Abstract
We present the results of a comparative gene expression analysis of 15 metastases (10 regressing and 5 progressing) obtained from 2 melanoma patients with mixed response following different forms of immunotherapy. Whole genome transcriptional analysis clearly indicate that regression of melanoma metastases is due to an acute immune rejection mediated by the upregulation of genes involved in antigen presentation and interferon mediated response (STAT-1/IRF-1) in all the regressing metastases from both patients. In contrast, progressing metastases showed low transcription levels of genes involved in these pathways. Histological analysis showed T cells and HLA-DR positive infiltrating cells in the regressing but not in the progressing metastases. Quantitative expression analysis of HLA-A, B and C genes on microdisected tumoral regions indicate higher HLA expression in regressing than in progressing metastases. The molecular signature obtained in melanoma rejection appeared to be similar to that observed in other forms of immune-mediated tissue-specific rejection such as allograft, pathogen clearance, graft versus host or autoimmune disease, supporting the immunological constant of rejection. We favor the idea that the major factor determining the success or failure of immunotherapy is the nature of HLA Class I alterations in tumor cells and not the type of immunotherapy used. If the molecular alteration is reversible by the immunotherapy, the HLA expression will be upregulated and the lesion will be recognized and rejected. In contrast, if the defect is structural the MHC Class I expression will remain unchanged and the lesion will progress.
Keywords: melanoma, metastasis, immunotherapy, rejection, HLA, IRF-1
Different protocols of melanoma immunotherapy have been used during the last decade, including interferon alpha 2b, interleukin 2 and 12 or more specific ones, such as autologous vaccination with naked peptides, dendritic cells, or plasmid DNA, or adoptive transfer of tumor-specific cytotoxic T cells.1–3 Active specific immunization with tumor antigen has been shown to generate immune T cells capable of recognizing antigenic peptides presented on tumor cells.3 However, successful activation of tumor-specific T cells does not correlate with tumor response and only a small percentage of patients demonstrate objective tumor regression.4,5
It is already known that melanoma and other tumors develop sophisticated immune evasion mechanisms6 that can explain the failure of current immunotherapies. One important tumor escape mechanism is represented by the loss or downregulation of major histocompatibility complex (MHC) Class I antigens in tumor cells that have been frequently observed in a variety of human malignancies derived from HLA Class I positive epithelia.7,8
A small proportion of melanoma patients develop a mix response after therapy, when some metastatic lesions regress, while other progress in the same patient.9,10 These patients are extremely valuable since they allows us to consider solely the tumor’s determining factors in immune responsiveness excluding variation in the genetic background of different patients or external variables affecting the potency of identical treatments provided at different times.11 The information obtained can help to explain the molecular basis of tumor immune rejection and the tumor mechanism of immune escape.
Our laboratory has had the opportunity to analyze two of such mixed-response melanoma patients after different immunotherapy treatments. In particular, we obtained five metastases (three progressing and two regressing) from Patient 1 (M1)12 and ten metastases (two progressing and eight regressing) from Patient 2 (M2).13 To date, no comparative analysis of simultaneous lesions with different clinical evolution has been performed at the global transcript level.
We present in this article the whole genome transcriptional analysis of various lesions obtained from two melanoma patients and related it with HLA expression data. Our results clearly indicate for the first time that the rejection pattern in melanoma lesions from both patients is the same and include activation of genes involved in antigen presentation, immune rejection and interferon pathways. In contrast, these particular groups of genes are downregulated in progressing melanoma lesions. The final consequence of this activation is a clear difference in HLA Class I cell surface expression that could explain why some lesions are rejected and others are not.
Material and Methods
Patients, clinical protocols and tumor specimens
We studied 15 metastases from two patient treated with immunotherapy.14 Both patients showed a mixed response to treatment: some lesions disappeared, others reduced the size, some metastases grew bigger and some new lesions appeared. Six subcutaneous metastases were removed from Patient 1 after 35 days with M-VAX treatment and sent to our laboratory.12 In this study one of the regressing metastases was not analyzed analyzed due to lack of material. Two of the remaining metastases (designated M1-VAX.R1 and M1-VAX.R2) were regressing at the time of excision, whereas the other three were progressing (designated M1-VAX.P1, M1-VAX.P2, M1-VAX.P3). We obtained ten metastases from Patient 2,13 five subcutaneous lesions were obtained after 10 months with alpha-INF-2b treatment and five lymph node metastases were obtained 10 days after finishing a 6-week M-VAX vaccination treatment. We obtained one progressing and four regressing lesions after each treatment from this patient. Samples obtained after IFNα-2b treatment were designated as M2-INF.R1, M2-INF.R2, M2-INF.R3, M2-INF.R4 and M2-INF.P1. Samples obtained after M-VAX were named M2-VAX.R1, M2-VAX.R2, M2-VAX.R3, M2-VAX.R4 and M2-VAX.P1. The detailed clinical history of the patients was previously described.12,13 Samples were obtained from the Department of Dermatology, Hospital Virgen de la Macarena, Sevilla, Spain. Informed consents and approval of the research protocol by the institutional review board were obtained. All metastases were removed from the patients at the same time after immunotherapy and snap-frozen in liquid nitrogen-cooled iso-pentane. Tumor response was established according to PET and CT scans.
