Abstract
To characterize human immunodeficiency virus (HIV-1) strains circulating in the Northern region of Colombia in South America, sequences of the viral envelope C2V3C3 region were obtained from patients with different high-risk practices. Close to 60% of the sequences were predicted to belong to macrophage-tropic viruses, according to the positions of acidic amino acids and putative N-linked glycosylation sites. This is in agreement with the fact that most of the patients were recently diagnosed individuals. Phylogenic analysis then allowed assignment of all 35 samples to subtype B viruses. This same subtype was found in previous studies carried out in other Colombian regions. This study thus expands previous analyses with previously missing data from the Northern region of the country. The number and the length of the sequences examined also help to provide a clearer picture of the prevailing situation of the present HIV epidemics in this country.
Genetic variability in human immunodeficiency virus (HIV-1) is due to mutations of the viral genome (nucleotide substitutions, deletions, or insertions) resulting from polymerase errors, as well as genomic recombinations between viral genomes. Characterization of the circulating viral strains is one critical factor, among many others, for monitoring HIV pandemics, vaccine design, and HIV treatment outcome. To date four HIV-1 groups have been reported: the M (main), the O (outlier), the N (non-M, non-O), and the P (putative) group. While all four groups are found in equatorial Africa, the M group is the most widely spread around the world and comprises nine subtypes (clades) designed A, B, C, D, F, G, H, J, and K.1 The O group is mainly confined to Cameroon and surrounding areas.2 The N group has a very low prevalence since all documented cases have been found in Cameroon.3,4 Finally, thus far, only two group P strains have been described. The first one was reported to be found in a Cameroonian woman living in France. The second was identified in an HIV-seropositive male hospital patient in Cameroon.5,6 The complexity of HIV subtype characterization has increased owing to the identification of intersubtype recombinants. These recombinant forms, classified as circulating recombinant forms (CRF) and unique recombinant forms (URF), are considered as a major driving force in the generation of viral diversity.7
In the main (M) group, subtype C accounts for nearly half of all worldwide infections.1 Subtype B causes 11% of infections but its epidemic is more widely and evenly spread around the world than other subtypes. It dominates in North America, the Caribbean, Latin America, Western and Central Europe, and Australia and it circulates with subtype A in Eastern Europe and Central Asia. The other subtypes belonging to the M group have a more limited geographic localization depending of their geographic site of introductions and expansion thereafter: subtype A in East Africa, Eastern Europe, and Central Asia, with the remainder in West and Central Africa and South and Southeast Asia; subtype F in Latin America, Central Africa, Eastern Europe, and Central Asia; and subtype G in West and Central Africa and Western and Central Europe.1 In South America, Brazil is the region where most molecular characterization studies have been performed. An epidemic by subtype B viruses was initially reported but, more recently, the presence of multiple subtypes was described with an apparent increase of new infections with subtype C viruses.8
Previous studies of the prevalence of HIV clades in Colombia showed that viruses belonging to subtype B are most often found. Nevertheless, in the few studies where sequencing methods were used, only a limited number of samples were examined.9,10 They were carried out in different geographic regions of the country but they never included the port of Barranquilla and its metropolitan area where different HIV subtypes might be present.9–11
In the present study, to better characterize current HIV-1 strains circulating in the metropolitan area of Barranquilla, Colombia, 80 samples were obtained from HIV-1-positive individuals. The samples were obtained from HIV-infected patients after obtaining their informed consent. Initial determination of HIV-positive status of these patients was performed by ELISA (Abbott Laboratories) with further confirmation by Western Blot Assay (GS HIV-1 Western Blot kit, Bio-Rad Laboratories blood virus division, Redmond, WA). Blood samples were collected in tubes with EDTA (18 mg K2 EDTA/10 ml; Vacutainer Brand Ref. 366643) and maintained at 4°C until DNA extraction, which was routinely performed on the same day.
