Abstract
Intrinsically disordered proteins are important in signaling, regulation, and translocation. Understanding their diffusion under physiologically relevant conditions will yield insight into their functions. We used NMR to quantify the translational diffusion of a globular and a disordered protein in dilute solution and under crowded conditions. In dilute solution, the globular protein chymotrypsin inhibitor 2 (CI2, 7.4 kDa) diffuses faster than the disordered protein α-synuclein (14 kDa). Surprisingly, the opposite occurs under crowded conditions; α-synuclein diffuses faster than CI2, even though α-synuclein is larger than CI2. These data show that shape is a key parameter determining protein diffusion under crowded conditions, adding to the properties known to be affected by macromolecular crowding. The results also offer a clue about why many signaling proteins are disordered.
Keywords: 19F NMR, Crowding, Diffusion, Disordered proteins
Intrinsically disordered proteins lack a stable tertiary structure.1,2 In bacteria, between 6% and 33% of proteins are disordered and 35 – 51% are disordered in eukaryotes.3 Despite the lack of well-defined structure, disordered proteins are involved in numerous cellular functions, including regulation, signaling, and translocation.1–7 Bioinformatics studies show that more than 70% of signaling proteins are unstructured.4 Diffusion is the major mode of macromolecular transport in cells, and therefore is expected to play a vital role in signaling.
The cellular interior, however, is an exceptionally complex environment where macromolecules can reach concentrations of 300 g/L and occupy 30% of the volume.8,9 This crowded environment is vastly different from the dilute, idealized conditions used in most biophysical studies. Fluorescence results show that cytoplasm of Escherichia coli slows the translational diffusion of a globular protein, green florescent protein, 10-fold compared to dilute solution.10,11 Results from in vitro experiments using synthetic polymers also show that crowding can dramatically decrease the translational diffusion of test globular proteins.12–14 However, there is no quantitative information about disordered protein diffusion under the crowded conditions.
The measurement of translational diffusion in cells has been dominated by fluorescence methods.15,16 Despite progress in site-specific protein labeling in living cells, most such studies are still limited to green fluorescent protein.17 Nuclear magnetic resonance spectroscopy (NMR) is a noninvasive approach that can assess protein biophysics in cells and under crowded in vitro conditions.18 Here, we use NMR to quantify the translational diffusion of the disordered 14-kDa Parkinson’s disease related protein α-synuclein in dilute solution and in 300 g/L solutions of glycerol, the synthetic polymers polyvinylpyrrolidone 40 (PVP, 40 kDa), and Ficoll 70 (a 70 kDa cross-linked sucrose polymer), and the globular proteins lysozyme (15 kDa), and bovine serum albumin (BSA, 67 kDa), and then compared the diffusion coefficients to those from parallel studies on the 7.4-kDa globular protein chymotrypsin inhibitor 2 (CI2).19,20
We used 19F NMR because of its high sensitivity, spectral simplicity, and large chemical shift range21–23 to assess the diffusion of α-synuclein. The protein was labeled with 3-fluorotyrosine.22 Translational diffusion was measured with the Dbppste sequence.24 Spectra of the protein in 300 g/L BSA solution as a function of gradient strength are shown in Figure 1. The slope of plots like the one shown in Figure 2 gives the translational diffusion coefficient (The uncertainties from repeating the experiment are less than 0.4 × 10−11 m2/s.). The linearity of the plot indicates that proteolysis of α-synuclein is not a problem, consistent with other investigations of α-synuclein under crowded conditions.25 The linearity of all the plots analyzed for this report is also consistent with simple- rather than sub-diffusion.26
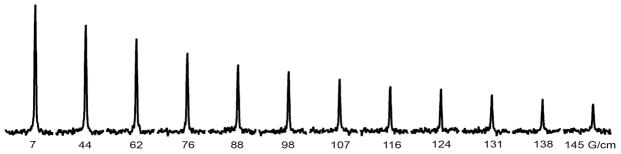
Figure 1.