Tumor cells microdissection
Cryopreserved 6-μm-thick tissue sections were fixed in 70% ethanol and stained with a 0.05% wt/vol solution of toluidine blue and microdissected using a Laser micromanipulator (PALM Microlaser Systems, ZEISS). Microdissected fragments were collected in PALM Adhesive Caps.
Total RNA isolation and amplification
Total RNA (tRNA) from whole sections and micro-dissected frozen tumor cells were isolated using the miRNeasy Mini kit (QIAGEN, Westburg, Leusden, The Netherlands). The quality of tRNA was tested with Agilent Bioanalyzer 2000 (Agilent Technologies, Santa Clara, USA). For gene expression studies, tRNA obtained from the whole frozen section was amplified into antisense RNA (aRNA) as previously described.15 Reference for human arrays consisted of pooled PBMCs from four normal donors. Human reference total RNA was also amplified into antisense RNA.
Microarray performance and data processing
Array quality was documented as previously described.16 Both reference and test aRNA were directly labeled using ULS aRNA Fluorescent Labeling kit (Kreatech) with Cy3 for reference and Cy5 for test samples and cohybridized at 45 °C for 18 hr into 36k whole genome human array.15 After incubation, the arrays were washed and stained in the fluidics station using GeneChip® Hybridization, Wash, and Stain Kit (Affymetrix, Santa Clara, USA). The data were uploaded to the mAdb databank http://nciarray.nci.nih.gov, further analyzed using BRBArray-Tools developed by the Biometric Research Branch, National Cancer Institute http://linus.nci.nih.gov/BRBArrayTools.html17 and clustered by TreeView software.18,19 Unsupervised analysis was used for class discovery using the stanford cluster program applying standard gene filtering parameters (80% gene presence across all experiments and at least 1.5-fold ratio change) and Treeview program for visualization. Average gene ratios were normalized and displayed according to uncentered correlation algorithms. Class comparison was performed using parametric unpaired Student’s t test to identify differentially expressed genes among progressing and regressing tumors. Validation by quantitative polymerase chain reaction (q-PCR) of the gene sets were not performed due to the fact that we have previously shown the present method for RNA amplification is robust and yields results comparable to those obtained by qPCR.16 Moreover, salient gene products were validated at the protein expression level by immunostaining. Gene function interpretation was based on GeneOntology software, while pathway analysis was based on Ingenuity Pathways Analysis (IPA) software. Primary microarray data are available in NCBI’s Gene Expression Omnibus public database (microarray platform, GPL7088; microarray data, GSE26383).
Quantitative RT-PCR of isolated tumor cells
RT products from microdissected metastases were analyzed for the expression of HLA-A, B and C loci by quantitative PCR. To control for variations in the amounts of RNA, G6PDH and HPRT were tested as a housekeeping gene. All PCR reactions were performed in a Light Cycler instrument using LC-FastStart DNA master probes kit and LC-FastStart DNA SYBR Green I Kit (Roche Diagnostics, Manheim, Germany). For G6PDH and HPRT we used commercial kits (Roche Diagnostics, Manheim, Germany). Amplification reactions of HLA loci were described previously.13
Immunohistological analysis
Immunohistological analysis was performed using Novolink® polymer detection system kit (Novocastra, Newcastle, United Kingdom). For HLA Class II staining we used GRB-1 mAb (anti-HLA-DR). Infiltration pattern was studied with, OKT8 mAb (anti CD8) OKT3 mAb (anti CD3) (hybridomes obtained from ATTC, Manassas, VA) and RPA-T4 mAb (anti CD4), (BD Biosciences Pharmingen, Belgium).
