Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that act as post-transcriptional gene regulators and have been shown to regulate many biological processes including embryonal development, cell differentiation, apoptosis, and proliferation. Variations in the expression of certain miRNAs have been linked to a wide range of human diseases – especially cancer – and the diversity of miRNA targets suggests that they are involved in various cellular networks. Several tools have been developed to control the function of individual miRNAs and have been applied to study their biogenesis, biological role, and therapeutic potential; however, common methods lack a precise level of control that allows for the study of miRNA function with high spatial and temporal resolution. Light-activated miRNA antagomirs for mature miR-122 and miR-21 were developed through the site-specific installation of caging groups on the bases of selected nucleotides. Installation of caged nucleotides led to complete inhibition of the antagomir-miRNA hybridization and thus inactivation of antagomir function. The miRNA-inhibitory activity of the caged antagomirs was fully restored upon decaging through a brief UV irradiation. The synthesized antagomirs were applied to the photochemical regulation of miRNA function in mammalian cells. Moreover, spatial control over antagomir activity was obtained in mammalian cells through localized UV exposure. The presented approach enables the precise regulation of miRNA function and miRNA networks with unprecedented spatial and temporal resolution using UV irradiation and can be extended to any miRNA of interest.
Introduction
MicroRNAs (miRNAs) are a recently discovered class of non-coding RNAs that regulate gene expression in a sequence specific fashion by binding the 3′ untranslated regions (UTRs) of target mRNAs,1 although recently, it has also been discovered that miRNA translational silencing can occur through targeting of 5′ UTRs.2 MiRNAs are single stranded RNA molecules of 21–23 nucleotides that down-regulate gene function by inhibiting translation, accelerating the degradation of the mRNA, or mediating deadenylation of the mRNA.1, 3 After transcription from the genome, miRNAs undergo several post-transcriptional processing steps via a dedicated miRNA pathway to produce mature miRNAs. Since miRNAs can bind to the 3′ UTR of target mRNAs with imperfect complementarity, each miRNA can target many different mRNA transcripts,4 and it is estimated that miRNAs control more than 30% of all genes and are involved in almost every genetic pathway.5 Biological processes that are regulated by miRNAs include embryonal development, cell differentiation, apoptosis, and proliferation.6 Targets of miRNAs range from signalling proteins, metabolic enzymes, and transcription factors to RNA binding proteins.7 The diversity of miRNA targets suggest that they are involved in every aspect of cellular circuitry, including signal transduction, gene regulatory, metabolic, and protein interaction networks.7 The aberrant expression of certain miRNAs has been linked to a wide range of human diseases, including cancer, cardiovascular diseases, immune disorders, and viral infections. 8–12
Due to the extensive involvement of miRNAs in biological processes and human diseases, several regulatory tools to control the function of individual miRNAs have been developed and have been applied to study their biogenesis, biological role, and therapeutic potential.13–16 Amongst these tools are miRNA antisense oligonucleotides,17, 18 miRNA sponges or decoys,19 miRNA expression vectors,20 and small molecule regulators.21, 22 The most common tools are miRNA antisense oligonucleotides, known as antagomirs, which are chemically modified oligonucleotides that are complementary to a miRNA and act as competitive inhibitors of the target mRNA binding sequence. The chemical modifications that are commonly introduced into antagomir structures in order to render the oligonucleotides more resistant to degradation in a cellular environment, to increase binding affinity, and to improve overall activity include 2′ sugar modifications such as 2′-O-methyl (2′OMe), 2′-O-methoxyethyl, 2′-fluoro, and the addition of a bridging methylene group between the 2′-O and the 4′-C of the ribose ring (locked nucleic acid, LNA). In addition, phosphorothioate bonds are typically included in the RNA backbone.13, 23, 24
In order to investigate miRNA function in a spatial and temporal context, antagomir reagents that can be activated with high spatio-temporal resolution are desirable. Light represents an external control element that can be regulated with high precision and has previously been applied to the control of various biological processes.25–29 Light-control of oligonucleotide function has been achieved through the installation of caging groups on nucleobases, thereby disrupting Watson-Crick base-pairing until the caging groups are removed through irradiation with UV light. 30–36 An alternative approach to the photochemical regulation of oligonucleotide hybridization is the synthesis of hairpin structures containing a light-cleavable linker. After UV irradiation, an inhibitor strand is released, leading to the activation of an antisense agent and its silencing of cellular RNA function.37–41
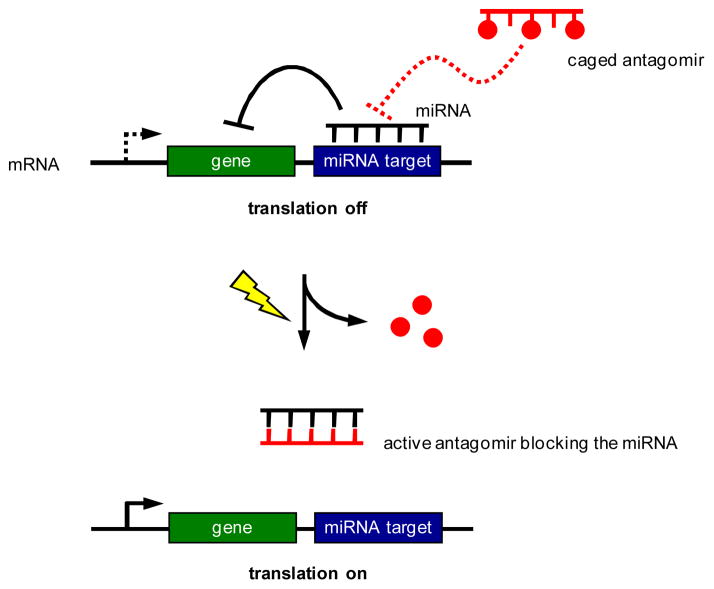
Here, we are applying the nucleobase-caging methodology to the photochemical control of antagomir and miRNA function. As shown in Scheme 1, a miRNA recognizes a (partially) complementary target sequence downstream of a gene and inhibits its expression. A nucleobase-caged antagomir has no effect on miRNA-mediated gene silencing, until it is activated through photochemical removal of the caging groups. The activated antagomir then binds to the miRNA and blocks its function, leading to the light-activation of gene expression. The gene of interest can be any endogenous gene or genetic circuit that is under control of a particular miRNA; or it can be an exogenous reporter gene, e.g., luciferase or EGFP, engineered to respond to a particular cellular miRNA.