19F spectra of 1 mM 15N-enriched, 3-fluorotyrosine labeled α-synuclein in 300-g/L bovine serum albumin [pH 7.4, 200 mM potassium phosphate buffer, 1 mM EDTA, 0.5 mM, 25 °C] with increasing gradient strength. Materials and Methods: Chicken lysozyme, BSA, glycerol, Ficoll and PVP were purchased from Sigma-Aldrich and used without further purification. For α-synuclein experiments, glycerol, PVP and Ficoll were dissolved in 50 mM potassium phosphate buffer, 1 mM EDTA, 0.5 mM phenylmethanesulfonyl fluoride (PMSF), pH 7.4, whereas lysozyme and BSA were dissolved in 200 mM potassium phosphate buffer, 1 mM EDTA, and 0.5 mM PMSF, pH 7.4. The final concentration of CI2 and α-synuclein was ~1 mM. 19F data were acquired without decoupling on a Varian Inova 600 spectrometer equipped with a 19F-{1H} z-gradient probe. Conventional 5-mm NMR tubes were used. Gradient strengths ranged from 5.4 G/cm to 129.1 G/cm and the diffusion delay was 0.15 s in solutions containing Ficoll and PVP. Gradient strengths ranged from 5.4 G/cm to 118.4 G/cm and the diffusion delay was 0.08 s in solutions containing lysozyme and BSA.
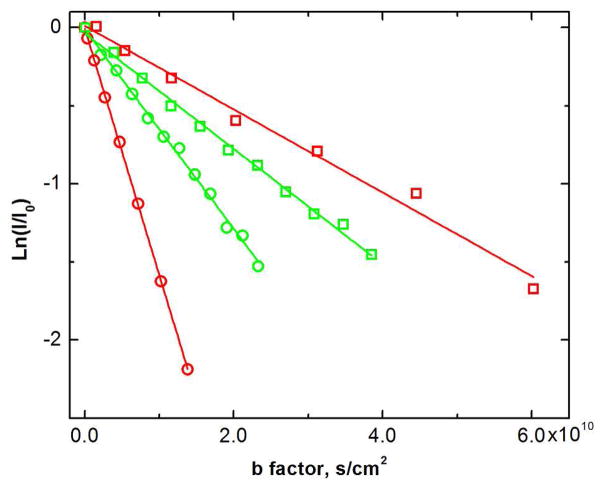
Figure 2.
Translational diffusion of CI2 (red)19,20 and α-synuclein (green) in dilute solution (circles) and in a 300 g/L solution of bovine serum albumin (squares). b factor = (γGd)2Δ where γ is the gyromagnetic ratio, G is the gradient strength, d is the duration of the gradient pulse, and Δ is the delay between gradient pulses.27
To assess the veracity of 19F NMR as a probe, we measured the diffusion coefficient 19F-labeled CI222 and α-synuclein in dilute solution (pH 7.4, 25 °C). The value for CI2, 16.0 × 10−11 m2/s, is consistent with that obtained from 15N enrichment (15.51 × 10−11 m2/s).19,20 Our value for 19F-labeled α-synuclein, 7.8 ×10−11 m2/s, is reasonable agreement with the value of 5.7 ×10−11 m2/s obtained at 10 °C,28 given that the properties of disordered proteins depend more heavily on conditions than those of globular proteins. For instance, the hydrated radius of α-synuclein increases by 8 Å between 10 °C and 35 °C.25 In summary our comparisons suggest that 19F is a reliable tool for measuring diffusion.
