Abstract
Background
Chinese bayberry (Myrica rubra Sieb. and Zucc.) is a subtropical evergreen tree originating in China. It has been cultivated in southern China for several thousand years, and annual production has reached 1.1 million tons. The taste and high level of health promoting characters identified in the fruit in recent years has stimulated its extension in China and introduction to Australia. A limited number of co-dominant markers have been developed and applied in genetic diversity and identity studies. Here we report, for the first time, a survey of whole genome shotgun data to develop a large number of simple sequence repeat (SSR) markers to analyse the genetic diversity of the common cultivated Chinese bayberry and the relationship with three other Myrica species.
Results
The whole genome shotgun survey of Chinese bayberry produced 9.01Gb of sequence data, about 26x coverage of the estimated genome size of 323 Mb. The genome sequences were highly heterozygous, but with little duplication. From the initial assembled scaffold covering 255 Mb sequence data, 28,602 SSRs (≥5 repeats) were identified. Dinucleotide was the most common repeat motif with a frequency of 84.73%, followed by 13.78% trinucleotide, 1.34% tetranucleotide, 0.12% pentanucleotide and 0.04% hexanucleotide. From 600 primer pairs, 186 polymorphic SSRs were developed. Of these, 158 were used to screen 29 Chinese bayberry accessions and three other Myrica species: 91.14%, 89.87% and 46.84% SSRs could be used in Myrica adenophora, Myrica nana and Myrica cerifera, respectively. The UPGMA dendrogram tree showed that cultivated Myrica rubra is closely related to Myrica adenophora and Myrica nana, originating in southwest China, and very distantly related to Myrica cerifera, originating in America. These markers can be used in the construction of a linkage map and for genetic diversity studies in Myrica species.
Conclusion
Myrica rubra has a small genome of about 323 Mb with a high level of heterozygosity. A large number of SSRs were identified, and 158 polymorphic SSR markers developed, 91% of which can be transferred to other Myrica species.
Background
Chinese bayberry is an important commercial horticultural crop. It has been cultivated for more than 7,000 years in southern China, but is little known elsewhere. The production area is currently 340,000 ha, with an annual production of 1.1 million tons. The plant is diploid (2n = 16), generally dioecious, with the female plants cultivated for fruit [1], growing well on poor soils due to the association of nitrogen-fixing bacteria with the root system. It is rich in anthocyanins exhibiting a wide range of pharmacological properties, such as anti-inflammatory, antitumor and antioxidative effects [2].
There are four species within the genus Myrica in China, namely Myrica rubra Sieb. & Zucc., M. esculenta Buch.-Ham., M. nana Cheval., and M. adenophora Hance. M. rubra is widely distributed, with many local cultivars in the Zhejiang, Jiangsu, Fujian and Guangdong provinces and a few from Guizhou, Yunnan and Hunan provinces. The best known cultivars are Biqi and Dongkui, both from the Zhejiang province. Although there are abundant germplasm resources, studies on genetics and breeding of the species are still in their infancy. Molecular marker technology is a popular tool for breeding and genetic research, and with the construction of a genomic library, 13 polymorphic microsatellite loci have been developed in M. rubra[3] and 11 from an expressed sequence tag library [4]. Recently, 12 primer pairs have been temporarily developed by ISSR-suppression PCR [5] with GSG (GT)6 as the primer for enriching microsatellite sequences. Reports on the genetic diversity in Chinese bayberry using SSR markers have also recently been published [6,7], but the number of markers for Chinese bayberry is limited.
The reproducibility, multiallelicism, co-dominance, relative abundance and good genome coverage of SSR markers have made them one of the most useful tools for genetic diversity and linkage mapping. Genomic SSRs and EST-SSRs, considered complementary to plant genome mapping, have been reported in many fruit crops, such as walnut [8], cherry [9], apricot [10] and coconut [11]. EST-SSRs are useful for genetic analysis, but their relatively low polymorphism and the high possibility of no gene-rich regions in the genome are limitations to their use. In contrast, genomic SSRs are highly polymorphic and tend to be widely distributed throughout the genome, resulting in better map coverage [12].
With genetic maps serving as the basis for future positional gene cloning, making map-based cloning and marker-assisted selection possible, the development of more SSRs is essential. As sequencing technologies advance, whole-genome shotgun (WGS) sequences are becoming increasingly available. These DNA sequences are valuable resources for SSR development in many plant species, such as rice [13] and papaya [14]. In addition, they can be used to evaluate the frequency and distribution of different types of SSRs in the genome, and even help to estimate genome size and characters such as heterozygosis and repeats.
As a way of reducing the cost of genotyping research, Schuelke [15] proposed a method for fluorescent dye labelling of PCR fragments with a sequence-specific forward primer: the universal fluorescent-labelled M13(-21) primer, at the 5’ end, acts as the forward primer in a ‘one-tube’ reaction. As this method allows for high-throughput genetic analyses, with a high number of microsatellite markers widely used, we considered the possibility of using this approach for multiplex PCR, to improve the efficiency and save costs.
In this study, we mined and validated 158 SSR markers and describe their application for understanding the genetic relationship among 29 Chinese bayberry accessions and other Myrica species. These markers are useful for genotyping and genetic diversity analysis and linkage mapping of Myrica rubra and other Myrica species.
