Abstract
A region with a high risk for esophageal squamous cell carcinoma (ESCC) in northeast of Iran was identified more than three decades ago. Previous studies suggest that hereditary factors play a role in the high incidence of cancer in the region. Polymorphisms of several genes have been associated with susceptibility to esophageal cancer in various populations, but these have not been studied in Iran. We selected 22 functional variants (and 130 related tagSNPs) from 15 genes which previously have been suggested to be associated with an increased risk of ESCC. We genotyped a primary set of samples from 451 Turkmen (197 cases and 254 controls). Seven of 152 variants were associated with ESCC at the P = 0.05 level; these SNPs were then studied in a validation set of 1668 cases and controls (Turkmen and non-Turkmen) under dominant and recessive models. In the joint sample set, five variants, from five different genes, showed significant associations with ESCC at the P = 0.05 level. For one variant, in ADH1B, the association was strong and was present in both Turkmen and non-Turkmen. The histidine allele at codon 48 of ADH1B gene was associated with a significantly decreased risk of ESCC under a recessive model (OR = 0.41, 95%, CI = 0.19 to 0.49; P = 4×10−4). For four additional variants, an association was present in the Turkmen subgroup, but the statistical significance of these was less compelling than for ADH1B. Two variants showed deleterious effects and two were protective. The G allele of the c.870A>G variant of CCND1 gene was associated with a 1.5-fold increased risk of ESCC under the recessive model (OR = 1.50, 95% CI = 1.14 to 2.16, P = 0.02) and the A allele of the rs1625895 variant of TP53 gene was associated with a 1.5-fold increased risk of ESCC under a dominant model (OR = 1.54, 95% CI = 1.21 to 4.07, P = 0.005). The C allele of the rs886205 variant of ALDH2 was associated with a decreased risk of ESCC under a recessive model (OR = 0.58, 95% CI = 0.34 to 0.87, P = 0.02) and the A allele of the rs7087131 variant of MGMT was associated with a decreased risk of ESCC under the recessive model (OR = 0.26, 95% CI = 0.05 to 0.49, P=0.01). These results confirm that genetic predisposition to ESCC plays a role in high incidence of this cancer among Turkmens who live in northeast of Iran.
Keywords: Esophageal squamous cell carcinoma, Turkmen population, ADH1B, ALDH2, MGMT, TP53, CCND1
Introduction
Worldwide, 462,000 new cases of esophageal cancer are diagnosed annually [1]. 75% of affected individuals die within a year of diagnosis [2] and the five-year survival rate is only 10–16% [1]. The incidence of esophageal cancer varies greatly between populations and across geographic regions. Gonbad City in northeastern Iran, which is inhabited mainly by Turkmen, is a region with a very high risk of esophageal cancer, but the causative factors remain elusive. The annual incidence rate exceeds 100/100,000 [3, 4], although the rate has declined somewhat over the last three decades [5]. Esophageal cancer has two main histological types; squamous cell carcinoma and adenocarcinoma. Esophageal squamous cell carcinomas (ESCC) constitute more than 90% of esophageal cancers among Turkmen [6]. We have recently shown that, among Turkmen, there is a strong familial component to esophageal cancer [7]. The first gene found to contribute to the burden of esophageal cancer in Iran was BRCA2 [8]. Protein-truncating mutations in BRCA2 are associated with an increased risk of esophageal cancer among Turkmen. However, because only 8% of the cases in the region carry a deleterious BRCA2 mutation [8] it is likely that other genes are involved.
Turkmen are believed to descend from Turkic tribes who migrated from the Altai Mountains on the border of China and Mongolia to northern Iran [9]. A population at high risk for ESCC is also present in China [10] and the two high-risk populations might share common susceptibility genes. Based on this hypothesis, we considered that the genes which have been previously reported to be associated with ESCC in the high-risk Chinese population [2, 11] were candidate genes among Turkmen.
We studied 15 different genes in cases and controls from Turkmen and non-Turkmen in the North of Iran. These genes are involved in DNA repair, cell cycle control and the metabolism of alcohol, folate and carcinogens.
Materials and Methods
Study Subject
This report is a component of a study of upper gastrointestinal (UGI) cancers [6, 12] which is underway in Gonbad, the second largest city of Golestan province, in northeastern Iran. The project is directed by the Digestive Disease Research Center (DDRC) of the Tehran University of Medical Sciences, in collaboration with the National Cancer Institute (NCI), the International Agency for Research on Cancer (IARC) and the University of Toronto. Cases and controls were collected between August 1, 2001 and May 15, 2008. The diagnosis of ESCC was confirmed for all cases by upper gastrointestinal endoscopy and by pathologic evaluation of tumour biopsies. The study protocol was approved by the ethics committee of the Digestive Disease Research Center of Tehran University of Medical Sciences.
Cases
A total of 746 cases was enrolled in this study. There were 281 Turkmen and 211 non-Turkmen ESCC cases from Gonbad city. Non-Turkmen included Persians (n = 114), Turks (n = 59), Sistanies (n = 28), Balouches (n = 9) and Kurds (n = 10). In addition, we enrolled 245 Turkish cases from Ardabil, a city in northwestern Iran with intermediate rates of ESCC [13]. The mean age at diagnosis was 63.6 years (range 25 to 89 years); 380 cases (50.9%) were male and 366 cases (49.1%) were female. Epidemiologic data included smoking status, use of opium and alcohol and a family history of cancer. The primary set of cases consisted of the first 197 Turkmen who were available at the time of testing. The remaining 549 cases were part of the validation set.
Controls
1373 controls were recruited to the study: 244 were Turks from Ardabil city, 811 were Turkmen from Gonbad city and 318 were non-Turkmen from Gonbad city [Persians (n = 133), Turks (n = 78), Sistanies (n = 79), Balouches (n = 19) and Kurds (n = 9)]. 898 controls were patients referred to local hospitals for a reason other than cancer and 475 controls were healthy individuals enrolled from the Golestan cohort study [12]. Controls had no personal history of cancer. The mean age for controls was 55.2 years (range 24 to 90 years); 700 (51.0%) were male and 673 (49.0%) were female. Charactersistics of the 746 cases and 1373 controls are presented in Table 1.
Table 1.
Subject characteristics of the ESCC cases and controls
| Variable | Cases | Controls | P-Value | |
|---|---|---|---|---|
| Total Number, n | 746 | 1373 | --- | |
| Age, Mean (range) | 63.6 (25–89) | 55.2 (24–90) | 1.3E-43 | |
| Gender, n (%) | Male | 380 (51) | 700 (51) | 1.0 |
| Female | 366 (49) | 673 (49) | ||
| Ethnicity, n (%) | Turkmen | 281 (38) | 811 (59) | 2.3E-7 |
| Turk | 304 (41) | 322 (23) | ||
| Persian | 114 (15) | 133 (10) | ||
| Sistani | 28 (4) | 79 (6) | ||
| Balooch | 9 (1) | 19 (1.4) | ||
| Kurd | 10 (1) | 9 (0.7) | ||
| Ever Smoker, n (%) | Yes | 200 (28) | 339 (25) | 0.14 |
| No | 516 (72) | 1018 (75) | ||
| Ever Opium User, n (%) | Yes | 158 (29) | 239 (20) | 3.5E-5 |
| No | 382 (71) | 943 (80) | ||
Candidate genes
Genes were selected based on literature review. Four genes were involved in DNA repair ERCC2 (XPD), XRCC1, hOGG1 and MGMT; four genes were involved in cell cycle control: CCND1 (Cyclin D1), CDKN2A (p16), TP53 (p53) and TMPRSS11A (ECRG1); four genes coded for carcinogen metabolizing enzymes: CYP1A1, CYP2E1, GSTP1and NAT2; two coded for alcohol metabolizing enzymes: ADH1B and ALDH2; and one coded for a folate metabolizing enzyme: MTHFR. The role of these genes and their variants in esophageal cancer has been reviewed [2, 11].
For each gene, at least one dysfunctional variant has been reported in the literature to be associated with ESCC [2, 11, and 14]. These variants include Asp312Asn (rs1799793) and Lys751Gln (rs13181) from ERCC2 gene, Arg399Gln (rs25487) from XRCC1 gene; Ser326Cys (rs1052133) from hOGG1 gene; Leu74Phe (rs12917) and Lys178Arg (rs2308327) from MGMT gene; c.870A>G (rs9344) from CCND1 gene; Ala148Thr (rs3731249) from CDKN2A gene; Arg72Pro (rs1042522) from TP53 gene; Arg293Gln (rs353163) from TMPRSS11A (ECRG1) gene; Ile462Val (rs1048943) from CYP1A1 gene; c.-1019C>T (rs2031920) from CYP2E1; Ile105Val (rs1695) from GSTP1 gene; Tyr94Tyr (rs1041983), Ile114Thr (rs1801280), Leu161Leu (rs1799929), Arg197Gln (rs1799930) and Gly286Glu (rs1799931) from NAT2 gene; Arg48His (rs1229984) from ADH1B gene; Glu487Lys (rs671) from ALDH2 gene; and Ala222Val (rs1801133) and Glu429Ala (rs1801131) from MTHFR gene.
