Abstract
The aim of this study was to elucidate the prevalence of transmitted drug-resistant (TDR) mutations and reverse transcriptase (RT) thumb subdomain polymorphisms in CRF01_AE and CRF07_BC virus among newly diagnosed, therapy-naive HIV-1 patients in Guangdong Province, China. One hundred and sixty-four samples were collected in the Guangzhou Eighth People's Hospital. The entire protease gene and 300 codons of the entry part of the reverse transcriptase were amplified and sequenced. Furthermore, genotypic drug resistance, polymorphisms, and their phylogeny were analyzed. According to eligibility criteria, seven samples were excluded, and 119 of 157 (75.8%) samples (84 CRF01_AE and 35 CRF07_BC) were amplified and sequenced successfully. The prevalence of TDR identified in the present study was 6.7% [8/119, 95% confidence interval (CI) 1.8–11.6%]. Three major resistance mutations, K103N, M184V, and Y188L, each of which caused more than one drug resistance, appeared in only two patients; the prevalence [1.7 % (2/119)] was relatively low. Until now, this is the first observation of the five newly identified accessory mutations, V35T, K43E, V60I, K122E, and E203D, and seven thumb subdomain polymorphisms, A272P, K277R, K281R, T286A, E291D, V292I, and I293V, in the RT gene in China. These findings provide useful information for guidance on the antiretroviral therapy (ART) policy in China where therapeutic options are still limited.
Introduction
HIV-1 infection has spread throughout China since the first HIV-1 case was identified in 1985, and the number of HIV-1 patients keeps rising. In Guangdong Province, the incidence of HIV-1 patients in 2011 increased 17.7%, compared with that in 2010.1 Sexual contact was the major risk factors for HIV-1 transmission in Guangdong Province, and the estimated prevalence was 56.5%, including 15.2% of men who have sex with men (MSM).1 In addition, among the HIV-1 patients, 81.4% were 20–49 years old.1 Therefore, it is urgent to monitor HIV-1 infection in Guangdong. In recent years, the distribution of HIV-1 subtypes has changed in China. Previous studies reported that CRF_BC (typically CRF07_BC) accounted for 45%, whereas subtype B' accounted for 38% and CRF01_AE accounted for 15%.2 Today, the numbers of HIV-1 CRF01_AE infections are also increasing in some provinces of China, such as Guangxi Pingxiang (88%)3 and Hainan (84%).4 It is possible that CRF07_BC and CRF01_AE have replaced subtype B' and become the most prevalent strains in China.
In recent years, expanded access to highly active antiretroviral therapy (HAART) for HIV-1 patients has significantly reduced the rate of AIDS-related morbidity and mortality.5 According to the ‘‘Four Frees and One Care’’ program launched by the Chinese government, since 2003 many HIV-1 patients who reside in China could acquire free antiretroviral therapy (ART). Transmitted drug-resistant (TDR) HIV-1 variants have been detected in therapy-naive HIV-1 patients since the massive scale-up of ART in China.2 A recent study reported that the prevalence of mutations conferring resistance to at least one antiretroviral (ARV) medicine was 4.5% among therapy-naive HIV-1 MSM in Liaoning province.6 However, TDR was also detected among therapy-naive patients in other Asian countries. In Japan, the prevalence of TDR increased from 5.9% in 2003 to 8.3% in 2008 in therapy-naive HIV-1 patients7; it increased to 5% in Vietnam,8 3% in Korea,9 and 4.3% in South Korea.10 Though the prevalence of TDR was relatively lower in China, it could threaten the public health of Chinese, since it could compromise the effectiveness of virologic suppression and successful treatment.
In this study, TDR prevalence was detected among newly diagnosed, therapy-naive HIV-1 patients infected with CRF01_AE and CRF07_BC variants in Guangdong province. Additionally, several newly identified accessory mutations and RT thumb subdomain polymorphisms were identified for the first time in China.
