Abstract
Aims
To determine if in vitro production of IL-22 and IL-17 correlated with resolution of HCV infection.
Materials & methods
Human peripheral blood cells isolated from a well-defined cohort of resolved and chronic HCV-infected subjects were used to measure HCV-, influenza- and mitogen-activated T-cell proliferation. In addition, IL-22 and IL-17 production was measured via ELISAs and flow cytometry.
Results
Resolved HCV subjects had a significantly higher T-cell proliferative response to recombinant NS3 protein compared with chronic HCV subjects. Resolved subjects had a dose-dependent IL-22 response to recombinant NS3 compared with chronic HCV subjects.
Conclusion
IL-22 production is associated with antigen-specific induction of CD4 + T cells in individuals that resolved HCV infection, suggesting a potential role for IL-22 in HCV clearance.
Keywords: hepatitis C virus, human PBMC, IL-17, IL-22, T cell
HCV is a global epidemic with over 100 million people infected with the virus worldwide [1]. Approximately 80% of individuals infected with HCV are unable to spontaneously resolve the infection, which can potentially lead to liver cirrhosis and hepatocellular carcinoma. However, approximately 20% of individuals infected with HCV are able to spontaneously clear the virus [2–6]. Therefore, the knowledge gained from examining resolved HCV subjects’ immune responses to HCV antigens in comparison with chronically infected HCV subjects could provide insight into the mechanisms used by the human immune system to clear HCV, thereby potentially revealing new targets and/or platforms to treat HCV infection.
HCV comprises a separate genus, Hepacivirus, in the family Flaviviridae [7]. The virus is a single-stranded, positive-sense RNA virus, that encodes ten proteins. In particular, the NS3 protein serves as both a helicase and a serine pro-tease, therefore making this protein critical for HCV replication. Furthermore, HCV has been detected in peripheral blood mononuclear cells (PBMCs), as well as the spleen, brain and other tissues of HCV infected individuals; however, the liver is the major site of HCV replication.
An effective adaptive immune response against HCV is dependent on the breadth and magnitude of the HCV-specific CD4+ T-helper (Th) cell responses against the HCV NS3 protein [8]. Furthermore, NS3 viral peptides contain numerous immunogenic epitopes recognized by human T cells [9–16]. CD4+ T-cell activation is dependent on the T-cell receptor recognizing a specific peptide (epitope) bound to MHC molecules expressed on the surface of APCs. The expression and interaction of co-receptors on the surface of APCs, in conjunction with the cytokines present in the extracellular milieu, are important factors in the activation and differentiation of CD4+ T cells.
The function of the CD4+ T-cell subset is to direct or ‘help’ other immune cells, thereby leading to an effective cell-mediated immune response against pathogens. CD4+ T cells are phenotypically characterized by the type of cytokines they secrete, along with the expression of specific transcription factors. The predominant model for the role of CD4+ T-cell responses in HCV clearance was the Th1 (viral clearance) – Th2 (viral persistence) paradigm [17,18]. However, recent studies have identified at least six different CD4+ T-cell subsets, including Th1, Th2, Tregs, Th17, Th22 and Th9 cells. Patients with chronic HCV have been shown to have higher serum levels of IL-10 and IL-4, suggestive of either Tregs and/or Th2 cell involvement in HCV persistence [19,20]. The role of Th17 cells in HCV pathogenesis is largely unknown. Interestingly, IL-17/IL-22-producing T cells were found at a high frequency in the livers of patients with chronic HCV infection, indicating that IL-22 and IL-17 could be involved in HCV pathogenesis [21]. However, as with IL-17 cytokine production, the physiological function of IL-22 in viral clearance is not well characterized.
IL-22 is produced by a variety of leukocytes, including Th17, Th22, γδ, NK, NKT and lymphoid tissue-inducer cells [22]. Conversely, the IL-22 receptor (IL-22R) is expressed almost exclusively on non-hematopoietic cells, including cells of the pancreas, intestine, lung, kidney and liver [23,24]. The function of IL-22 has been demonstrated to be both pathogenic and protective depending on the microenvironment. For example, IL-22 amplifies the TNF-α, IFN-γ and/or IL-17 proinflammatory response, potentially leading to ankylosing spondylitis and rheumatoid arthritis in humans [25,26]. However, IL-22 produced alone can provide protection and promote tissue regeneration. For example, murine models of liver inflammation induced by concanavalin A, which induces a T-cell mediated hepatitis in mice, suggested that IL-22 provided protection to hepatocytes [27–29]. Furthermore, IL-22 could have antiviral effects. For example, Misse et al. demonstrated a potential association between HIV-1 resistance and IL-22 [30]. Individuals resistant to continual exposure to HIV-1 had increased IL-22 serum levels and a higher number of IL-22-producing T cells after non-specific activation with anti-CD3/CD28, when compared with control groups [30]. Furthermore, single nucleotide polymorphisms (SNPs) in the IL22 gene of individuals were associated with viral clearance and treatment in HCV patients [31]. In addition, IL22 mapped to a locus that contains the genes that are potentially associated with Theiler’s murine encephalomyelitis virus (TMEV) clearance in mice [32]. B.10 mice, a mouse strain resistant to TMEV persistence, had SNPs in the promoter region of IL22, leading to an increase in mRNA for IL-22 [32]. These SNPs were compared with SJL/J mice, which are unable to clear TMEV, and this comparison suggested that increased IL-22 production in the B.10 (viral clearing) mice could potentially be important for viral clearance.