Three slow metabolizing alleles of M1, M2 and M3 were reported for NAT2 gene. Variants Ile114Thr and Leu161Leu constitute allele M1; variants Arg197Gln and Tyr94Tyr constitute allele M2; and variant Gly286Glu is named allele M3 [15].
tagSNP Selection
Within populations, it is believed that SNPs are distributed along chromosomes in haplotype blocks and therefore genotyping of one or more SNPs which are characteristic of a haplotype block will capture the important information for the additonal SNPs in that block. These SNPS are in linkage disequilibrium with the genotyped SNPs, and are called tagSNPs. Genotype data from the Chinese population of the HapMap project [16] (Rel21/Phase II) was used for selecting tagSNPs to cover the genomic region of each gene, including 5 kbp in each 5’ and 3’end to cover the regulatory regions of the genes. Haploview software [17] was used for selecting tagSNps. Aggressive tagging with using 2- and 3-marker haplotypes, LOD score threshold of 3.0 for multi-marker testing and r2 threshold of 0.8 were employed for tagging procedure. Genomic regions of all 15 candidate genes encompass 705 kbp of the human genome. There are 635 SNPs with MAF of more than 1% in Chinese population of HapMap project. 141 tagSNP were selected to capture 594 SNPs (94% of the total common SNPs) with mean r2 of o.98 in 152 tests [Table 2].
Table 2.
The number of tagSNPs genotyped in this study along with the number of the SNPs captured with these tagSNPs for each gene
| No. | Gene | Chromosome | Size (kbp)* | Number of Genotyped tagSNPs, n |
Number of Captured SNPs, n (%)† |
Mean Max r2 |
|---|---|---|---|---|---|---|
| 1 | ERCC2 | 19 | 29 | 8 | 18/20 (90) | 0.97 |
| 2 | ADH1B | 4 | 10 | 3 | 4/4 (100) | 1.00 |
| 3 | ALDH2 | 12 | 53 | 10 | 22/23 (95) | 0.95 |
| 4 | CCND1 | 11 | 23 | 2 | 2/2 (100) | 1.00 |
| 5 | CDKN2A | 9 | 18 | 6 | 8/8 (100) | 0.99 |
| 6 | CYP1A1 | 15 | 16 | 1 | 1/1 (100) | 1.00 |
| 7 | CYP2E1 | 10 | 22 | 6 | 27/34 (79) | 0.97 |
| 8 | ECRG1 | 4 | 63 | 9 | 49/54 (90) | 0.99 |
| 9 | GSTP1 | 11 | 13 | 4 | 12/12 (100) | 0.99 |
| 10 | MGMT | 10 | 310 | 49 | 321/332 (96) | 0.95 |
| 11 | MTHFR | 1 | 30 | 8 | 35/36 (97) | 0.99 |
| 12 | NAT2 | 8 | 20 | 10 | 40/46 (86) | 0.94 |
| 13 | OGG1 | 3 | 27 | 5 | 5/5 (100) | 1.00 |
| 14 | TP53 | 17 | 29 | 6 | 7/14 (50) | 1.00 |
| 15 | XRCC1 | 19 | 42 | 14 | 43/44 (97) | 0.93 |
| Total | 705 | 141 | 594/635 (94) | 0.98 | ||
The size of the gene including 5 kbp at each end for covering gene regulatory regions
In this column the number of the SNPs with MAF >1% identified in the Chinese population of the HapMap project and the proportion of these SNPs which is captured by the genotyped tagSNPs in this study are shown for each gene.
SNP Genotyping
We initially tested the 152 SNPs in a primary set of 197 cases and 254 controls. From the 141 tagSNPs selected for study, 11 were among the dysfunctional variants above mentioned and 130 were additional. All SNPs were genotyped in germline DNA of all samples of the primary set of cases and controls. iPLEX chemistry on a MALDI-TOF MassARRAY system (Sequenom Inc., San Diego, CA, USA) was used for genotyping the 152 SNPs in eight reactions. The procedures were performed according to the manufacturer’s standard protocol. SNPs which showed association with ESCC in the primary set of samples (p < 0.05) were combined and a new assay design was performed to genotype them in a single reaction. ESCC-associated SNPs identified in the primary set of samples were genotyped in the germline DNA of the validation set of samples. 10% blinded quality control samples were included in each plate; the concordance rates ranged from 98% to 100%. The average genotyping call rate was 98% (ranging from 95% to 100%). Two ESCC-associated SNPs were failed for genotyping in the primary set of samples; they were genotyped using TaqMan assay on ABI 7500 fast real-time system (Applied Biosystems Co., Foster City, CA, USA).
Statistical Analysis
The permutation version of the exact test was done to test for Hardy–Weinberg Equilibrium. All the genotypes first compared in the primary sample set and then the identified ESCC-associated variants were genotyped in the validation sample set. Haplotype analysis using expectation maximization algorithm [18] was utilized for comparing SNPs tagged by multiple markers. The significance level of α = 0.05 was used for all comparisons. Genotyping results of the initially ESCC-associated variants in the primary and validation sample sets were combined to make a joint sample set. Genotypes of the cases and controls in the joint sample set were assessed in Turkmen and non-Turkmen. All case-control comparisons were adjusted for age and ethnicity, using multivariate logistic regression and the adjusted P-values and odds ratios (OR) were reported. The p-values of the comparisons in the final analysis were not adjusted for multiple testing.
In order to exclude the possibility that there were technical errors in genotyping between the typing of subjects in the primary and validation sets, we re-genotyped all the seven variants genotyped in the validation set in the primary sample set using the genotyping assay used for the validation sample set. The concordance rate of the genotyping results was 97%.
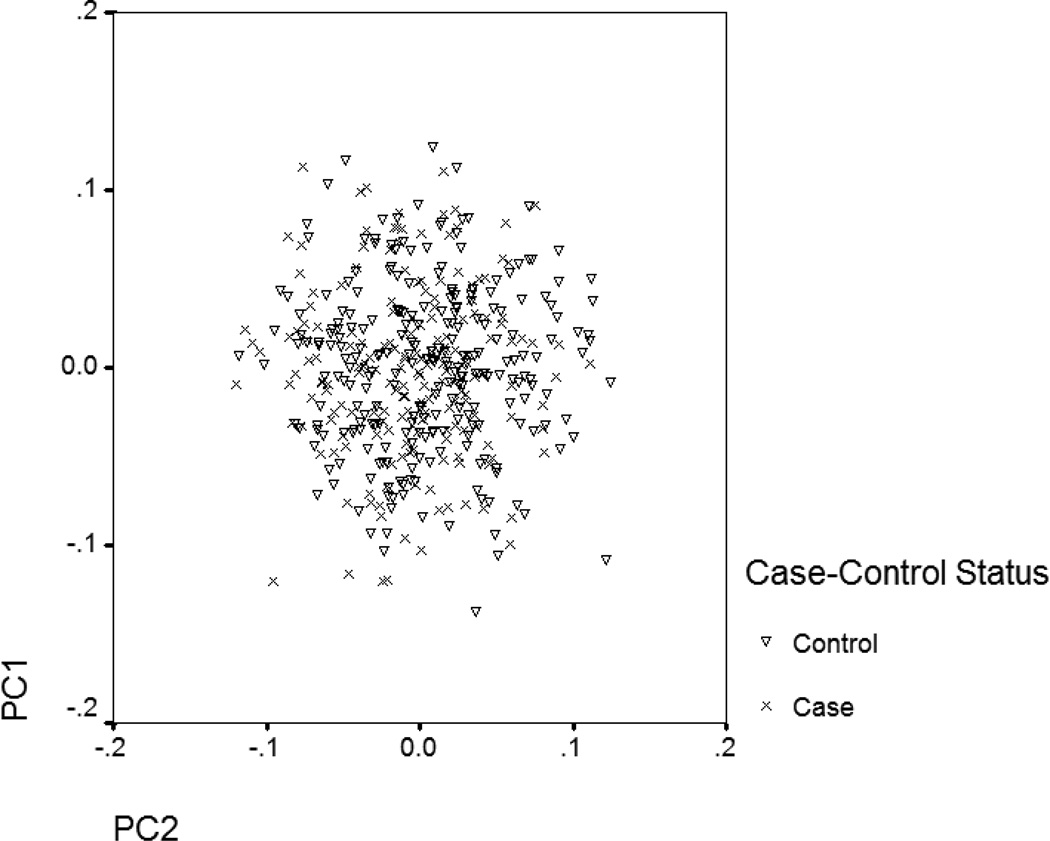
The possibility that there is spurious population stratification in the Turkmen was addressed by principle component analysis using the Eigenstrat method [19] for identifying any population stratification in the primary sample set which we had genotypes of 130 tagSNPs plus 22 dysfunctional variants for them. By plotting the Eigenvectors of the first two principle components with the largest Eigenvalues for the cases and the controls [figure 1], it appears that population stratification was not present. All analyses were done by SNP Variation Suit V.7 (Golden Helix Inc., Bozeman, MT, USA).
