Abstract
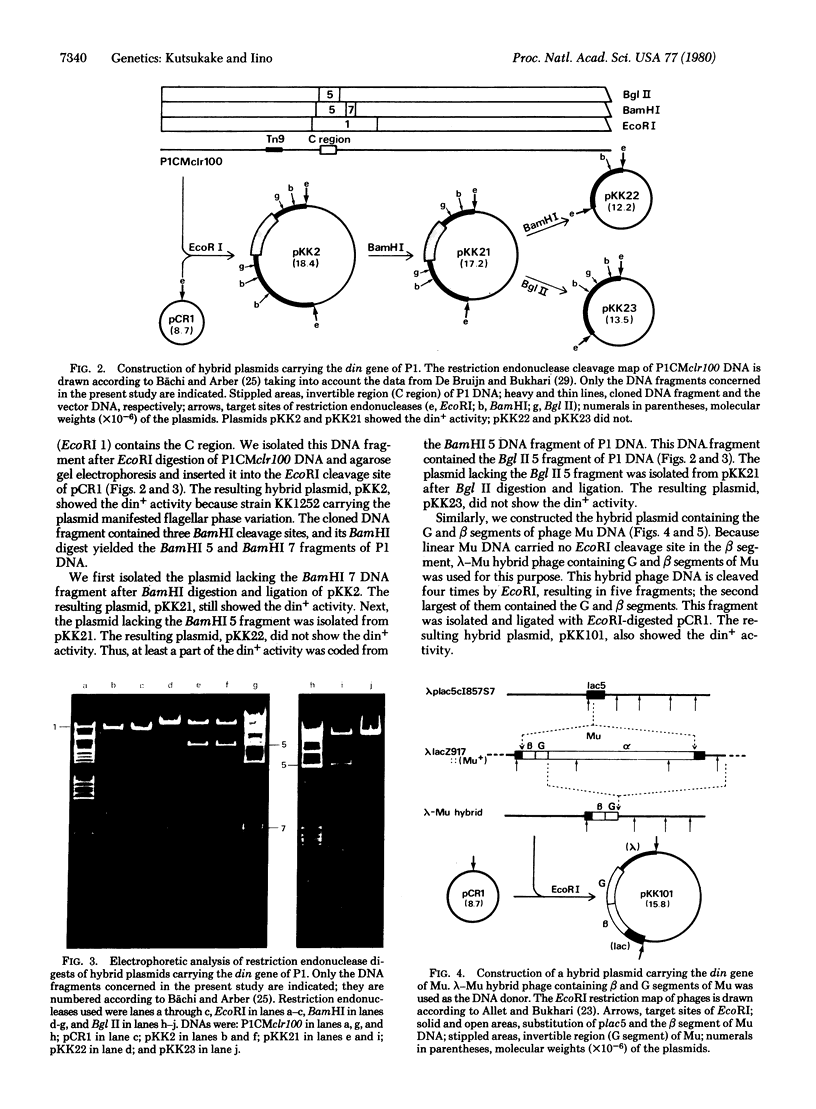
Prophages P1 and Mu produces a trans-acting factor possessing the din+ activity which catalyzes the inversion of the specific DNA segment responsible for flagellar phase variation of Salmonella, din mutants were isolated from PICMclr100 phage by selecting phages that did not suppress the yh2 mutation of Salmonella in prophage state. No inversion loop structure was detected among DNA forms arising after denaturation and rehybridization of DNAs extracted from the din mutants. The DNA fragment containing C region of P1 was cloned on a plasmid vector, pCR1. The resulting hybrid plasmid, pKK2, was shown to possess the din+ activity: the vh2 mutant of Salmonella harboring the plasmid changed the flagellar phase. From analysis of the plasmid by use of BamHI and Bgl II, the din gene specifying the din+ activity was located near or within the C region of P1. It is highly plausible that the din gene of P1 was also involved in the inversion of the C region. Similarly, the DNA fragment containing the G and beta segments of Mu was cloned on pCR1. The resulting hybrid plasmid, pII101, also possessed the din+ activity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I., Allet B. Plaque-forming lambda-Mu hybrids. Virology. 1975 Jan;63(1):30–39. doi: 10.1016/0042-6822(75)90367-0. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I., Ambrosio L. The invertible segment of bacteriophage Mu DNA determines the adsorption properties of Mu particles. Nature. 1978 Feb 9;271(5645):575–577. doi: 10.1038/271575a0. [DOI] [PubMed] [Google Scholar]
- Bächi B., Arber W. Physical mapping of BglII, BamHI, EcoRI, HindIII and PstI restriction fragments of bacteriophage P1 DNA. Mol Gen Genet. 1977 Jun 24;153(3):311–324. doi: 10.1007/BF00431596. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Bukhari A. I. The invertible DNA segments of coliphages Mu and P1 are identical. Virology. 1976 Oct 1;74(1):242–248. doi: 10.1016/0042-6822(76)90148-3. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Kahmann R., Kamp D. Electron microscopic characterization of DNAs of non-defective deletion mutants of bacteriophage Mu. J Mol Biol. 1977 Jul 15;113(4):591–609. doi: 10.1016/0022-2836(77)90224-8. [DOI] [PubMed] [Google Scholar]
- Covey C., Richardson D., Carbon J. A method for the deletion of restriction sites in bacterial plasmid deoxyribonucleic acid. Mol Gen Genet. 1976 May 7;145(2):155–158. doi: 10.1007/BF00269587. [DOI] [PubMed] [Google Scholar]
- De Bruijn F. J., Bukhari A. I. Analysis of transposable elements inserted in the genomes of bacteriophages Mu and P1. Gene. 1978 Jul;3(4):315–331. doi: 10.1016/0378-1119(78)90041-0. [DOI] [PubMed] [Google Scholar]
- Enomoto M., Stocker B. A. Integration, at hag or elsewhere, of H2 (phase-2 flagellin) genes transduced from Salmonella to Escherichia coli. Genetics. 1975 Dec;81(4):595–614. doi: 10.1093/genetics/81.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H., Yamaguchi S., Iino T. Studies on H-O variants in Salmonella in relation to phase variation. J Gen Microbiol. 1973 May;76(1):127–134. doi: 10.1099/00221287-76-1-127. [DOI] [PubMed] [Google Scholar]
- Howe M. M., Bade E. G. Molecular biology of bacteriophage mu. Science. 1975 Nov 14;190(4215):624–632. doi: 10.1126/science.1103291. [DOI] [PubMed] [Google Scholar]