In the current study, we demonstrated that PBMCs isolated from resolved HCV subjects had a significantly higher T-cell proliferative response to recombinant NS3 (rNS3) protein. Interestingly, the cell culture supernatants, collected after PBMC activation with the rNS3 protein, showed a significantly higher IL-22 level in the resolved HCV subjects compared with the chronic HCV subjects. In addition, chronic and resolved HCV subjects did not have increased levels of IL-17, suggesting that memory Th17 cells are not activated by rNS3. Flow cytometric analyses of PBMCs stimulated with rNS3 provided evidence of CD4+ T cells producing IL-22. Taken together, these results suggest that IL-22 production is associated with antigen- specific induction of CD4+ T cells in individuals that resolved HCV infection.
Patients, materials & methods
Patient samples
Blood was collected in acid citrate dextrose and PBMCs were isolated over Lymphocyte Separation Medium (GE-Healthcare, NJ, USA) and preserved in liquid nitrogen. Quantitative RT-PCR and HCV genotyping were performed at ARUP laboratories (UT, USA). Chronic HCV subjects used in this study were genotype 1a (Table 1). If the subjects had no detectable viral load, serum was screened for HCV antibodies by recombinant immunoblot assay (ARUP laboratories). These studies have been reviewed and approved by the University of Utah Institutional Review Board.
Table 1.
Demographics of subjects used in this study.
| Subjects | Gender | Age (years) | HCV genotype | HCV quantification (IU/ml) | Anti-HCV antibody† |
|---|---|---|---|---|---|
| C1 | Female | 33 | 1a | 0.2 × 105 | N/A |
| C2 | Female | 59 | 1a | 20 × 105 | N/A |
| C3 | Female | 61 | 1a | 1.2 × 105 | N/A |
| C4 | Female | 54 | 1a | 20 × 105 | N/A |
| C5 | Male | 56 | 1a | 1.7 × 105 | N/A |
| C6 | Female | 59 | 1a | 10 × 105 | N/A |
| C7 | Male | 56 | 1a | 0.6 × 105 | N/A |
| C8 | Female | 61 | 1a | 10 × 105 | N/A |
| R9 | Male | 56 | N/A | BLD | Positive |
| R10 | Male | 53 | N/A | BLD | Positive |
| R11 | Female | 31 | N/A | BLD | Positive |
| R12 | Male | 64 | N/A | BLD | Positive |
| R13 | Male | 57 | N/A | BLD | Positive |
| R14 | Female | 41 | N/A | BLD | Positive |
| R15 | Male | 50 | N/A | BLD | Positive |
| R16 | Male | 40 | N/A | BLD | Positive |
| H17 | Female | N/A | N/A | BLD | Negative |
| H18 | Female | N/A | N/A | BLD | Negative |
| H19 | Male | N/A | N/A | BLD | Negative |
| H20 | Male | N/A | N/A | BLD | Negative |
All subjects’ sera were screened for HCV antibodies, except for patients that had HCV RNA detectable by RT-PCR, as described in the ‘Patients, materials and methods’ section.
BLD: Below level of detection; C: Chronic; H: Healthy; IU: Infectious unit; N/A: Not assessed; R: Resolved.
Cell culture & media
PBMCs were cultured in RPMI 1640 tissue culture medium (BioWhittaker, ME, USA) supplemented with 25 mM HEPES, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 5 μg/ml gentamycin (all from Mediatech Cellgro, VA, USA), 10 U/ml heparin sodium (Fisher Scientific, PA, USA) and 10% pure human serum (Atlanta Biologicals, GA, USA). Cells incubator for were cultured in a 37°C, 5% CO2 the indicated amount of time.
Antigens & mitogens
The rNS3 protein was expressed and purified as previously described [33]. The recombinant influenza virus H3 (A/Phillipines/1992) antigen and H5 (A/Vietnam/2004) antigen were obtained from Protein Sciences (CT, USA). Phytohemagglutinin (PHA, Phaseolus vulgaris; Sigma, MO, USA) and influenza antigens were suspended in dimethyl sulfoxide, 0.1% and RPMI-1640.
T-cell proliferation assays
PBMCs (1 × 105 cells/well), in 200 μl complete medium containing 10% human serum, were plated in round-bottomed 96-well plates and incubated at 37°C, 5% CO2, for either 4 or 6 days, as indicated, pulsed overnight with 1 μCi/well of tritiated (3H)-thymidine (PerkinElmer, MA, USA), and harvested onto glass fiber filters (PerkinElmer) for measurement of radiolabel incorporation by liquid scintillation spectroscopy. Results are presented as the mean counts per minute (CPM) ± standard error of the mean of triplicate cultures. Proliferation data from PBMCs stimulated with recombinant influenza virus H3 antigen was transformed using a previously described algorithm: log10ΔCPM = log10 [Xexp−Xbkg]. To account for individual variation between subjects, NS3 antigen proliferation data from PBMCs stimulated with at least three different concentrations (0.1, 1 and 10 μg/ml) were averaged together and the mean background was subtracted. Subsequently, data was transformed onto a log scale [34]. The delta (Δ) CPM value was the mean experimental proliferative response minus the mean background.
ELISA
Cell culture supernatants were assayed for IL-22 and IL-17 cytokine levels by sandwich ELISA according to the manufacturer’s instructions (R&D Systems, Inc., MN, USA). Briefly, medium-binding 96-well microtiter plates (Corning Costar, NY, USA) were coated with capture antibody in phosphate-buffered saline (Thermo Fisher Scientific, Inc., MA, USA) overnight at 4°C. The plates were washed five times with wash buffer: phosphate-buffered saline + 0.01% Tween-20 (Thermo Fisher Scientific, Inc.). Supernatants obtained from cell cultures (100 μl/well) were added for 2 h at room temperature. The plates were washed five times in wash buffer and detection antibody was added according to the instructions. Europium-coupled StreptAvidin (200 ng/ml; PerkinElmer) was added for 1 h and then the plates were washed five times with wash buffer. Enhancing Solution (PerkinElmer) was added to detect fluorescence. Fluorescence was determined by a Victor2 Fluorometer (Wallac, Finland). Experimental values were determined by comparing the optical densities with a standard curve derived from recombinant proteins. Negative controls consisted of background levels of PBMCs in media alone and positive controls were PHA-activated PBMCs. Each sample was run in duplicate.