Materials and Methods
Study subjects and specimens
This study was conducted among patients attending the Outpatient Clinic of the Guangzhou Eighth People's Hospital between March 2009 and April 2011. Referred based on the criteria defined by the WHO for surveillance of HIV-1 TDR, newly diagnosed, therapy-naive HIV-1 patients were enrolled.11 Eligibility criteria included age above 18 years, never exposed to antiretroviral medicines (ARVs) (refer to the record of the medical chart review), no WHO clinical stage 4 event, HIV RNA viral load more than 1000 copies/ml, and laboratory evidence of newly acquired HIV-1 infection [confirmed by an HIV-1 RNA test (One Step HIV-1 SYBR real-time RT-PCR Kit, DaAn Gene Diagnostic Center of Sun Yat-sen University, Guangzhou, China; limit of detection is 200 copies/ml) or Western blot test of p24 antigen].11 Exclusion criteria included age below 18 years, previous ARVs used, documented WHO clinical stage 4 event, HIV RNA viral load less than 1000 copies/ml, and HIV-1 RNA test or Western blot test of p24 antigen was negative.11 Information on demographics and risk behavior for HIV-1 patients was recorded during the medical chart review and personal interview. CD4 T cell count was also determined at study entry. All patients provided written informed consent. The Guangzhou Eighth People's Hospital Ethics Committee approved the study.
RNA extraction, RT-PCR, and sequencing
HIV-1 RNA was extracted from 200 μl of HIV-1-positive plasma using the DaAn Gene nucleotide extraction kit (DaAn Gene Diagnostic Center of Sun Yat-sen University, Guangzhou, China) according to the manufacturer's instructions. A 1.3-kp region of the pol gene, covering the whole protease and an entire part of the reverse transcriptase gene, was reverse transcribed and amplified from the extracted RNA with one-step reverse transcriptase polymerase chain reaction (RT-PCR) (Takara, Dalian, China) and nested PCR (Takara, Dalian, China). The forward primer WMA26-TTGGAAATGTGGAAAGGAAGGAC-3 (HXB2 location 2028–2050) and the reverse primer RT21-CTGTATTTCTGCTATTAAGTCTTTTGATGGG-3 (HXB2 location 3509–3539) were used in one-step RT-PCR with cycling conditions of reverse transcript reaction at 50°C for 30 min and 94°C for 5 min, followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 2 min, and a final extension at 72°C for 7 min.12 For the nested PCR, the forward primer PRO1 5-CAGAGCCAACAGCCCCACCA-3 (HBX2 location 2147–2166) and the reverse primer RT20 5-CTGCCAGTTCTAGCTCTGCTTC-3 (HBX2 location 3441–3462) were used.12 The product of the one-step RT-PCR (5 μl) was used as the template for nested PCR amplification. The cycling conditions were 94°C for 5 min, followed by 45 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 7 min. Both one-step RT-PCR and nested PCR amplification were carried out in a thermal cycler (ABI 9700, USA). Furthermore, PCR products were detected by agarose gel electrophoresis and the expected amplicons were excised and extracted from the gel using the Takara spin gel extraction kit (Takara, Dalian, China). Moreover, DNA sequencing was performed using BigDye Terminators v3.1 on an ABI3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). All PCR products were sequenced in both directions.
Drug-resistant mutations of HIV-1 isolates
Protease (PR) and reverse transcriptase (RT) drug resistance mutations were determined based on the last updated (December 2010) guidelines from the International AIDS Society Resistance Testing-USA panel. The drug resistance interpretation was obtained from the Stanford Resistance Database tool: HIValg Program Version 6.0.11 (http://hivdb.stanford.edu/pages/algs/HIValg.html). This tool compares the input sequences with sequences of HIV-1 subtype B isolates shown to confer resistance to ARVs.