Next, we compared the diffusion of CI2 to that of α-synuclein in dilute solution and in a 300 g/L solution of the small-molecule co-solute, glycerol (Figure 3). CI2 diffuses faster for three reasons. First, CI2 has a smaller molecule weight. Second, CI2 is folded and compact, making its hydrodynamic radius smaller than that of α-synuclein.25 Third, when the radius of a protein is comparable to or greater than the radius of the co-solute (e.g., glycerol), a larger protein will diffuse more slowly.19,20,29 In addition, the slowing of CI2 diffusion by glycerol is consistent with the Stokes-Einstein equation.20
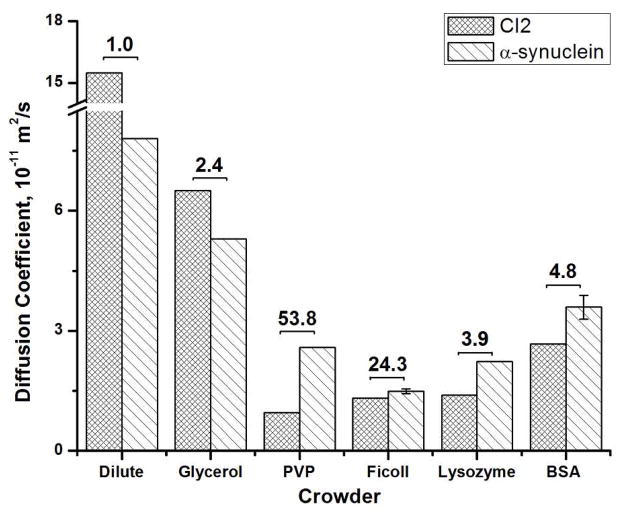
Figure 3.
Histogram showing the translational diffusion coefficients of CI2 and α-synuclein in dilute solution and 300 g/L solutions of glycerol, PVP, Ficoll 70, lysozyme and bovine serum albumin at 25 °C. Error bars represent the standard deviation of the mean from three trials. The corresponding macroviscosity (in cP) is indicated at the top of each entry. The CI2 data have been published.19,20
We then studied macromolecular crowding. Compared to dilute solution, CI2 and α-synuclein diffusion is slowed 5- to 10-fold in solutions crowded by PVP, Ficoll, BSA and lysozyme (Figure 3). The slowing by protein crowders is consistent with studies of green fluorescent protein in E. coli,10,11 suggesting that our data have some biological relevance. Circular dichroism and NMR data collected under crowded conditions rule out large crowding-induced conformational changes of CI2 and α-synuclein as the cause of the slowing.30–36
Synthetic polymers impede the diffusion of both CI2 and α-synuclein more than the protein crowders (Figure 3). The explanation is that the synthetic polymers overlap to form a mesh that impedes translation,37 but proteins, being quasi-spherical, cannot overlap and offer less of an impediment. In summary, CI2 and α-synuclein diffuse faster in crowded protein solutions than they do in crowded synthetic polymer solutions.
The key observation of this study is that macromolecular crowding affects the diffusion of the disordered and the globular protein differently (Figure 3). While CI2 diffusion is slowed 5- to 10-fold, α-synuclein diffusion is slowed only 3- to 6-fold, despite the fact that α-synuclein has a larger molecular weight. The consequence is that α-synuclein diffusion is now faster than CI2 diffusion; the opposite of what is observed in dilute solution and in glycerol. These results do not arise from a structural change because CI2 remains compact20 and α-synuclein remains collapsed30,33–36 under crowded conditions. We suggest that α-synuclein adopts a different diffusion strategy under crowded conditions. For instance, we know that the cellular environment dampens that the rotational diffusion of disordered proteins less than the rotational diffusion of globular proteins.33 This relative increase in rotation motion for disordered proteins arises because they possess more internal (segmental) motion. Perhaps the inherent internal motion of disordered proteins is converted to translational motion under crowded conditions.
Our results may help explain an observation about FlgM, an intrinsically disordered protein that regulates flagella and chemotaxis genes in E. coli and Salmonella typhimurium. Specifically, the disorder would facilitate its export from the cell via the narrow central channel of the flagella filament.38,39 More generally, this facilitation of diffusion would decrease the apparent size of disordered signaling proteins, making it easier for them to shuttle through the small cavities between macromolecules inside the crowded cell.
Acknowledgments
This research was supported by the National Institutes of Health (5DP1OD783) and the National Science Foundation (MCB-1051819). We thank Marc ter Horst and Gregory Young for spectrometer maintenance, and Elizabeth Pielak for helpful comments. GJP thanks Science Foundation Ireland for an E.T.S Walton Visitor Award.