Results
Regressing metastases display upregulation of antigen presentation and immune rejection patterns
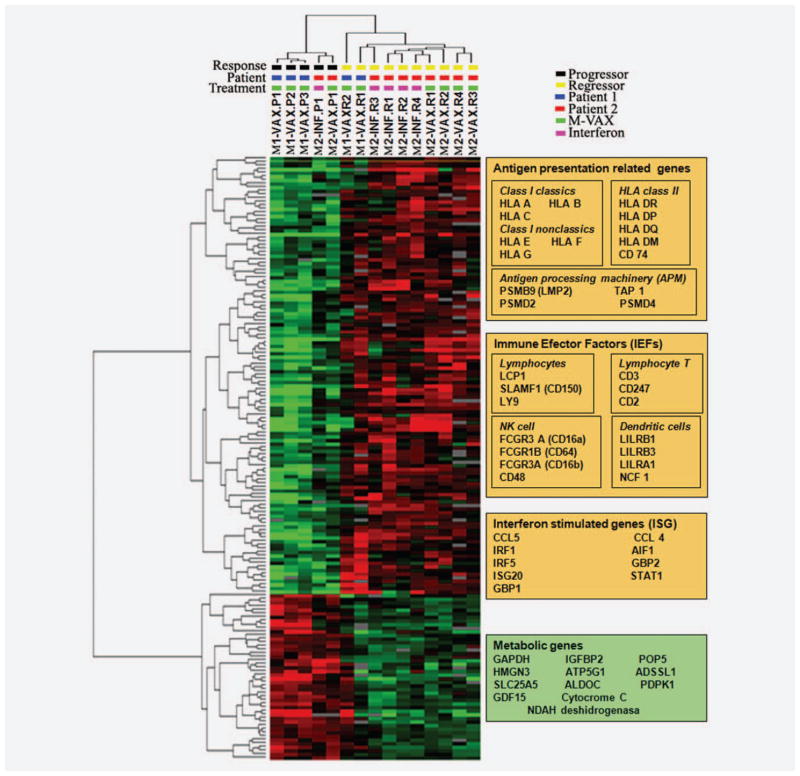
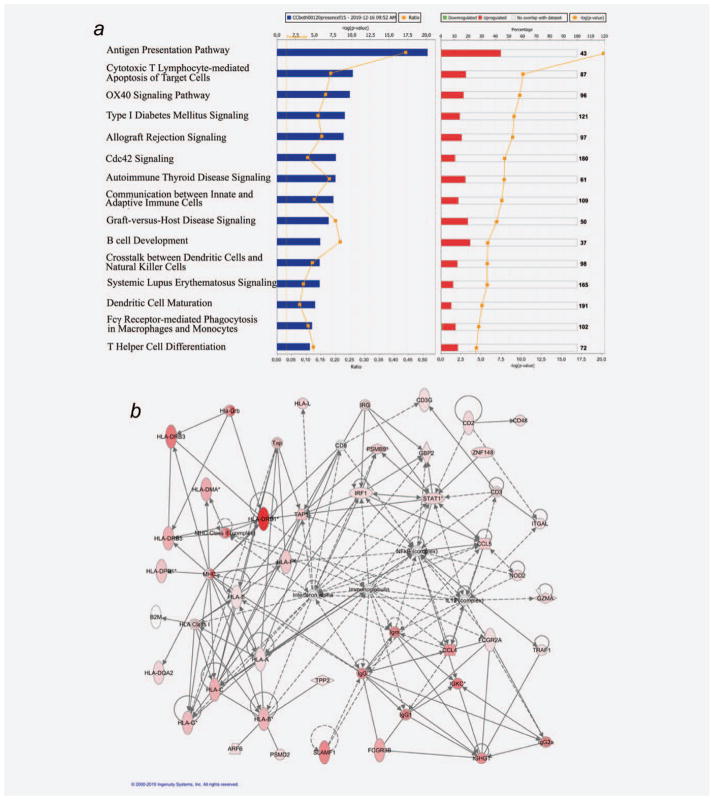
Unsupervised analysis clustering lesions according to its general profile expression showed that metastases are first grouped according to the patient and second according to the response (Supporting Information Fig. S1). To understand the different response between progressing and regressing metastases from both patients we perform a high stringency class comparison between the progressing and regressing metastases from both patients using a nominal cut-off significance level of p ≤ 0.001. This analysis identified 167 genes differentially expressed between regressing and non regressing lesions—most of which were associated with antigen presentation function and acute immune response (Supporting Information Table S1; Fig. 1). To gain a more comprehensive portrait of the transcriptional signatures associated with rejection, we performed gene enrichment analysis comparing regressing and nonregressing lesions at a nominal significance cut-off p-value ≤ 0.01. This analysis identified 540 transcripts differentially expressed between the two phenotypes (Fig. 1). Expression pattern in regressing metastases is related to a release of interferon gamma from infiltrating T lymphocytes. The upregulated genes (IRF-1, STAT-1 AIF-1, CCL5, GBP1, GBP2…) are mainly involved in Type II interferon response. We observed this pattern in metastases obtained after MVAX and even after Type I interferon alpha 2b treatment. The enriched data set (540 transcripts) was analyzed by ingenuity pathway analysis (IPA) software to obtain the molecular pathways differentially expressed in regressing versus progressing melanoma lesions. The top 15 most significantly altered pathways found among the ~400 included in IPA were upregulated in regressing metastases and were related to mechanisms associated with immune-mediated tissue-specific destruction (Fig. 2a). These pathways include cytotoxic T-lymphocyte apoptosis of target cells, allograft rejection signaling and different autoimmune disorders. Figure 2b shows the top ranking self-organizing network by IPA based on this data set. Interaction network clearly demonstrated upregulation of IRF and antigen presentation networks in regressing metastases. Interesting we did not find any downregulated gene in this network, demonstrating the consistency of the results.