Total genomic DNA was extracted from the buffy coat separated by density gradient using the MasterPure DNA Purification Kit for Blood Version II (EPICENTRE Biotechnologies) according to the manufacturer's procedure. DNA samples were then subjected to polymerase chain reaction (PCR) amplification using a nested PCR protocol in a Perkin Elmer 480 Thermal Cycler. For amplification of the HIV env gene, well-conserved primer sequences were chosen.12 First round reactions were conducted using the ED3 (5′-TTAGGCATCTCCTATGGCAGGAAGAAGCGG) primer, corresponding to position 5956–5985 of the HIV-1 HXB-2 genome (GenBank accession no. K03455) and ED-14 (TCTTGCCTGGAGCTGTTTGATGCCCCAGAC) position 7960–7931. These primers allow amplification of a 2.0-kbp fragment of the HIV-1 env gene. For the second round PCR, the set of inner primers ED-31(5′-CCTCAGCCATTACACAGGCCTGTCCAAAG) position 6816-6844 and ED-33(5′-TTACAGTAGAAAAATTCCCCTC) position 7359-7380 was used to amplify the 500 bp of the HIV-1 env C2V3C3 region.
The first round of amplification was carried out in a final volume of 50 μl containing Tris–HCl 10 mM pH 8.3, MgCl2 1.5 mM, 200 μM of each dNTP, 10 pmol of each primer, and 2.5 units of ampliTaq polymerase (Biolase DNA polymerase, BIOLINE). First round PCR reaction included 1 μg of target DNA. Amplification conditions in the first PCR were three cycle (94°C for 1 min, 64°C for 1 min, 72°C for 1 min); 37 cycles (94°C for 15 s, 64°C for 45 s, 72°C for 1 min) followed by a final incubation at 72°C for 5 min. Concentrations in the second round PCR mix were the same as that for the first round. They were carried out in a final volume of 100 μl and included 2 μl of the first round reaction. In the second amplification the conditions were three cycles (94°C for 1 min, 55°C for 1 min, 72°C for 1 min); 37 cycles (94°C for 15 s, 55°C for 45 s, 72°C for 1 min) followed by a final incubation at 72°C for 5 min. The expected DNA fragment was obtained in 42 samples, as detected by agarose gel electrophoresis followed by ethidium bromide staining, while there was no visible PCR product for 38 samples, despite repeated attempts and the use of alternative PCR protocols.
The reasons for these negative results were not further investigated but it was observed that patients with low viremia levels accounted for the majority in whom gene amplification was unsuccessful. These patients were treated with antiretroviral therapy at the time of their inclusion in the study. The analysis was then pursued on the 42 positive samples obtained. PCR products were recovered by ethanol precipitation and directly submitted to sequencing in both directions using primers ED31 and ED33 with the ABI PRISM Big Dye Terminator kit (Applied Biosystems, Foster City, CA) using the Applied Biosystems 3130 Genetic Analyzer.