Phylogenetic analysis
HIV-1 subtypes were classified according to the phylogenetic analysis of the pol sequence (whole protease and entire part of the reverse transcriptase gene). The sequences obtained were edited by ContigExpress software (Invitrogen), and then were aligned by the BioEdit program (North Carolina State University, Raleigh, NC: http://www.mbio.ncsu.edu/bioedit/bioedit.html). Alignments also included the reference sequences representing HIV-1 genetic circulating recombinant forms obtained from the Los Alamos National Laboratory (http://hiv-web.lanl.gov). A phylogenetic tree was constructed using the MEGA program (Molecular Evolutionary Genetic Analysis Software, Version 4.0). The reliability of genetic distance was evaluated using the bootstrap test (1025 bootstrap replicates).
Statistical methods
Statistical Package for the Social Sciences (SPSS, 16.0) (Chicago, IL) was used to analyze the data in the present study. TDR prevalence was estimated with a 95% CI based on the binomial distribution. We use the Chi-square test for comparing categorical data and Kruskal–Wallis or Student's t-test for investigating continuous data. The difference was considered statistically significant when p<0.05.
Results
Patient characteristics
One hundred and sixty-four newly diagnosed HIV-1 patients were enrolled locally from March 2009 to April 2011. Seven patients who did not meet the eligibility criteria were excluded, including three of five patients infected with the CRF01_AE virus who were less than 18 years old; the other two patients were exposed to ARVs prior to plasma sample collection. Two patients infected with the CRF07_BC virus were diagnosed as HIV-1 seropositive before 2009. Based on RT-PCR and sequencing results, 119/157 (75.8%) samples (included 84 CRF01_AE and 35 CRF07_BC) were amplified and sequenced successfully. Baseline characteristics, except for mean viral load did not differ between the patients who were infected with CRF01_AE and CRF07_BC virus (Table 1).
Table 1.
Characteristics of Patients Infected with CRF01_AE and CRF07_BC Virus in This Study
| Total | CRF01_AE | CRF_07BC | p | |
|---|---|---|---|---|
| Patients | 119 | 84 (70.6%) | 35 (29.4%) | |
| Sex | 0.516 | |||
| Female | 49 (41.2%) | 33 (39.3%) | 16 (45.7%) | |
| Male | 70 (58.8%) | 51 (60.7%) | 19 (54.3%) | |
| Age—mean years (SD) | 43.6 (12.9) | 43.1 (12.9) | 44.9 (13.0) | 0.581 |
| Risk factor | 0.520 | |||
| Heterosexual contact | 61 (51.3%) | 49 (58.3%) | 12 (34.3%) | |
| MSM | 5 (4.2%) | 3 (3.6%) | 2 (5.7%) | |
| IDU | 49 (41.1%) | 29 (34.5%) | 20 (57.1%) | |
| Blood transfusion | 2 (1.7%) | 1 (1.2%) | 1 (2.9%) | |
| Unknown | 2 (1.7%) | 2 (2.4%) | 0 (0%) | |
| Viral load (log10 copies/ml) [median (range)] | 5.12 (3.94–5.18) | 5.12 (4.16–5.31) | 4.26 (3.44–4.35) | 0.000 |
| CD4 count (cells/ml) [median (range)] | 213 (92–318) | 223 (92–338) | 192 (87–263) | 0.489 |
MSM, men who have sex with men; IDU, intravenous drug user; SD, standard deviation.
Resistance-associated mutations of the pol gene
No major drug-resistant mutations to protease inhibitors (PIs) were detected in the present study. One or more TDR associated with reverse transcriptase inhibitors (RTIs) were identified in eight of 119 patients, yielding an estimated TDR prevalence of 6.7% with a 95% CI 1.8%–11.6% (Table 2). The prevalence of TDR associated with nucleoside reverse transcriptase inhibitors (NRTIs) and nonnucleoside reverse transcriptase inhibitors (NNRTIs) was 4.2% (5 of 119) and 5.9 % (7 of 119), respectively. In addition, prevalences of 7.1% (6/84) and 5.7% (2/35) of TDR associated with RTIs were identified in CRF01_AE and CRF07_BC, respectively. Nine different TDR, T69S, T69N, K70Q, V75L, K101Q, K103N, V179E, M184V, and Y188L, were identified. Among them, mutations K103N, M184V, and Y188L were major RTI resistance mutations, each of which caused at least one drug resistance, and were found in only two patients. The prevalence [1.7% (2/119)] of major TDR was relatively low. The demographic and virological characteristics of the eight patients are shown in Table 2.
