Abstract
Roseobacter clade bacteria (RCB) are abundant in marine bacterioplankton worldwide and central to pelagic sulfur cycling. Very little is known about their abundance and function in marine sediments. We investigated the abundance, diversity and sulfur oxidation potential of RCB in surface sediments of two tidal flats. Here, RCB accounted for up to 9.6% of all cells and exceeded abundances commonly known for pelagic RCB by 1000-fold as revealed by fluorescence in situ hybridization (FISH). Phylogenetic analysis of 16S rRNA and sulfate thiohydrolase (SoxB) genes indicated diverse, possibly sulfur-oxidizing RCB related to sequences known from bacterioplankton and marine biofilms. To investigate the sulfur oxidation potential of RCB in sediments in more detail, we analyzed a metagenomic fragment from a RCB. This fragment encoded the reverse dissimilatory sulfite reductase (rDSR) pathway, which was not yet found in RCB, a novel type of sulfite dehydrogenase (SoeABC) and the Sox multi-enzyme complex including the SoxCD subunits. This was unexpected as soxCD and dsr genes were presumed to be mutually exclusive in sulfur-oxidizing prokaryotes. This unique gene arrangement would allow a metabolic flexibility beyond known sulfur-oxidizing pathways. We confirmed the presence of dsrA by geneFISH in closely related RCB from an enrichment culture. Our results show that RCB are an integral part of the microbial community in marine sediments, where they possibly oxidize inorganic and organic sulfur compounds in oxic and suboxic sediment layers.
Keywords: geneFISH, Roseobacter , sediment, sulfur oxidation
Introduction
Marine Roseobacter clade bacteria (RCB) are one of the most abundant bakterioplanktonic groups in oceans worldwide and are particularly important in organic sulfur cycling (Gonzalez and Moran, 1997; Buchan et al., 2005; Wagner-Döbler and Biebl, 2006). RCB are free-living, but they are also often found in epibiotic biofilms on macro-algae and various invertebrates (Wagner-Döbler and Biebl, 2006). Moreover, RCB are the dominant primary colonizers of submerged surfaces (Dang and Lovell, 2002). Accordingly, their 16S rRNA gene sequences and isolates have been frequently recovered from biofilms of deep-sea and coastal sediments (Buchan et al., 2005; Wagner-Döbler and Biebl, 2006; Brinkhoff et al., 2008; Sass et al., 2010). However, little is known about RCB in marine sediments. In 16S rRNA gene libraries from marine sediments, RCB accounted for 2–15% of all clones (Buchan et al., 2005). In the only quantitative study, RCB accounted for 3–11% of all 16S rRNA genes in bulk DNA from coastal sediments (Gonzalez et al., 1999), but their cell abundance in sediments is unknown.
RCB are metabolically heterogeneous and are capable of, for example, anoxygenic phototrophy, aromatic and organosulfur degradation. In water columns, RCB and SAR11 are the main groups involved in the demethylation of dimethylsulfoniopropionate and in routing it to the food web or to other compounds such as the climatically active DMS (Gonzalez et al., 1999; Howard et al., 2006; Curson et al., 2011). In addition, a few cultivated strains oxidize inorganic sulfur compounds such as sulfite, sulfide and thiosulfate (Sorokin, 1995; Gonzalez et al., 1999; Sass et al., 2010).
The detection and phylogeny of the soxB gene in the Sox multi-enzyme (SOX) pathway for thiosulfate oxidation in environmental polymerase chain reaction (PCR)-based libraries and genome sequences confirmed the involvement of RCB in sulfur oxidation (Meyer et al., 2007; Moran et al., 2007; Newton et al., 2010). Sox genes occur in various phylogenetic clades, including many Proteobacteria and Chlorobi (Petri et al., 2001; Meyer et al., 2007). The majority of these organisms encode a complete SOX pathway, whereas only a few groups such as some phototrophic and chemotrophic bacteria lack the subunits sulfur dehydrogenase SoxCD (Meyer et al., 2007; Frigaard and Dahl, 2009; Gregersen et al., 2011). These SoxCD-lacking organisms oxidize thiosulfate or sulfide and transiently form zero-valence sulfur, which is further oxidized to sulfite via the rDSR pathway (Hensen et al., 2006; Grimm et al., 2008). In RCB such as Roseobacter denitrificans, the complete SOX pathway including SoxCD is present; however, the rDSR pathway has not been found yet (Moran et al., 2007, www.roseobase.org).
In coastal sediments, large amounts of hydrogen sulfide are released during degradation of organic matter and re-oxidized to sulfate at the sediment surface (Jansen et al., 2009; Kamyshny and Ferdelman, 2010). Generally, Gamma- and Epsilonproteobacteria are considered as dominant sulfur-oxidizing organisms at marine sediment surfaces (Sievert et al., 2008; Lenk et al., 2011). In previous studies, we recovered alphaproteobacterial DsrAB sequences, which indicated that also yet unknown Alphaproteobacteria may be important to sulfur oxidation in coastal water columns and sediments (Lavik et al., 2009; Lenk et al., 2011). In this study, we investigated whether alphaproteobacterial RCB, some of which are known to oxidize sulfur compounds in the water column, could also play a role in sulfur oxidation in coastal sediments. We first studied the environmental abundance and diversity of RCB by comparative analysis of 16S rRNA and soxB genes in tidal sediments from the German Wadden Sea. Using fluorescence in situ hybridization (FISH), we quantified RCB in different types of marine sediments. Furthermore, we linked the 16S rRNA genes of uncultured RCB with sulfur oxidation genes using metagenomics, single geneFISH (Moraru et al., 2010) and enrichment cultures of sulfur-oxidizing RCB.
Materials and methods
Sampling
The sediment was sampled in two tidal sites located in the East and in the North Frisian German Wadden Sea. Sediment cores from the Janssand intertidal sand flat (53°44′07″N, 007°41′57″E) were taken during repeated sampling campaigns in April 2005, August 2007 and May 2009 for catalyzed reporter deposition-FISH (CARD-FISH) and 16S rRNA gene libraries (Lenk et al., 2011), and in June 2009 for cell sorting and 16S rRNA gene cloning (this study). Sediment from a silty tidal site of Koenigshafen at the island of Sylt (55.02544°N, 8.4317°E) was sampled in October 2008 for the enrichment culture and in October 2010 for the RCB-specific 16S rRNA gene library and CARD-FISH analysis. For a biogeographic survey of RCB, we also analyzed sediment samples of diverse origin (see Supplementary Table 1). Sediment processing is detailed in Supplementary Methods.