ABBREVIATIONS
- CI2
chymotrypsin inhibitor 2
- PVP
polyvinylpyrrolidone
References
- 1.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic Disorder and Protein Function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 2.Wright PE, Dyson HJ. Intrinsically Unstructured Proteins: Re-Assessing the Protein Structure-Function Paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 3.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic Protein Disorder in Complete Genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 4.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic Disorder in Cell-Signaling and Cancer-Associated Proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 5.Sigalov AB. Protein Intrinsic Disorder and Oligomericity in Cell Signaling. Mol Biosyst. 2010;6:451–461. doi: 10.1039/b916030m. [DOI] [PubMed] [Google Scholar]
- 6.Uversky VN. What Does It Mean to Be Natively Unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 7.Dunker AK, Obradovic Z. The Protein Trinity--Linking Function and Disorder. Nat Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerman SB, Trach SO. Estimation of Macromolecule Concentrations and Excluded Volume Effects for the Cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman SB, Minton AP. Macromolecular Crowding: Biochemical, Biophysical, and Physiological Consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 10.Konopka MC, Shkel IA, Cayley S, Record MT, Weisshaar JC. Crowding and Confinement Effects on Protein Diffusion in Vivo. J Bacteriol. 2006;188:6115–6123. doi: 10.1128/JB.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slade KM, Baker R, Chua M, Thompson NL, Pielak GJ. Effects of Recombinant Protein Expression on Green Fluorescent Protein Diffusion in. Escherichia coli Biochemistry. 2009;48:5083–5089. doi: 10.1021/bi9004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banks DS, Fradin C. Anomalous Diffusion of Proteins Due to Molecular Crowding. Biophys J. 2005;89:2960–2971. doi: 10.1529/biophysj.104.051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dauty E, Verkman AS. Molecular Crowding Reduces to a Similar Extent the Diffusion of Small Solutes and Macromolecules: Measurement by Fluorescence Correlation Spectroscopy. J Mol Recognit. 2004;17:441–447. doi: 10.1002/jmr.709. [DOI] [PubMed] [Google Scholar]
- 14.Kozer N, Kuttner YY, Haran G, Schreiber G. Protein-Protein Association in Polymer Solutions: From Dilute to Semidilute to Concentrated. Biophys J. 2007;92:2139–2149. doi: 10.1529/biophysj.106.097717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bacia K, Kim SA, Schwille P. Fluorescence Cross-Correlation Spectroscopy in Living Cells. Nat Methods. 2006;3:83–89. doi: 10.1038/nmeth822. [DOI] [PubMed] [Google Scholar]
- 16.Verkman AS. Diffusion in Cells Measured by Fluorescence Recovery after Photobleaching. Methods Enzymol. 2003;360:635–648. doi: 10.1016/s0076-6879(03)60132-1. [DOI] [PubMed] [Google Scholar]
- 17.Chen I, Ting AY. Site-Specific Labeling of Proteins with Small Molecules in Live Cells. Curr Opin Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Pielak GJ, Li C, Miklos AC, Schlesinger AP, Slade KM, Wang GF, Zigoneanu IG. Protein Nuclear Magnetic Resonance under Physiological Conditions. Biochemistry. 2009;48:226–234. doi: 10.1021/bi8018948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Wang Y, Pielak GJ. Translational and Rotational Diffusion of a Small Globular Protein under Crowded Conditions. J Phys Chem B. 2009;113:13390–13392. doi: 10.1021/jp907744m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Li C, Pielak GJ. Effects of Proteins on Protein Diffusion. J Am Chem Soc. 2010;132:9392–9397. doi: 10.1021/ja102296k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Lutz EA, Slade KM, Ruf RA, Wang G, Pielak GJ. 19F-NMR Studies of α-Synuclein Conformation and Fibrillation. Biochemistry. 2009;48:8578–8584. doi: 10.1021/bi900872p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Wang GF, Wang Y, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RA, Mehl RA, Pielak GJ. Protein 19F NMR in Escherichia coli. J Am Chem Soc. 2010;132:321–327. doi: 10.1021/ja907966n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams SP, Haggie PM, Brindle KM. 19F NMR Measurements of the Rotational Mobility of Proteins in Vivo. Biophys J. 1997;72:490–498. doi: 10.1016/S0006-3495(97)78690-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, Chen A, Johnson CS. An Improved Diffusion-Ordered Spectroscopy Experiment Incorporating Bipolar-Gradient Pulses. J Magn Reson Ser A. 1995;115:260–264. [Google Scholar]