Table 2.
Clinical Information and Resistance-Related Amino Acid Substitutions of Patients Who Harbored Primary Resistance to Antiretroviral Drugs
| |
|
|
|
|
|
|
Surveillance drug resistance |
||
|---|---|---|---|---|---|---|---|---|---|
| Access no. | Age (years) | Sex | Risk factor/diagnosis (year) | CD4 cell count (cells/ml) | HIV-1 RNA load (log10 copies/ml) | Subtype | PI | NRTI | NNRTI |
| JN848881 | 64 | Female | Hetero/2011 | 78 | 5.81 | CRF01_AE | T69S | V179E | |
| JN848848 | 42 | Male | Hetero/2009 | 61 | 5.33 | CRF01_AE | T69N, V75L | ||
| JN848901 | 61 | Male | Hetero/2010 | 196 | 4.92 | CRF01_AE | K101Q, K103N | ||
| JN848853 | 55 | Male | Hetero/2009 | 75 | 5.40 | CRF01_AE | T69N | ||
| JN848847 | 47 | Female | Hetero/2010 | 100 | 3.52 | CRF01_AE | K70Q | ||
| JN848906 | 51 | Male | Hetero/2010 | 358 | 5.44 | CRF01_AE | V179E | ||
| JN848940 | 44 | Female | IDU/2009 | 339 | 3.72 | CRF 07_BC | L10V | M184V | V179E, Y188L |
| JN848924 | 39 | Male | IDU/2009 | 80 | 3.79 | CRF 07_BC | L33I | V179D | |
Hetero, heterosexual; IDU, intravenous drug user; PI, protease inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; Access no., GenBank accession number(s).
Polymorphisms of reverse transcriptase
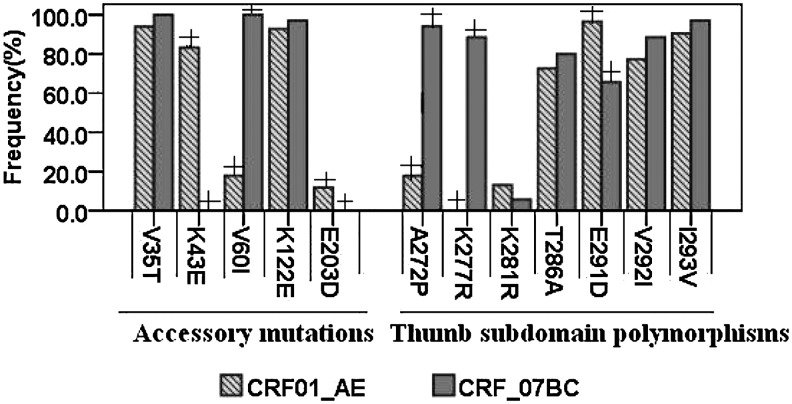
Five newly identified polymorphisms, V35T, K43E, V60I, K122E, and E203D, were found (Fig. 1). Among them, polymorphism frequencies of K43E, V60I, and E203D were different between CRF01_AE and CRF07_BC strains (p<0.05). Otherwise, there was no significant different between V35T and K122E (p<0.05) in the two recombinant types in the study. This is the first observation on thumb subdomain polymorphisms A272P, K277R, K281R, T286A, E291D, V292I, and I293V in China (Fig. 1). The frequencies of A272P, K277R, and E291D were statistically significant between CRF01_AE and CRF07_BC (p<0.05). It is worth mentioning that K277R was found in none of the CRF01_AE sequences, but in 88.6% of the CRF07_BC sequences.
FIG. 1.