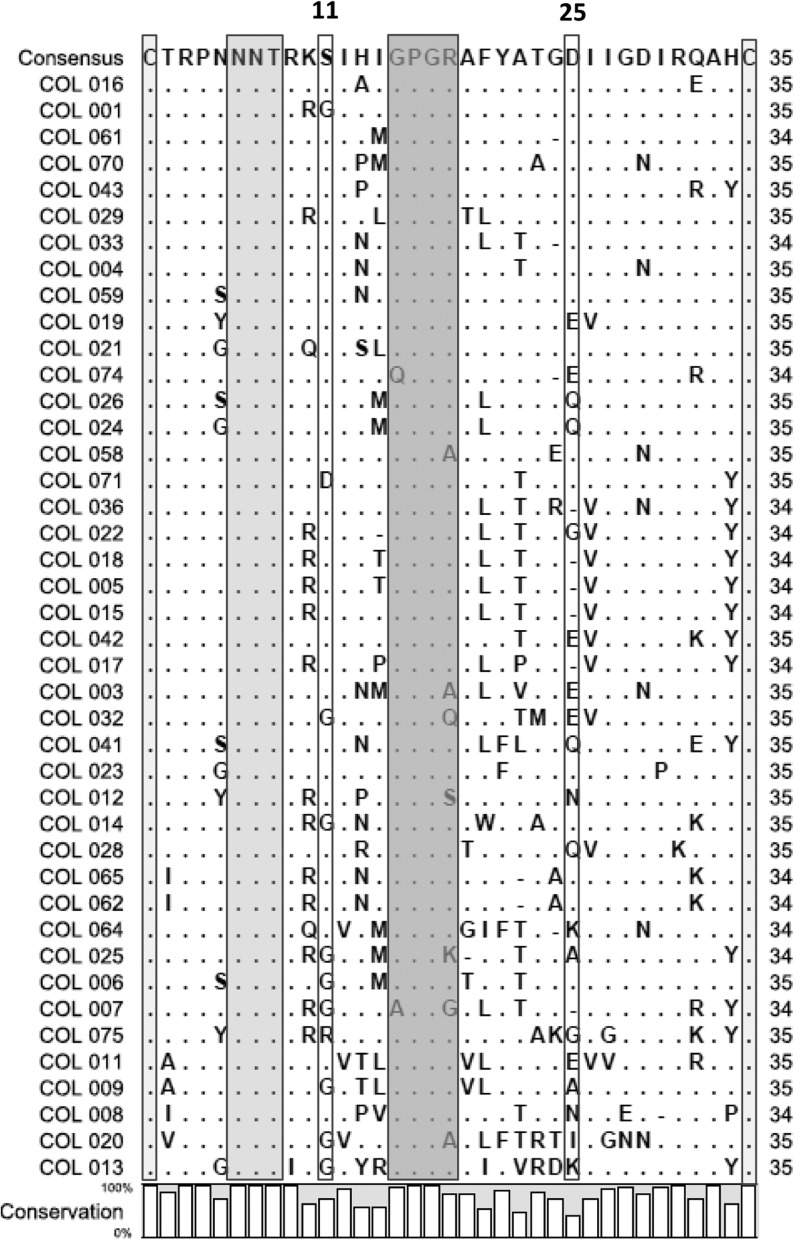
The Four Peaks program was first used to open sequence chromatograms for visual inspection of sequence quality and to recover the amino acid sequences of the V3 region in the correct reading frame. All 42 sequences were then aligned with each other. The alignment indicated that 16 (38%) of 42 V3 domain sequences had 34 amino acid residues whereas 26 (62%) had 35. All the V3 sequences are framed by the highly conserved cysteines (C), responsible for the loop structural configuration (Fig. 1).
FIG. 1.
Alignments of the putative protein sequences of the env V3 loop region of the 42 sequenced Colombian HIV-1 strains. Dots indicate identity to the consensus sequence generated from these 42 sequences, as indicated, and dashes indicate gaps in alignments. The cysteines framing the V3 loop are identified by the light gray shaded box. The potential N-linked glycosylation site is identified by the gray shaded box. The crown sequence, GPGR, is indicated by the dark gray box. The positions of acid residues, associated with binding to CCR5, are identified by the colorless boxes (positions 11 and 25).
The four amino acids in the 15 to 18 residues forming the tip of the V3 loop were quite well conserved while the GPGR motif was the most common and was found in 35 (83%) of the studied V3 loop sequences. In the COL058, COL003, and COL020 sequences, the basic residue arginine (R) was replaced by alanine (A). In the COL032, COL012, COL025, and COL007 sequences, this same residue was replaced by glutamine (Q), serine (S), lysine (K), and glycine (G) residues, respectively. Finally, the first glycine (G) residue was replaced by glutamine and alanine (A) in the COL074 and COL007 sequences, respectively (Fig. 1).