Figure 1.
expression pattern clustering of melanoma metastases obtained from comparing regressing versus progressing metastases (p = 0.001). Genes upregulated in regressing metastases are represented in red, genes overexpressed in progressing metastases are represented in green. Most of the genes upregulated in regressing metastases belong to the immune response. Key genes involved in immunological rejection (antigen presentation, immune effectors factors and interferon stimulates genes) are included in the orange box, metabolism genes upregulated in progressing metastases are included in the green box.
Figure 2.
Class comparison between progressing and regressing metastases in both patients. (a) Top 15 first canonical pathways ranking according to significance level (Fisher exact test −log p-value) based on the 540 identified genes using gene enrichment (p = 0.01) analysis. Blue bar represent the p-value of the comparison and yellow line represents the change ratio of the pathway. Red bar represent the percentage of genes upregulated inside each pathway in regressing lesions meanwhile green bars represent downregulated ones, yellow line in this graphic represents the p-value of the comparison. All the most significant pathways belong to the immune response and are upregulated in regressing lesions. (b) Antigen presentation plus IRF-1 self organizing network according to IPA analysis based on the 540 identified genes derived from the low stringency analysis. Upregulated genes in regressing metastases are represented in red. Fold-change of the gene is represented by color intensity. The figure shows a consistent activation of the immune genes inside regressing lesions. The analysis did not identify any downregulated gene in the network.
We found that progressing metastases have upregulate genes involve in cellular metabolism (Fig. 1). This pattern is due to the cellular growth of tumor cells. We did not find markers of inhibitory immune cells, as regulatory T cells (Tregs) or myeloid derived suppressor cells (MDSC), in progressing metastases (Supporting Information Table S1). The only escape mechanism detected by whole transcriptional analysis is the downregulation of HLA Class I molecules.
To determine the major role of the immune rejection against the regressing lesions in each patient separately, we compared progressing versus regressing lesions inside each patient. The results are showed in Supporting Information Figure S2. Both patients showed parallel differences between the two groups of metastases. Most significantly altered IPA canonical pathways were similar in both patients and maintain the same pathways observed when we compared all metastases together (Supporting Information Figs. S2 a and c), including antigen presentation, cytotoxic T-lymphocyte apoptosis of target cells, allograft rejection signaling and different autoimmune disorders. Antigen presentation, immune infiltration and IRF activation networks were mostly involved in discriminating responding from non responding lesions in spite of the different patient genetic background and different treatments (Supporting Information Figs. S2 b and d).
Chromosomal distribution of genes differentially expressed between progressing and regressing metastases demonstrated a great enrichment for transcripts located in Chromosome 6 (Supporting Information Fig. S3), where antigen presentation and many inflammation genes are located.
The results clearly showed an acute immune response in regressing metastases, with most transcripts associated with immune rejection response being upregulated. These included activation of antigen presentation, interferon stimulated genes (ISGs), immune effector function related genes (IEFs) and several chemokines (Fig. 2 and Supporting Information Fig. S2, network analysis).
To investigate the differences between regressing metastases after interferon and autologous vaccination we compared regressing metastases from Patient 2 after interferon versus those after M-VAX. IPA canonical pathways analyses using the significant genes from the high stringency comparison showed a borderline significant upregulation of antigen presentation in M-VAX treated regressing lesions. We found no other significant different pathways between regressing lesions after interferon and M-VAX (Supporting Information Table S1; Fig. S4). Therefore, there were not major differences in the regressing pathway response after interferon and M-VAX treatment.