Newly identified accessory mutations and thumb subdomain polymorphisms. Histograms represent the frequencies of the indicated mutations among CRF01_AE and CRF07_BC sequences. Frequencies were expressed as percentages of the total CRF01_AE (N=84) or CRF07_BC (N=35) samples examined, and compared with the χ2 test or Fisher's exact test. “+” denoted statistically significant (p<0.05).
Phylogenetic tree reconstructed
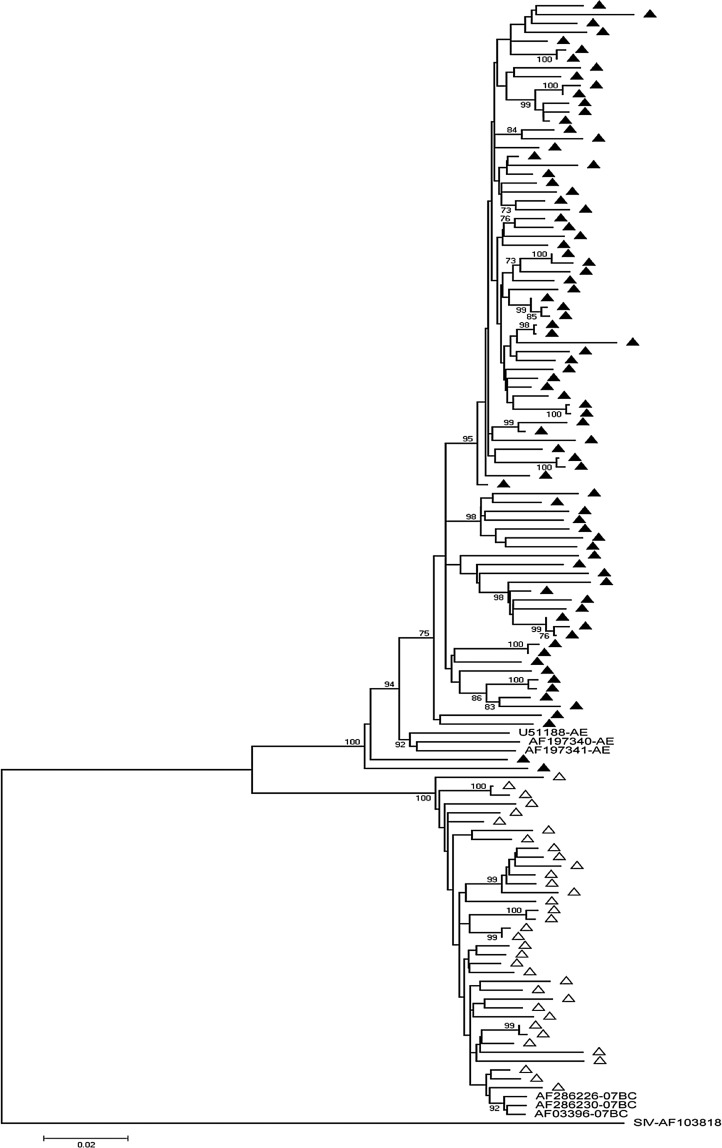
Eighty-four CRF01_AE and 35 CRF07_BC samples were amplified and sequenced successfully. A 1.3-kb pol sequence for every sample was analyzed and a phylogenetic tree was constructed using the neighbor-joining method (Fig. 2). The bootstrap value was 1025 replicates.
FIG. 2.
The phylogenetic tree was created based on the neighbor-joining method and the Kimura two-parameter distance estimation method with the protease (PR) and reverse transcriptase (RT) sequences of HIV-1 obtained from newly diagnosed drug-naive patients in this study. The reference sequences of recombinant types used in the comparative phylogenetic analysis were CRF01_AE: U51188, AF197340, and AF197341; CRF_07BC: AF286226, AF03396, and AF286230. The simian immunodeficiency virus sequence AF103818 was used as the outgroup sequence. ▲ represents the sequences of CRF01_AE; △ represents the sequences of CRF_07BC. The bootstrap values (1025 replicates) over 70% are marked at the major branches of the tree. The scale bar represents 0.02 nucleotide substitution per site between the sequences.