Figure 1.
Eigenvectors of the first two principle components with the largest Eigenvalues have been plotted for the primary sample set using genotyping data of 152 SNPs.
Results
Initially, we genotyped 152 SNPs on a primary set of 197 case and 254 controls. These represent the first samples to be collected in the course of the project and were all Turkmen. These SNPs included 22 functional variants of 15 genes and 130 tagSNPs from these genes. The results of the 17 dysfunctional variants and three slow metabolizing alleles of NAT2 gene in the primary set of 451 samples are shown in table 3. The frequencies of the minor alleles of all variants except one (ALDH2 Glu487Lys) were greater than 1%. All were in Hardy-Weinberg equilibrium.
Table 3.
Genotypes of the dysfunctional variants on the 197 cases and 254 controls of the primary sample set
| Variant | Gene | MAF* | Cases, n (%) | Controls, n (%) | OR† | P-Value† |
|---|---|---|---|---|---|---|
| Arg48His | ADH1B | 0.21 | ||||
| Arg/Arg | 129 (66.5) | 145 (57.3) | 1.00 | 1.00 | ||
| Arg/His | 59 (30.5) | 96 (37.9) | 0.66 | 0.04 | ||
| His/His | 6 (3.0) | 12 (4.8) | 0.70 | 0.17 | ||
| Dominant | 0.64 | 0.03 | ||||
| Recessive | 0.61 | 0.33 | ||||
| Glu487Lys | ALDH2 | 0.01 | ||||
| Glu/Glu | 196 (99.5) | 248 (98.4) | 1.00 | 1.00 | ||
| Glu/Lys | 1 (0.5) | 4 (1.6) | 0.28 | 0.22 | ||
| Lys/Lys | 0 (0.0) | 0 (0.0) | --- | --- | ||
| c.870A>G | CCND1 | 0.48 | ||||
| A/A | 53 (27.2) | 79 (31.6) | 1.00 | 1.00 | ||
| A/G | 83 (42.6) | 116 (46.4) | 1.09 | 0.72 | ||
| G/G | 59 (30.2) | 55 (22.0) | 1.30 | 0.04 | ||
| Dominant | 1.27 | 0.26 | ||||
| Recessive | 1.61 | 0.03 | ||||
| Ala148Thr | CDKN2A | 0.03 | ||||
| Ala/Ala | 186 (95.9) | 235 (94.0) | 1.00 | 1.00 | ||
| Ala/Thr | 8 (4.1) | 15 (6.0) | 0.71 | 0.45 | ||
| Thr/Thr | 0 (0.0) | 0 (0.0) | --- | --- | ||
| Ile462Val | CYP1A1 | 0.13 | ||||
| Ile/Ile | 149 (75.6) | 186 (74.1) | 1.00 | 1.00 | ||
| Ile/Val | 47 (23.8) | 61 (24.3) | 0.98 | 0.92 | ||
| Val/Val | 1 (0.6) | 4 (1.6) | 0.50 | 0.18 | ||
| Dominant | 0.96 | 0.86 | ||||
| c.-1019C>T | CYP2E1 | 0.05 | ||||
| C/C | 175 (91.1) | 225 (90.4) | 1.00 | 1.00 | ||
| C/T | 15 (7.8) | 24 (9.6) | 0.76 | 0.42 | ||
| T/T | 2 (1.1) | 0 (0.0) | --- | --- | ||
| Dominant | 0.83 | 0.59 | ||||
| Arg293Gln | ECRG1 | 0.28 | ||||
| Arg/Arg | 103 (52.8) | 130 (52.0) | 1.00 | 1.00 | ||
| Arg/Gln | 70 (35.9) | 101 (40.4) | 0.84 | 0.39 | ||
| Gln/Gln | 22 (11.3) | 19 (7.6) | 1.24 | 0.21 | ||
| Dominant | 0.96 | 0.84 | ||||
| Recessive | 1.69 | 0.12 | ||||
| Asp312Asn | ERCC2 | 0.37 | ||||
| Asp/Asp | 77 (40.1) | 104 (41.8) | 1.00 | 1.00 | ||
| Asp/Asn | 87 (45.3) | 115 (46.2) | 1.06 | 0.77 | ||
| Asn/Asn | 28 (14.6) | 30 (12.0) | 1.15 | 0.39 | ||
| Dominant | 1.14 | 0.50 | ||||
| Recessive | 1.27 | 0.40 | ||||
| Lys751Gln | ERCC2 | 0.36 | ||||
| Lys/Lys | 77 (39.1) | 99 (39.3) | 1.00 | 1.00 | ||
| Lys/Gln | 94 (47.7) | 119 (47.2) | 1.06 | 0.77 | ||
| Gln/Gln | 26 (13.2) | 34 (13.5) | 1.02 | 0.90 | ||
| Dominant | 1.06 | 0.76 | ||||
| Recessive | 1.03 | 0.93 | ||||
| Ile105Val | GSTP1 | 0.28 | ||||
| Ile/Ile | 103 (52.6) | 126 (50.4) | 1.00 | 1.00 | ||
| Ile/Val | 77 (39.3) | 105 (42.0) | 0.90 | 0.63 | ||
| Val/Val | 16 (8.1) | 19 (7.6) | 0.99 | 0.96 | ||
| Dominant | 0.90 | 0.58 | ||||
| Recessive | 1.07 | 0.84 | ||||
| Lys178Arg | MGMT | 0.14 | ||||
| Lys/Lys | 163 (83.2) | 213 (84.9) | 1.00 | 1.00 | ||
| Lys/Arg | 30 (15.3) | 35 (13.9) | 1.14 | 0.64 | ||
| Arg/Arg | 3 (1.5) | 3 (1.2) | 1.17 | 0.71 | ||
| Dominant | 1.14 | 0.62 | ||||
| Leu84Phe | MGMT | 0.09 | ||||
| Leu/Leu | 142 (72.4) | 185 (74.0) | 1.00 | 1.00 | ||
| Leu/Phe | 53 (27.0) | 63 (25.2) | 1.02 | 0.92 | ||
| Phe/Phe | 1 (0.6) | 2 (0.8) | 0.76 | 0.65 | ||
| Dominant | 1.03 | 0.90 | ||||
| Glu429Ala | MTHFR | 0.40 | ||||
| Glu/Glu | 74 (75.6) | 94 (74.1) | 1.00 | 1.00 | ||
| Glu/Ala | 92 (23.8) | 109 (24.3) | 1.03 | 0.88 | ||
| Ala/Ala | 29 (0.6) | 47 (1.6) | 0.86 | 0.30 | ||
| Dominant | 0.94 | 0.75 | ||||
| Recessive | 0.73 | 0.23 | ||||
| Ala222Val | MTHFR | 0.25 | ||||
| Ala/Ala | 102 (52.3) | 151 (60.4) | 1.00 | 1.00 | ||
| Ala/Val | 79 (40.5) | 84 (33.6) | 1.47 | 0.09 | ||
| Val/Val | 14 (7.2) | 15 (6.0) | 1.16 | 0.46 | ||
| Dominant | 1.44 | 0.10 | ||||
| Recessive | 1.10 | 0.81 | ||||
| Ser326Cys | OGG1 | 0.32 | ||||
| Ser/Ser | 91 (46.4) | 124 (49.4) | 1.00 | 1.00 | ||
| Ser/Cys | 76 (38.8) | 102 (40.6) | 1.03 | 0.87 | ||
| Cys/Cys | 29 (14.8) | 25 (10.0) | 1.20 | 0.24 | ||
| Dominant | 1.10 | 0.62 | ||||
| Recessive | 1.36 | 0.30 | ||||
| Arg72Pro | TP53 | 0.36 | ||||
| Arg/Arg | 77 (39.3) | 108 (43.0) | 1.00 | 1.00 | ||
| Arg/Pro | 90 (45.9) | 116 (46.2) | 1.12 | 0.59 | ||
| Pro/Pro | 29 (14.8) | 27 (10.8) | 1.17 | 0.31 | ||
| Dominant | 1.18 | 0.39 | ||||
| Recessive | 1.36 | 0.29 | ||||
| Arg399Gln | XRCC1 | 0.37 | ||||
| Lys/Lys | 78 (39.1) | 94 (39.3) | 1.00 | 1.00 | ||
| Lys/Gln | 89 (47.7) | 120 (47.2) | 0.93 | 0.71 | ||
| Gln/Gln | 26 (13.2) | 35 (13.5) | 1.00 | 0.98 | ||
| Dominant | 0.91 | 0.62 | ||||
| Recessive | 1.02 | 0.94 | ||||
| Gly286Glu(M3) | NAT2 | 0.08 | ||||
| Gly/Gly | 165 (84.2) | 209 (83.6) | 1.00 | 1.00 | ||
| Gly/Glu | 29 (14.8) | 39 (15.6) | 0.86 | 0.57 | ||
| Glu/Glu | 2 (1.0) | 2 (0.8) | 0.90 | 0.84 | ||
| Dominant | 0.86 | 0.56 | ||||
| M1 ‡ | NAT2 | |||||
| wt/wt | 97 (51.9) | 130 (53.3) | 1.00 | 1.00 | ||
| wt/m1 | 77 (41.2) | 93 (38.1) | 1.12 | 0.58 | ||
| m1/m1 | 13 (6.9) | 21 (8.6) | 0.92 | 0.67 | ||
| Dominant | 1.07 | 0.72 | ||||
| Recessive | 0.90 | 0.56 | ||||
| M2 ‡ | NAT2 | |||||
| wt/wt | 91 (46.7) | 114 (45.8) | 1.00 | 1.00 | ||
| wt/m2 | 84 (43.1) | 102 (41.0) | 1.01 | 0.97 | ||
| m2/m2 | 20 (10.2) | 33 (13.2) | 0.91 | 0.54 | ||
| Dominant | 0.96 | 0.83 | ||||
| Recessive | 0.90 | 0.50 |
Minor Allele Frequency
P-value and odds ratio (OR) are adjusted for age using multivariate logistic regression analysis.