Intracellular cytokine staining
PBMC cultures were stained 48 h after stimulation with rNS3 in a dose-dependent manner. Brefeldin A (BD Bioscience, CA, USA) was added to the cultures according to the manufacturer’s instructions for the last 4 h prior to antibody staining; however, no further activation was performed. Extracellular surface staining was performed using a combination of anti-CD4-pacific blue, anti-CD8-Amcyan, anti-CD3-PEcy7 and 7-amino-actinomycin D (BD Bioscience). Cells were washed and then fixed and permeabilized with BD Cytofix/Cytoperm™ buffer (BD Bioscience), washed in BD Perm/Wash™ buffer (BD Bioscience) and stained with 0.5 μg/ml anti-IL-17F-APC and anti-IL-22-PE (R&D Systems, Inc.) for 45 min at 4°C. Cells were washed and stored until analyzed by flow cytometry.
Results
T-cell proliferative response to HCV NS3 proteins
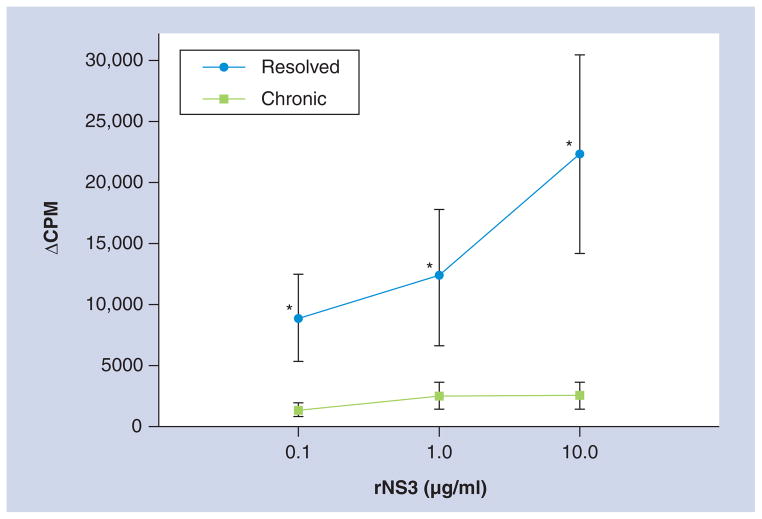
rNS3 protein, for which the protein sequence was based on HCV genotype 1a, was added into culture with PBMCs, and the T-cell proliferation was measured by 3H-thymidine uptake assays. The cohort of subjects included resolved patients and those chronically infected with HCV genotype 1a (Table 1). Resolved HCV subjects were nonviremic, as determined by PCR, and were HCV antibody-positive (Table 1). Unfortunately, resolved individuals were HCV RNA-negative by PCR, excluding the possibility of determining the HCV genotype. However, all of the resolved subjects had a dose-dependent T-cell proliferative response to rNS3, suggesting memory T cells specific for HCV were present in these individuals. HCV viral quantification in chronic HCV subjects was broad, ranging in viral titer from 0.6 × 105 IU/ml to 20 × 105 IU/ml (Table 1). As we have observed previously [18,35], significantly lower T-cell proliferation was measured in chronic HCV subjects versus resolved (Figure 1 & Table 2). T-cell responses to non-HCV antigens, influenza antigens (H3 and H5) and the mitogen PHA, were compared with those against rNS3 (Table 2). To be noted, the proliferation values of rNS3-activated PBMCs in Table 2 are the mean average of the three rNS3 concentrations that were used in Figure 1, and chronic HCV subjects did have a proliferative response to rNS3. Interestingly, as demonstrated in previous studies, the T-cell proliferative response to influenza antigens (in subjects that had a greater than twofold response over background) and PHA were not significantly different between chronic and resolved subjects (Table 2). These results suggested that chronic HCV subjects were able to process, recognize and present antigen similarly to resolved HCV subjects [33,35]. Importantly, noninfected negative control individuals (Table 1; H17–H20) had no T-cell proliferative responses when incubated with rNS3 (data not shown). All patients used in this study had a significant T-cell response to PHA, indicating that the PBMCs were viable (Table 2 ; data not shown). Therefore, HCV antigen-specific T-cell responses are significantly different from responses to other viral antigens, suggesting that HCV is able to specifically modulate the human immune response.
Figure 1. Dose-dependent T-cell response to recombinant NS3.
Peripheral blood mononuclear cells from HCV chronically infected (n = 8) and resolved subjects (n = 8) at 1 × 105 cells/well performed in triplicate were incubated with HCV rNS3 protein at 0.1, 1 or 10 μg/ml. 3H-thymidine uptake was measured at day 5. Background was subtracted from each culture condition, which is represented by ΔCPM.
*p < 0.05, Student’s paired t test.
CPM: Counts per minute; rNS3: Recombinant NS3.
Table 2.
Chronic and resolved HCV subjects’ T-cell proliferative responses to recombinant HCV NS3 protein, influenza antigens (H3 and H5) and phytohemagglutinin.
| Antigen | Chronic (n = 8) | Resolved (n = 8) | Significance |
|---|---|---|---|
| NS3† (log ΔCPM) | 3.6 ± 0.2 | 4.2 ± 0.2 | p < 0.05 |
| H3‡ | 3.9 ± 0.2 | 3.9 ± 0.2 | ns |
| H5‡ | 3.3 ± 0.3 | 3.5 ± 0.3 | ns |
| PHA | 4.8 ± 0.3 | 4.9 ± 0.06 | ns |
Values are representative of the log mean of three concentrations (0.1, 1 and 10 μg/ml) of recombinant NS3-induced T-cell proliferative response minus background (log ΔCPM). Results are expressed as mean ± standard error of the mean of proliferation. Peripheral blood mononuclear cells were added in triplicate at 1 × 105 cells/well.