Enrichment of RCB
RCB were enriched from sediment sampled in October 2008 (site Koenigshafen, Island of Sylt, Germany). Initially, 2.5 ml of sediment from the anoxic layer of 3–4 cm depth were inoculated into 50 ml sulfate-free, sodium bicarbonate-buffered, artificial seawater medium (Widdel and Bak, 1992) containing 1 mℳ sulfide, 5 mℳ acetate and 5 mℳ nitrate. The headspace contained a N2/CO2 (80/20 (v v−1)) atmosphere. After 1 month of incubation, the presence of nitrate reducers was indicated by gas formation. Subsequently, RCB were further enriched by repeated Media Processing Node series. FISH revealed high relative abundance of cells targeted by probe ROS537 (>50%) in the highest positive dilution (10−6). This culture was used for substrate tests (details are given in Supplementary Figure 5).
CARD-FISH on sediment samples and enrichment cultures
The in situ abundance of RCB was assessed by CARD-FISH using probe ROS537 (Gonzalez and Moran, 1997; Eilers et al., 2001) following an established protocol (Ishii et al., 2004). Probe match analysis against the SILVA SSU Ref. database release 102 revealed 92% coverage for sequences of the marine Roseobacter clade. Only 3% of the sequences targeted by ROS537 were non-target organisms. RCB were counted along three vertical sediment profiles sampled in April 2005, August 2007 and May 2009. Probe ALF968 (Glöckner et al., 1999) specific for Alphaproteobacteria was applied for double hybridizations. CARD-FISH on sediments of Sylt and enrichment cultures are detailed in Supplementary Methods.
Flow cytometry sorting of RCB and amplification of 16S rRNA gene
Surface sediment (0–3 cm) of Janssand site was sampled in June 2009 and preserved for FISH. Subsamples were sonicated (Lenk et al., 2011) and cells were purified via density gradient centrifugation (Fazi et al., 2005). Purified cell fractions were hybridized via CARD-FISH with probe ROS537 and then sorted using a MoFlo flow cytometer (Cytomation Inc., Fort Collins, CO, USA) according to Sekar et al. (2004). Hybridized (ROS537-positive) cells were selected based on green fluorescent signals and side angle light scatter (SSC) by plotting SSC versus green fluorescence in a bivariant dot plot diagram. A total of 133 000 target cells were sorted and concentrated on a 0.2 μm polycarbonate membrane filter. To check the purity of the sorted cell fraction, a subsample was counterstained with 4′,6-diamidino-2-phenylindole (1 μg ml−1) and the relative abundance of non-hybridized cells was determined. Amplification of the 16S rRNA gene was performed from filter pieces (Sekar et al., 2004). PCR, cloning and sequencing of the amplification product was performed as described previously (Lenk et al., 2011).
Fosmid library screening and sequence analysis
For metagenomic analysis, we screened two fosmid libraries that were established from Janssand sediment of 5–12 cm depth (Mussmann et al., 2005) and from 490 cm sediment depth (K Bischof, unpublished) for the presence of dsrAB and soxB. We used primer pairs DSR1F/DSR4R (Loy et al., 2009), rDSR240F/rDSR808R (Lenk et al., 2011) and soxB432F/1446B (Petri et al., 2001) according to PCR conditions previously published. After screening, selected clones were chosen for full-length sequencing of the insert (∼40 kb). The fosmid insert sequences were determined by a shotgun approach. Sequencing approach, assembly and annotation are detailed in Supplementary Methods.
Simultaneous in situ hybridization of dsrA gene (geneFISH) and 16S rRNA
A dsrA targeting polynucleotide probe ds285 RCB (359 bp, GC content 65%) was designed based on the dsrAB sequence in fosmid ws101A12 using the PolyPro software (Moraru et al., 2011). The target region of the dsrA probe showed 92.3% sequence identity to the alphaproteobacterial dsrA phylotype that was derived from the sulfidic enrichment. It displayed 66.3% sequence identity to the gammaproteobacterial dsrA phylotype detected in the enrichment. Fosmid copies were extracted from Escherichia coli clone cultures using the Spin Miniprep Kit (Qiagen, Hilden, Germany) and served as template for the synthesis of the double-stranded DNA polynucleotide (dsDNA probe). Dig-dUTP-labeled polynucleotide probes were synthesized using the PCR Dig Probe Synthesis Kit (Roche, Diagnostics, Mannheim, Germany). Primers dsr1-F, 5′-GAAGTATCCCGAGTCGAAGG-3′ and dsr1-R, 5′-GCGCCGGGCGGTGCATCTC-3′ targeting dsrA of fosmid ws101A12 were used for synthesis at 56 °C annealing temperature. The template and the primers applied for synthesis of probe NonPolyPro350 are described elsewhere (Moraru et al., 2010). GeneFISH was performed according to Moraru et al. (2010). Details with regard to the protocol are given in Supplementary Methods.
Nucleotide sequences
The sequence data from this study have been submitted to the GenBank databases under accession No. JQ256774–JQ256779 (enrichment culture dsrA, dsrB, soxC, soxD), JQ256780–JQ256790 (fosmid clones), JQ256791–JQ256838 (16S rRNA gene) and JQ256839–JQ256904 (soxB).
Results
Abundance of RCB in marine sediments
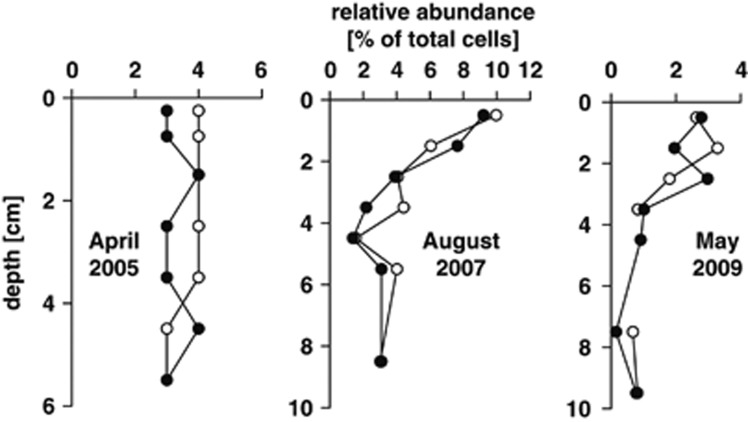
We quantified RCB in surface sediments of the Janssand site in different seasons using probe ROS537 for CARD-FISH. Relative abundances in the upper 2 cm varied substantially between the summer and spring samples (Figure 1). Maximum relative and total abundances were reached in August 2007, where RCB accounted for up to 9.6% of all cells (2.5 × 108 cells ml−1). In deeper, permanently anoxic, sulfidic layers down to 9 cm RCB made up 0.8% to 3.1% of all cells. Double hybridizations supported the specific detection of RCB, as all ROS537-hybridized cells also showed signals with probe ALF968 targeting Alphaproteobacteria (Figure 2). We also quantified RCB in other coastal sediments including Sylt sediments (Supplementary Table 1). Here, RCB constituted approximately up to 8% of all cells in the upper 1 cm of the sediment. In contrast, they made up <0.5% in 7–8 cm depth. RCB also occurred in other coastal habitats such as North and Baltic Sea sediments, where they accounted for approximately 2% of all cells, but RCB were not detected in significant numbers in hydrothermal (Guaymas Basin), arctic (Svalbard) or subtropical (Bermuda, Elba) sediments (Supplementary Table 1).