Scheme 1.
Regulation of gene expression through photocontrol of endogenous miRNAs with nucleobase-caged antagomirs.
Results and Discussion
In order to demonstrate that light-activated antagomirs can inhibit miRNA function, microRNA miR-122 and miR-21 were selected as targets. MicroRNA miR-122 is a liver specific miRNA42 that is required by the hepatitis C virus for replication and infectious virus production.43 It was reported that knockdown of miR-122 with antagomirs resulted in a decrease in HCV RNA replication in human liver cells43 and reduced HCV levels in chronically infected primates.44 The miRNA miR-21 has been linked to several human malignancies and over expression of miR-21 has been observed in glioblastomas, breast, pancreatic, cervical, colorectal, ovarian, lung, and hepatocellular cancers.45 MiR-21 functions as an anti-apoptotic factor in cancer cells and a dependence of tumor growth on miR-21 presence has been demonstrated in a mouse model.46 The involvement of miR-122 and miR-21 in human malignancies and the promising in vivo results make these miRNAs potential targets for the development of fundamentally new therapeutics and the development of new photochemical tools to study these miRNAs could provide additional information on their involvement in diseases.
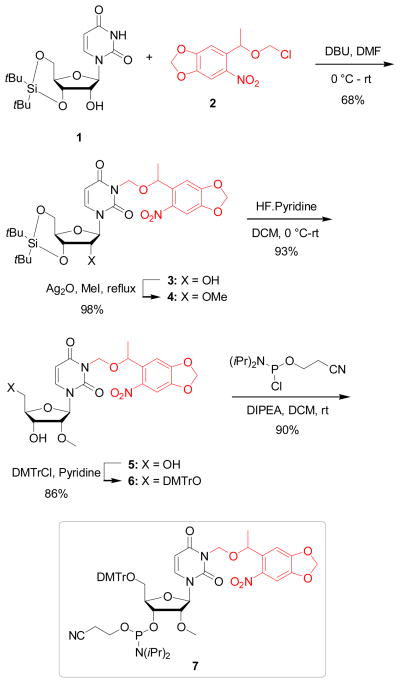
To develop light-activated antagomirs, we designed perfectly complementary oligonucleotides with 2′OMe modified nucleotides and phosphorothioate backbones targeting mature miR-21 and miR-122. In order to site-specifically install a light-removable caging group, a 2′-OMe NPOM (6-nitropiperonyloxymethyl)-caged uridine phosphoramidite (7) was synthesized (Scheme 2). 3′,5′-O-(Di-t-butylsilanediyl)uridine47 (1) was synthesized in one step and was subsequently reacted with NPOM chloride48 (2) in the presence of DBU in DMF to furnish the corresponding NPOM-caged uridine 3 in 68% yield. NPOM chloride (2) was synthesized in three steps from commercially available 6-nitropiperonal.48 The NPOM caged uridine 3 was heated under reflux in methyl iodide in the presence of Ag2O49 to deliver the corresponding 2′OMe derivative 4 in 98% yield. Treatment of 4 with HF-pyridine at 0° C in DCM yielded the NPOM caged 2′OMe uridine 5 in 93%. Protection of the 5′-OH group of 5 with DMTrCl in pyridine resulted the corresponding DMT-protected caged compound 6 in 68% yield. Finally, 6 was activated with 2-cyanoethyl N,N-diisopropylchlorophosphoramidite to give the desired phosphoramidite 7 in 90% yield.
Scheme 2.
Synthesis of the NPOM-caged 2′OMe U phosphoramidite 7.
Non-caged and caged 2′OMe phosphorothioate antagomirs containing the 2′OMe NPOM-caged uridine groups were synthesized for mature miR-21 and miR-122 (Table 1). The synthesis was conducted on an Applied Biosystems Model 394 automated DNA/RNA synthesizer using standard β-cyanoethyl phosphoramidite chemistry. The sulfurization was performed using a standard ABI synthesis cycle and the Beaucage sulfurizing reagent.50 The antagomirs were eluted from the solid-phase support with ammonium hydroxide and subsequently deprotected. The deprotected oligonucleotides were purified using Illustra NAP-10 columns by gravity flow through Sephadex G-25 DNA grade resin and a high purity of the final antagomirs was confirmed using PAGE analysis. The miR-21 caged antagomir was designed to contain four NPOM-caged 2′OMe uridine nucleotides whereas the miR-122 caged antagomir contains three 2′OMe NPOM-caged uridines (Table 1).