Proliferation data are representative of individuals who had a response greater than twofold over medium (control background) for influenza antigens.
CPM: Counts per minute; ns: Not significant; PHA: Phytohemagglutinin.
Th17 cells are not involved in HCV clearance
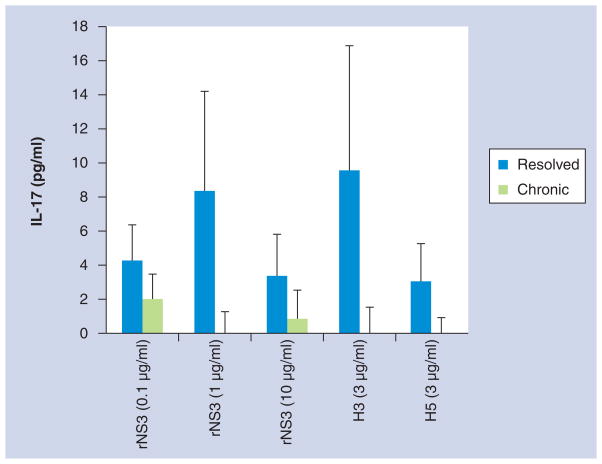
Recently, serum levels of IL-17 were demonstrated to not be significantly different in HCV-infected subjects in relation to HCV-related fibrosis [21]. However, IL-17 has been implicated in hepatic inflammation in patients with autoimmune hepatitis and hepatitis B [36,37]. Interestingly, preliminary studies, comparing gene expression profiles of subject C4’s PBMCs stimulated with rNS3 to subject C4’s PBMCs with no stimulation, demonstrated that the stimulated PBMCs had a greater than tenfold increase in IL22 and IL17 gene expression (microarray data not shown). In an effort to translate the microarray results obtained from one subject to a larger cohort of subjects, PBMCs from both chronic and resolved subjects were stimulated with rNS3 at rising doses (0.1, 1, 10 μg/ml), H3 (3 μg/ml), H5 (3 μg/ml) and PHA (2 μg/ml) (Figure 1 & Table 2). Cell culture supernatants were collected 48 h after antigen stimulation and IL-17 levels were measured by ELISA. Data was normalized by subtracting the cytokine levels detected from PBMCs treated with medium alone. The IL-17 cytokine levels induced by rNS3 were not significantly different between chronic and resolved subjects (Figure 2). However, there was a significant difference in the resolved (507 ± 104 pg/ml) and chronic (66 ± 33 pg/ml) PHA-induced IL-17 cytokine levels (data not shown). Furthermore, the IL-17 levels were not significantly different between chronic and resolved subjects in the H3 or H5 responses (Figure 2). Therefore, IL-17 cytokine production induced by rNS3 was not significantly different between chronic and resolved subjects, although mitogen-stimulated PBMCs from subjects chronically infected with HCV had a significantly lower level of IL-17 production than the HCV-resolved subjects.
Figure 2. Production of IL-17 was not significantly different between resolved and chronic HCV subjects.
Peripheral blood mononuclear cells from resolved (n = 8) and chronic (n = 8) subjects were incubated with either medium, rNS3 (0.1, 1 and 10 μg/ml), H3 (3 μg/ml), H5 (3 μg/ml) or phytohemagglutinin (2 μg/ml). Cell culture supernatants were collected 48 h after the addition of antigens, mitogen or medium. Data are normalized to medium.
rNS3: Recombinant NS3.
HCV antigen-specific induction of IL-22 cytokine production in T cells
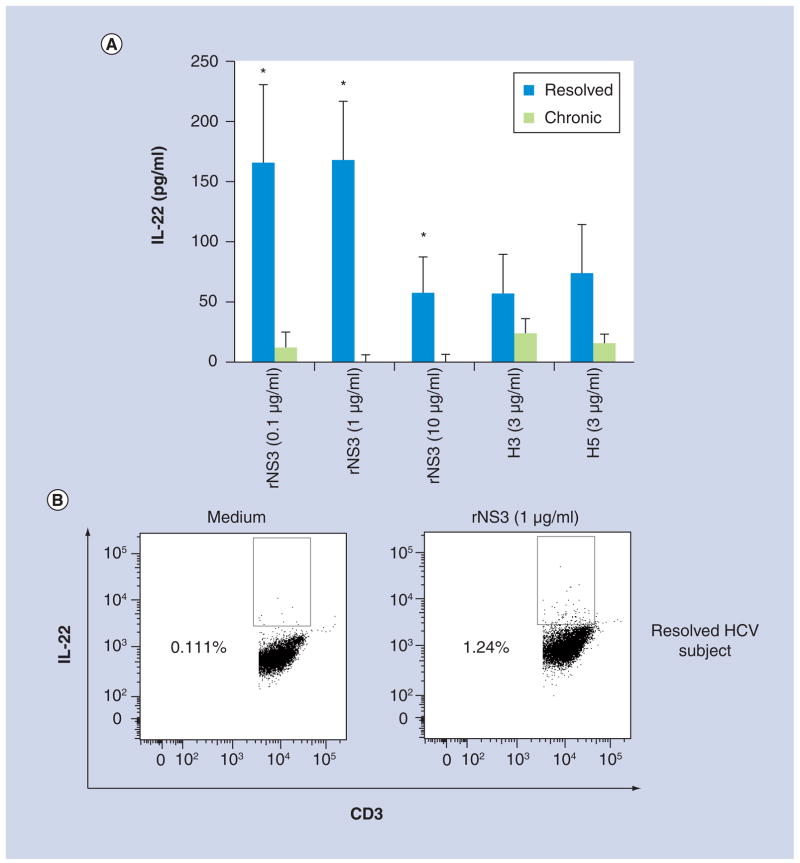
To determine if NS3 induced an IL-22 response in chronic and/or resolved HCV subjects, PBMCs were incubated with either medium, rNS3 at rising doses (0.1, 1, 10 μg/ml), H3 (3 μg/ml), H5 (3 μg/ml) or PHA (2 μg/ml), supernatants were collected 48 h after antigen stimulation and IL-22 levels were measured by ELISA. Data was normalized by subtracting the cytokine levels detected from PBMCs treated with medium alone. Interestingly, the IL-22 cytokine levels in the cell culture supernatants were significantly higher in PBMCs from resolved subjects stimulated with rNS3 in a dose-dependent manner, compared with chronic subjects (Figure 3a). Similar to the IL-17 ELISA results (above), the PHA-induced IL-22 cytokine response was significantly higher in resolved subjects (2533 ± 390 pg/ml) in comparison with chronic subjects (405 ± 281 pg/ml; data not shown). The IL-22 levels were not significantly different between chronic and resolved subjects in the H3 or H5 responses (Figure 3a). To determine if T cells were the source of the IL-22, flow cytometry was used to measure the number of CD3+ IL-22+ cells (Figure 3b). The representative flow cytometric data of a resolved HCV subject suggests that CD3+ T cells are producing IL-22; however, statistical analysis was not performed due to the low number of subjects tested.