- 25.McNulty BC, Tripathy A, Young GB, Charlton LM, Orans J, Pielak GJ. Temperature-Induced Reversible Conformational Change in the First 100 Residues of Alpha-Synuclein. Protein Sci. 2006;15:602–608. doi: 10.1110/ps.051867106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shakhov A, Valiullin R, Kärger J. Tracing Molecular Propagation in Dextran Solutions by Pulsed Field Gradient NMR. J Phys Chem Lett. 2012;3:1854–1857. doi: 10.1021/jz300734m. [DOI] [PubMed] [Google Scholar]
- 27.Le Bihan D, Breton E. Imagerie De Diffusion in-Vivo Par Résonance Magnétique Nucléaire. C R Acad Sci. 1985;301:1109–1112. [Google Scholar]
- 28.Bodner CR, Dobson CM, Bax A. Multiple Tight Phospholipid-Binding Modes of Alpha-Synuclein Revealed by Solution NMR Spectroscopy. J Mol Biol. 2009;390:775–790. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dix JA, Verkman AS. Crowding Effects on Diffusion in Solutions and Cells. Annu Rev Biophys. 2008;37:247–263. doi: 10.1146/annurev.biophys.37.032807.125824. [DOI] [PubMed] [Google Scholar]
- 30.McNulty BC, Young GB, Pielak GJ. Macromolecular Crowding in the Escherichia coli Periplasm Maintains Alpha-Synuclein Disorder. J Mol Biol. 2006;355:893–897. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 31.Miklos AC, Li C, Sharaf NG, Pielak GJ. Volume Exclusion and Soft Interaction Effects on Protein Stability under Crowded Conditions. Biochemistry. 2010;49:6984–6991. doi: 10.1021/bi100727y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miklos AC, Sarkar M, Wang Y, Pielak GJ. Protein Crowding Tunes Protein Stability. J Am Chem Soc. 2011;133:7116–7120. doi: 10.1021/ja200067p. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Charlton LM, Lakkavaram A, Seagle C, Wang G, Young GB, Macdonald JM, Pielak GJ. Differential Dynamical Effects of Macromolecular Crowding on an Intrinsically Disordered Protein and a Globular Protein: Implications for in-Cell NMR Spectroscopy. J Am Chem Soc. 2008;130:6310–6311. doi: 10.1021/ja801020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morar AS, Olteanu A, Young GB, Pielak GJ. Solvent-Induced Collapse of Alpha-Synuclein and Acid-Denatured Cytochrome C. Protein Sci. 2001;10:2195–2199. doi: 10.1110/ps.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, Perovic I, Chittuluru J, Kaganovich A, Nguyen LT, Liao J, Auclair JR, Johnson D, Landeru A, Simorellis AK, Ju S, Cookson MR, Asturias FJ, Agar JN, Webb BN, Kang C, Ringe D, Petsko GA, Pochapsky TC, Hoang QQ. A Soluble Alpha-Synuclein Construct Forms a Dynamic Tetramer. Proc Natl Acad Sci U S A. 2011;108:17797–17802. doi: 10.1073/pnas.1113260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zigoneanu IG, Pielak GJ. Interaction of Alpha-Synuclein and a Cell Penetrating Fusion Peptide with Higher Eukaryotic Cell Membranes Assessed by 19F NMR. Mol Pharm. 2012;9:1024–1029. doi: 10.1021/mp200615m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubinstein M, Colby RH. Polymer Physics. Oxford University Press; USA: 2003. [Google Scholar]
- 38.Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-Terminal Half of the Anti-Sigma Factor, Flgm, Becomes Structured When Bound to Its Target, Sigma 28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 39.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing Structural Intermediates in Bacterial Flagellar Assembly by Export of a Negative Regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]