Immunohistological validation of immune infiltrates shows a large T-cell population inside regressing tumors
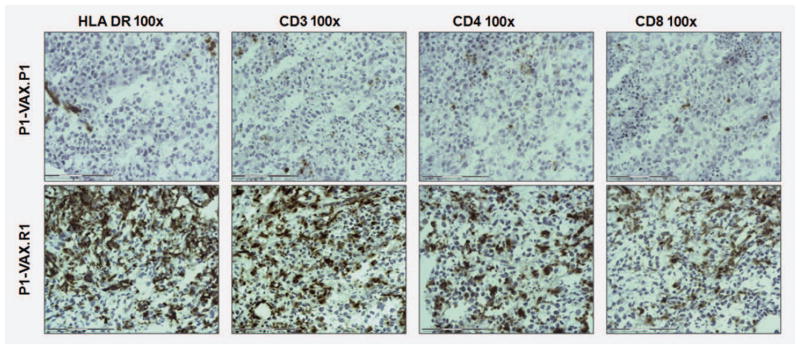
To confirm the enhancement of immune activity in regressing lesions in both patients, we studied the immune infiltrate in cryopreserved tissue sections. Immunostaining with antibodies against HLA Class II DR indicated that both progressing and regressing tumor cells were negative for HLA Class II expression, while the immune infiltrate showed strong positive staining (Fig. 3). There was a dramatic difference in the number of infiltrating cells between phenotypes; progressing lesions showed low number of infiltrating cells, and vice versa for regressing lesions. Differences in HLA Class II expression documented by microarray transcriptional analysis of bulk mRNA were not associated with enhanced tumor cell expression, but rather with the enrichment of the tumor environment with immune cells. The composition of the infiltrate in regressing metastases consisted mostly in CD3+ T cells (80% of CD4+ and 20% of CD8 + cells), (Fig. 3).
Figure 3.
Immunoperoxidase staining of frozen sections of metastases: (a) tumor cells from both progressing and regressing metastases do not express HLA-DR. The staining shows a high degree of HLA-DR positive cellular infiltration in regressing metastasis; in contrast, the progressing lesions have very little infiltration. (b) CD3 staining corresponds to T lymphocyte infiltration (c and d). The number of CD4 positive cells in the infiltrate is higher than the number of CD8 cells.
Microdissected isolated tumor cells from progressing metastases showed a reduced expression of HLA Class I A, B and C loci
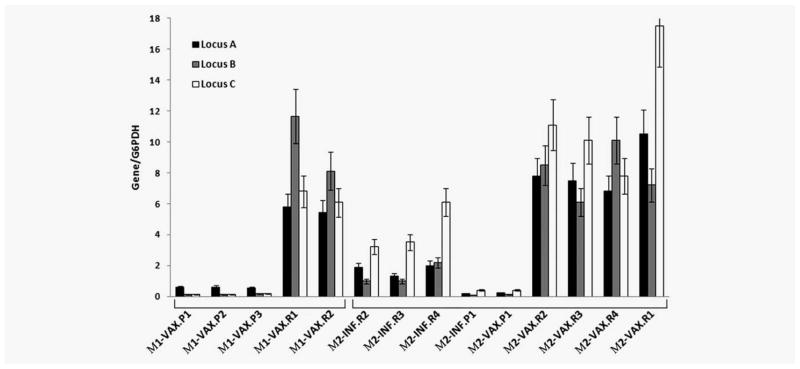
For the analysis of tumor HLA Class I expression we isolated cancer cells from frozen tumor samples using laser microdissection to restrict the analysis to tumor cells only and avoid sample contamination with stroma and infiltrating cells. Quantitative RT-PCR analysis of isolated tumor cells obtained from progressing metastases in Patient 1 demonstrated low mRNA levels for HLA-A locus transcripts with residual transcription of HLA-B and -C loci (Fig. 4). In contrast, cancer cells from regressing lesions from the same patient showed high transcription levels of HLA-A, HLA-B and HLA-C. Similar correlation we have previously reported in Patient 2,13 in which high mRNA levels of all three HLA loci were observed in regressing metastases and only residual levels in progressing lesions. Here we present data on both Patients 1 and 2 to better illustrate the similarity of the observed correlation between low HLA Class I transcriptional expression level and the tendency of the lesions to progress. In Patient 2 the HLA levels were higher in post M-VAX regressing lesions compared to the lesion after IFN therapy. These data are in agreement with the array results. The mRNA levels were also consistent with immunohistochemical staining of HLA Class I molecules described previously.12,13
Figure 4.
Normalized mRNA levels in tumor cells isolated by laser microdissection from melanoma patients. The progressing metastases (P) obtained from both patients showed low residual levels of HLA-A, -B and -C loci. In contrast, the regressing tumor cells (R) displayed a high transcription of the tree HLA loci. Regressing metastases obtained after M-VAX treatment in patient M2 showed higher HLA loci expression than those obtained after interferon α 2b.
Discussion
We have observed that the differential activation of genes involved in acute inflammatory processes (Fig. 2) between regressing and progressing melanoma lesions obtained after immunotherapy are dramatic. Among 30,000 studied genes in regressing metastases we mostly detected upregulation of the genes related to the immune rejection with a great enrichment for transcripts located in Chromosome 6, where antigen presentation and many inflammation related genes are located. We propose that these differences are due to efficient tumor recognition and elimination of regressing metastases by the activated immune system “triggered” by HLA Class I upregulation. However, the systemic immunotherapy cannot induce the regression of all lesions. Because of the major differences in HLA expression, we propose that progressing lesions are not recognized by immune cells due to irreversible alterations in HLA Class I.