Figure 3. T-cell production of IL-22 was significantly different between resolved and chronic HCV subjects.
(A) Peripheral blood mononuclear cells from resolved (n = 8) and chronic (n = 8) subjects were incubated with either medium, rNS3 (0.1, 1 and 10 μg/ml), H3 (3 μg/ml), H5 (3 μg/ml) or phytohemagglutinin (2 μg/ml). Cell culture supernatants were collected 48 h after the addition of antigens, mitogen or medium. Data are normalized to medium. (B) Representative flow cytometry dot plots of CD3+ IL-22+ expression in a resolved HCV subject’s cells at 48 h post-antigen stimulation. Flow cytometry data are representative of four different experiments.
*p < 0.05, Student’s paired t test.
rNS3: Recombinant NS3.
CD4+ T cells produce IL-22
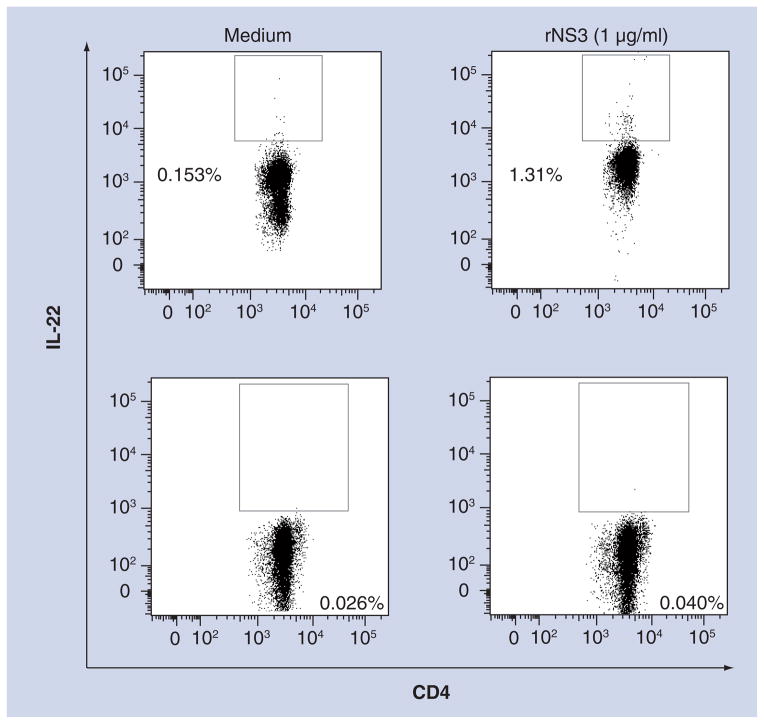
To determine whether CD4+ T cells were producing IL-22, we measured intracellular cytokine staining by flow cytometry. The flow cytometric dot plots showing increased numbers of CD4+ IL-22+ cells in the resolved HCV subject’s PBMCs activated by rNS3 (1 μg/ml) suggested that CD4+ T cells comprised at least one source of IL-22 in these cultures (Figure 4). Although the IL-22 background levels are higher in the resolved HCV subject in comparison with the chronic HCV subject, the testing of more subjects is needed to determine if clearance of HCV and background levels of IL-22 are related. Furthermore, CD8+ T cells had no detectable IL-22 from PBMCs stimulated with rNS3 (1 μg/ml) in resolved and chronic HCV subjects (data not shown).
Figure 4. CD4+ T cells express IL-22 in resolved HCV subjects.
Representative flow cytometric dot plots of CD4+ IL-22+ expression. Data are representative of three different experiments. Top panels are dot plots of peripheral blood mononuclear cells from a resolved HCV subject. Bottom panels are dot plots of peripheral blood mononuclear cells from a chronic HCV subject. Gating was determined by both medium samples (left panels) and fluorescence minus one including the isotype control (data not shown).
rNS3: Recombinant NS3.
Discussion
The quality of the CD4+ T-cell response has been found to be critical in the clearance of HCV [9,11,38–40]. In agreement with previous work, our study demonstrates attenuated T-cell proliferative responses to rNS3 in chronic HCV subjects along with intact influenza memory and mitogen-activated T-cell proliferative responses, suggesting that HCV is detected by the immune system in chronically infected subjects; however, HCV is able to evade clearance, possibly through mechanisms such as antigen-specific induction of Tregs and/or CD8+ T-cell exhaustion (Figure 1 & Table 2) [33,35]. Taken together, these results suggest that HCV may be specifically modulating some immune responses consistent with viral persistence in HCV-infected individuals.