Figure 1.
Relative abundance of marine RCB determined by CARD-FISH (probe ROS537) in vertical sediment profiles in April 2005, August 2007 and May 2009. Sediment core A, white circles; sediment core B, black circles.
Figure 2.
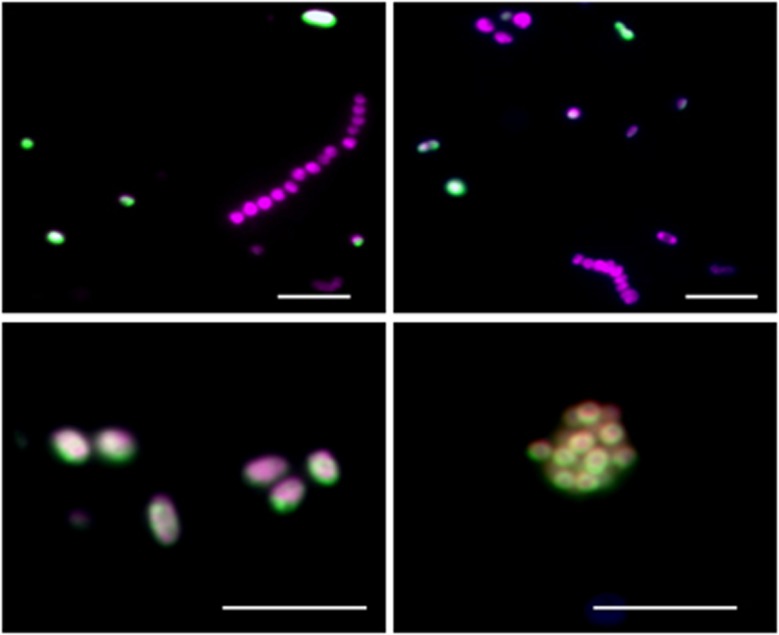
Epifluorescence microscopy images of marine RCB in Janssand sediments. For all images, green (Alexa 488): cells that were identified by RCB-specific probe ROS537; red (Alexa 594): Alphaproteobacteria that hybridized with the general probe ALF968. Only RCB are simultaneously stained in green and red. The scale bar corresponds to 5 μm.
16S rRNA and soxB gene diversity of sedimentary RCB
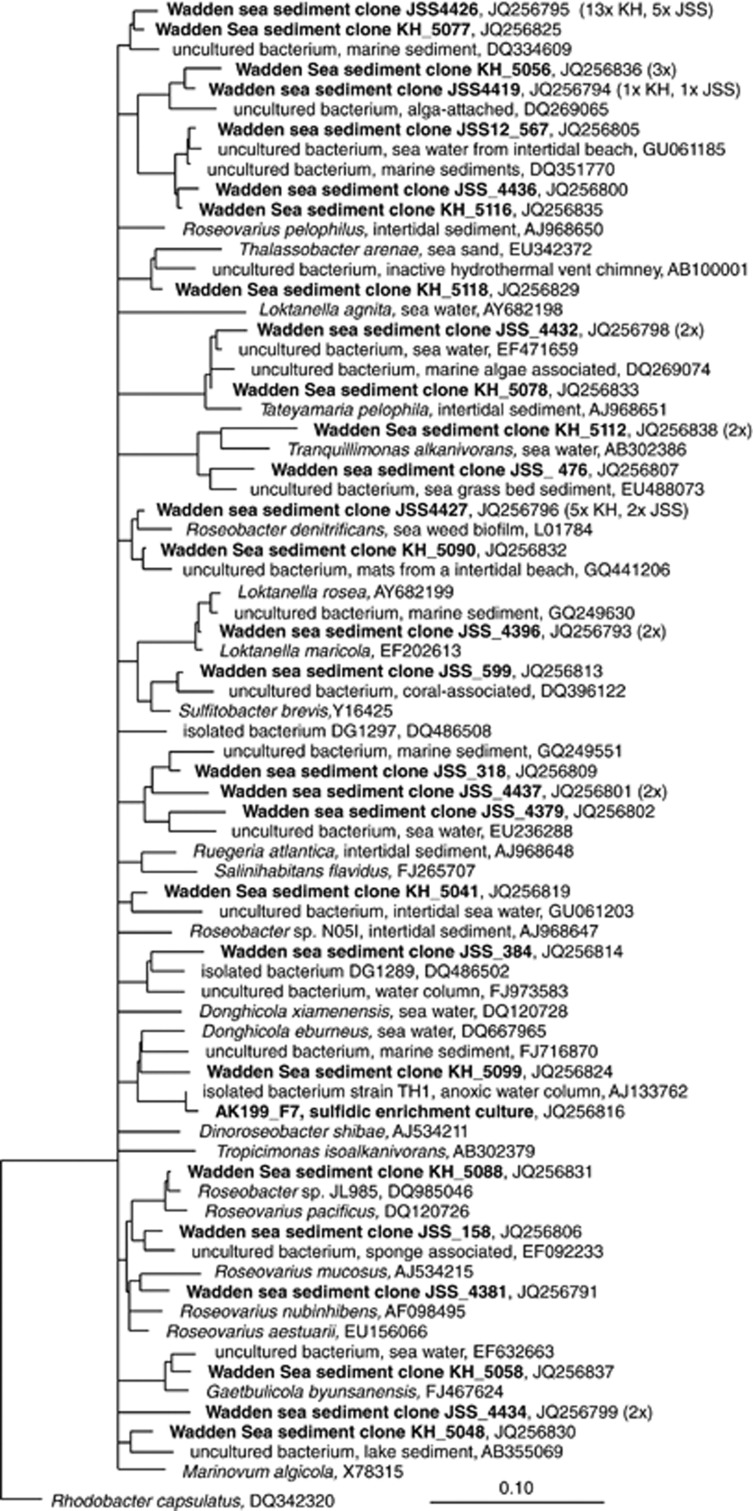
To study the diversity of RCB, we constructed and sequenced 16S rRNA gene libraries from both Janssand and Sylt sediments and a soxB library from Janssand sediment. In previously generated, Bacteria-targeted 16S rRNA gene libraries from Janssand sediments (Lenk et al., 2011), out of 458 analyzed clones, 10 sequences related to RCB were recovered. To study the RCB diversity in more detail, we generated a 16S rRNA gene library from probe ROS537-hybridized cells that we mechanically enriched from sediment samples by fluorescence-activated cell sorting. The sorted cells served as template for PCR and subsequent cloning and sequencing. Out of 30 clones, 14 sequences grouped with RCB. Despite a sorting purity of 97%, the non-RCB sequences were diverse but displayed several mismatches to probe ROS537 and likely originated from contaminating cells in the sorted cell fraction. Biased cell lysis and PCR probably favored amplification of non-target cells over hybridized RCB cells. To recover specifically RCB sequences from the Sylt sediments, we used probe ROS537 as a reverse primer and combined it with a general forward primer for PCR. Here, we recovered 34 partial, RCB-related sequences. In total, we obtained 58 RCB-related 16S rRNA sequences from the two tidal sediments. These sequences were highly diverse (79–99% sequence identity (SI)) and affiliated with sequences of cultured and uncultured organisms from biofilms, pelagic and benthic habitats (Figure 3). The closest cultured relatives were among the genera Sulfitobacter, Thalassobacter, Roseobacter and others. Clones JSS_4432 and KH_5078 grouped with Tateyamaria pelophila (97% SI), a facultatively anaerobic, sulfite and thiosulfate oxidizing strain that was previously isolated from the study site (Sass et al., 2010).