It has been suggested that acidic amino acids at some critical positions within the V3 loop are determinant for viral tropism and coreceptor usage. Localization of acid residues in position 11 and/or 25 of the V3 region is especially associated with CCR5 utilization. This phenotype can be strongly influenced by the presence of N-linked glycosylation sites in the V3 region.13 According to our analysis of the N-linked glycosylation pattern and the presence of acidic amino acids, 23 of the V3 sequences (57%) were predicted to belong to macrophage-tropic viruses using CCR5 as the coreceptor for viral entry. The acidic amino acid more frequently found in such R5 viruses was aspartate (D) with a frequency of 74% at amino acid position 25, although only once at amino acid position 11. The high frequency of macrophage-tropic viruses is also in agreement with the fact that the samples were obtained from recently diagnosed individuals. However, surprisingly, a patient who received his diagnosis 8 years before was still exhibiting infection with a R5 virus (Fig. 1).
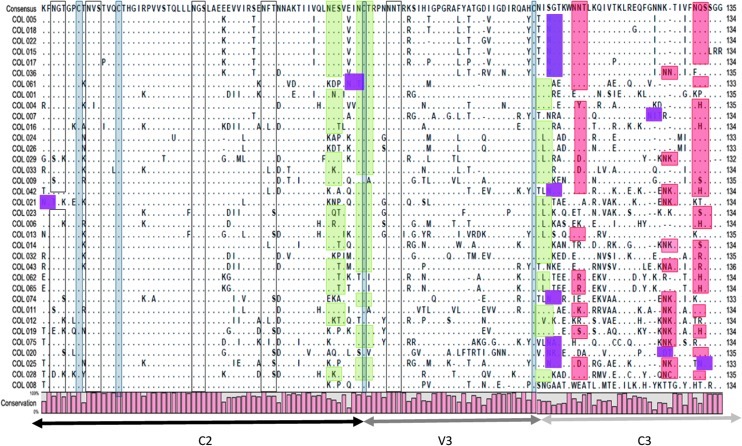
The whole C2V3C3 sequence encompassing 136 amino acids was then examined in more detail for phylogenetic analysis (Figs. 2 and 3). The amino acid sequences deduced from C2V3C3 nucleotide sequences were aligned for 35 samples where sequencing results were of sufficient quality to encompass this whole region. The deduction of the C2V3C3 sequence allowed analysis of the distribution and conservation of N-linked glycosylation sites in this region.
FIG. 2.
Alignments of the env C2V3C3 region of the 35 Colombian HIV-1 strains. Dots indicate identity to the consensus sequence generated from these 35 sequences, as indicated, and dashes indicate gaps in alignments. The conserved putative N-linked glycosylation sites are identified by empty boxes while the less conserved putative sites are highlighted in green. The least conserved sites are identified by orange boxes often followed by new putative sites highlighted in magenta. The four cysteines are identified by gray boxes.
FIG. 3.
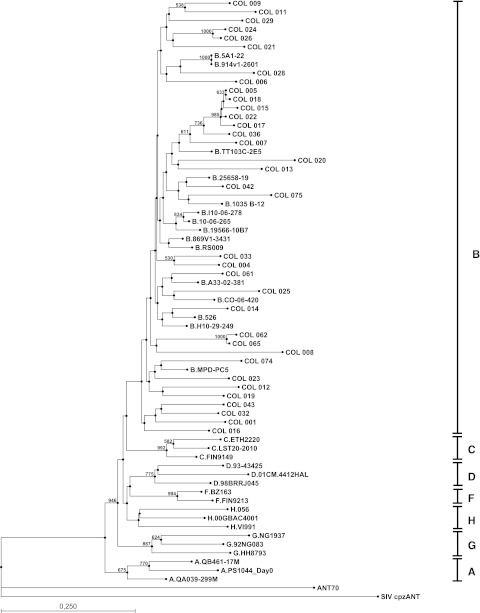
Phylogenetic analysis of the 35 sequenced HIV-1 env C2V3C3 region from Colombian samples and representative HIV-1 strains from groups M, subtypes A to J, and O (ANT70), taken from the Los Alamos sequence database; SIVcpzANT was used as the outgroup sequence. Trees were constructed by the neighbor-joining method and branch nodes supported by bootstrap values above 500 are indicated. Horizontal lines are drawn to proportion.