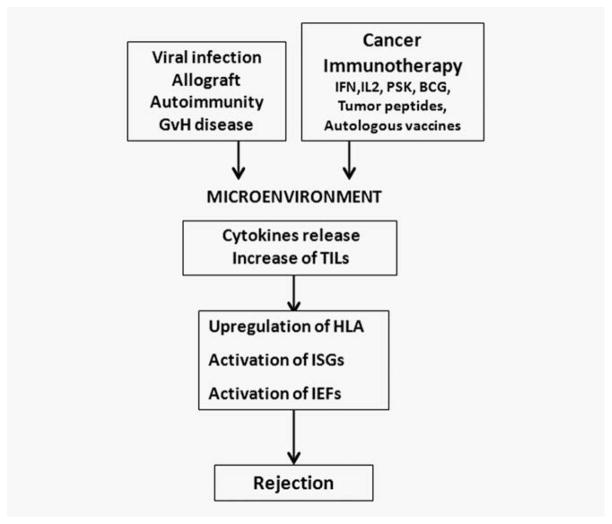
The transcriptional pattern expressed by regressing metastases was quite similar, independently of the type of immunotherapy used. This suggests that the mechanisms leading to tumor rejection converge in a unique pathway (Fig. 5). In this context, the expression pattern of regressing lesions was also similar to that observed after imiquimod or IL-2 administration in basal cell carcinoma,20,21 allograft rejection,22 graft versus host disease, autoimmunity or acute infection resulting in clearance of pathogen.23 This has been postulated as the existence of an immunologic constant of rejection.24,25 The molecular mechanisms used to eliminate all of them converge in a final pathway consisting of the expression of antigen presentation, ISGs and IEFs. The expression pattern that we have found is ultimately an interferon Type II signature most probably mediated by a release of interferon-gamma by tumor infiltrating T cells, even when the melanoma patient was treated with interferon alpha2b.
Figure 5.
A common pathway of T cell mediated rejection is showed in different pathological entities.
Immunohistochemistry staining of tumors showed the correlation between tumor infiltration and the expression of genomic transcripts implicated in tumor rejection (ISGs and IEFs). Several studies have described the presence of tumor-antigen specific cytotoxic T lymphocytes in the tumor microenvironment.23 Nevertheless, these cells are unable to eliminate tumor cells.23 In this study, immune cells potentially induced or activated by the immunotherapy can recognize and eliminate lesions that previously were low immunogenic and invisible for the immune system. However, this immunomodulation occurred only in a subset of metastases, while another group of lesions continued to grow in spite of systemic activation of immune surveillance mechanisms. Therefore, intratumoral factors are most likely to be responsible for the failure of the immune response within the progressing metastases. Genome transcriptional analysis indicates that antigen presentation is the most critical pathway to determine tumor regression. Previous studies12,13 and microdissected tumor from both patients confirm that specific HLA-A, B and C high expression is associated with immune infiltrate and tumor regression, meanwhile a low or negative HLA Class I transcription correlate with tumor progression (Table 1). Interestingly, we did not find differences in markers corresponding to regulatory T cells (Tregs) or myeloid derived suppressor cells (MDSC) when both types of metastases were compared.
Table 1.
Correlation of HLA class I surface expression with tumor response after immunotherapy
| Patient Treatment Lesions | M-1
|
M-2
|
||||
|---|---|---|---|---|---|---|
| Autologous vaccination + BCG
|
Interferon α 2b
|
Autologous vaccination + BCG
|
||||
| Regressing | Progressing | Regressing | Progressing | Regressing | Progressing | |
| HLA expression | +++ | +/− | +++ | +/− | +++ | +/− |
|
| ||||||
| Upregulated pathways | Antigen presentation | Metabolic pathways | Antigen presentation | Metabolic pathways | Antigen presentation | Metabolic pathways |
|
| ||||||
| ISGs | ISGs | ISGs | ||||
|
| ||||||
| IEFs | IEFs | IEFs | ||||
|
| ||||||
| Rejection | Yes | No | Yes | No | Yes | No |
Our observations suggest that immunotherapy promotes a modification of tumor microenvironment, leading to a release of immune stimulating factors by immune infiltrating cells. This immune stimulation will lead to upregulation of HLA expression in tumor cells with reversible alterations of HLA Class I expression (“Soft” lesions) and, subsequently, these tumor cells will be recognized and destroyed by the antigen-specific T cells. These T cells, as they recognize tumor cells, produce more proinflammatory factors such as IFN-γ, IL-2, TNF-α and GM-CSF.26 This, in turn, triggers a positive and self perpetuating feedback between tumor and immune cells until tumor rejection occurs. In contrast, if cancer cells bear irreversible defects in HLA Class I genes (“Hard” lesions), antigen presentation remains defective after immunotherapy impairing the amplification of the immune response in situ and promoting their escape from immune recognition.26
Our prediction is that “soft” and “hard” HLA Class I tumor lesions will coexist during the natural history of tumor development. However after immunotherapy tumor cells with “soft” lesion will upregulate antigen presentation and can be rejected. In contrast, those tumor cells bearing “hard” and irreversible genetic defects will prevail and progress to kill the host.27 This clearing would lead to the selection of a tumor variant with additional alteration of HLA Class I expression.28,29
To the best of our knowledge, this is the first comparison of the transcriptional profile of melanoma metastases with different responses after immunotherapy in the same patient, showing the “molecular image” of tumor rejection. Our results strongly suggest that the genetic makeup of individual tumor cells is a major factor determining immune responsiveness. In particular the capacity to modulate tumor HLA Class I expression.