Although the role of IL-22 in viral clearance is not well-defined, evidence suggests that IL-22 does not act directly on HCV replication [41]. However, IL-22 has been found to upregulate the production of acute-phase serum proteins such as serum amyloid A, α1-anti-chymotrypsin and haptoglobin in human hepatoma cell lines, suggesting that IL-22 production can lead to a proinflammatory response [42], which might be thought to be conducive to viral elimination. In HCV-infected individuals, Foster et al. found a greater number of intrahepatic lymphocytes secreting IL-22 and IL-17 when compared with lymphocytes from peripheral blood, suggesting that IL-17/IL-22 could be involved in HCV pathogenesis within the liver [21]. Furthermore, liver biopsies of chronically infected HBV and HCV patients were demonstrated to have a significantly higher number of IL-22+ cells when compared with healthy control liver biopsies, suggesting that IL-22+ cell abundance increased in the liver during HCV infection [43,44]. Although such previous studies provide valuable insight into which cells in the liver could be involved in HCV clearance, there are no studies describing HCV antigen-specific induction of IL-22. In this study, human PBMCs stimulated with rNS3 yielded a dose-dependent IL-22 response in resolved HCV subjects (Figure 3a). Importantly, in this study, nonstimulated PBMCs from resolved HCV subjects had a higher IL-22 background in comparison with PBMCs from chronically infected subjects (data not shown), suggesting T cells in the periphery of chronically infected subjects are not producing more background IL-22 in comparison to T cells obtained from resolved subjects. Therefore, the lack of response from chronic subjects is not due to the presence of a higher basal level of IL-22. In addition, the IL-22 production by PBMCs incubated with either H3 or H5 influenza antigens showed an increase in IL-22 production in the resolved subjects, but this was not significantly higher in comparison to chronic HCV subjects (Figure 3a). Mitogen-activated T cells had a significant difference in IL-22 production in resolved versus chronic subjects, even though there was no difference in PHA-induced T-cell proliferation, suggesting that HCV infection could have a broader effect on T-cell responses than previously thought. However, further investigation into the levels of IFN-γ and IL-2 produced by CD4+ T cells in conjunction with IL-22-producing cells would provide more insight into the activation level of these cell types. Therefore, HCV infection may potentially modulate immune responses, leading to persistent infection and possibly chronic liver disease.
IL-22R is expressed almost exclusively on non-hematopoietic cells, including epithelial cells, hepatic stellate cells and hepatocytes, suggesting that IL-22’s effect is tissue-specific [45]. Ligation of the IL-22R on non-hematopoietic cells by IL-22 leads to the modulation of a variety of genes encoding for molecules involved in chemotaxis, proliferation, innate immunity and inflammation (reviewed in [46]). For example, murine liver models and hepatocyte cell lines have provided evidence for a therapeutic benefit to exogenous recombinant IL-22. More specifically, previous studies using a murine stress-induced liver damage model that simulates ischemia and reperfusion injury leading to acute liver inflammation had shown a benefit from T-cell-derived IL-22 in hepatocyte regeneration [47]. Furthermore, it was postulated that IL-22 could provide a potential therapeutic option to prevent ischemia–reperfusion injury in transplant recipients due to STAT3 activation in a murine model [47]. The importance of STAT3 activation by IL-22 has been demonstrated by activation-induced phosphorylation of STAT3 in hepatocytes and thymic epithelium, which initiated hepatocyte proliferation and thymopoiesis [29,48,49]. It has been suggested that STAT3 regulates an antiviral response by inducing a strong potent immune response without leading to immunopathology [50]. In addition, IL-22 has been demonstrated to inhibit liver fibrosis by targeting hepatic stellate cells, suggesting IL-22 inhibits liver fibrosis [45]. However, in the case of chronically infected viral hepatitis subjects, the long-term effect of IL-22 stimulation within the liver could potentially play a role in hepatic carcinogenesis [43]. Therefore, further investigation into the role of IL-22 in the liver is clearly needed. Taken together, we hypothesize that IL-22 might have a role in facilitating a liver microenvironment that is conducive for viral clearance due to IL-22 inducing hepatocyte proliferation and activation of the immune system in an antigen-specific manner. Further investigation is underway to identify specific HCV epitopes that could potentially induce CD4+ IL-22. Identifying HCV-specific epitopes would allow for further investigation into the potential correlation of an antiviral effect of IL-22 in the liver and T-cell responses measured in the peripheral blood.
Conclusion
HCV modulates anti-HCV T-cell proliferation in vitro. Although CD4+ T cells are critical in HCV clearance, mechanisms employed by HCV to avoid detection by these cells are most likely multifactorial. In this study, we compared T-cell production of IL-17 and IL-22 cytokines in resolved and chronic HCV subject’s PBMCs when stimulated with an HCV antigen. We demonstrated a significance difference in the IL-22 produced by resolved HCV subjects. Therefore, HCV-specific induction of in vitro IL-22 correlates with individuals that have cleared HCV, suggesting a potential anti-viral and/or protective effect of IL-22. Clearly, further investigation of the role of IL-22 in HCV clearance is warranted based on the significant difference in the production of this cytokine in resolved versus chronic HCV subjects.
Executive summary.
Resolved HCV subjects’ peripheral blood mononuclear cell (PBMC) proliferative response to recombinant NS3 was significantly higher in comparison with chronically infected HCV subjects’ PBMC proliferative response.
Resolved HCV subjects’ PBMCs stimulated with rNS3 had significantly higher IL-22 cytokine production, thereby correlating HCV clearance to production of IL-22 by T cells.
Further characterization of CD4+ IL-22+ T cells specific for HCV antigens in the peripheral blood and in the liver would strengthen the observation that IL-22 is important in HCV clearance.
