Abstract
We report a high frequency of drug resistance mutations among patients with unusual insertions or deletions at the β3–β4 hairpin-loop-coding region of HIV-1 subtype C reverse transcriptase, during failure of first-line antiretroviral therapy containing only reverse transcriptase inhibitors in Chennai, India.
Treatment of HIV-1-infected individuals with antiretroviral (ARV) drugs is highly effective in inhibiting viral replication, thereby, increasing both the duration and the quality of life.1 The development of viral resistance to ARV drugs is a primary cause of treatment failure.2 Extensive genetic diversity is inherent in HIV because the highly error-prone reverse transcriptase (RT) enzyme creates a population of variants from which drug-resistant strains can be selected during treatment.3 With the increasing complexity of the ARV regimens, novel mutational patterns conferring high-level resistance to nucleoside and nonnucleoside RT inhibitors have been identified in viral isolates. Among them, insertions and deletions in the β3–β4 hairpin-loop-coding region of HIV-1 RT have been identified in heavily treated patients.4 Insertions of one, two, or several residues appear to have a significant impact on nucleoside analogue resistance.5 In this study, we describe unusual amino acid (aa) insertions/deletions (indels) observed in β3–β4 hairpin-loop-coding region of HIV-1 RT among South Indian patients who are experiencing failure of first-line ARV therapy, which contains both nucleoside and nonnucleoside RT inhibitors.
The YRG Centre for AIDS Research and Education (YRG CARE) is a nonprofit medical and research institution in Chennai, India, that provides medical care to more than 15,000 HIV-infected individuals. All patients were treated according to WHO treatment guidelines. Plasma viral load monitoring was not standard of care. Patients were seen every 3 months or as clinically indicated. CD4 cell count monitoring was performed every 3–6 months. Data were collected under the approval of YRG CARE's free-standing institutional review board. Eligible patients were investigated between June 2007 and June 2011. These patients were ARV naive before initiation of therapy, who later underwent genotyping after immunologic or clinical failure of first-line therapy was identified. For genotyping, the RT region of the HIV-1 pol gene was amplified from cDNA reverse transcribed from plasma viral RNA (QIAGEN Inc., CA) by nested PCR.6 Nested products were bidirectionally sequenced on an automated ABI 3100-Avant genetic analyzer (Applied Biosystems, CA). Multiple sequence alignment SeqScape v2.5 software (Applied Biosystems, CA) generated consensus sequences were analyzed for mutations using the Stanford drug resistance database (www.hivdb.stanford.edu).7,8
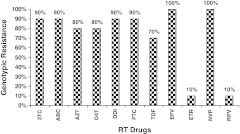
The demographics and mutation profiles for these patients with genomic rearrangements are shown in Table 1. Ten patients infected with HIV-1 subtype C (mean age 37 years±12) had unusual indels in the β3–β4 hairpin-loop-coding region of HIV-1 RT. Five patients had two aa insertion at RT codon 69, whereas three patients had one aa deletion at codon 69 and another two patients had one aa deletion at codon 67. All of the five patients with two aa insertions had at least two thymidine analog mutations (TAMs). Of the nucleoside RT inhibitor resistance mutations, M184V was the most common (70%), and TAMs were found in 60% of patients. Three of the ten patients had virus that remained sensitive (30%) to tenofovir. Though all the ten patients had NNRTI mutations, sensitivity to Etravirine and Rilpivirine was very high (100%) (Figure 1), as per genotypic drug resistance-associated mutations (IAS-USA and Stanford HIV Resistance Database v 6.1.1).7,8
Table 1.