Table 1.
Sequences of miR-21 and miR-122 and the synthesized antagomirs.a
| miRNA Target/Antagomir | Sequence 5′ → 3′ |
|---|---|
| mature miR-21 | UAGCUUAUCAGACUGAUGUUGA |
| miR-21 antagomir | mA*mU*mC*mA*mA*mC*mA*mU*mC*mA* mG*mU*mC*mU*mG*mA*mU*mA*mA*mG* mC*mU*mA |
| caged miR-21 antagomir | mA*mU*mC*mA*mA*mC*mA*mU*mC*mA* mG*mU*mC*mU*mG*mA*mU*mA*mA*mG* mC*mU*mA |
| mature miR-122 | UGGAGUGUGACAAUGGUGUUUG |
| miR-122 antagomir | mA*mC*mA*mA*mA*mC*mA*mC*mC*mA* mU*mU*mG*mU*mC*mA*mC*mA*mC*mU* mC*mC*mA |
| caged miR-122 antagomir | mA*mC*mA*mA*mA*mC*mA*mC*mC*mA* mU*mU*mG*mU*mC*mA*mC*mA*mC*mU* mC*mC*mA |
An asterisk indicates a phosphorothioate bond, an m indicates a 2′OMe modified nucleotide, and a bold mU denotes a NPOM-caged uridine nucleotide.
To investigate the photochemical control of antagomir activity in mammalian cell culture, two Renilla luciferase sensors for either mature miR-122 or miR-21 based on the psiCHECK-2 (Promega) reporter plasmid were used.21, 22 The psiCHECK-2 vector was selected because it contains both a Renilla luciferase as well as an independently transcribed firefly luciferase reporter gene, which can be used for normalization purposes in order to account for variation in transfection efficiency and cell viability. The complementary sequences of miR-122 or miR-21 were inserted downstream of the Renilla luciferase gene (Scheme 1). Thus, the presence of the mature miRNA will lead to a decrease in the Renilla luciferase signal enabling the detection of endogenous miRNA levels. An active antagomir would act as a competitive inhibitor of miRNA target binding and would lead to an increase in Renilla luciferase expression. The ability of the reporters to detect endogenous miR-21 or miR-122 was validated by transiently transfecting the generated psiCHECK constructs into Huh7 human hepatoma cells which have previously been shown to express high levels of miR-122 and miR-21.42, 51, 52 The specificity of the luciferase reporters for miR-122 and miR-21 were verified by transfection of the non-caged miR-122 and miR-21 antagomirs (Supporting Figures S1 and S2).
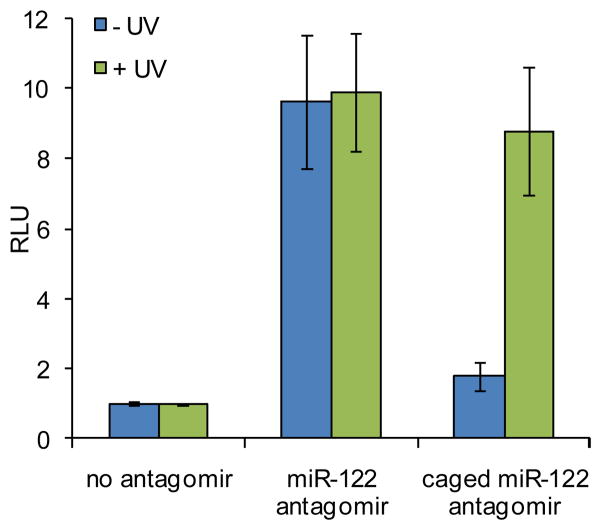
A Huh7 cell line that is stably transfected with the Renilla luciferase sensor for endogenous miR-122 was used to confirm the activity of the synthesized miR-122 antagomirs.53 In order to determine the best irradiation conditions for the caged antagomir, a time course experiment was conducted (Supporting Figure S3). The Huh7-psiCHECK-miR122 cells were transfected with the caged miR-122 antagomir in triplicate in a 96-well format. Following transfection, the cells were either kept in the dark or were irradiated at 365 nm for 10, 15, 30, 120, or 300 sec. After 48 h, the relative luciferase signal was measured using a Dual Luciferase Assay Kit (Promega). The Renilla luciferase was normalized to the firefly luciferase activity providing relative luminescence units (RLU). An irradiation time of 2 to 5 min efficiently activated miR-122 antagomir function (Supporting Figure S3). The Huh7-psiCHECK-miR122 cells were then transfected with either the non-caged or caged miR-122 antagomir in triplicate and were kept in the dark or irradiated at 365 nm for 5 min following transfection. After 48 h, the relative luciferase signal was measured. The non-caged miR-122 antagomir produced a 10-fold increase in RLU values relative to a transfection control containing no antagomir (Figure 1). Prior to UV irradiation, the caged miR-122 antagomir showed RLU values similar to the transfection control, indicating that the presence of three NPOM-caging groups on a 2′OMe U prevents the antagomir from inhibiting miR-122 function. After UV irradiation, the caging groups are removed and the decaged miR-122 antagomir effectively inhibits miR-122 and leads to a 9-fold increase in RLU.