Due to tissue-specific action by IL-22, further studies are needed to elucidate the function of this cytokine in the context of the liver microenvironment during HCV pathogenesis.
Future studies directed at understanding IL-22/IL-22R biology could potentially lead to novel therapeutic targets or strategies in multiple human diseases, including chronic HCV infection.
Acknowledgments
The authors would like to thank C Zabawa for technical assistance.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
RS Fujinami recieved funding from the Emma Mary Deland Foundation and DD Eckels is NIH funded (NIH 5R01AI047347-11). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Hepatitis C. global prevalence. Wkly Epidemiol Rec. 1997;72(46):341–344. [PubMed] [Google Scholar]
- 2.Conry-Cantilena C, Vanraden M, Gibble J, et al. Routes of infection, viremia, and liver disease in blood donors found to have hepatitis C virus infection. N Engl J Med. 1996;334(26):1691–1696. doi: 10.1056/NEJM199606273342602. [DOI] [PubMed] [Google Scholar]
- 3.Kenny-Walsh E. Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. Irish Hepatology Research Group. N Engl J Med. 1999;340(16):1228–1233. doi: 10.1056/NEJM199904223401602. [DOI] [PubMed] [Google Scholar]
- 4.Seeff LB, Miller RN, Rabkin CS, et al. 45-year follow-up of hepatitis C virus infection in healthy young adults. Ann Intern Med. 2000;132(2):105–111. doi: 10.7326/0003-4819-132-2-200001180-00003. [DOI] [PubMed] [Google Scholar]
- 5.Messick K, Sanders JC, Goedert JJ, Eyster ME. Hepatitis C viral clearance and antibody reactivity patterns in persons with haemophilia and other congenital bleeding disorders. Haemophilia. 2001;7(6):568–574. doi: 10.1046/j.1365-2516.2001.00559.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang M, Rosenberg PS, Brown DL, et al. Correlates of spontaneous clearance of hepatitis C virus among people with hemophilia. Blood. 2006;107(3):892–897. doi: 10.1182/blood-2005-07-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo QL, Richman KH, Han JH, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckels DD, Tabatabail N, Bian TH, et al. In vitro human Th-cell responses to a recombinant hepatitis C virus antigen: failure in IL-2 production despite proliferation. Hum Immunol. 1999;60(3):187–199. doi: 10.1016/s0198-8859(98)00111-6. [DOI] [PubMed] [Google Scholar]
- 9.Day CL, Lauer GM, Robbins GK, et al. Broad specificity of virus-specific CD4+ T-helper-cell responses in resolved hepatitis C virus infection. J Virol. 2002;76(24):12584–12595. doi: 10.1128/JVI.76.24.12584-12595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diepolder HM, Gerlach JT, Zachoval R, et al. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71(8):6011–6019. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlach JT, Diepolder HM, Jung MC, et al. Recurrence of hepatitis C virus after loss of virus-specific CD4+ T-cell response in acute hepatitis C. Gastroenterology. 1999;117(4):933–941. doi: 10.1016/s0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 12.Thimme R, Bukh J, Spangenberg HC, et al. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci USA. 2002;99(24):15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194(10):1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Zhu F, Sonderstrup G, Eckels DD. Recognition of endogenously synthesized HLA-DR4 restricted HCV epitopes presented by autologous EBV transformed B-lymphoblastoid cell line. Vaccine. 2005;23(7):951–962. doi: 10.1016/j.vaccine.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 15.Fuller MJ, Shoukry NH, Gushima T, et al. Selection-driven immune escape is not a significant factor in the failure of CD4 T cell responses in persistent hepatitis C virus infection. Hepatology. 2010;51(2):378–387. doi: 10.1002/hep.23319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckels DD, Zhou H, Bian TH, Wang H. Identification of antigenic escape variants in an immunodominant epitope of hepatitis C virus. Int Immunol. 1999;11(4):577–583. doi: 10.1093/intimm/11.4.577. [DOI] [PubMed] [Google Scholar]
- 17.Mosmann TR, Coffman RL. Th1 and Th2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 18.Eckels DD, Wang H, Bian TH, Tabatabai N, Gill JC. Immunobiology of hepatitis C virus (HCV) infection: the role of CD4 T cells in HCV infection. Immunol Rev. 2000;174:90–97. doi: 10.1034/j.1600-0528.2002.017403.x. [DOI] [PubMed] [Google Scholar]
- 19.Macdonald AJ, Duffy M, Brady MT, et al. CD4 T helper type 1 and regulatory T cells induced against the same epitopes on the core protein in hepatitis C virus-infected persons. J Infect Dis. 2002;185(6):720–727. doi: 10.1086/339340. [DOI] [PubMed] [Google Scholar]
- 20.Tsai SL, Liaw YF, Chen MH, Huang CY, Kuo GC. Detection of type 2-like T-helper cells in hepatitis C virus infection: implications for hepatitis C virus chronicity. Hepatology. 1997;25(2):449–458. doi: 10.1002/hep.510250233. [DOI] [PubMed] [Google Scholar]
- 21▪▪.Foster RG, Golden-Mason L, Rutebemberwa A, Rosen HR. Interleukin (IL)-17/IL-22-producing T cells enriched within the liver of patients with chronic hepatitis C viral (HCV) infection. Dig Dis Sci. 2012;57(2):381–389. doi: 10.1007/s10620-011-1997-z. Co-producing IL-22 and IL-17 T cells were enriched in the livers of HCV-infected patients. [DOI] [PubMed] [Google Scholar]
- 22.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- 23.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21(2):241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Tachiiri A, Imamura R, Wang Y, Fukui M, Umemura M, Suda T. Genomic structure and inducible expression of the IL-22 receptor alpha chain in mice. Genes Immun. 2003;4(2):153–159. doi: 10.1038/sj.gene.6363934. [DOI] [PubMed] [Google Scholar]