Demographics and Drug Resistance-Related Mutations Within the Polymerase Domain of HIV-1 RT, Associated with Insertions or Deletions of One or Two Amino Acids Between Codons 67 and 69 (β3–β4 Loop)
| S. No. | HIV-1 RT insertion/deletion | Age/sex | Mode of transmision | CD4+count cells/μl | HAART exposure (months) | NRTI mutations | NNRTI mutations |
|---|---|---|---|---|---|---|---|
| 1 | Dipeptide (SG) insertion at 69 | 32/M | HT | 90 | 64 | M41L, T69Si, M184V | Y188L |
| 2 | 38/M | HT | 171 | 57 | M41L, D67E, T69Si, L74V, M184V, T215Y | K103KN, V108I, Y181C, H221Y | |
| 3 | Dipeptide (SS) insertion at 69 | 38/M | HT | 318 | 54 | M41L, T69Si, M184V, T215Y | Y188L, G190A |
| 4 | Dipeptide (ST) insertion at 69 | 37/M | HT | 38 | 81 | M41L, T69Si, M184V, T215Y | A98AG, Y188L |
| 5 | Dipeptide (VT) insertion at 69 | 45/M | HT | 180 | 52 | M41L, D67N, T69Si, L74V, M184V, T215Y | A98G, L100IL, K101KPQT, K103N, V108IV |
| 6 | D67 deletion | 30/F | HT | 115 | 65 | D67d, T69G, K70R, M184V, T215F, K219Q | Y188L |
| 7 | 39/M | HT | 166 | 72 | M41L, D67d, T69G, K70R, M184V, T215FY, K219E | V90I, A98G, V108I, Y181C, G190A | |
| 8 | T69 deletion | 29/M | HT | 123 | 39 | K65R, T69d, Q151M | K103N, Y181C |
| 9 | 14/F | BT | 150 | 40 | K65R, D67N, T69d, Q151M | V90I, V106M, Y181C | |
| 10 | 36/M | HT | 180 | 48 | K65R, T69d | A98AG, E138A, G190E, H221Y |
HT, heterosexual transmission; BT, blood transmission; RT, reverse transcriptase; HAART, highly active antiretroviral therapy; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor.
FIG. 1.
Interpretation of genotypic resistance to HIV-1 reverse transcriptase (RT) drugs [nucleoside and nonnucleoside reverse transcriptase inhibitor (NRTI and NNRTI)]. 3TC, lamivudine; ABC, abacavir; AZT, zidovudine; d4T, stavudine; DDI, didanosine; FTC, emtricitabine; TDF, tenofovir; EFV, efavirenz; ETR, etravirine; NVP, nevirapine; RPV, rilpivirine.
Since insertions are usually found in viral strains isolated from heavily treated patients, it is not surprising to find them in combination with drug resistance-related mutations, particularly with TAMs such as M41L, L210W, or T215Y.9 T215Y appears to be strongly associated with the identified insertions (80% of the insertion-containing sequences have Tyr at position 215) whereas K65R and Q151M were strongly associated with a deletion at codon 69 compared to a deletion at codon 67. In line with earlier studies, the aa deletion at codon 67 was also associated with the T69G mutation in combination with TAMs, such as K70R, T215F, and K219E. It has been reported that T69G by itself does not affect nucleoside analogue sensitivity10,11; however, within a sequence background containing TAMs, it confers resistance with a loss in replicative capacity.5 The development of the D67 deletion was believed to compensate for the loss of replication capacity, rendering a virus that replicates efficiently while showing a high level of resistance to zidovudine.10
Resistance to RT inhibitors can be achieved through the accumulation of one or more mutations in the HIV-1 RT coding region.12,13 The underlying genetic variation of RT genes (e.g., polymorphisms) may drive preferential selection; however, no particular viral genetic background has been associated with the development of the identified resistance mutations.5 Other virus-related genetic interactions such as the inherent replication rate and overall population diversity may influence the preferential selection of resistance-associated mutations. In addition, host-related factors, such as drug metabolism and immune system recognition of particular epitopes, may also influence strain selection.14 Further studies of these factors are necessary and important to elucidate the conditions by which certain genetic features in HIV strains are selected. Current evidence suggests that a large portion of HIV-1 subtype C isolated with either insertions or deletions in the β3–β4 hairpin-loop-coding region of RT contains one or more TAMs, exhibiting a high level of resistance to multiple nucleoside analogues. With increasing numbers of HIV-infected patients initiating ARV therapy and not being monitored for virologic failure, these insertions and deletions are likely to become a common occurrence, thereby, compromising the efficacy of future regimens.