M1: combination of Ile114Thr and Leu161Leu variants; M2: combination of Arg197Gln and Tyr94Tyr variants
In addition, we selected 130 tagSNPs of 15 different genes on the primary set of samples. These SNPs had not previously been studied in ESCC. Eight tagSNPs had minor allele frequencies (MAF) of less than 0.01 and three tagSNPs were not in Hardy-Weinberg equilibrium. These 11 tagSNPs were excluded from the analysis. The remaining 119 tagSNPs were analyzed in the primary set of 197 cases and 254 controls.
Two functional variants showed associations with ESCC in the primary sample set with p-values less than 0.05 (the Arg48His variant from ADH1B gene and c.870A>G from CCND1). Five tagSNPs, from five different genes, also showed significant associations (p-values <0.05) in the primary set. These seven SNPs (two functional variants and five tagSNPs) were then genotyped in the validation set of samples. The results reported below are based on the combined data set (primary and validation set). Results were analyzed separately for Turkmen and non-Turkmen and for the two ethnic groups combined. Results were reported in the form of odds ratios for the three genotype classes, with the most common homozygous genotype being the reference class. Results were reported assuming dominant and recessive models.
The strongest association was observed with the His allele of the Arg48His variant of ADH1B gene. This allele was associated with a decreased risk of ESCC under a recessive model [table 4]. Compared to those who carried no His allele, who carried two His alleles were at 0.63 times the risk (P = 0.0002).
Table 4.
Genotyping results of the Arg48His variant of ADH1B gene on the joint sample set
| Turkmens, n (%) | Non-Turkmen, n (%) | Total samples, n (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (281) | Controls (811) | OR* | P-Value* | Cases (465) | Controls (562) | OR* | P-Value* | Cases (746) | Controls (1373) | OR* | P-Value* | |
| Arg/Arg | 190 (67.6) | 488 (60.2) | 1.00 | 1.00 | 300 (64.5) | 339 (60.3) | 1.00 | 1.00 | 490 (65.7) | 827 (60.2) | 1.00 | 1.00 |
| Arg/His | 80 (28.5) | 288 (35.5) | 0.72 | 0.03 | 152 (32.7) | 183 (32.5) | 0.92 | 0.58 | 232 (31.1) | 471 (34.3) | 0.84 | 0.10 |
| His/His | 8 (2.8) | 34 (4.2) | 0.72 | 0.11 | 13 (2.8) | 39 (6.9) | 0.57 | 0.0006 | 21 (2.8) | 73 (5.3) | 0.63 | 0.0002 |
| Dominant | 0.69 | 0.02 | 0.81 | 0.13 | 0.77 | 0.01 | ||||||
| Recessive | 0.59 | 0.19 | 0.33 | 0.0007 | 0.41 | 0.0004 | ||||||
P-value and odds ratio (OR) were adjusted for age in Turkmens and for age and ethnicity for Non-Turkmens and total samples using multivariate logistic regression analysis.
The G allele of the c.870A>G variant of CCND1 gene was associated with an increased risk of ESCC under both additive and recessive models. The association was limited to Turkmen [table 5]. Those who were homozygotes for the G allele had 1.5 fold higher chance of having ESCC in comparison to A/A and G/A carriers of this variant (P = 0.02). Three tagSNPs from ALDH2 (rs886205), MGMT (rs7087131) and TP53 (rs1625895) genes were associated with increased risks of ESCC in Turkmen only [table 6].
Table 5.
Genotyping results of the c.870A>G variant of CCND1 gene on the joint sample set
| Turkmens, n (%) | Non-Turkmen, n (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Cases (281) | Controls (811) | OR* | P-Value* | Cases (465) | Controls (562) | OR* | P-Value* | |
| A/A | 81 (28.8) | 270 (33.3) | 1.00 | 1.00 | 130 (27.9) | 164 (29.2) | 1.00 | 1.00 |
| A/G | 126 (44.8) | 376 (46.4) | 1.05 | 0.77 | 238 (51.2) | 290 (51.6) | 0.90 | 0.51 |
| G/G | 72 (25.6) | 161 (19.9) | 1.23 | 0.03 | 97 (20.9) | 107 (19.0) | 0.99 | 0.91 |
| Dominant | 1.20 | 0.26 | 0.93 | 0.65 | ||||
| Recessive | 1.50 | 0.02 | 1.07 | 0.67 | ||||
P-value and odds ratio (OR) were adjusted for age in Turkmens and for age and ethnicity in non-Turkmens using multivariate logistic regression analysis.
Table 6.
Genotyping results of the five initially ESCC-associated tagSNPs on the joint sample set
| variant | Gene | Turkmens, n (%) | Non-Turkmen, n (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases (281) | Controls (811) | OR* | P-Value* | Cases (465) | Controls (562) | OR* | P-Value* | ||
| rs886205 | ALDH2 | ||||||||
| T/T | 141 (50.2) | 355 (43.8) | 1.00 | 1.00 | 247 (53.1) | 316 (56.2) | 1.00 | 1.00 | |
| T/C | 113 (40.2) | 344 (42.4) | 0.85 | 0.31 | 176 (37.8) | 205 (36.5) | 1.05 | 0.73 | |
| C/C | 24 (8.5) | 106 (13.1) | 0.73 | 0.01 | 40 (8.6) | 41 (7.3) | 1.11 | 0.43 | |
| Dominant | 0.78 | 0.09 | 1.08 | 0.57 | |||||
| Recessive | 0.58 | 0.02 | 1.20 | 0.46 | |||||
| rs7087131 | MGMT | ||||||||
| G/G | 203 (72.2) | 533 (65.7) | 1.00 | 1.00 | 351(75.5) | 424 (75.4) | 1.00 | 1.00 | |
| G/A | 72 (25.6) | 246 (30.3) | 0.74 | 0.07 | 99 (21.3) | 124 (22.1) | 0.92 | 0.60 | |
| A/A | 3 (1.1) | 27 (3.3) | 0.49 | 0.007 | 13 (2.8) | 13 (2.3) | 1.06 | 0.77 | |
| Dominant | 0.68 | 0.02 | 0.94 | 0.67 | |||||
| Recessive | 0.26 | 0.01 | 1.12 | 0.78 | |||||
| rs1625895 | TP53 | ||||||||
| G/G | 167 (59.4) | 546 (67.3) | 1.00 | 1.00 | 263 (56.6) | 310 (55.2) | 1.00 | 1.00 | |
| G/A | 97 (34.5) | 231 (28.5) | 1.51 | 0.01 | 173 (37.2) | 209 (37.2) | 0.98 | 0.87 | |
| A/A | 15 (5.3) | 30 (3.7) | 1.36 | 0.08 | 27 (5.8) | 43 (7.6) | 0.98 | 0.89 | |
| Dominant | 1.54 | 0.005 | 0.98 | 0.86 | |||||
| Recessive | 1.65 | 0.15 | 0.95 | 0.86 | |||||
| rs6552135 | ECRG1 | ||||||||
| A/A | 99 (35.6) | 244 (30.8) | 1.00 | 1.00 | 146 (32.4) | 164 (29.9) | 1.00 | 1.00 | |
| A/G | 114 (41.0) | 372 (46.9) | 0.70 | 0.08 | 209 (46.3) | 269 (49.0) | 0.92 | 0.57 | |
| G/G | 65 (23.4) | 177 (22.3) | 0.96 | 0.68 | 96 (21.3) | 116 (21.1) | 0.98 | 0.84 | |
| Dominant | 0.77 | 0.09 | 0.94 | 0.65 | |||||
| Recessive | 1.13 | 0.47 | 1.01 | 0.94 | |||||
| rs3916874 | ERCC2 | ||||||||
| G/G | 194 (69.3) | 514 (63.9) | 1.00 | 1.00 | 328 (70.7) | 377 (67.4) | 1.00 | 1.00 | |
| G/C | 75 (26.8) | 262 (32.6) | 0.74 | 0.07 | 124 (26.7) | 162 (29.0) | 0.91 | 0.53 | |
| C/C | 11 (3.9) | 28 (3.5) | 1.04 | 0.85 | 12 (2.6) | 20 (3.6) | 0.75 | 0.13 | |
| Dominant | 0.77 | 0.09 | 0.85 | 0.27 | |||||
| Recessive | 1.17 | 0.69 | 0.55 | 0.12 | |||||
P-value and odds ratio (OR) were adjusted for age in Turkmens and for age and ethnicity in non-Turkmens using multivariate logistic regression analysis.