Discussion
The present study provided a description of the prevalence of TDR and genetic polymorphisms of HIV-1 among newly diagnosed HIV-1 patients in Guangdong Province. The overall prevalence of TDR was 6.7% (8/119), representing 7.1% (6/84) for CRF01_AE and 5.7% (2/35) for CRF07_BC, even though the patients had never been exposed to ARVs. In previous surveys of HIV-1 drug resistance, a TDR prevalence rate of 4.5% was found in 217 samples from ARV therapy-naive MSM in Liaoning6; in a nationwide investigation on TDR in ARV therapy-naive patients in China,2 the prevalence rate was found to be 3.8%. However, the TDR prevalence rates in the present study were relatively higher. It is noteworthy that the TDR prevalence rates were different among the studies, although most of the HIV/AIDS patients had the same opportunity to acquire free ART in China.
We presumed that several factors could contribute to the differences, such as subtype distribution and risk behaviors.13 It is known that HIV-1 is characterized by a highly genetic diversity in distinct subtypes. However, CRF01_AE and CRF07_BC were the major subtypes in the present study, which were different from previous studies (Fig. 2). Therefore, It is possible that TDR were different among subtypes. Otherwise, TDR would be associated with risk behaviors, such as sexual contact and injecting drug use.14,15 More than 95% of the patients were infected with HIV-1 through sexual contact or injecting drug use in this study (Table 1). Another possible explanation was that the increasing occurrence of TDR was related to the massive scale-up of ART in China. However, the prevalence of TDR maintained a low trend in the present study.
Present findings agreed with the results reported in other Asian countries.9,10 No major PI drug resistance mutations were identified in the study. As we know, PIs were not used extensively in China. We inferred that PIs in combination with RTIs (i.e., zidovudine, didanosine, nevirapine, stavudine, and lamivudine) could benefit HIV-1 patients in China.
Several new mutations were found in the RT gene: V35T, K43E, V60I, K122E, and E203D (Fig. 1). It is possible that an accumulation of these mutations increased the numbers of thymidine analog-associated mutations (TAMs) and NNRTI mutations.16 The copresence of V35T, K43E, V60I, K122E, and E203D, combined with TAM-1 (i.e., M41L, L210W, and T215Y) or other polymorphic mutations, might contribute to the high levels of resistance to nucleoside analogs.17 However, single accessory mutations had no effect on the susceptibility to ARVs. For example, Huigen et al. found that the K43E mutation alone did not increase the levels of resistance, but showed a compensatory role when combined with E40F.18 A study by Cane et al. showed that K43E was detected in only 1% of HIV-1 patients with no TAMs but in 24% of samples with four or more TAMs,16 whereas in the present study the mutation was seen in 83.3% of newly diagnosed, therapy-naive HIV-1 patients who were infected with CRF01_AE, and in none of the CRF07_BC patients. It is possible that the K43E change was a subtype-specific polymorphic mutation that was found more frequently in CRF01_AE than CRF07_BC. However, the resistance mechanism of these newly identified accessory mutations was not exactly elaborated.
Most previous studies analyzed the RT sequence information but were restricted only to RT residues 1 to 240. In this study, the polymorphisms on codons 1–300 were identified in the RT gene, which included the p66 thumb subdomain. The p66 thumb subdomain plays a role in DNA polymerization by making important interactions with the minor groove of the template primer through α-helices H (residues 255–268) and I (residues 278–286).19,20 The frequencies of T286A, V292I, and I293V were identified to be relatively higher in both CRF01_AE and CRF07_BC (Fig. 1). These results were similar to the results of Garriga and his colleagues.17 They reported that mutation frequencies of T286A, V292I, and I293V were higher in naive patients than in patients who failed treatment. However, all of their sequences were from subtype B HIV-1 isolates. Thus, we presumed that T286A, V292I, and I293V would be polymorphisms in some subtypes. Additionally, results showed that the prevalence of A272P, K277R, and E291D in CRF01_AE was 15.9%, 0%, and 96.6%, respectively, and in CRF07_BC was 94.3%, 88.6%, and 65.7%, respectively (Fig. 1). Obviously, mutations A272P and K277R were mainly found in CRF07_BC. However, Garriga et al.19 reported that mutations A272P and K277R were more prevalent in patients who failed treatment with abacavir/stavudine than in naive patients; the prevalence of E291D decreased in treated patients. Accordingly, we presumed that A272P and K277R were polymorphisms in CRF07_BC, but not in CRF01_AE. Furthermore, it is possible that combinations of RTIs in some subtypes such as CRF01_AE would cause A272P and K277R. Further investigations would be necessary to determine whether those RT thumb subdomain polymorphisms in different HIV-1 subtypes were related to RTI therapy failure.