Figure 1.
Light-activation of miR-122 inhibition and subsequent luciferase expression by decaging of a miR-122 antagomir in Huh7 cells. Cells were transfected with the caged and non-caged miR-122 antagomir, and UV irradiation (365 nm, 25 W) for 5 min efficiently activated antagomir function. The error bars represent standard deviations from three independent experiments.
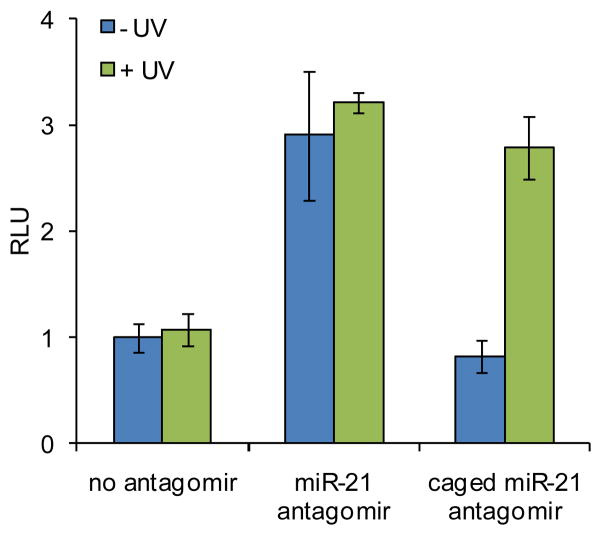
The miR-21 antagomirs were assayed using a Huh7 cell line transiently transfected with the Renilla luciferase sensor for endogenous miR-21. In order to determine the best irradiation conditions for decaging, a time course experiment was performed (Supporting Figure S4). The Huh7 cells were co-transfected with the psiCHECK-miR21 plasmid and the caged miR-21 antagomir at 10 pmol in triplicate in a 96-well format. Following transfection, the cells were kept in the dark or irradiated at 365 nm for 10, 15, 30, 120, or 300 sec. After 48 h, the relative luciferase signal was measured using a Dual Luciferase Assay Kit (Promega). An irradiation time of 2 to 5 min efficiently activated miR-21 antagomir function (Supporting Figure S4). Huh7 cells were then co-transfected with the psiCHECK-miR21 plasmid and either the non-caged or caged miR-122 antagomir in triplicate and were kept in the dark or irradiated at 365 nm for 5 min. The relative luciferase signal was measured after 48 h. The non-caged miR-21 antagomir produced a 3-fold increase in RLU values relative to a transfection control containing no antagomir (Figure 2). Prior to decaging, the caged miR-21 antagomir showed RLU values equal to the transfection control, indicating that the presence of the four NPOM-caging groups prevents that antagomir from inhibiting miR-21 function. After UV irradiation, the decaged miR-21 antagomir provides a 3-fold increase in RLU values, equal to that of the non-caged antagomir, demonstrating that the miR-21 inhibitory activity is fully restored.
Figure 2.
Light-activation of miR-21 inhibition and subsequent luciferase expression by decaging of a miR-21 antagomir in Huh7 cells. Cells were transfected with the caged and non-caged miR-21 antagomir, and UV irradiation (365 nm, 25 W) for 5 min efficiently activated antagomir function. The error bars represent standard deviations from three independent experiments.
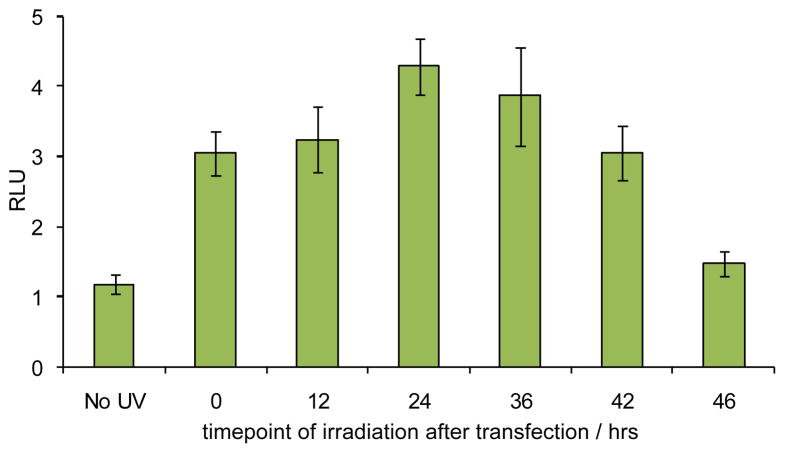
Light-activated antagomirs are unique tools that could allow for the temporal control over miRNA function. The ability to control antagomir activity and thereby inhibit miRNA function in a temporal fashion was investigated using the caged miR-21 antagomir. Huh7 cells were co-transfected with the psiCHECK-miR21 reporter plasmid and the caged miR-21 antagomir. Following transfection, the cells were kept in the dark and then irradiated at 0, 12, 24, 36, 42, and 46 h. The cells were incubated for a total of 48 h post transfection, as in previous experiments, and luciferase activity was measured. Prior to decaging, the caged miR-21 antagomir showed RLU values equal to the transfection control (Figure 3). After UV irradiation immediately following transfection (0 h), the decaged miR-21 antagomir provides a 3-fold increase in RLU values, demonstrating that based on comparison with Figure 2 the miR-21 inhibitory activity is fully restored. Similar results were obtained when the cells were irradiated 12 h post transfection. Interestingly, when UV irradiation was administered 24 and 36 h following transfection, an unexpected further increase in RLU values were obtained. We hypothesize that this observation could potentially be caused by an increase in antagomir stability prior to decaging, a limited lifetime of the target protein (Renilla luciferase) generated after miR-21 silencing, establishment of a compensatory effect by the cells in the case of constitutively active antagomirs (early irradiation) by upregulation of miR-21 expression, or a combination thereof. Irradiations at 42 and 46 h post transfection lead to decreasing RLU values, as there is not sufficient time for Renilla luciferase production following the inhibition of miR-21.