Discussion
We studied 22 different dysfunctional variants of 15 genes which were previously associated with ESCC. Notably, the odds ratios reported for these variants in the earlier studies were typically 2.0 and above [11]. Given that the frequencies of the minor alleles of these variants in our population were almost all above 0.1 [table 3], the power of our study to detect odds ratios above 2.0 exceeded 90%. However, of the 22 variants tested, only two polymorphisms (ADH1B Arg48His and CCND1 c.870A>G) showed significant associations at the p = 0.05 level in the entire data set and for only one of these (ADH1B Arg48His) was the association compelling.
The His allele of the Arg48His variant of ADH1B gene was associated with a decreased risk of ESCC in both Turkmen and non-Turkmen under a recessive model [table 5]. ADH1B is one of seven different genes which encode five different classes of alcohol dehydrogenase (ADH) enzymes. These enzymes are involved in the oxidation of alcohol groups of a variety of substrates. ADH class I, which is responsible for the oxidation of ethanol to acetaldehyde, is a homodimer or heterodimer of three related subunits (α, β and γ) encoded by ADH1A, ADH1B and ADH1C respectively [20]. All ADH enzymes use NAD+/NADH as their oxidizing cofactor. The arginine residue at codon 48 of ADH1B is in close contact with its cofactor and seems to have a crucial role on modulating the protein’s ethanol oxidizing capability. When the arginine is replaced by histidine at this codon, the enzyme activity increases by 70–80 fold [20]. In a rodent model, ADH1B, which has proline at this codon, is not capable of oxidizing ethanol [21].
In previous studies, the Arg allele of ADH1B has been consistently reported to increase the risk of ESCC by 1.6- to 4-fold among alcohol drinkers, compared to individuals with two His alleles [2, 11, 22]. The suggested mechanism is that alcohol and acetaldehyde persist for a longer time in the circulation of individuals with the Arg allele, who are slow metabolizers. These individuals tend to experience less flushing upon exposure to alcohol and are prone to alcoholism [23]. We confirmed that the presence of the Arg allele at codon 48 of the ADH1B protein increases the risk of ESCC by 2.4 fold in comparison to two His alleles (p = 4 × 10−4). Considering that only 4.5 % of our study subjects were alcohol drinkers, this finding suggests that the association is not due to metabolism of alcohol per se. Our observation is contrary to other studies which report no association between Arg48His variant of the ADH1B gene and the risk of ESCC among non-drinkers [22, 24, and 25]. Most studies which showed the association of Arg48His variant of ADH1B with ESCC in alcohol drinkers also report an association between the Glu487Lys variant of ALDH2 and esophageal cancer [11]. The ALDH2 protein with lysine residue at codon 487 is catalytically inactive and can not metabolize acetaldehyde, a known carcinogen [23]. We did not observe this association.
Our data suggests that, in Northern Iran, ESCC may be related to an environmental factor other than ethanol, but that is also metabolized by ADH1B. 1-methypyrene (1-MP) might be the environmental factor. 1-MP is a carcinogenic polycyclic aromatic hydrocarbon (PAH) which is metabolized to 1-hydroxymethylpyrene (1-HMP) by CYP450 enzymes. 1-HMP could be activated by sulphotransferases (SULTs) to 1- Sulfooxymethylpyrene (1-SMP), which is capable of forming DNA adducts, but most of 1-HMP is inactivated by ADH1B and excreted in urine and does not form DNA adducts [26, 27].
Polycyclic aromatic hydrocarbons (PAH) have been shown to be esophageal carcinogens in both animal and human studies. In a two-year feeding study in mice, a dose-response relationship was shown between benzo[a]pyrene intake and ESCC incidence [28]. Occupational exposure to PAHs in Sweden was reported to double the risk of ESCC [29]. Other lines of evidence which support the contribution of PAHs in the pathogenesis of ESCC come from a high-risk region in China, and include high urinary levels of PAH metabolites in residents [30], increased concentrations of PAHs in cooked and uncooked foods in the region [28] and the presence of PAH-DNA adducts in ESCC tissue samples of patients [31].
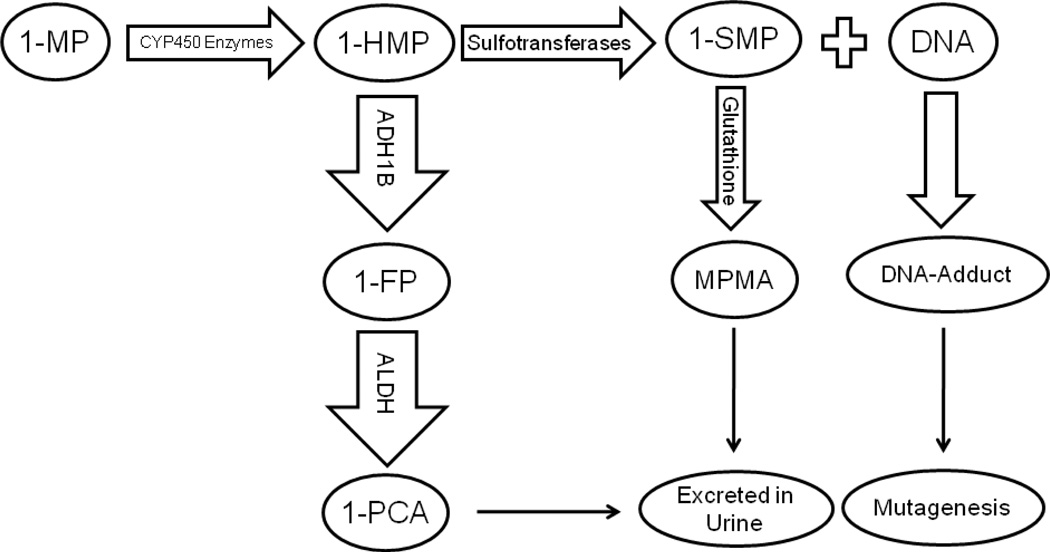
PAHs require metabolic activation to become carcinogens. Purely aromatic PAHs, such as benzo[a]pyrene, are activated via the diol-epoxide pathway [32], but alkylated PAHs, such as 1-MP, are metabolized by an alternative pathway named the aralkylating hydrocarbon pathway [33]. In this pathway [figure 2], 1-MP is hydroxylated to 1-HMP by cytochrome P450 enzymes such as CYP1A1 [34]. 1-HMP subsequently forms reactive sulphuric acid ester (1- SMP) by the action of sulphotransferases [35, 36]. 1- SMP forms DNA adducts and has been shown to be mutagenic in rats [33] and in Salmonella Typhimurium [37]. 1- SMP is also reacts with glutathione and is excreted in urine as methylpyrenyl mercapturic acid (MPMA). The urine MPMA constitutes only 2% of the administrated 1-HMP to rats; 80% of it is excreted as free 1-pyrenylcarboxylic acid (1-PCA) and its glucuronic acid or other conjugates [26]. 1-PCA is an inactivate metabolite of 1-HMP which has been oxidized by ADH1B [27]. Therefore ADH1B competes with 1-SMP for the metabolism of 1-HMP and ADH1B activity decreases the formation of DNA-adducts. If ADH1B is the major enzyme for the inactivation of 1-HMP, then other substrates of ADH1B are expected to interfere with inactivation of 1-HMP and increase the amount of its activated metabolite. This has been exhibited by concurrent administration of 1-HMP and ethanol to rats, thereby enhancing the hepatic level of PAH-DNA adducts by 15-fold [26]. The competition of ethanol with 1-HMP for oxidation by ADH1B could also explain the synergistic carcinogenic effect of alcohol and cigarette smoke (which is rich in 1-MP, the precursor of 1-HMP) [27].