Acknowledgments
Grant sponsor: Fondo de Investigaciones Sanitarias (FIS); Grant numbers: CP03/0111, PI 08/1265; Grant sponsor: Proyecto de investigacion MEC I + D; Grant numbers: SAF 2007-63262, SAF 2010-20273; Grant sponsor: Red Genomica del Cancer; Grant number: RETIC RD 06/020; Grant sponsor: Plan Andaluz de Investigacion; Grant numbers: CTS 143, CTS-695; Grant sponsor: Proyectos de Excelencia; Grant numbers: CTS-03952, CVI-04740; Grant sponsor: Integrated European Cancer Immunotherapy; Grant numbers: OJ2004/C158, 518234
The authors thank Jingo Zhao for technical assistance and Dr. Harvey Alter for his helpful advice.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Terando AM, Faries MB, Morton DL. Vaccine therapy for melanoma: current status and future directions. Vaccine. 2007;25:4–16. doi: 10.1016/j.vaccine.2007.06.033. [DOI] [PubMed] [Google Scholar]
- 2.Kirkwood JM, Tarhini AA, Panelli MC, Moschos SJ, Zarour HM, Butterfield LH, Gogas HJ. Next generation of immunotherapy for melanoma. J Clin Oncol. 2008;26:3445–55. doi: 10.1200/JCO.2007.14.6423. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg SA, Dudley ME. Adoptive cell therapy for the treatment of patients with metastatic melanoma. Curr Opin Immunol. 2009;21:233–40. doi: 10.1016/j.coi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geldmacher A, Freier A, Losch FO, Walden P. Therapeutic vaccination for cancer immunotherapy: antigen selection and clinical responses. Hum Vaccine. 2011;7S:115–9. doi: 10.4161/hv.7.0.14573. [DOI] [PubMed] [Google Scholar]
- 5.Klebanoff CA, Acquavella N, Yu Z, Restifo NP. Therapeutic cancer vaccines: are we there yet? Immunol Rev. 2011;239:27–44. doi: 10.1111/j.1600-065X.2010.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Adv Immunol. 2006;90:51–81. doi: 10.1016/S0065-2776(06)90002-9. [DOI] [PubMed] [Google Scholar]
- 7.Garrido F, Algarra I. MHC antigens and tumour escape from immune surveillance. Adv Cancer Res. 2001;83:117–58. doi: 10.1016/s0065-230x(01)83005-0. [DOI] [PubMed] [Google Scholar]
- 8.Garrido F, Ruiz-Cabello F, Cabrera T, Pérez-Villar JJ, López-Botet M, Duggan-Keen M, Stern PL. Implications for immunosurveillance of altered HLA class I phenotypes in human tumours. Immunol Today. 1997;18:89–95. doi: 10.1016/s0167-5699(96)10075-x. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10:909–15. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruit WH, van Ojik HH, Brichard VG, Escudier B, Dorval T, Dréno B, Patel P, van Baren N, Avril MF, Piperno S, Khammari A, Stas M, et al. Phase 1/2 study of subcutaneous and intradermal immunization with a recombinant MAGE-3 protein in patients with detectable metastatic melanoma. Int J Cancer. 2005;117:596–604. doi: 10.1002/ijc.21264. [DOI] [PubMed] [Google Scholar]
- 11.Yuan J, Page DB, Ku GY, Li Y, Mu Z, Ariyan C, Gallardo HF, Roman RA, Heine AI, Terzulli SL, Ritter E, Gnjatic S, et al. Correlation of clinical and immunological data in a metastatic melanoma patient with heterogeneous tumor responses to ipilimumab therapy. Cancer Immunol. 2010;10:1. [PMC free article] [PubMed] [Google Scholar]
- 12.Cabrera T, Lara E, Romero JM, Maleno I, Real LM, Ruiz-Cabello F, Valero P, Camacho FM, Garrido F. HLA class I expression in metastatic melanoma correlates with tumor development during autologous vaccination. Cancer Immunol Immunother. 2007;56:709–17. doi: 10.1007/s00262-006-0226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carretero R, Romero JM, Ruiz-Cabello F, Maleno I, Rodriguez F, Camacho FM, Real LM, Garrido F, Cabrera T. Analysis of HLA class I expression in progressing and regressing metastatic melanoma lesions after immunotherapy. Immunogenetics. 2008;60:439–47. doi: 10.1007/s00251-008-0303-5. [DOI] [PubMed] [Google Scholar]
- 14.Berd D. M-VAX: an autologous, hapten-modified vaccine for human cancer. Expert Rev Vaccines. 2004;3:521–7. doi: 10.1586/14760584.3.5.521. [DOI] [PubMed] [Google Scholar]