Figure 3.
Temporal control over miR-21 inhibition and subsequent luciferase expression by decaging of a miR-21 antagomir in Huh7 cells. Cells were transfected with the caged miR-21 antagomir, and were irradiated (365 nm, 5 min) at 0, 12, 24, 36, 42, and 46 hours post transfection. The error bars represent standard deviations from three independent experiments.
The ability to control miRNA function with light-activated antagomirs was further demonstrated by constructing an EGFP sensor for endogenous miR-21 through inserting the complementary sequence of miR-21 downstream of an EGFP gene in the pEGFP-C2 plasmid. Thus, the presence of mature miR-21 will lead to a decrease in the EGFP expression (Scheme 1). The ability of the reporter to detect endogenous miR-21 was validated by transiently transfecting the generated pEGFP-C2-miR21 construct into Huh7 cells. The pEGFP-C2 construct without the miR-21 target site was transfected as a positive control and fluorescence was imaged on a Zeiss Axio Observer inverted microscope. Transfection of the pEGFP-C2-miR21 construct showed almost complete silencing of EGFP expression due to endogenous miR-21.
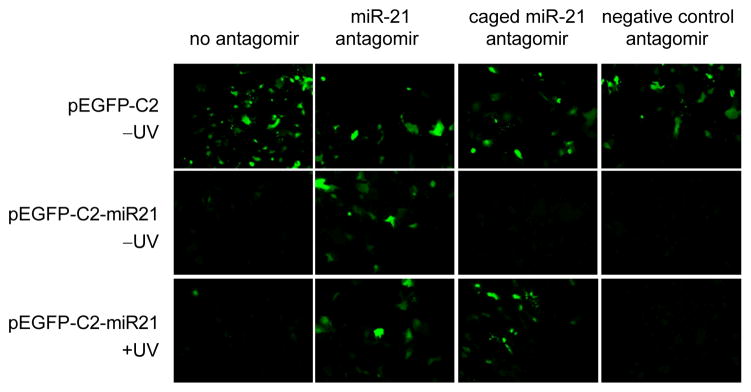
The construct was then used to assay the synthesized miR-21 non-caged and caged antagomirs. The transfection conditions were optimized for the co-transfection of the reporter plasmid and antagomirs and an irradiation time of 5 min was utilized based on the time course experiment performed using the luciferase reporter. Huh7 cells were co-transfected with either the parent pEGFP-C2 or pEGFP-C2-miR21 construct and the caged and non-caged miR-21 antagomirs. In addition, the miR-122 antagomir was used as a negative control. Following transfection, the cells were irradiated at 365 nm for 5 min, and fluorescence was imaged after 48 h (Figure 4). When the pEGFP-C2-miR21 plasmid was co-transfected with the non-caged, positive control miR-21 antagomir the EGFP expression was restored, indicating that the antagomir effectively inhibits miR-21 function. As expected, the negative control antagomir had no effect on EGFP expression. Prior to irradiation, the miR-21 caged antagomir is inactive and EGFP expression is silenced, as in case of the nonfunctional negative control antagomir. However, upon irradiation at 365 nm for 5 min, the caging groups are removed and the decaged miR-21 antagomir regains miR-21 inhibitory activity as shown by an increase in EGFP expression to the same extend as observed with the non-caged, positive control miR-21 antagomir.
Figure 4.
Light-activation of EGFP expression by decaging of a miR-21 antagomir in Huh7 cells transiently transfected with an EFGP sensor for miR-21 function. UV irradiation (365 nm, 5 min) restored antagomir activity and inhibited miR-21 function. The miR-122 antagomir was used as a negative control since it shows no crossreactivity with the reporter.