Figure 3.
Consensus tree of alphaproteobacterial 16S rRNA gene sequences from intertidal sediments of Janssand (JSS) and Sylt (KH). Bar indicates 10% sequence divergence.
To test whether the detected RCB at Janssand have the potential to oxidize inorganic sulfur compounds, we screened a soxB gene library for RCB-related sequences. In total, 66 soxB sequences were recovered, which grouped into 35 operational taxonomic units (Supplementary Figure 1). The majority of sequences (41 sequences, 17 operational taxonomic units) affiliated with Alphaproteobacteria. Among these, 39 sequences (15 operational taxonomic units) consistently clustered with SoxB sequences of cultured RCB. Similar to the results obtained with the 16S rRNA genes, the RCB-associated SoxB sequences were highly diverse (46–96% amino-acid SI). They were only distantly related to sequences of known cultured representatives such as Oceanibulbus indolifex (JSS043, 86% SI) and sulfite-oxidizing Silicibacter pomeroyi (JSS037, 82% SI).
Metagenomic analysis of sulfur-oxidizing bacteria in a tidal sediment
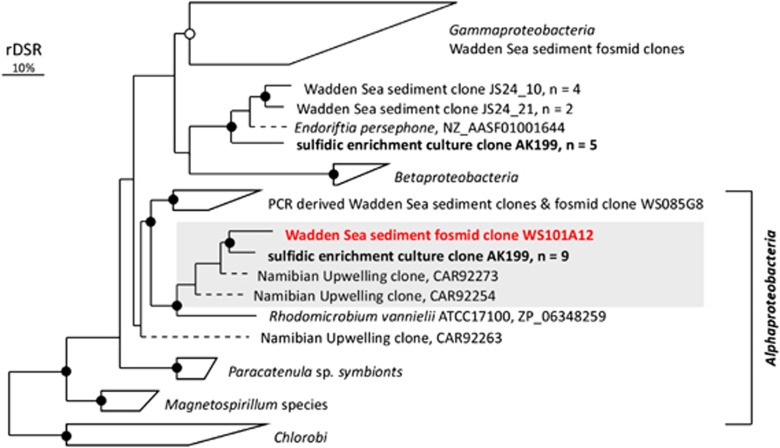
To reveal genomic and metabolic features of sulfur-oxidizing microbes, in particular of RCB, we screened fosmid clones for genome fragments encoding dsrAB or soxB. Out of ∼24 000 fosmids, we identified 11 fosmids of interest with insert sizes ranging from 30 to 43 kb. Eight fosmids carried the dsrAB gene and two fosmids carried the soxB gene, while one fosmid (ws101A12) carried both genes (Figure 5 and Supplementary Table 2). The deduced DsrAB or SoxB amino-acid sequences of eight fosmids grouped with Gammaproteobacteria (Supplementary Table 3). The derived SoxB sequences of two fosmids including fosmid ws101A12 affiliated with Alphaproteobacteria (Figure 5 and Supplementary Figure 1). The DsrAB sequence derived from fosmid clone ws101A12 was most closely related to sequences of uncultured, alphaproteobacterial sulfur-oxidizing prokaryotes (SOP) detected in sulfidic waters off the coast of Namibia (84% sequence identity SI) (Figure 4). The closest cultured relative was Rhodomicrobium vannielli (78% SI), a photoautotrophic alphaproteobacterium. The SoxB sequence of fosmid ws101A12 affiliated with the marine Roseobacter clade (Supplementary Figure 1). It had the highest sequence identity to the SoxB sequence of Sagittula stellata (86% SI). Furthermore, the comparison of intrinsic nucleotide signatures of this genomic fragment using the TaxSOM software (Weber et al., 2011) provided additional evidence for a Roseobacter clade-related affiliation of ws101A12 and supported Oceanicola batsensis as closest known relative (data not shown).
Figure 4.
Phylogenetic reconstruction (RaxML) of DsrAB protein sequences encoded on Wadden Sea sediment fosmids from Janssand and DsrAB sequences retrieved from the sulfidic enrichment culture AK199 from Koenigshafen (in bold). Gray shading indicates sequences likely originating from members of the marine Roseobacter clade. Bootstrapping values are indicated with >70% (closed circles) and >50% (open circles) RAxML bootstrap support. The bar indicates 10% sequence divergence.