- 25.Eyerich S, Eyerich K, Pennino D, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119(12):3573–3585. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nograles KE, Zaba LC, Guttman-Yassky E, et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol. 2008;159(5):1092–1102. doi: 10.1111/j.1365-2133.2008.08769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27▪▪.Wahl C, Wegenka UM, Leithauser F, Schirmbeck R, Reimann J. IL-22-dependent attenuation of T cell-dependent (ConA) hepatitis in herpes virus entry mediator deficiency. J Immunol. 2009;182(8):4521–4528. doi: 10.4049/jimmunol.0802810. IL-22 was protective in a murine model of acute hepatitis. [DOI] [PubMed] [Google Scholar]
- 28.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Karow M, Flavell RA. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27(4):647–659. doi: 10.1016/j.immuni.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29▪.Radaeva S, Sun R, Pan HN, Hong F, Gao B. Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology. 2004;39(5):1332–1342. doi: 10.1002/hep.20184. IL-22 promoted hepatocyte survival by activation of the STAT3 signaling pathway in a murine acute hepatitis model. [DOI] [PubMed] [Google Scholar]
- 30▪.Misse D, Yssel H, Trabattoni D, et al. IL-22 participates in an innate anti-HIV-1 host-resistance network through acute-phase protein induction. J Immunol. 2007;178(1):407–415. doi: 10.4049/jimmunol.178.1.407. Implicates the production of IL-22 in viral pathogenesis due to HIV-1-resistant humans having higher levels of IL-22-producing T cells. [DOI] [PubMed] [Google Scholar]
- 31▪▪.Hennig BJ, Frodsham AJ, Hellier S, et al. Influence of IL-10RA and IL-22 polymorphisms on outcome of hepatitis C virus infection. Liver Int. 2007;27(8):1134–1143. doi: 10.1111/j.1478-3231.2007.01518.x. Polymorphisms in the IL22 gene were associated with HCV outcome. [DOI] [PubMed] [Google Scholar]
- 32.Levillayer F, Mas M, Levi-Acobas F, Brahic M, Bureau JF. Interleukin 22 is a candidate gene for Tmevp3, a locus controlling Theiler’s virus-induced neurological diseases. Genetics. 2007;176(3):1835–1844. doi: 10.1534/genetics.107.073536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urbani S, Amadei B, Tola D, et al. PD-1 expression in acute hepatitis C virus (HCV) infection is associated with HCV-specific CD8 exhaustion. J Virol. 2006;80(22):11398–11403. doi: 10.1128/JVI.01177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cusick MF, Wang S, Eckels DD. In vitro responses to avian influenza H5 by human CD4 T cells. J Immunol. 2009;183(10):6432–6441. doi: 10.4049/jimmunol.0901617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cusick MF, Schiller JJ, Gill JC, Eckels DD. Hepatitis C virus induces regulatory T cells by naturally occurring viral variants to suppress T cell responses. Clin Dev Immunol. 2011 doi: 10.1155/2011/806061. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao L, Tang Y, You Z, et al. Interleukin-17 contributes to the pathogenesis of autoimmune hepatitis through inducing hepatic interleukin-6 expression. PLoS One. 2011;6(4):e18909. doi: 10.1371/journal.pone.0018909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang JY, Zhang Z, Lin F, et al. Interleukin-17-producing CD4+ T cells increase with severity of liver damage in patients with chronic hepatitis B. Hepatology. 2010;51(1):81–91. doi: 10.1002/hep.23273. [DOI] [PubMed] [Google Scholar]
- 38.Grakoui A, Shoukry NH, Woollard DJ, et al. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302(5645):659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 39.Schulze Zur Wiesch J, Ciuffreda D, Lewis-Ximenez L, et al. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med. 2012;209(1):61–75. doi: 10.1084/jem.20100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleming VM, Harcourt G, Barnes E, Klenerman P. Virological footprint of CD4+ T-cell responses during chronic hepatitis C virus infection. J Gen Virol. 2010;91(Pt 6):1396–1406. doi: 10.1099/vir.0.017699-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dambacher J, Beigel F, Zitzmann K, et al. The role of interleukin-22 in hepatitis C virus infection. Cytokine. 2008;41(3):209–216. doi: 10.1016/j.cyto.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Dumoutier L, Van Roost E, Colau D, Renauld JC. Human interleukin-10-related T cell-derived inducible factor: molecular cloning and functional characterization as an hepatocyte-stimulating factor. Proc Natl Acad Sci USA. 2000;97(18):10144–10149. doi: 10.1073/pnas.170291697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park O, Wang H, Weng H, et al. In vivo consequences of liver-specific interleukin-22 expression in mice: implications for human liver disease progression. Hepatology. 2011;54(1):252–261. doi: 10.1002/hep.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng D, Kong X, Weng H, et al. Interleukin-22 promotes proliferation of liver stem/progenitor cells in mice and patients with chronic hepatitis B virus infection. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.03.044. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong X, Feng D, Wang H, et al. Interleukin-22 induces hepatic stellate cell senescence and restricts liver fibrosis. Hepatology. 2012 doi: 10.1002/hep.25744. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12(5):383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 47.Chestovich PJ, Uchida Y, Chang W, et al. Interleukin-22: implications for liver ischemia-reperfusion injury. Transplantation. 2012;93(5):485–492. doi: 10.1097/TP.0b013e3182449136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudakov JA, Hanash AM, Jenq RR, et al. Interleukin-22 drives endogenous thymic regeneration in mice. Science. 2012;336(6077):91–95. doi: 10.1126/science.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren X, Hu B, Colletti LM. IL-22 is involved in liver regeneration after hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2010;298(1):G74–G80. doi: 10.1152/ajpgi.00075.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem. 2006;281(20):14111–14118. doi: 10.1074/jbc.M511797200. [DOI] [PubMed] [Google Scholar]