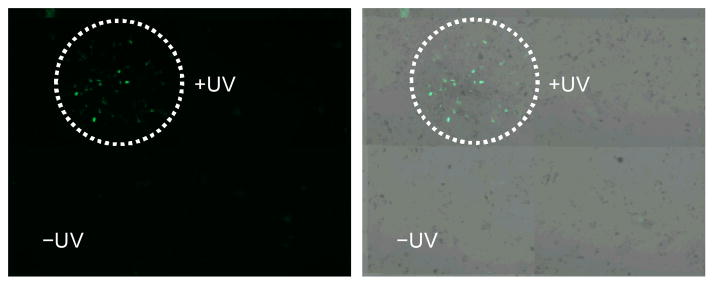
Based on the successful light-activation of the caged miR-21 antagomir, the ability to control miR-21 function in a spatial fashion was explored in tissue culture. A monolayer of Huh7 cells was transfected with the pEGFP-C2-miR21 construct and the caged miR-21 antagomir. Following transfection, localized UV irradiation (365 nm, 5 min) was conducted using a UV LED fiber optics probe. Cells were incubated for 48 h and fluorescence was imaged using a 10× objective and a 2×2 tile scan (Figure 5). EGFP expression was only observed within the irradiated area and no EGFP expression was visible in the non-irradiated area, thus demonstrating spatial control over antagomir activity and thereby miR-21 function.
Figure 5.
Spatial activation of EGFP expression by decaging of a miR-21 antagomir in a localized fashion. Huh7 cells were co-transfected with an EGFP sensor for miR-21 function and the caged miR-21 antagomir (100 pmol). Cells were irradiated at 365 nm using a LED fiber optics probe (Prizmatix) and were imaged after 48 hrs on a Zeiss Axio Observer inverted microscope (10× objective, 2×2 tile scan). The EGFP channel and corresponding bright field images are shown.
Conclusions
In summary, we have developed light-activated antagomirs for mature miR-122 and miR-21 through the site-specific installation of caging groups on the bases of selected nucleotides. Installation of three or four caged nucleotides led to complete inactivation of the antagomir-miRNA hybridization and thus inactivation of antagomir function. The miRNA-inhibitory activity of the caged antagomirs was fully restored upon decaging through a brief UV irradiation. These synthesized antagomirs were then applied to the photochemical regulation of miRNA function in mammalian cells. The ability to control antagomir activity and thereby miRNA function with light was demonstrated using both a Renilla luciferase reporter and a fluorescent EGFP reporter. In addition, temporal control over miRNA function was achieved using a caged miR-21 antagomir. Moreover, spatial control over antagomir activity was obtained in Huh7 cells through localized UV exposure. The presented approach enables the precise deactivation of miRNA function using UV irradiation and can be extended to any miRNA of interest. Applications may include the study of miRNAs involved in cell-to-cell signaling54, 55, 56 and signal transduction57, 58 by inhibiting miRNA function with single-cell resolution and the study of miRNAs involved in the cell cycle59, 60, 61 by inactivating miRNA function at a specific time point. These caged antagomirs can be used as tools to further explore the spatial and temporal role of specific miRNAs in cellular networks, embryonal development, and human diseases.
Experimental Section
Synthesis and purification of antagomirs
Oligonucleotides with 2′OMe bases and phosphorothioate linkages were synthesized on an Applied Biosystems Model 394 automated DNA/RNA synthesizer using standard β-cyanoethyl phosphoramidite chemistry. The antagomirs were synthesized on a 0.2 μM scale with solid-phase supports obtained from Glen Research. Reagents for automated DNA synthesis were also obtained from Glen Research. The NPOM-2′OMe uridine phosphoramidite was resuspended in anhydrous acetonitrile to a concentration of 0.1 M. Standard synthesis cycles provided by Applied Biosystems were used with 15 minute coupling times for all bases. The sulfurization step was performed using the Beaucage sulfurizing reagent 3H-1,2-Benzodithiole-3-one-1,1-dioxide at 0.05 M in acetonitrile. Oligonucleotides were eluted from the solid-phase supports with ammonium hydroxide and deprotected at 65 °C for 16 hrs. Deprotection reactions were purified using Illustra NAP-10 columns by gravity flow through Sephadex G-25DNA grade resin and the antagomirs were eluted in water. Poly-acrylamide gel analysis was used to confirm the purity of the final antagomirs.
Photochemical activation of miR-122 and miR-21 antagomir function in mammalian cells
Huh7-psiCHECK-miR122 or Huh7 cells were seeded at 10,000 cells per well in white clear-bottom 96-well plates (BD Falcon). After an overnight incubation, the Huh7-psiCHECK-miR122 cells were transfected with the miR-122 antagomirs (10 or 50 pmol; 50 or 250 nM) using X-tremeGENE siRNA transfection reagent (1.5 μL/well; Roche) in Opti-Mem media (Invitrogen) at 37 °C for 4 h. The Huh7 cells were transfected with the psiCHECK-miR21 plasmid (250 ng/well) and the miR-21 antagomirs (10 pmol; 50 nM) using X-tremeGENE siRNA transfection reagent (1.0 μL/well) in Opti-Mem media at 37 °C for 4 h. The media was removed and replaced with PBS (50 μL/well) and the cells were either kept in the dark or irradiated on a transilluminator for 5 min (365 nm, 25 W) in triplicate. The PBS was replaced with standard DMEM growth media (supplemented with 10% FBS and 2% penicillin/streptomycin) and the cells were incubated at 37 °C for 48 h. The media was then removed and the cells were lysed and assayed with a Dual Luciferase Assay Kit (Promega). The ratio of Renilla to firefly luciferase expression was calculated for each of the triplicates, the data was averaged, and standard deviations were determined.