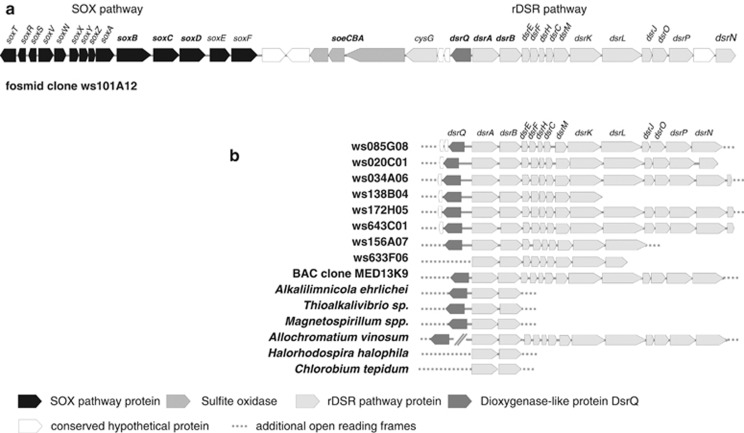
Gene content on fosmid clone ws101A12
The 35 kb insert of fosmid ws101A12 contained 37 open reading frames (ORFs) (Figure 5 and Supplementary Table 2). In 18 out of 37 predicted proteins, the closest homologs were found in genome sequences of known RCB. This fosmid clone harbored a gene set of 14 genes in the Sox multi-enzyme pathway (soxTRSVWXYZABCDEF, ORFs 1–14) including the sulfur dehydrogenase SoxCD encoding genes (ORF 11, ORF 12). All deduced protein sequences displayed highest sequence identity to sox genes of marine RCB (Supplementary Table 2). In addition, the same metagenomic fragment encoded a nearly complete rDSR operon (dsrABEFHCMKLJOPN, ORFs 24–35, ORF 37). Of these, seven deduced protein sequences displayed significant sequence similarity to homologs in Alphaproteobacteria (ORFs 24–26, 29, 32–34). Between the rDSR and SOX operons, three subunits of a putative sulfite dehydrogenase were encoded (soeCBA, ORFs 17–19). The deduced proteins showed highest sequence similarity to homologs among RCB (62–82% SI; Supplementary Table 2) including the SoeABC subunits of Ruegeria pomeroyi DSS-3 (YP_168752-54).
Figure 5.
(a) Genomic organization of sox, soe and dsr loci in fosmid clone ws101A12. (b) Gene neighborhood of the dsrAB locus encoded on Wadden Sea sediment fosmid clones and in genomes of selected SOP.
Comparison of the dsr locus of fosmid ws101A12 with the gammaproteobacterial fosmid clones and available SOP genomes pointed at the presence of previously uncharacterized ORFs (ORFs 21–23) that are consistently located upstream of dsrA (Figure 5). ORFs 21–22 are conserved hypothetical proteins of which homologs were found in a few SOP. The deduced protein sequences of ORF 23 weakly matched a taurine catabolism dioxygenase (Supplementary Table 2), but displayed sequence motifs typical for dioxygenases (Supplementary Figure 2). Comparative genome analysis of the dsr loci revealed that the ORF23 homologs are located upstream of dsrAB in nearly all our fosmid clones and in most (facultatively) aerobic chemotrophic SOP (Figure 5). In contrast, homologs are not associated with the dsr locus in the genomes of the sulfur-oxidizing endosymbionts Ruthia magnifica and Vesicomyosocius oktuanii and of the anaerobic phototrophs A. vinosum and Halorhodospira halophila. It is moreover absent in the genomes of the strictly anaerobic, phototrophic Chlorobi.
Enrichment and molecular characterization of rDSR-encoding RCB
To confirm the hypothesis that the dsr locus in fosmid ws101A12 indeed belonged to RCB, we screened for RCB-related 16S rRNA and dsrAB genes in a sulfidic enrichment culture from anoxic sediments. After repeated transfers, RCB dominated the enrichment and accounted for >50% of all cells, whereas Gamma- and Epsilonproteobacteria were less abundant (Supplementary Figure 3). From this enrichment culture, bacterial 16S rRNA and dsrAB genes were amplified, cloned and sequenced. Consistent with our FISH results, 16S rRNA gene sequences (55 clones) affiliated with RCB, Gamma- and Epsilonproteobacteria. Only a single RCB phylotype was identified that most closely affiliated with the nitrate-reducing strain TH1 (98% SI), and with Donghicola eburneus, an acetate-oxidizing aerobe (96% SI). A sequence obtained from a denaturing gradient gel electrophoresis analysis with RCB specific primers matched the clone library-derived sequence, which strongly supported the presence of only one RCB phylotype in the enrichment (Supplementary Figure 4).
Two different DsrAB phylotypes were identified in the enrichment culture. One phylotype was most closely related to the DsrAB encoded on the RCB-affiliated fosmid clone ws101A12 (93% SI). The second phylotype affiliated with gammaproteobacterial DsrAB sequences previously retrieved from Janssand sediments (85% SI). To test whether the RCB in the enrichment also possess soxCD genes, we amplified a fragment using novel primers that target soxC at the 5′ end and soxD at the 3′ end. We recovered a fragment of approximately 1130 bp in size that was directly sequenced and had the highest nucleotide sequence identity to soxC of fosmid ws101A12 (83% SI). The corresponding soxD sequence displayed 73% nucleic acid sequence identity to soxD of fosmid ws101A12 and 76% to soxD of R. pomeroyi. As no pure culture of the dsr-encoding RCB organism could be recovered, we monitored their growth in the enrichment upon supply with organic and inorganic sulfur compounds and organic acids. Under nitrate-respiring conditions, the addition of inorganic sulfur compounds stimulated growth of RCB. Strongest growth of RCB was observed with dimethylsulfoxide and taurine under aerobic conditions (Supplementary Figure 5). As other sulfur-oxidizing bacteria were present in the enrichment, we cannot completely rule out that the observed RCB relied on metabolites released by these organisms.
In situ localization of dsrA in RCB cells in the enrichment using geneFISH
As a 16S rRNA gene was absent on the metagenomic fragment and as we could not obtain a pure RCB culture from the enrichment, we combined 16S rRNA CARD-FISH with gene-targeted FISH (Moraru et al., 2010). To link directly the RCB identity and the dsrA gene at the single-cell level, we designed a polynucleotide probe dsr285_RCB (Supplementary Figure 6) that targeted the alphaproteobacterial dsrA phylotype of fosmid ws101A12 (Supplementary Table 4) and hybridized it to the sulfidic enrichment culture. The sequence identity between the probe and the target dsrA sequence was 91.9%, which is sufficient to ensure specific binding (Moraru et al., 2011). GeneFISH of dsrA clearly localized the gene in those cells that were simultaneously hybridized with the 16S rRNA targeting probe ROS537 (Figure 6). Because of known methodological issues (Moraru et al., 2010), only approximately 50% of all RCB were targeted by the polynucleotide probe. Cells not hybridized to probe ROS537 also gave no dsrA geneFISH signals. The second DsrAB phylotype in the enrichment belonged to Gammaproteobacteria and displayed 66.3% sequence identity to the geneFISH probe. Therefore, it was not targeted by probe dsr285_RCB (Supplementary Table 4). Hybridization with the negative control probe NonPolyPr350 (Supplementary Figure 6) resulted in background level unspecific binding and showed that the probe did not bind unspecifically to cellular components (Figure 6). We also attempted to quantify the abundance of dsrA-encoding RCB in surface sediment. However, the low detection efficiency of geneFISH in sediment samples due to high particle background hampered any further in situ quantification.