Figure 2.
The Suggested Metabolic Pathway for 1- Methylpyrene (1-MP). 1-MP is first metabolized to 1-Hydroxymethypyrene (1-HMP) by CYP450 enzymes. The Majority (~80%) of 1-HMP is detoxified by alcohol dehydrogenase (ADH1B) to 1-Formylpyrene (1-FP) and then to 1-Pyrenyl carboxylic acid (1-PCA) by aldehyde dehydrogenase (ALDH). 1-PCA and its glucoronic acid conjugates are excreted in urine. Some of (~20%) 1-HMP is transformed to 1-Sulfooxymethylpyrene (1-SMP) of which the majority form DNA- adducts and tiny of it (~2%) detoxified by nucleophile glutathione and form Methylpyrenyl mercapturic acid (MPMA).
It is possible that individuals with two slow-metabolizing Arg alleles at codon 48 of ADH1B gene do not inactivate 1-HMP as efficiently as those who carry one or more His alleles at this codon, thus more 1-HMP will be available for conversion to 1-SMP by sulphutransferases and consequently, more DNA-adducts will be formed [figure 2]. The high urinary levels of 1-hydroxypyrene (1-OHP) which characterize exposure to PAHs in the normal population of this region of Iran support our hypothesis. The median urine level of 1-OHP was reported to be 4.2 pmol/ml in Iran [38] – this is almost 18 times higher than the mean 1-OHP urine level in American office workers [39]. Although 1-OHP is a metabolite of pyrene, rather than of 1-MP, PAHs exist as a mixture in the environment and pyrene is always present in the mixture [40]. The urinary level of 1-OHP as a pyrene metabolite is representative of exposure to a mixture of PAHs, which could also include 1-MP.
There was no interaction between Arg48His variant and smoking or opium use observed in our study. Also the 1-OHP urine level of those who are neither smokers nor opium users was shown to be very high in this region [38]. Therefore, neither smoking nor opium is likely to be the source of 1-MP in our study subjects. However, charcoal-broiled red meat is rich in PAHs that could include 1-MP [41]. Turkmen have ready access to red meat and, in this region, eating charcoal-broiled red meat is common.
The second functional variant associated with ESCC in this study was c.870A>G of the Cyclin D1 (CCND1) gene. Among Turkmen, the G allele of this variant increased the risk of ESCC by 1.5-fold, under a recessive model. The CCND1 protein initiates cell cycle progression and promotes cell proliferation through cyclin-dependent kinases (CDK4 and CDK6) by inactivation of retinoblastoma protein (RB) [42, 43]. Tumours of several different types, including esophagus, may over-express CCND1 [44]. Alternative splicing of CCND1 at the exon 4 and intron 4 boundaries results in the formation of two different transcripts, CCND1a and CCND1b [45]. CCND1b has enhanced transforming activity, in comparison to CCND1a [46] and overexpression of CCND1b has been shown in esophageal tumor cells [47]. The c.870A>G variant, which is located at the end of exon 4, is believed to be associated with alternative splicing of CCND1 between transcripts a and b. Although some studies reported the A allele at this codon favors the formation of transcript b [45, 48], others have detected no association [49, 50]. A recent study described the preferential formation of transcript b for genes with the G allele [51]. The association of c.870A>G variant and different cancer types is also controversial [52, 53]. In some studies, the A allele is associated with an increased risk of cancer and in others the G allele is the high-risk variant [52, 53]. These discrepancies suggest that there are cis- or trans-acting modifiers which, in combination with the c.870A>G variant, determine the splicing of CCND1 [52].
In addition to these two functional variants, tagSNPs from ALDH2 (rs886205), MGMT (rs7087131) and TP53 (rs1625895) were also associated with ESCC in Turkmen. Aldehyde dehydrogenase 2 (ALDH2) catalyzes the conversion of acetaldehyde to acetate. Acetaldehyde is the major carcinogenic metabolite of ethanol. The rs886205 variant is the substitution of T by C nucleotide at the 5’ untranslated region of Exon 1 at the promoter region of ALDH2. A transfection assay showed that the expression activity of the T allele is approximately 50% less than the C allele in hepatoma cells [54], also the acetaldehyde- and ethanol-induced gene expression of genes with the T allele was lower than those with the C allele [55]. These reports are consistent with our finding that the C allele (minor allele) of this variant decreases the risk of ESCC under a recessive model (OR = 0.58, p = 0.02); possibly by increasing the expression of ALDH2, which metabolizes the aldehyde group of carcinogens like acetaldehyde.
The rs7087131 variant of MGMT was also found to be associated with ESCC. O6-methylguanine is formed by alkylating agents such as nitrosamines, which are associated with an increased risk of ESCC [56]. O6-methylguanine has been found in the epithelial cells of the esophagus of individuals living in the high-risk region of China [57]. Unrepaired O6-methylguanine tends to pair with thymine and introduces G:C to A:T transitions during cell replication [58]. O6-methylguanine-DNA methytransferase (MGMT) repairs O6-methylguanine by transferring the methyl group to a cysteine residue at codon 145 of the enzyme [59]. Binding of methyl group to the cytosine residues of MGMT is an irreversible reaction and after repairing each single O6-methylguanine, the protein is ubiquitylated and degraded and needs to be replaced by a new protein. Therefore, it is expected that cells with low expression of MGMT can not repair O6-methylguanine efficiently. Low expression of MGMT in cells can result from either promoter hypermethylation of the gene or by cis-acting nucleotide polymorphisms which change the binding sites of key regulatory factors [60]. A group of SNPs at the 5’ half of the gene were described to be in association with MGMT activity in white blood cells. [60]. The rs7087131 variant is also located at the 5’ half of the MGMT gene in the intronic region between exons 2 and 3 and directly or indirectly might affect expression level of the gene and contribute to the risk of ESCC in Turkmens. In our study, the substitution of G by A nucleotide in this variant decreased the risk of ESCC in Turkmens under a recessive model (OR = 0.26, P=0.01).
The last tagSNP identified in this study to be associated with ESCC in Turkmen was the rs1625895 variant of TP53. TP53 is a tumor suppressor protein which is activated by phosphorylation following a genotoxic event and prevents the cell from proceeding from G1 to S phase of the cell cycle [42]. This variant is the result of a nucleotide substitition (G to A) in the intronic region of TP53 gene (between exons 6 and 7). In our study, the A allele of this variant was associated with a 1.5-fold increase in the risk of ESCC under a dominant model (p = 0.005). This variant has been described in association with colorectal [61] and nasopharyngeal [62] cancer. Recently, one study reported that the rs1625895 variant is associated with a higher level of micronuclei in the blood cells of individuals exposed to vinyl chloride [63].
We observed more evidence for a genetic component for ESCC in Turkmen, compared to non-Turkmens, consistent with previous report of a high incidence of ESCC among Turkmen [64] and our previous study which showed strong familial component to esophageal cancer among Turkmen [7]. The different associations in Turkmen and non-Turkmen could be due to chance, but could also be the result of social and environmental differences in the populations. Second, these variants may be indirectly associated with ESCC, and the responsible variants might be in linkage disequilibrium with these variants in Turkmen, but not in other groups. Third, genetic modifier variants may be required to increase cancer risk and the relevant genetic modifier alleles may be prevalent in Turkmen, but not in non-Turkmen.
In conclusion, this study extends our earlier observations that genetic factors play an important role in the high incidence of esophageal cancer in the northeast of Iran. A strong and significant association was seen with the Arg48His allele of ADH1B gene. Interestingly, the variant is related to the risk of ESCC in alcohol non-drinkers, suggesting that other carcinogens might be causative. It is important that these observations be confirmed in larger series of patients, ideally with reference to exposure data and incorporating data from other biomarkers, such as DNA adducts. To date, we have strong evidence that both BRCA2 and ADH1B contribute to the burden of esophageal cancer in Northern Iran, but the known variants in these two genes are not sufficient to explain the high incidence of esophageal cancer in the region, or the high familial relative risk, and further studies are needed.