In conclusion, the present study showed a low prevalence of TDR for newly diagnosed, therapy-naive HIV-1 patients. From the study, we could see the models of HIV-1 drug-resistant mutations for newly diagnosed, therapy-naive HIV-1 patients. The data would be helpful in understanding the drug resistance HIV-1 strains and determining better treatment for HIV-1 patients living in China. Hence, continued surveillance is needed for newly diagnosed HIV/AIDS patients to understand drug resistance HIV-1 transmission. In addition, the present findings showed that the prevalence of polymorphisms was different between CRF01_AE and CRF07_BC. The relationship between polymorphisms and drug resistance in circulating recombinant types was not completely elucidated. Thus, further work focused on elaborating the effects of polymorphic mutations on drug resistance would be valuable.
Sequence Data
GenBank accession numbers for the sequences obtained in our study are as follows: CRF01_AE sequences, JN848837–JN848920; CRF07_BC sequences, JN848921–JN848955.
Acknowledgments
We are grateful to all the patients who participated in the present study. We thank the doctors in Guangzhou Eighth People's Hospital for collecting samples. This work was supported by the China national special funds for prevention and treatment of major infection disease-AIDS and viral hepatitis (Grant 2008ZX10001-013).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Department of Public Health, Guangdong province. The report of HIV/AIDS prevention and control work in 2011. http://zwgk.gd.gov.cn/006940132/201112/t20111206_294678.html? [Apr 12;2011 ]. http://zwgk.gd.gov.cn/006940132/201112/t20111206_294678.html? (in Chinese).
- 2.Liao L. Xing H. Shang H, et al. The prevalence of transmitted antiretroviral drug resistance in treatment-naive HIV-infected individuals in China. J Acquir Immune Defic Syndr. 2010;53:10–14. doi: 10.1097/QAI.0b013e3181c7d363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laeyendecker O. Zhang GW. Quinn TC, et al. Molecular epidemiology of HIV-1 subtypes in southern China. J Acquir Immune Defic Syndr. 2005;38:356–362. [PubMed] [Google Scholar]
- 4.Deng W. Fu P. Bao LL, et al. Molecular epidemiological tracing of HIV-1 outbreaks in Hainan island of southern China. AIDS. 2009;23:977–985. doi: 10.1097/QAD.0b013e328329217d. [DOI] [PubMed] [Google Scholar]
- 5.Li JY. Li HP. Li L, et al. Prevalence and evolution of drug resistance HIV-1 variants in Henan, China. Cell Res. 2005;15:843–849. doi: 10.1038/sj.cr.7290356. [DOI] [PubMed] [Google Scholar]
- 6.Zhao B. Han XX. Dai D, et al. New trends of primary drug resistance among HIV Type 1-infected men who have sex with men in Liaoning Province, China. AIDS Res Hum Retroviruses. 2011;27:1–4. doi: 10.1089/AID.2010.0119. [DOI] [PubMed] [Google Scholar]
- 7.Hattori J. Shiino T. Gatanaga H, et al. Trends in transmitted drug-resistant HIV-1 and demographic characteristics of newly diagnosed patients: Nationwide surveillance from 2003 to 2008 in Japan. Antiviral Res. 2010;88:72–79. doi: 10.1016/j.antiviral.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen HT. Duc NB. Shrivastava R, et al. HIV drug resistance threshold survey using specimens from voluntary counselling and testing sites in Hanoi, Vietnam. Antivir Ther. 2008;13:115–121. [PubMed] [Google Scholar]