Acknowledgments
We are most grateful to the clinical and laboratory staff at YRG Centre for AIDS Research and Education, VHS, Chennai, India, for their facilitation of the study. We would like to thank the Fogarty AIDS International Training and Research Program (AITRP), Brown University, Providence, RI, for their financial support, thus providing us with an opportunity to present the poster at the XVIII International AIDS Conference, 2010. Work by D.S. was supported by grants from the National Institutes of Health: AI69432, AI043638, MH62512, MH083552, AI077304, AI36214, AI047745, AI74621, and AI080353, and the James B. Pendleton Charitable Trust.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Thompson MA. Aberg JA. Cahn P. Montaner JS. Rizzardini G. Telenti A. Gatell JM. Günthard HF. Hammer SM. Hirsch MS. Jacobsen DM. Reiss P. Richman DD. Volberding PA. Yeni P. Schooley RT the International AIDS Society-USA. Antiretroviral treatment of adult HIV infection: 2010 recommendations of the International AIDS Society-USA panel. JAMA. 2010;304(3):321–333. doi: 10.1001/jama.2010.1004. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch MS. Günthard HF. Schapiro JM. Brun-Vézinet F. Clotet B. Hammer SM. Johnson VA. Kuritzkes DR. Mellors JW. Pillay D. Yeni PG. Jacobsen DM. Richman DD. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47(2):266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 3.Rambaut A. Posada D. Crandall KA. Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 4.Huigen MC. de Graaf L. Eggink D. Schuurman R. Müller V. Stamp A. Stammers DK. Boucher CA. Nijhuis M. Evolution of a novel 5-amino-acid insertion in the beta3-beta4 loop of HIV-1 reverse transcriptase. Virology. 2007;364(2):395–406. doi: 10.1016/j.virol.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 5.Menéndez-Arias L. Matamoros T. Cases-González Insertions and deletions in HIV-1 reverse transcriptase: Consequences for drug resistance and viral fitness. Curr Pharm Des. 2006;12(15):1811–1825. doi: 10.2174/138161206776873608. [DOI] [PubMed] [Google Scholar]
- 6.Saravanan S. Vidya M. Balakrishanan P, et al. Evaluation of two human imunodeficiency virus-1 genotyping systems: ViroSeq 2.0 and an in-house method. J Virol Methods. 2009;159:211–216. doi: 10.1016/j.jviromet.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson VA. Brun-Vézinet F. Clotet B. Günthard HF. Kuritzkes DR. Pillay D. Schapiro JM. Richman DD. Update of the drug resistance mutations in HIV-1: December 2010. Top HIV Med. 2010;18(5):156–163. [PubMed] [Google Scholar]
- 8.Liu TF. Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42(11):1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross L. Parkin N. Chappey C. Fisher R. St Clair M. Bates M, et al. Phenotypic impact of HIV reverse transcriptase M184I/V mutations in combination with single thymidine analog mutations on nucleoside reverse transcriptase inhibitor resistance. AIDS. 2004;18:1691–1696. doi: 10.1097/01.aids.0000131355.44834.e4. [DOI] [PubMed] [Google Scholar]
- 10.Imamichi T. Berg SC. Imamichi H. Lopez JC. Metcalf JA. Falloon J. Lane HC. Relative replication fitness of a high-level 3′-azido-3′-deoxythymidine-resistant variant of human immunodeficiency virus type 1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr to Gly) at codon 69. J Virol. 2000;74:10958–10964. doi: 10.1128/jvi.74.23.10958-10964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winters MA. Merigan TC. Variants other than aspartic acid at codon 69 of the human immunodeficiency virus type 1 reverse transcriptase gene affect susceptibility to nucleoside analogs. Antimicrob Agents Chemother. 2001;45:2276–2279. doi: 10.1128/AAC.45.8.2276-2279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarafianos SG. Das K. Clark AD., Jr Ding J. Boyer PL. Hughes SH, et al. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with b-branched amino acids. Proc Natl Acad Sci USA. 1999;96:10027–10032. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deval J. White KL. Miller MD. Parkin NT. Courcambeck J. Halfon P, et al. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J Biol Chem. 2004;279:509–516. doi: 10.1074/jbc.M308806200. [DOI] [PubMed] [Google Scholar]
- 14.Manosuthi W. Butler DM. Chantratita W. Sukasem C. Richman DD. Smith DM. Patients infected with HIV type 1 subtype CRF01_AE and failing first-line nevirapine- and efavirenz-based regimens demonstrate considerable cross-resistance to etravirine. AIDS Res Hum Retroviruses. 2010;26(6):609–611. doi: 10.1089/aid.2009.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]