Temporal control over miR-21 antagomir function in mammalian cells
Huh7 cells were seeded at 10,000 cells per well in white clear-bottom 96-well plates (BD Falcon). After an overnight incubation, the cells were transfected with the psiCHECK-miR21 plasmid (250 ng/well) and the caged miR-21 antagomir (10 pmol; 50 nM) using X-tremeGENE siRNA transfection reagent (1.0 μL/well; Roche) in Opti-Mem media (Invitrogen) at 37 °C for 4 h. The media was removed and replaced with standard DMEM growth media (supplemented with 10% FBS and 2% penicillin/streptomycin). The cells were irradiated on a transilluminator for 5 min (365 nm, 25 W) in triplicate at 0, 12, 24, 36, 42, or 46 h post transfection in PBS (50 μL/well). The PBS was replaced with standard DMEM growth media and the cells were incubated at 37 °C for a total of 48 h following transfection. The media was then removed and the cells were lysed and assayed with a Dual Luciferase Assay Kit (Promega). The ratio of Renilla to firefly luciferase expression was calculated for each of the triplicates, the data was averaged, and standard deviations were determined.
EGFP activation through photochemical control of miR-21 function
Huh7 cells were seeded at 10,000 cells per well in black clear-bottom 96-well plates (BD Falcon) treated with poly-D-lysine. After an overnight incubation, the cells were transfected with either the pEGFP-C2 plasmid or the pEGFP-C2-miR21 plasmid (250 ng/well) and either the miR-21 antagomir, the miR-122 antagomir, or the caged miR-21 antagomir (10 pmol; 50 nM) using X-tremeGENE siRNA transfection reagent (1.0 μL/well; Roche) in Opti-Mem media (Invitrogen) at 37 °C for 4 h. The media was removed and replaced with PBS (50 μL/well) and the cells were either kept in the dark or irradiated on a transilluminator for 5 min (365 nm, 25 W). The PBS was replaced with standard DMEM growth media (supplemented with 10% FBS and 2% penicillin/streptomycin) and the cells were incubated at 37 °C for 48 h. The media was then removed and replaced with DMEM without phenol red and the cells were imaged on a Zeiss Axio Observer inverted microscope using a 10× objective and filter set 38 (excitation 470/40, emission 525/50).
Spatial control of miR-21 function in mammalian cells
Huh7 cells were seeded at 100,000 cells per well in a 6-well plate (BD Falcon) treated with poly-D-lysine. After a 48 h overnight incubation, the cells were co-transfected with the pEGFP-C2-miR21 plasmid (2.5 μg/well) and the caged miR-21 antagomir (100 pmol; 500 nM) using X-tremeGENE siRNA transfection reagent (10 μL/well; Roche) in Opti-Mem media (Invitrogen) at 37 °C for 4 h. The media was removed and replaced with PBS (1 mL/well) and the cells were irradiated from the top using a LED fiber optics probe (Prizmatix) for 5 min (365 nm). The PBS was replaced with standard DMEM growth media (supplemented with 10% FBS and 2% penicillin/streptomycin) and the cells were incubated at 37 °C for 48 h. The media was then removed and replaced with DMEM without phenol red and the cells were imaged on a Zeiss Axio Observer inverted microscope using a 10× objective, a 2×2 tile scan, and filter set 38 (excitation 470/40, emission 525/50).
Supplementary Material
Acknowledgments
This research was supported in part by the Teva USA Scholars Grants Program, administered by the American Chemical Society Office of Research Grants, and by a Research Scholar Grant (120130-RSG-11-066-01-RMC) from the American Cancer Society. CMC acknowledges an NIH/NCSU Molecular Biotechnology Traineeship (2T32GM008776-11).