- 9.Kim CO. Chin BS. Han SH, et al. Low prevalence of drug-resistant HIV-1 in patients newly diagnosed with early stage of HIV infection in Korea. Tohoku J Exp Med. 2008;216:259–265. doi: 10.1620/tjem.216.259. [DOI] [PubMed] [Google Scholar]
- 10.Choi JY. Kim EJ. Park YK. Lee JS. Kim SS. National survey for drug-resistant variants in newly diagnosed antiretroviral drug-naive patients with HIV/AIDS in South Korea: 1999–2005. J Acquir Immune Defic Syndr. 2008;49:237–242. doi: 10.1097/QAI.0b013e318188a919. [DOI] [PubMed] [Google Scholar]
- 11.Bennett DE. Myatt M. Bertagnolio S, et al. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008;13:25–36. [PubMed] [Google Scholar]
- 12.Han X. Zhang M. Dai D, et al. Genotypic resistance mutations to antiretroviral drugs in treatment-naive HIV/AIDS patients living in Liaoning Province, China: Baseline prevalence and subtype-specific difference. AIDS Res Hum Retroviruses. 2007;23:357–364. doi: 10.1089/aid.2006.0094. [DOI] [PubMed] [Google Scholar]
- 13.Goldsamt LA. Clatts MC. Parker MM, et al. Prevalence of sexually acquired antiretroviral drug resistance in a community sample of HIV-positive men who have sex with men in New York City. AIDS Patient Care STDS. 2011;25:287–293. doi: 10.1089/apc.2011.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yerly S. Junier T. Gayet-Ageron A, et al. The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS. 2009;23:1415–1423. doi: 10.1097/QAD.0b013e32832d40ad. [DOI] [PubMed] [Google Scholar]
- 15.Parker MM. Gordon D. Reilly A, et al. Prevalence of drug-resistant and nonsubtype B HIV strains in antiretroviral-naive, HIV-infected individuals in New York State. AIDS Patient Care STDS. 2007;21:644–652. doi: 10.1089/apc.2006.0172. [DOI] [PubMed] [Google Scholar]
- 16.Cane PA. Green H. Fearnhill E. Dunn D the UK collaborative group on HIV Drug Resistance. Identification of accessory mutations associated with high-level resistance in HIV-1 reverse transcriptase. AIDS. 2007;21:447–455. doi: 10.1097/QAD.0b013e3280129964. [DOI] [PubMed] [Google Scholar]
- 17.Svicher V. Sing T. Santoro MM, et al. Involvement of novel human immunodeficiency virus type 1 reverse transcriptase mutations in the regulation of resistance to nucleoside inhibitors. J Virol. 2006;80:7186–7198. doi: 10.1128/JVI.02084-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huigen MC. van Ham PM. de Graaf L. Kagan RM. Boucher CA. Nijhuis M. Identification of a novel resistance (E40F) and compensatory (K43E) substitution in HIV-1 reverse transcriptase. Retrovirology. 2008;5:20–31. doi: 10.1186/1742-4690-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garriga C. Pérez-Elías MJ. Delgado R, et al. HIV-1 reverse transcriptase thumb subdomain polymorphisms associated with virological failure to nucleoside drug combinations. J Antimicrob Chemother. 2009;64:251–258. doi: 10.1093/jac/dkp200. [DOI] [PubMed] [Google Scholar]
- 20.Betancor G. Puertas MC. Nevot M, et al. Mechanisms involved in the selection of HIV-1 reverse transcriptase thumb subdomain polymorphisms associated with nucleoside analogue therapy failure. Antimicrob Agents Chemother. 2010;54:4799–4811. doi: 10.1128/AAC.00716-10. [DOI] [PMC free article] [PubMed] [Google Scholar]