Acknowledgments
We thank Dr. Jafar Bashiri from the case-control study center (Aras Clinic) in Ardabil city, Ms. Safura Kor from the case-control study center (Atrak Clinic) and Mrs. Goharshad Goglani and Mrs. Mina Bahrami from cohort study center in Gonbad city for helping with study subject recruitment. We also appreciate the kind help of Cheryl Crozier from Analytical Genetics Technology Centre of the University of Toronto and Alexandre Belisle from the Genotyping Platform of the McGill University and Genome Quebec Innovation Centre for helping in performing the genetic tests of this study. This study was supported by grants from the Canadian Institutes of Health Research (CIHR) and the Digestive Disease Research Center (DDRC), Tehran University of Medical Sciences (TUMS).
Contributor Information
MR Akbari, Digestive Disease Research Center, Tehran University of Medical Sciences, Tehran, Iran; Women’s College Research Institute, University of Toronto, Toronto, Canada; Institute of Medical Science, Faculty of Medicine, University of Toronto, Toronto, Canada.
R Malekzadeh, Digestive Disease Research Center, Tehran University of Medical Sciences, Tehran, Iran.
R Shakeri, Digestive Disease Research Center, Tehran University of Medical Sciences, Tehran, Iran.
D Nasrollahzadeh, Digestive Disease Research Center, Tehran University of Medical Sciences, Tehran, Iran.
M Foumani, Women’s College Research Institute, University of Toronto, Toronto, Canada.
Y Sun, Women’s College Research Institute, University of Toronto, Toronto, Canada.
A Pourshams, Digestive Disease Research Center, Tehran University of Medical Sciences, Tehran, Iran.
M Sotoudeh, Digestive Disease Research Center, Tehran University of Medical Sciences, Tehran, Iran.
P Boffetta, International Agency for Research on Cancer, Lyon, France.
SM Dawsey, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland, USA.
P Ghadirian, Epidemiology Research Unit Research Centre, CHUM- Hôtel-Dieu, University of Montreal, Montreal, Canada.
SA Narod, Women’s College Research Institute, University of Toronto, Toronto, Canada.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Xing D, Tan W, Lin D. Genetic polymorphisms and susceptibility to esophageal cancer among Chinese population (review) Oncol Rep. 2003;10:1615–1623. [PubMed] [Google Scholar]
- 3.Kmet J, Mahboubi E. Esophageal cancer in the Caspian littoral of Iran: initial studies. Science. 1972;175:846–853. doi: 10.1126/science.175.4024.846. [DOI] [PubMed] [Google Scholar]
- 4.Saidi F, Sepehr A, Fahimi S, et al. Oesophageal cancer among the Turkomans of northeast Iran. Br J Cancer. 2000;83(9):1249–1254. doi: 10.1054/bjoc.2000.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semnani S, Sadjadi A, Fahimi S, et al. Declining incidence of esophageal cancer in the Turkmen Plain, eastern part of the Caspian littoral of Iran: a retrospective cancer surveillance. Cancer Detect Prev. 2006;30:14–19. doi: 10.1016/j.cdp.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Islami F, Kamangar F, Aghcheli K, et al. Epidemiologic features of upper gastrointestinal tract cancers in Northeastern Iran. Br J Cancer. 2004;90:1402–1406. doi: 10.1038/sj.bjc.6601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbari MR, Malekzadeh R, Nasrollahzadeh D, et al. Familial risks of esophageal cancer among the Turkmen population of the Caspian littoral of Iran. Int J Cancer. 2006;119:1047–1051. doi: 10.1002/ijc.21906. [DOI] [PubMed] [Google Scholar]
- 8.Akbari MR, Malekzadeh R, Nasrollahzadeh D, et al. Germline BRCA2 mutations and the risk of esophageal squamous cell carcinoma. Oncogene. 2008 Feb 21;27(9):1290–1296. doi: 10.1038/sj.onc.1210739. [DOI] [PubMed] [Google Scholar]
- 9.Sumer F. Oguzlar (Turkmenler) fifth edition. Istanbul: Turk Dunyasi Arastimalari Yakf; 1999. [Google Scholar]
- 10.Ke L. Mortality and incidence trends from esophageal cancer in selected geographic areas of China CIRCA 1970–90. Int J Cancer. 2002;102:271–274. doi: 10.1002/ijc.10706. [DOI] [PubMed] [Google Scholar]
- 11.Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and esophageal cancer risk. Int J Cancer. 2007;121(8):1643–1658. doi: 10.1002/ijc.23044. [DOI] [PubMed] [Google Scholar]
- 12.Pourshams A, Saadatian-Elahi M, Nouraie M, et al. Golestan cohort study of oesophageal cancer: feasibility and first results. Br J Cancer. 2005 Jan 17;92(1):176–181. doi: 10.1038/sj.bjc.6602249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadjadi A, Malekzadeh R, Derakhshan MH, Sepehr A, Nouraie M, Sotoudeh M, et al. Cancer occurrence in Ardabil: results of a population-based cancer registry from Iran. Int J Cancer. 2003;107(1):113–118. doi: 10.1002/ijc.11359. [DOI] [PubMed] [Google Scholar]
- 14.Bugni JM, Han J, Tsai MS, Hunter DJ, Samson LD. Genetic association and functional studies of major polymorphic variants of MGMT. DNA Repair (Amst) 2007;6(8):1116–1126. doi: 10.1016/j.dnarep.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Morita S, Yano M, Tsujinaka T, et al. Association between genetic polymorphisms of glutathione S-transferase P1 and N-acetyltransferase 2 and susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 1998;79(5):517–520. doi: 10.1002/(sici)1097-0215(19981023)79:5<517::aid-ijc12>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 16.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Fallin D, Schork NJ. Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data. Am J Hum Genet. 2000;67(4):947–959. doi: 10.1086/303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- 21.Höög JO, Hedberg JJ, Strömberg P, Svensson S. Mammalian alcohol dehydrogenase - functional and structural implications. J Biomed Sci. 2001;8(1):71–76. doi: 10.1007/BF02255973. [DOI] [PubMed] [Google Scholar]
- 22.Hashibe M, McKay JD, Curado MP, et al. Multiple ADH genes are associated with upper aerodigestive cancers. Nat Genet. 2008;40(6):707–709. doi: 10.1038/ng.151. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama A, Omori T. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and risk for esophageal and head and neck cancers. Jpn J Clin Oncol. 2003;33(3):111–121. doi: 10.1093/jjco/hyg026. [DOI] [PubMed] [Google Scholar]
- 24.Yang SJ, Wang HY, Li XQ, et al. Genetic polymorphisms of ADH2 and ALDH2 association with esophageal cancer risk in southwest China. World J Gastroenterol. 2007;13(43):5760–5764. doi: 10.3748/wjg.v13.i43.5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee CH, Lee JM, Wu DC, et al. Carcinogenetic impact of ADH1B and ALDH2 genes on squamous cell carcinoma risk of the esophagus with regard to the consumption of alcohol, tobacco and betel quid. Int J Cancer. 2008;122(6):1347–1356. doi: 10.1002/ijc.23264. [DOI] [PubMed] [Google Scholar]
- 26.Ma L, Kuhlow A, Glatt H. Ethanol enhances the activation of 1-hydroxymethylpyrene to DNA adduct-forming species in the rat. Polycyclic Aromatic Compounds. 2002;22:933–946. [Google Scholar]
- 27.Kollock R, Meinl W, Schneider H, et al. Efficient oxidation of promutagenic hydroxymethylpyrenes by cDNA-expressed human alcohol dehydrogenase ADH2 and its inhibition by various agents. Biochem Pharmacol. 2008;75(2):527–537. doi: 10.1016/j.bcp.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 28.Roth MJ, Guo-King W, Lewin KJ, et al. High levels of carcinogenic polycyclic aromatic hydrocarbons present within food from Linxian, China may contribute to that region’s high incidence of oesophageal cancer. Eur J Cancer. 1998;34:757–758. doi: 10.1016/s0959-8049(97)10071-5. [DOI] [PubMed] [Google Scholar]
- 29.Gustavsson P, Jakobsson R, Johansson H, Lewin F, Norell S, Rutkvist LE. Occupational exposures and squamous cell carcinoma of the oral cavity, pharynx, larynx, and oesophagus: a case-control study in Sweden. Occup Environ Med. 1998;55(6):393–400. doi: 10.1136/oem.55.6.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth MJ, Qiao Y, Rothman N, et al. High urine 1-hydroxypyrene glucuronide concentrations in Linxian, China, an area of high risk for squamous oesophageal cancer. Biomarkers. 2001;6:381–386. doi: 10.1080/13547500110044780. [DOI] [PubMed] [Google Scholar]
