K, Coward E, Jonassen I. Fast sequence clustering using a suffix array algorithm. Bioinformatics. 2003;19:1221. doi: 10.1093/bioinformatics/btg138. [DOI] [PubMed] [Google Scholar]
- 36.Ptitsyn A, Hide W. CLU: a new algorithm for EST clustering. BMC Bioinformatics. 2005;6(Suppl. 2):S3. doi: 10.1186/1471-2105-6-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hazelhurst S, Lipták Z. KABOOM! A new suffix-array based algorithm for clustering expression data. Bioinformatics. 2011;27:3348–55. doi: 10.1093/bioinformatics/btr560. [DOI] [PubMed] [Google Scholar]
- 38.Suzek BE, Huang HZ, McGarvey P, et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–8. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 39.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 40.Ghodsi M, Liu B, Pop M. DNACLUST: accurate and efficient clustering of phylogenetic marker genes. BMC Bioinformatics. 2011;12:271. doi: 10.1186/1471-2105-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao E, Jiang T, Kaloshian I, et al. SEED: efficient clustering of next-generation sequences. Bioinformatics. 2011;27:2502–9. doi: 10.1093/bioinformatics/btr447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun S, Chen J, Li W, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res. 2011;39:D546–51. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu S, Zhu Z, Fu L, et al. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer F, Paarmann D, D'Souza M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markowitz VM, Ivanova NN, Szeto E, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–8. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rusch DB, Halpern AL, Sutton G, et al. The sorcerer II global ocean sampling expedition: northwest atlantic through eastern tropical pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:U480–7. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–6. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Y, Cai Y, Huse SM, et al. A large-scale benchmark study of existing algorithms for taxonomy-independent microbial community analysis. Brief Bioinform. 2012;13:107–21. doi: 10.1093/bib/bbr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Y, Cai Y, Liu L, et al. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res. 2009;37:e76. doi: 10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huse SM, Welch DM, Morrison HG, et al. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–98. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kunin V, Engelbrektson A, Ochman H, et al. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–23. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 54.Quince C, Lanzen A, Curtis TP, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–41. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 55.Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–9. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quince C, Lanzen A, Davenport RJ, et al. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qu W, Hashimoto S, Morishita S. Efficient frequency-based de novo short-read clustering for error trimming in next-generation sequencing. Genome Res. 2009;19:1309–15. doi: 10.1101/gr.089151.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao X, Palmer LE, Bolanos R, et al. EDAR: an efficient error detection and removal algorithm for next generation sequencing data. J Comput Biol. 2010;17:1549–60. doi: 10.1089/cmb.2010.0127. [DOI] [PubMed] [Google Scholar]
- 60.Medvedev P, Scott E, Kakaradov B, et al. Error correction of high-throughput sequencing datasets with non-uniform coverage. Bioinformatics. 2011;27:i137–41. doi: 10.1093/bioinformatics/btr208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kao WC, Chan AH, Song YS. ECHO: a reference-free short-read error correction algorithm. Genome Res. 2011;21:1181–92. doi: 10.1101/gr.111351.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li R, Yu C, Li Y, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 63.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li R, Zhu H, Ruan J, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–72. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niu B, Zhu Z, Fu L, et al. FR-HIT, a very fast program to recruit metagenomic reads to homologous reference genomes. Bioinformatics. 2011;27:1704–5. doi: 10.1093/bioinformatics/btr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ye Y, Choi JH, Tang H. RAPSearch: a fast protein similarity search tool for short reads. BMC Bioinformatics. 2011;12:159. doi: 10.1186/1471-2105-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li WZ, Jaroszewski L, Godzik A. Sequence clustering strategies improve remote homology recognitions while reducing search times. Protein Eng. 2002;15:643–9. doi: 10.1093/protein/15.8.643. [DOI] [PubMed] [Google Scholar]
- 70.Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cole JR, Wang Q, Cardenas E, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ogata H, Goto S, Sato K, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rychlewski L, Jaroszewski L, Li WZ, et al. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–41. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yooseph S, Li W, Sutton G. Gene identification and protein classification in microbial metagenomic sequence data via incremental clustering. BMC Bioinformatics. 2008;9:182. doi: 10.1186/1471-2105-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li W. Analysis and comparison of very large metagenomes with fast clustering and functional annotation. BMC Bioinformatics. 2009;10:359. doi: 10.1186/1471-2105-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–30. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–11. [PubMed] [Google Scholar]