Figure 6.
Epifluorescence microscopy images of RCB encoding the dsrAB gene from sulfidic enrichment culture. Dual color hybridization of the 16S rRNA of RCB stained in green (Alexa 488 tyramide) and the dsrA targeting polynucleotide probe stained in red (Alexa 594 tyramide). Overlay images demonstrate that the signal for dsrA (red, a3 and a4) colocalizes with the 16S rRNA FISH signal for RCB (green, a4). Other bacteria in the enrichment (Epsilon- and Gammaproteobacteria) are stained with 4′,6-diamidino-2-phenylindole and are not fluorescent (a1). In addition, no fluorescent signals were obtained from hybridization of the NonPoly350 probe (b3) to the enrichment culture, which indicates the absence of nonspecific binding (b1–b4). a1, b1: DNA stains (4′,6-diamidino-2-phenylindole). a2: FISH signals of probe ROS537 targeting 16S rRNA (RCB specific). b2: FISH signals of probe GAM42a targeting 16S rRNA (Gammaproteobacteria). a3: FISH signals of dsrA gene-targeted probe. b3: negative control probe NonPoly350. a4: overlay images of 16S rRNA-FISH and dsrA-FISH. b4: overlay images of 16S rRNA-FISH and negative control probe NonPoly350. Scale bar corresponds to 5 μm.
Discussion
Abundance and diversity of RCB in marine sediments
The marine Roseobacter clade often accounts for a substantial fraction of coastal bacterioplankton, but so far it was unknown, how abundant they are in coastal sediments. Here, we provide evidence that RCB also reach high total abundances in marine sediments, which outnumbered RCB in bacterioplankton by up to three orders of magnitude (Eilers et al., 2000; Alderkamp et al., 2006). Our results complement early findings of Gonzalez and Moran (1999) and corrobate that RCB are also an integral part of the microbial community in coastal surface sediments.
Using several methods, we explored the RCB diversity in two tidal sediments to look for groups specific to marine sediments. We obtained 58 16S rRNA gene sequences indicative of a large diversity within the marine RCB group in tidal sediments. In several diversity studies, RCB-related sequences were recovered from sediments (Buchan et al., 2005), but an analysis of available sequences did not reveal a dedicated RCB group that is exclusively found in sediments. Most of our sequences were related to those from bacterioplankton and algae or invertebrate biofilms, which rather mirrors the flexibility of RCB to thrive as free-living or attached to surfaces. In support of this, pelagic RCB were found to rapidly colonize submerged surfaces in marine waters (Dang and Lovell, 2002). Moreover, we detected RCB in high numbers in permanently anoxic layers and also enriched a facultatively anaerobic RCB from anoxic sediments. Previous results indeed showed that planktonic RCB in the open North Sea can assimilate glucose both under aerobic and anaerobic conditions (Alonso and Pernthaler, 2005). Taken together, these results strongly suggest that many pelagic RCB are able to survive and grow even in anoxic marine sediments.
Some RCB encode both the rDSR and the complete SOX pathway
Thiosulfate is a key intermediate in sulfur cycling in marine sediments (Jørgensen, 1990). As we recovered many and diverse soxB sequences related to RCB, we propose that RCB are important thiosulfate-oxidizing bacteria in Janssand tidal sediments. This is consistent with published data and genomic surveys indicating that known RCB encode the SOX pathway including soxCD subunits but not the rDSR pathway (Meyer et al., 2007, Moran et al., 2007). Moreover, in RCB the SOX pathway also appears to be involved in the oxidation of sulfur intermediates during breakdown of dimethylsulfoniopropionate (Rinta-Kanto et al., 2011).
Here, we provide evidence that some RCB also possibly employ the rDSR pathway for sulfur oxidation. First, we identified a RCB-related metagenomic fragment that harbored a nearly complete set of genes of both the SOX and the rDSR pathway. Second, we enriched a RCB population in a sulfidic, nitrate-respiring culture, in which we simultaneously visualized the 16S rRNA and the dsrA gene in single cells. Congruent phylogenies of DsrAB and 16S rRNA and denaturing gradient gel electrophoresis analysis supported the presence of only one dsrAB-encoding RCB in the enrichment culture. This organism most likely also encoded SoxCD as we could also amplify a soxCD gene fragment from the enrichment culture that was closely related to soxCD on fosmid ws101A12.
Extended sulfur oxidation potential of RCB from tidal sediments
The co-occurrence of dsr and soxCD loci is novel, as both are mutually exclusive in known SOP genomes (Meyer et al., 2007; Frigaard and Dahl, 2009; Gregersen et al., 2011). In soxCD-lacking organisms, the rDSR pathway substitutes the sulfur dehydrogenase activity of SoxCD and the sulfane–sulfur transiently accumulates as elemental sulfur before it is further oxidized to sulfite (Hensen et al., 2006; Grimm et al., 2008). In contrast, a complete pathway including SoxCD allows the direct oxidation of thiosulfate to sulfate (Friedrich et al., 2001; Rother et al., 2001). It was presumed that the rDSR pathway conserves more energy than the SOX pathway (Gregersen et al., 2011), which would explain the loss of soxCD in all yet described rDSR-encoding SOP.
However, we propose that such an extended genomic repertoire provides an increased metabolic flexibility. A fast, direct oxidation of thiosulfate using only the SOX pathway including SoxCD is beneficial, when electron donor and acceptor are available in excess. In turn, the rDSR pathway likely catalyzes the oxidization of elemental sulfur that has been formed from ambient sulfide or thiosulfate under electron acceptor-limiting conditions (Van den Ende and Van Gemerden, 1993); for instance, during tidal fluctuations (Jansen et al., 2009). Unfortunately, the enrichment culture could not be maintained in our lab. Hence, it remains to be shown under which conditions these RCB employ the SOX and rDSR pathways.
A novel subunit in the proteobacterial DSR operon
On fosmid ws101A12, we identified a previously undescribed ORF (ORF 23) that is located upstream of dsrAB in most of our dsr-encoding metagenomic fragments and in several SOP genomes. Such a conserved gene neighborhood and the typical dioxygenase motifs among many different phyla are strongly indicative of a catalytic activity in the rDSR pathway. Thus we suggest the designation ‘dsrQ' for this ORF. The actual function of the deduced protein is unclear, but different scenarios are conceivable. The DsrQ protein may catalyze the release of sulfite during oxygenolytic breakdown of intracellular (Franz et al., 2009) or ambient sulfonates, similar to the function of taurine dioxygenase in E. coli (Eichhorn et al., 1997). This is supported by the fact that the derived proteins of ORFs 17–19 are homolog to a recently proposed novel-type sulfite dehydrogenase SoeABC (Lehmann et al., 2012). Mutation studies have shown that this enzyme is crucially involved in sulfite oxidation and in taurine degradation by R. pomeroyi (Lehmann et al., 2012; S. Lehmann, personal communication). Moreover, homologs enzymes are possibly involved in dimethylsulfoniopropionate breakdown by RCB (Rinta-Kanto et al., 2011).