Footnotes
Electronic Supplementary Information (ESI) available: Synthetic protocols and analytical data for 3–7, cloning procedures, cell culture details, and supporting figures. See DOI: 10.1039/b000000x/
References
- 1.Carthew R. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Lytle JR, Yario TA, Steitz JA. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djuranovic S, Nahvi A, Green R. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pillai RS. RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Appasani K. MicroRNAs: from basic science to disease biology. Cambridge University Press; Cambridge: 2008. [Google Scholar]
- 7.Janga SC, Vallabhaneni S. Adv Exp Med Biol. 2011;722:59–74. doi: 10.1007/978-1-4614-0332-6_4. [DOI] [PubMed] [Google Scholar]
- 8.Tong AW, Nemunaitis J. Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 9.Sevignani C, Calin G, Siracusa L, Croce C. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Port JD, Sucharov C. J Cardiovasc Pharmacol. 2010;56:444–453. doi: 10.1097/FJC.0b013e3181f605b6. [DOI] [PubMed] [Google Scholar]
- 11.Lindsay MA. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Cullen BR. Genes Dev. 2011;25:1881–1894. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esau C. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Veedu R, Wengel J. Chem Biodivers. 2010;7:536–542. doi: 10.1002/cbdv.200900343. [DOI] [PubMed] [Google Scholar]
- 15.Brown B, Naldini L. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 16.Deiters A. AAPS J. 2010;12:51–60. doi: 10.1208/s12248-009-9159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meister G, Landthaler M, Dorsett Y, Tuschl T. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krützfeldt J, Rajewsky N, Braich R, Rajeev K, Tuschl T, Manoharan M, Stoffel M. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 19.Ebert MS, Neilson JR, Sharp PA. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gumireddy K, Young D, Xiong X, Hogenesch J, Huang Q, Deiters A. Angew Chem Int Ed Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young D, Connelly C, Grohmann C, Deiters A. J Am Chem Soc. 2010;132:7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, Sall A, Yang D. Int J Mol Sci. 2008;9:978–999. doi: 10.3390/ijms9060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grünweller A, Hartmann R. BioDrugs. 2007;21:235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 25.Riggsbee CW, Deiters A. Trends Biotechnol. 2010;28:468–475. doi: 10.1016/j.tibtech.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deiters A. Chembiochem. 2010;11:47–53. doi: 10.1002/cbic.200900529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deiters A. Curr Opin Chem Biol. 2009;13:678–686. doi: 10.1016/j.cbpa.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young DD, Deiters A. Org Biomol Chem. 2007;5:999–1005. doi: 10.1039/b616410m. [DOI] [PubMed] [Google Scholar]
- 29.Fenno L, Yizhar O, Deisseroth K. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prokup A, Hemphill J, Deiters A. J Am Chem Soc. 2012;134:3810–3815. doi: 10.1021/ja210050s. [DOI] [PubMed] [Google Scholar]
- 31.Govan JM, Lively MO, Deiters A. J Am Chem Soc. 2011;133:13176–13182. doi: 10.1021/ja204980v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young D, Lively M, Deiters A. J Am Chem Soc. 2010;132:6183–6193. doi: 10.1021/ja100710j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young DD, Lusic H, Lively MO, Yoder JA, Deiters A. Chembiochem. 2008;9:2937–2940. doi: 10.1002/cbic.200800627. [DOI] [PubMed] [Google Scholar]
- 34.Young DD, Edwards WF, Lusic H, Lively MO, Deiters A. Chem Commun (Camb) 2008:462–464. doi: 10.1039/b715152g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi KB, Vlachos A, Mikat V, Deller T, Heckel A. Chem Commun (Camb) 2012;48:2746–2748. doi: 10.1039/c2cc16654b. [DOI] [PubMed] [Google Scholar]
- 36.Mikat V, Heckel A. RNA. 2007;13:2341–2347. doi: 10.1261/rna.753407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shestopalov IA, Sinha S, Chen JK. Nat Chem Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- 38.Tang X, Maegawa S, Weinberg ES, Dmochowski IJ. J Am Chem Soc. 2007;129:11000–11001. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- 39.Tang X, Swaminathan J, Gewirtz AM, Dmochowski IJ. Nucleic Acids Res. 2008;36:559–569. doi: 10.1093/nar/gkm1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CL, Li WH, Olson AJ, Chen JK. J Am Chem Soc. 2009;131:13255–13269. doi: 10.1021/ja809933h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng G, Cochella L, Liu J, Hobert O, Li WH. ACS Chem Biol. 2011;6:1332–1338. doi: 10.1021/cb200290e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esau C, Davis S, Murray S, Yu X, Pandey S, Pear M, Watts L, Booten S, Graham M, McKay R, Subramaniam A, Propp S, Lollo B, Freier S, Bennett C, Bhanot S, Monia B. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Jopling C, Yi M, Lancaster A, Lemon S, Sarnow P. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 44.Lanford R, Hildebrandt-Eriksen E, Petri A, Persson R, Lindow M, Munk M, Kauppinen S, Ørum H. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong A, Nemunaitis J. Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 46.Medina PP, Nolde M, Slack FJ. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 47.FURUSAWA K, UENO K, KATSURA T. Chemistry Letters. 1990:97–100. [Google Scholar]
- 48.Lusic H, Deiters A. Synthesis-Stuttgart. 2006:2147–2150. [Google Scholar]
- 49.AKIYAMA T, NISHIMOTO H, OZAKI S. Bulletin of the Chemical Society of Japan. 1990;63:3356–3357. [Google Scholar]
- 50.IYER R, EGAN W, REGAN J, BEAUCAGE S. Journal of the American Chemical Society. 1990;112:1253–1254. [Google Scholar]
- 51.Kida K, Nakajima M, Mohri T, Oda Y, Takagi S, Fukami T, Yokoi T. Pharm Res. 2011;28:2467–2476. doi: 10.1007/s11095-011-0473-y. [DOI] [PubMed] [Google Scholar]
- 52.Chen HL, Huang JY, Chen CM, Chu TH, Shih C. PLoS One. 2012;7:e34116. doi: 10.1371/journal.pone.0034116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Connelly CM, Thomas M, Deiters A. J Biomol Screen. 2012 doi: 10.1177/1087057112439606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 55.Iguchi H, Kosaka N, Ochiya T. Commun Integr Biol. 2010;3:478–481. doi: 10.4161/cib.3.5.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Cui Q, Yu Z, Purisima EO, Wang E. Mol Syst Biol. 2006;2:46. doi: 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inui M, Martello G, Piccolo S. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 59.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 60.Lizé M, Klimke A, Dobbelstein M. Cell Cycle. 2011;10:2874–2882. doi: 10.4161/cc.10.17.17181. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Blelloch R. Cancer Res. 2009;69:4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.