These oligosaccharide addition sites are required for the structure/function of the protein and are shown in Asn-X-Ser or Asn-X-Thr contexts. High conservation was found in the potential N-linked glycosylation sites localized in the C2 region (positions 234–236, 241–243, 262–264, and 276–278) and within the V3 loop (position 301–303). Compared to the N-linked glycosylation sites localized in the C2 region, those close to the V3 loop (positions 289–291, 295–297, and 331–333) are less conserved. The less conserved N-linked glycosylation sites are localized in the C3 region (positions 338–340, 356–358, and 362–364). A compensation was found in some sequences when an N-linked glycosylation site is lost (positions 331–333) by the generation of another potential glycosylation sequence close to the previous one. A possible selective pressure can be the cause of the conservation of the oligosaccharides addition sites in V3 (Fig. 2).
Four highly conserved cysteine residues were found through the C2V3C3 region. They included the two cysteines that bordered the V3 loop at positions 296 and 330 (numbers corresponding to the positions in the HIV-1-HXB2 genome). The other two were localized at positions 239 and 247.
Samples were subtyped by phylogenetic analysis of env corresponding to the deduced protein sequence of the C2V3C3 region. Multiple sequence alignment was obtained as before with addition of reference sequences taken from the Los Alamos HIV sequence database (http://www.hiv.lanl.gov/content/sequences/HIV/mainpage.html). The following reference sequences were used: subtype A: PS1044_Day0, QB461-17M, QA039-299M; subtype B: 5A1-22, 914v1-2601, TT103C-2E5, 25658-19, 1035 B-12, I10-06-278, 10-06-265, 19566-10B7, 869V1-3431, RS009, A33-02-381, CO-06-420, 562, H10-29-249, MPD-PC5; subtype C: ETH2220, LST20-2010, FIN9149; subtype D: 93-43425, 98BRRJ045, 01CM.4412HAL; subtype F: BZ163, FIN9213; subtype G: 92NG083, NG1937, HH8793; subtype H: 056, 00GBAC4001, VI991; The group O reference sequence ANT70 and the outgroup sequence SIV cpzANT were also used as controls.
Phylogenetic trees were constructed by the neighbor-joining method and bootstrap analyses were performed with 1000 resamplings of the original sequence data. Our results show clearly that the entire 35 analyzed samples cluster with the sequences belonging to subtype B (Fig. 3). This same subtype was found in all studies carried out to date in other Colombian regions, perhaps as a result of only one or a few points of HIV introduction and expansion in this country. Nevertheless, the phylogenetic tree shows a divergence in the majority of bootstrap values suggesting a change in the behavior of the national epidemic; the sequences from patients COL 005, COL 018, COL 015, COL 022, and COL 017 clustered with a high bootstrap value (985), suggesting an epidemiological link between these sequences. Because the samples were randomly obtained in the Metropolitan area of Barranquilla, and these patients are unrelated, this suggests a recent outbreak of some unusual subtype.
Our study expands past analyses by providing previously missing data from the Northern region of the country. The number and the length of the envelope sequence examined, compared to previous studies,9–11 also provide a clearer picture of the prevailing situation of the HIV epidemic in Colombia at the present time.