- Howe M. M., Schumm J. W., Taylor A. L. The S and U genes of bacteriophage mu are located in the invertible G segment of mu DNA. Virology. 1979 Jan 15;92(1):108–124. doi: 10.1016/0042-6822(79)90218-6. [DOI] [PubMed] [Google Scholar]
- Iino T. A Stabilizer of Antigenic Phases in Salmonella Abortus-Equi. Genetics. 1961 Nov;46(11):1465–1469. doi: 10.1093/genetics/46.11.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp D., Kahmann R., Zipser D., Broker T. R., Chow L. T. Inversion of the G DNA segment of phage Mu controls phage infectivity. Nature. 1978 Feb 9;271(5645):577–580. doi: 10.1038/271577a0. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Iino T. A trans-acting factor mediates inversion of a specific DNA segment in flagellar phase variation of Salmonella. Nature. 1980 Apr 3;284(5755):479–481. doi: 10.1038/284479a0. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J, Iino T. Phase Variation in Salmonella. Genetics. 1956 Sep;41(5):743–757. doi: 10.1093/genetics/41.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Otsubo E., Deonier R. C., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. V. ilv+ Deletion mutants of F14. J Mol Biol. 1974 Nov 15;89(4):585–597. doi: 10.1016/0022-2836(74)90037-0. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Takeda Y., Nishimura A., Suzuki H., Inouye M., Hirota Y. Synthetic ColE1 plasmids carrying genes for cell division in Escherichia coli. Plasmid. 1977 Nov;1(1):67–77. doi: 10.1016/0147-619x(77)90009-9. [DOI] [PubMed] [Google Scholar]
- Pearce U. B., Stocker B. A. Phase variation of flagellar antigens in Salmonella: abortive transduction studies. J Gen Microbiol. 1967 Nov;49(2):335–349. doi: 10.1099/00221287-49-2-335. [DOI] [PubMed] [Google Scholar]
- Rosner J. L. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972 Jun;48(3):679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- Silverman M., Zieg J., Hilmen M., Simon M. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc Natl Acad Sci U S A. 1979 Jan;76(1):391–395. doi: 10.1073/pnas.76.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Iino T. In vitro synthesis of phase-specific flagellin of Salmonella. J Mol Biol. 1973 Nov 25;81(1):57–70. doi: 10.1016/0022-2836(73)90247-7. [DOI] [PubMed] [Google Scholar]
- Velten J., Fukada K., Abelson J. In vitro construction of bacteriophage lambda and plasmid DNA molecules containing DNA fragments from bacteriophage T4. Gene. 1976;1(1):93–106. doi: 10.1016/0378-1119(76)90009-3. [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Inokuchi H., Ozeki H. Excision and duplication of su3+-transducing fragments carried by bacteriophage phi 80. I. Novel structure of phi 80sus2psu3+ DNA molecule. J Virol. 1976 Jun;18(3):1016–1023. doi: 10.1128/jvi.18.3.1016-1023.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun T., Vapnek D. Electron microscopic analysis of bacteriophages P1, P1Cm, and P7. Determination of genome sizes, sequence homology, and location of antibiotic-resistance determinants. Virology. 1977 Mar;77(1):376–385. doi: 10.1016/0042-6822(77)90434-2. [DOI] [PubMed] [Google Scholar]
- Zieg J., Hilmen M., Simon M. Regulation of gene expression by site-specific inversion. Cell. 1978 Sep;15(1):237–244. doi: 10.1016/0092-8674(78)90098-3. [DOI] [PubMed] [Google Scholar]
- Zieg J., Silverman M., Hilmen M., Simon M. Recombinational switch for gene expression. Science. 1977 Apr 8;196(4286):170–172. doi: 10.1126/science.322276. [DOI] [PubMed] [Google Scholar]