- 15.Wang E, Miller L, Ohnmacht GA, Liu E, Marincola FM. High fidelity mRNA amplification for gene profiling using cDNA microarrays. Nat Biotech. 2000;17:457–9. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 16.Jin P, Zhao Y, Ngalame Y, Panelli MC, Nagorsen D, Monsurró V, Smith K, Hu N, Su H, Taylor PR, Marincola FM, Wang E. Selection and validation of endogenous reference genes using a high throughput approach. BMC Genomics. 2004;5:55. doi: 10.1186/1471-2164-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Worschech A, Kmieciak M, Knutson KL, Bear HD, Szalay AA, Wang E, Marincola FM, Manjili MH. Signatures associated with rejection or recurrence in HER-2/neu-positive mammary tumors. Cancer Res. 2008;68:2436–46. doi: 10.1158/0008-5472.CAN-07-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-array tools. Cancer Inform. 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panelli MC, Stashower ME, Slade HB, Smith K, Norwood C, Abati A, Fetsch P, Filie A, Walters SA, Astry C, Aricó E, Zhao Y, et al. Sequential gene profiling of basal cell carcinomas treated with imiquimod in a placebo-controlled study defines the requirements for tissue rejection. Genome Biol. 2007;8:R8. doi: 10.1186/gb-2007-8-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panelli MC, Wang E, Phan G, Puhlmann M, Miller L, Ohnmacht GA, Klein HG, Marincola FM. Gene-expression profiling of the response of peripheral blood mononuclear cells and melanoma metastases to systemic IL-2 administration. Genome Biol. 2002;3:1–17. doi: 10.1186/gb-2002-3-7-research0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saint-Mezard P, Berthier CC, Zhang H, Hertig A, Kaiser S, Schumacher M, Wieczorek G, Bigaud M, Kehren J, Rondeau E, Raulf F, Marti HP. Analysis of independent microarray datasets of renal biopsies identifies a robust transcript signature of acute allograft rejection. Transpl Int. 2009;22:293–302. doi: 10.1111/j.1432-2277.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- 23.Pilla L, Patuzzo R, Rivoltini L, Maio M, Pennacchioli E, Lamaj E, Maurichi A, Massarut S, Marchianò A, Santantonio C, Tosi D, Arienti F, et al. A phase II trial of vaccination with autologous, tumor-derived heat-shock protein peptide complexes Gp96, in combination with GM-CSF and interferon-alpha in metastatic melanoma patients. Cancer Immunol Immunother. 2006;55:958–68. doi: 10.1007/s00262-005-0084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marincola FM, Wang E, editors. Immunologic signatures of rejection. New York: Springer-Verlag; 2010. [Google Scholar]
- 25.Wang E, Worschech A, Marincola FM. The immunologic constant of rejection. Trends Immunol. 2008;29:256–62. doi: 10.1016/j.it.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Aptsiauri N, Carretero R, Garcia-Lora A, Real LM, Cabrera T, Garrido F. Regressing and progressing metastatic lesions: resistance to immunotherapy is predetermined by irreversible HLA class I antigen alterations. Cancer Immunol Immunother. 2008;57:1227–33. doi: 10.1007/s00262-008-0532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrido F, Cabrera T, Aptsiauri N. Hard and soft lesions underlying the HLA class I alterations in cancer cells: implications for immunotherapy. Int J Cancer. 2010;127:249–56. doi: 10.1002/ijc.25270. [DOI] [PubMed] [Google Scholar]
- 28.Garrido F, Cabrera T, Concha A, Glew S, Ruiz-Cabello F, Stern P. Natural history of HLA expression during tumor development. Immunol Today. 1993;14:491–9. doi: 10.1016/0167-5699(93)90264-L. [DOI] [PubMed] [Google Scholar]
- 29.Carretero R, Cabrera T, Gil H, Saenz-Lopez P, Maleno I, Aptsiauri N, Cozar JM, Garrido F. Bacillus Calmette-Guerin immunotherapy of bladder câncer induces selection of human leukocyte antigen class I-deficient tumor cells. Int J Cancer. 2011;129:839–46. doi: 10.1002/ijc.25733. [DOI] [PubMed] [Google Scholar]