- 31.van Gijssel HE, Divi RL, Olivero OA, et al. Semiquantitation of polycyclic aromatic hydrocarbon-DNA adducts in human esophagus by immunohistochemistry and the automated cellular imaging system. Cancer Epidemiol Biomarkers Prev. 2002;11(12):1622–1629. [PubMed] [Google Scholar]
- 32.Thakker DR, Yagi H, Levin W, Wood AW, Conney AH, Jerina DM. Polycyclic aromatic hydrocarbons: metabolic activation to ultimate carcinogens. In: Anders MW, editor. Bioactivation of Foreign Compounds. Academic Press Inc.; 1985. pp. 177–242. [Google Scholar]
- 33.Horn J, Flesher JW, Lehner AF. 1-Sulfooxymethylpyrene is an electrophilic mutagen and ultimate carcinogen of 1-methyl- and 1-hydroxymethylpyrene. Biochem Biophys Res Commun. 1996;228(1):105–109. doi: 10.1006/bbrc.1996.1623. [DOI] [PubMed] [Google Scholar]
- 34.Engst W, Landsiedel R, Hermersdörfer H, Doehmer J, Glatt H. Benzylic hydroxylation of 1-methylpyrene and 1-ethylpyrene by human and rat cytochromes P450 individually expressed in V79 Chinese hamster cells. Carcinogenesis. 1999;20(9):1777–1785. doi: 10.1093/carcin/20.9.1777. [DOI] [PubMed] [Google Scholar]
- 35.Surh Y-J, Miller JA. Roles of electrophilic sulfuric acid ester metabolites in mutagenesis and carcinogenesis by some polynuclear aromatic hydrocarbons. Chem-Biol Interact. 1994;92:351–362. doi: 10.1016/0009-2797(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 36.Glatt HR, Meinl W, Kuhlow A, Ma L. Metabolic formation, distribution and toxicological effects of reactive sulphuric acid esters. Nova Acta Leopoldina NF87. 2003;329:151–161. [Google Scholar]
- 37.Glatt HR, Seidel A, Harvey RG, Coughtrie MWH. Activation of benzylic alcohols to mutagens by human hepatic sulphotransferases. Mutagenesis. 1994;9:553–557. doi: 10.1093/mutage/9.6.553. [DOI] [PubMed] [Google Scholar]
- 38.Kamangar F, Strickland PT, Pourshams A, et al. High exposure to polycyclic aromatic hydrocarbons may contribute to high risk of esophageal cancer in northeastern Iran. Anticancer Res. 2005;25(1B):425–428. [PubMed] [Google Scholar]
- 39.Kang DH, Rothman N, Poirier MC, et al. Interindividual differences in the concentration of 1-hydroxypyrene-glucuronide in urine and polycyclic aromatic hydrocarbon-DNA adducts in peripheral white blood cells after charbroiled beef consumption. Carcinogenesis. 1995 May;16(5):1079–1085. doi: 10.1093/carcin/16.5.1079. [DOI] [PubMed] [Google Scholar]
- 40.Jongeneelen FJ. Methods for routine biological monitoring of carcinogenic PAH-mixtures. Sci Total Environ. 1997;199(1–2):141–149. doi: 10.1016/s0048-9697(97)00064-8. [DOI] [PubMed] [Google Scholar]
- 41.Rothman N, Poirier MC, Baser ME, et al. Formation of polycyclic aromatic hydrocarbon-DNA adducts in peripheral white blood cells during consumption of charcoal-broiled beef. Carcinogenesis. 1990 Jul;11(7):1241–1243. doi: 10.1093/carcin/11.7.1241. [DOI] [PubMed] [Google Scholar]
- 42.Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 43.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98(6):859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 44.Zheng Y, Shen H, Sturgis EM, et al. Cyclin D1 polymorphism and risk for squamous cell carcinoma of the head and neck: a case-control study. Carcinogenesis. 2001;22(8):1195–1199. doi: 10.1093/carcin/22.8.1195. [DOI] [PubMed] [Google Scholar]
- 45.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11(5):1005–1011. [PubMed] [Google Scholar]
- 46.Solomon DA, Wang Y, Fox SR, et al. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278(32):30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- 47.Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63(21):7056–7061. [PubMed] [Google Scholar]
- 48.Holley SL, Parkes G, Matthias C, et al. Cyclin D1 polymorphism and expression in patients with squamous cell carcinoma of the head and neck. Am J Pathol. 2001;159(5):1917–1924. doi: 10.1016/S0002-9440(10)63038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casson AG, Zheng Z, Evans SC, et al. Cyclin D1 polymorphism (G870A) and risk for esophageal adenocarcinoma. Cancer. 2005;104(4):730–739. doi: 10.1002/cncr.21229. [DOI] [PubMed] [Google Scholar]
- 50.Gupta VK, Feber A, Xi L, Pennathur A, Wu M, Luketich JD, Godfrey TE. Association between CCND1 G/A870 polymorphism, allele-specific amplification, cyclin D1 expression, and survival in esophageal and lung carcinoma. Clin Cancer Res. 2008;14(23):7804–7812. doi: 10.1158/1078-0432.CCR-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sathyan KM, Nalinakumari KR, Abraham T, Kannan S. CCND1 polymorphisms (A870G and C1722G) modulate its protein expression and survival in oral carcinoma. Oral Oncol. 2008;44(7):689–697. doi: 10.1016/j.oraloncology.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25(11):1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 53.Pabalan N, Bapat B, Sung L, Jarjanazi H, Francisco-Pabalan O, Ozcelik H. Cyclin D1 Pro241Pro (CCND1-G870A) polymorphism is associated with increased cancer risk in human populations: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2773–2781. doi: 10.1158/1055-9965.EPI-08-0169. [DOI] [PubMed] [Google Scholar]
- 54.Kimura Y, Nishimura FT, Abe S, Fukunaga T, Tanii H, Saijoh K. A Promoter Polymorphism in the ALDH2 Gene Affects Its Basal and Acetaldehyde/Ethanol-Induced Gene Expression in Human Peripheral Blood Leukocytes and HepG2 Cells. Alcohol Alcohol. 2009 Jan 14; doi: 10.1093/alcalc/agn123. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 55.Chou WY, Stewart MJ, Carr LG, et al. An A/G polymorphism in the promoter of mitochondrial aldehyde dehydrogenase (ALDH2): effects of the sequence variant on transcription factor binding and promoter strength. Alcohol Clin Exp Res. 1999;23(6):963–968. [PubMed] [Google Scholar]
- 56.Lu SH, Chui SX, Yang WX, Hu XN, Guo LP, Li FM. Relevance of N-nitrosamines to oesophageal cancer in China. IARC Sci Publ. 1991;(105):11–17. [PubMed] [Google Scholar]
- 57.Umbenhauer D, Wild CP, Montesano R, et al. O(6)-methyldeoxyguanosine in oesophageal DNA among individuals at high risk of oesophageal cancer. Int J Cancer. 1985;36(6):661–665. doi: 10.1002/ijc.2910360607. [DOI] [PubMed] [Google Scholar]
- 58.Snow ET, Foote RS, Mitra S. Base-pairing properties of O6-methylguanine in template DNA during in vitro DNA replication. J Biol Chem. 1984;259(13):8095–8100. [PubMed] [Google Scholar]
- 59.Olsson M, Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980;255(22):10569–10571. [PubMed] [Google Scholar]
- 60.Margison GP, Heighway J, Pearson S, et al. Quantitative trait locus analysis reveals two intragenic sites that influence O6-alkylguanine-DNA alkyltransferase activity in peripheral blood mononuclear cells. Carcinogenesis. 2005;26(8):1473–1480. doi: 10.1093/carcin/bgi087. [DOI] [PubMed] [Google Scholar]
- 61.Själander A, Birgander R, Athlin L, et al. P53 germ line haplotypes associated with increased risk for colorectal cancer. Carcinogenesis. 1995;16(7):1461–1464. doi: 10.1093/carcin/16.7.1461. [DOI] [PubMed] [Google Scholar]
- 62.Birgander R, Själander A, Zhou Z, Fan C, Beckman L, Beckman G. p53 polymorphisms and haplotypes in nasopharyngeal cancer. Hum Hered. 1996;46(1):49–54. doi: 10.1159/000154325. [DOI] [PubMed] [Google Scholar]
- 63.Qiu YL, Wang W, Wang T, et al. Genetic polymorphisms, messenger RNA expression of p53, p21, and CCND1, and possible links with chromosomal aberrations in Chinese vinyl chloride-exposed workers. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2578–2584. doi: 10.1158/1055-9965.EPI-07-2925. [DOI] [PubMed] [Google Scholar]
- 64.Mahboubi E, Kmet J, Cook PJ, Day NE, Ghadirian P, Salmasizadeh S. Oesophageal cancer studies in the Caspian littoral of Iran: the Caspian cancer registry. Br J Cancer. 1973;28:197–214. doi: 10.1038/bjc.1973.138. [DOI] [PMC free article] [PubMed] [Google Scholar]