Conclusions
We derived potential ecophysiological traits of RCB in coastal sediments from FISH counts, metagenomics and analyses of an enrichment culture. Some of these RCB possess an extended genetic repertoire for sulfur oxidation that is novel among RCB and all known sulfur-oxidizing microorganisms. Using the enrichment culture, we monitored growth of RCB by FISH upon supply of different electron donors and oxygen or nitrate as electron acceptors. Although RCB could have relied on metabolites released by other organisms in the enrichment culture, the observed growth of RCB on different sulfur compounds is consistent with our genomic data and supports a sulfur-based energy metabolism. Our substrate tests in the enrichment culture indicated a potential to grow on organic and inorganic sulfur compounds of the dsrAB-containing RCB phylotype. Such a unique metabolic flexibility is particularly useful in highly fluctuating environments, for example, at oxic–anoxic interfaces in tidal sediments and may allow occupying new ecological niches. These RCB may oxidize sulfur compounds via two different pathways and could overcome oxygen limitation by respiring nitrate. Unlike most known sulfur-oxidizing bacteria, they are probably not thioautotrophic as we did not observe any carbon fixation by RCB in a previous microautoradiography experiment using 14C bicarbonate (Lenk et al., 2011). This is in line with the general absence of CO2-fixing pathways in RCB (Moran et al., 2007; Newton et al., 2010). During lithoheterotrophic growth, the use of reduced inorganic sulfur compounds as additional energy source would provide a competitive advantage over autotrophic SOP (Sorokin, 2003; Moran et al., 2004) that are present in our tidal sediments (Lenk et al., 2011). Because of their ability to switch between planktonic and a surface-adapted lifestyle, RCB most likely persist in sediments by vertical migration, active settlement and growth, and may therefore contribute to the pelagic–benthic coupling. Considering their high cell abundances, RCB may significantly contribute to sulfur cycling not only in the marine water columns, but also in coastal surface sediments.
Acknowledgments
We thank A Krupke, C Probian, J Wulf, S Dyksma, K Bischof and K Zerjatke for excellent technical assistance. We are grateful to thank Felix Jansen, Manuel Kleiner, Juliane Wippler, Petra Pjevac and the MarMic students of class 2014 for providing sediment samples. We acknowledge the captains of ‘Spes Mea' and ‘Doris von Ochtum' for supporting the sampling campaigns. This work was funded by the German Science Foundation (DFG) during research group ‘Biogeochemistry of Tidal Flats' (JO 307/4, BO 1584/4) and the Max Planck Society.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (/http://www.nature.com/ismej)
Supplementary Material
References
- Alderkamp AC, Sintes E, Herndl GJ. Abundance and activity of major groups of prokaryotic plankton in the coastal North Sea during spring and summer. AME. 2006;45:237–246. [Google Scholar]
- Alonso C, Pernthaler J. Incorporation of glucose under anoxic conditions by bacterioplankton from coastal North Sea surface waters. Appl Environ Microbiol. 2005;71:1709–1716. doi: 10.1128/AEM.71.4.1709-1716.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhoff T, Giebel HA, Simon M. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol. 2008;189:531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- Brune DC. Sulfur oxidation by phototrophic bacteria. Biochim Biophys Acta. 1989;975:189–221. doi: 10.1016/s0005-2728(89)80251-8. [DOI] [PubMed] [Google Scholar]
- Buchan A, Gonzalez JM, Moran MA. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curson ARJ, Todd JD, Sullivan MJ, Johnston AWB. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Micro. 2011;9:849–859. doi: 10.1038/nrmicro2653. [DOI] [PubMed] [Google Scholar]
- Dang HY, Lovell CR. Numerical dominance and phylotype diversity of marine Rhodobacter species during early colonization of submerged surfaces in coastal marine waters as determined by 16S ribosomal DNA sequence analysis and fluorescence in situ hybridization. Appl Environ Microbiol. 2002;68:496–504. doi: 10.1128/AEM.68.2.496-504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn E, van der Ploeg JR, Kertesz MA, Leisinger T. Characterization of alpha-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J Biol Chem. 1997;272:23031–23036. doi: 10.1074/jbc.272.37.23031. [DOI] [PubMed] [Google Scholar]
- Eilers H, Pernthaler J, Glöckner FO, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers H, Pernthaler J, Peplies J, Glockner FO, Gerdts G, Amann R. Isolation of novel pelagic bacteria from the German bight and their seasonal contributions to surface picoplankton. Appl Environ Microbiol. 2001;67:5134–5142. doi: 10.1128/AEM.67.11.5134-5142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi S, Amalfitano S, Pernthaler J, Puddu A. Bacterial communities associated with benthic organic matter in headwater stream microhabitats. Environ Microbiol. 2005;7:1633–1640. doi: 10.1111/j.1462-2920.2005.00857.x. [DOI] [PubMed] [Google Scholar]
- Franz B, Gehrke T, Lichtenberg H, Hormes J, Dahl C, Prange A. Unexpected extracellular and intracellular sulfur species during growth of Allochromatium vinosum with reduced sulfur compounds. Microbiol Sgm. 2009;155:2766–2774. doi: 10.1099/mic.0.027904-0. [DOI] [PubMed] [Google Scholar]
- Friedrich CG, Rother D, Bardischewsky F, Quentmeier A, Fischer J. Oxidation of reduced inorganic sulfur compounds by bacteria: Emergence of a common mechanism. Appl Environ Microbiol. 2001;67:2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard NU, Dahl C. Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol. 2009;54:103–200. doi: 10.1016/S0065-2911(08)00002-7. [DOI] [PubMed] [Google Scholar]
- Glöckner FO, Fuchs BM, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JM, Kiene RP, Moran MA. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl Environ Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JM, Moran MA. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Bryant DA, Frigaard N-U. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front Microbiol 2. 2011;116:1–14. doi: 10.3389/fmicb.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm F, Franz B, Dahl C.2008Thiosulfate and sulfur oxidation in purple sulfur bacteriaIn: Dahl C, Friedrich CG (eds).Microbial Sulfur Metabolism Springer: Berlin; 101–116. [Google Scholar]
- Hensen D, Sperling D, Truper HG, Brune DC, Dahl C. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol Microbiol. 2006;62:794–810. doi: 10.1111/j.1365-2958.2006.05408.x. [DOI] [PubMed] [Google Scholar]
- Howard EC, Henriksen JR, Buchan A, Reisch CR, Buergmann H, Welsh R, et al. Bacterial taxa that limit sulfur flux from the ocean. Science. 2006;314:649–652. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- Ishii K, Mussmann M, MacGregor BJ, Amann R. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol Ecol. 2004;50:203–212. doi: 10.1016/j.femsec.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Jansen S, Walpersdorf E, Werner U, Billerbeck M, Bottcher ME, de Beer D. Functioning of intertidal flats inferred from temporal and spatial dynamics of O2, H2S and pH in their surface sediment. Ocean Dynam. 2009;59:317–332. [Google Scholar]
- Jørgensen BB. A thiosulfate shunt in the sulfur cycle of marine sediments. Science. 1990;249:152–154. doi: 10.1126/science.249.4965.152. [DOI] [PubMed] [Google Scholar]
- Kamyshny A, Ferdelman TG. Dynamics of zero-valent sulfur species including polysulfides at seep sites on intertidal sand flats (Wadden Sea, North Sea) Mar Chem. 2010;121:17–26. [Google Scholar]
- Lavik G, Stuhrmann T, Bruchert V, Van der Plas A, Mohrholz V, Lam P, et al. Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature. 2009;457:581–U586. doi: 10.1038/nature07588. [DOI] [PubMed] [Google Scholar]
- Lehmann S, Johnston AWB, Curson ARJ, Todd JD, Cook AM.2012SoeABC, a novel sulfite dehydrogenase in RoseobactersIn: Muyzer G, Stams AJM (eds)EMBO Workshop on Microbial Sulfur Metabolism 15–18 April 2012 (Abstract Book)Sieca Repro, Delft, p 29.