Acknowledgments
This work was supported by a grant from the Departamento Administrativo de Ciencia, Tecnología e Innovación, COLCIENCIAS, Colombia (Contract UNINORTE-COLCIENCIAS No. 418-2004). The authors thank Hospital General de Barranquilla and Medicina Integral IPS (Barranquilla, Colombia) for their cooperation in this study. We thank Isis Arias for expert technical assistance resulting in the smooth operation of our laboratories and Rosa Gómez-Castro and Miriam Torregrosa for excellent technical assistance with field work. We thank Dr. Joaquin Piñeros-Castillo for his kind cooperation. We thank Rodolfo Hernández and Carole Hardy for numerous helpful discussions. G.C.-A. also wishes to thank Virginie Brochu-Lafontaine, Roland Jabre, and Véronique Sandekian for their help during his brief stay in Montreal.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hemelaar J. Gouws E. Ghys PD, et al. WHO-UNAIDS Network for HIV isolation and characterization. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roques P. Robertson DL. Souquière S, et al. Phylogenetic analysis of 49 newly derived HIV-1 group O strains high viral diversity but no group M-liked subtype structure. Virology. 2002;302:259–273. doi: 10.1006/viro.2002.1430. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi J. McArthur CP. Vallari A, et al. HIV-1 group N: Evidence of ongoing transmission in Cameroon. AIDS Res Hum Retroviruses. 2006;22:453–457. doi: 10.1089/aid.2006.22.453. [DOI] [PubMed] [Google Scholar]
- 4.Vallari A. Bodelle P. Ngansop C, et al. Four new HIV-1 group N isolates from Cameroon: Prevalence continues to be low. AIDS Res Hum Retroviruses. 2010;26:109–115. doi: 10.1089/aid.2009.0178. [DOI] [PubMed] [Google Scholar]
- 5.Plantier J-C. Leoz M. Dickerson J-E, et al. New human immunodeficiency virus derived from gorillas. Nat Med. 2009;15:871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 6.Vallari A. Holzmayer V. Harris B, et al. Confirmation of putative HIV-1 group P in Cameroon. J Virol. 2011;85:1403–1407. doi: 10.1128/JVI.02005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomson M. Nájera R. Molecular epidemiology of HIV-1 variants in the global AIDS pandemic: An update. AIDS Rev. 2005;7:210–224. [PubMed] [Google Scholar]
- 8.Soares EA. Santos RP. Pellegrini JA, et al. Epidemiologic and molecular characterization of human immunodeficiency virus type 1 in southern Brazil. J Acquir Immune Defic Syndr. 2003;34:520–526. doi: 10.1097/00126334-200312150-00012. [DOI] [PubMed] [Google Scholar]
- 9.Navas MC. Letourneur F. Gomas F, et al. Analysis of the V3 loop sequences from 12 HIV type 1-infected patients from Colombia, South America. AIDS Res Hum Retroviruses. 1999;15:1141–1144. doi: 10.1089/088922299310430. [DOI] [PubMed] [Google Scholar]
- 10.Eyzaguirre L. Bautista CT. Ayala C, et al. First case of HIV type 1 subtype F among men who have sex with men in Colombia. AIDS Res Hum Retroviruses. 2006;22:808–811. doi: 10.1089/aid.2006.22.808. [DOI] [PubMed] [Google Scholar]
- 11.Sánchez GI. Bautista CT. Eyzaguirre L, et al. Molecular epidemiology of human immunodeficiency virus-infected individuals in Medellin, Colombia. Am J Trop Med Hyg. 2006;74:674–677. [PubMed] [Google Scholar]
- 12.Delwart EL. Herring B. Rodrigo AG, et al. Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. Genome Res. 1995;4:S202–S216. doi: 10.1101/gr.4.5.s202. [DOI] [PubMed] [Google Scholar]
- 13.Pollakis G. Kang S. Kliphuis A, et al. N-linked glycosylation of the HIV-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J Biol Chem. 2001;276:13433–13441. doi: 10.1074/jbc.M009779200. [DOI] [PubMed] [Google Scholar]