- Lenk S, Arnds J, Zerjatke K, Musat N, Amann R, Mussmann M. Novel groups of Gammaproteobacteria catalyse sulfur oxidation and carbon fixation in a coastal, intertidal sediment. Environ Microbiol. 2011;13:758–774. doi: 10.1111/j.1462-2920.2010.02380.x. [DOI] [PubMed] [Google Scholar]
- Loy A, Duller S, Baranyi C, Mussmann M, Ott J, Sharon I, et al. Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ Microbiol. 2009;11:289–299. doi: 10.1111/j.1462-2920.2008.01760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Imhoff JF, Kuever J. Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria—evolution of the Sox sulfur oxidation enzyme system. Environ Microbiol. 2007;9:2957–2977. doi: 10.1111/j.1462-2920.2007.01407.x. [DOI] [PubMed] [Google Scholar]
- Moran MA, Belas R, Schell MA, Gonzalez JM, Sun F, Sun S, et al. Ecological genomics of marine roseobacters. Appl Environ Microbiol. 2007;73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- Moraru C, Lam P, Fuchs BM, Kuypers MMM, Amann R. GeneFISH—an in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ Microbiol. 2010;12:3057–3073. doi: 10.1111/j.1462-2920.2010.02281.x. [DOI] [PubMed] [Google Scholar]
- Moraru C, Moraru G, Fuchs BM, Amann R. Concepts and software for a rational design of polynucleotide probes. Environ Microbiol Rep. 2011;3:69–78. doi: 10.1111/j.1758-2229.2010.00189.x. [DOI] [PubMed] [Google Scholar]
- Mussmann M, Richter M, Lombardot T, Meyerdierks A, Kuever J, Kube M, et al. Clustered genes related to sulfate respiration in uncultured prokaryotes support the theory of their concomitant horizontal transfer. J Bacteriol. 2005;187:7126–7137. doi: 10.1128/JB.187.20.7126-7137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4:784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- Petri R, Podgorsek L, Imhoff JF. Phylogeny and distribution of the soxB gene among thiosulfate-oxidizing bacteria. FEMS Microbiol Lett. 2001;197:171–178. doi: 10.1111/j.1574-6968.2001.tb10600.x. [DOI] [PubMed] [Google Scholar]
- Rinta-Kanto JM, Burgmann H, Gifford SM, Sun SL, Sharma S, del Valle DA, et al. Analysis of sulfur-related transcription by Roseobacter communities using a taxon-specific functional gene microarray. Environ Microbiol. 2011;13:453–467. doi: 10.1111/j.1462-2920.2010.02350.x. [DOI] [PubMed] [Google Scholar]
- Rother D, Henrich HJ, Quentmeier A, Bardischewsky F, Friedrich CG. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J Bacteriol. 2001;183:4499–4508. doi: 10.1128/JB.183.15.4499-4508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass H, Kopke B, Rutters H, Feuerlein T, Droge S, Cypionka H, et al. Tateyamaria pelophila sp nov., a facultatively anaerobic alphaproteobacterium isolated from tidal-flat sediment, and emended descriptions of the genus Tateyamaria and of Tateyamaria omphalii. Int J Syst Evol Microbiol. 2010;60:1770–1777. doi: 10.1099/ijs.0.013524-0. [DOI] [PubMed] [Google Scholar]
- Sekar R, Fuchs BM, Amann R, Pernthaler J. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol. 2004;70:6210–6219. doi: 10.1128/AEM.70.10.6210-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert SM, Hugler M, Taylor CD, Wirsen CO.2008Sulfur oxidation at deep-sea hydrothermal ventsIn: Dahl C, Friedrich CG (eds).Microbial Sulfur Metabolism Springer: Berlin; 238–258. [Google Scholar]
- Sorokin DY. Sulfitobacter pontiacus, gen-nov, sp. nov.—a new heterotrophic bacterium from the Black Sea, specialized on sulfite oxidation. Microbiology. 1995;64:295–305. [Google Scholar]
- Sorokin DY. Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology. 2003;72:641–653. [PubMed] [Google Scholar]
- Van den Ende FP, Van Gemerden H. Sulfide oxidation under oxygen limitation by a Thiobacillus thioparus isolated from a marine microbial mat. FEMS Microbiol Ecol. 1993;13:69–77. [Google Scholar]
- Wagner-Döbler I, Biebl H. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol. 2006;60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- Weber M, Teeling H, Huang SX, Waldmann J, Kassabgy M, Fuchs BM, et al. Practical application of self-organizing maps to interrelate biodiversity and functional data in NGS-based metagenomics. ISME J. 2011;5:918–928. doi: 10.1038/ismej.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F, Bak F.1992Gram-negative mesophilic sulfate-reducing bacteriaIn: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H (eds).The ProkaryotesVol. 12nd edn.Springer: New York; 3352–3378. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.