Results
Genome survey using whole genome shotgun data in Chinese bayberry
WGS generated 273,161 (>100 bp) high quality sequence reads from two DNA libraries (250 bp and 500 bp) of the androphyte individual ‘C2010-55’. We used 9.01 G raw data for K-mer analysis and heterozygous simulation. For the 17-mer frequency distribution (Figure 1), the peak of the depth distribution was about 22. The estimated genome size was 323 Mb, using the formula: genome size = k-mer count/peak of the kmer distribution. The minor peak at 1/2 altitude of the main peak indicates the high level of heterozygosity in this genome (Figure 1). A total of 739,969 contigs were assembled with a total sequence length of 255.7 Mb. The length of N50 was 295 bp in our assembly, and the longest contig and scaffold 7,593 and 127,008 bp, respectively.
Figure 1.
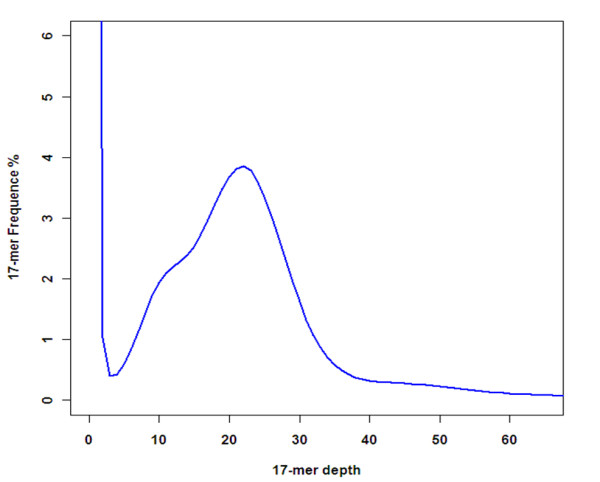
The distribution of 17-mer depth analysis based on whole genome shotgun data in Chinese bayberry.
Frequency distribution of different types of SSR markers
A total of 17,172 out of 273,161 scaffolds (6%) retrieved from the genome survey sequence contained 28,602 SSRs (Table 1), of which 5,401 contained more than one SSR, and 1,444 SSRs were present in compound format. Among the derived SSR repeats, the di-nucleotide was the most abundant repeat, accounting for 84.72% of total SSRs, followed by tri- (13.78%), tetra- (1.34%), penta- (0.12%), and hexa- (0.04%) nucleotides (Table 1). There was a large proportion of both dinucleotides and trinucleotides while the rest amounted to less 2%. The average frequency of occurrence was about 10.47% (Table 1).
Table 1.
Occurrence of SSRs in the Genome Survey of Chinese bayberry
| Type | Number | Proportion in all SSRs (%) | Frequency (%) |
|---|---|---|---|
| Dinucleotide |
24,233 |
84.72% |
8.87% |
| Trinucleotide |
3,941 |
13.78% |
1.44% |
| Tetranucleotide |
383 |
1.34% |
0.14% |
| Pentanucleotide |
35 |
0.12% |
0.013% |
| Hexanucleotide |
10 |
0.04% |
0.004% |
| Total | 28,602 | 100% | 10.47% |
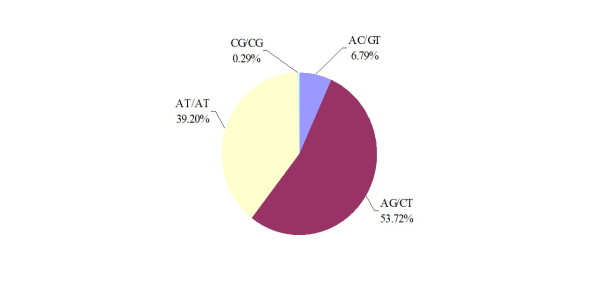
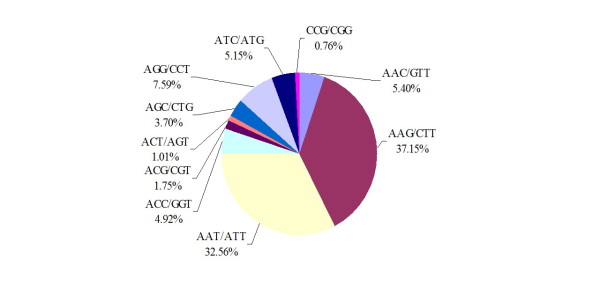
The SSR frequency of each motif is presented in Additional file 1. The SSR motif consists of 69 types. Among the repeat motifs of the dinucleotide, the AG/CT repeat was the most common, representing 53.72%, followed by 39.20% AT repeats (Figure 2), and the predominant motifs of trinucleotide (AAG/CTT and AAT/ATT) repeats accounted for 37.15% and 32.56%, respectively (Figure 3).
Figure 2.
Percentage of different motifs in dinucleotide repeats in Chinese bayberry genome.
Figure 3.
Percentage of different motifs in trinucleotide repeats in Chinese bayberry genome.
Polymorphism of SSR markers
We first designed and synthesised 600 SSR primer pairs from those scaffolds more than 2Kb long. The majority of SSR loci were dinucleotide repeats (597, 99.5%), and the remainder trinucleotide. Initially, the effectiveness of these primer pairs was detected in two cultivars (Biqi and Dongkui) and M. cerifera through denaturing PAGE (Polyacrylamide gel electrophoresis), and 581 (96.8%) of these were amplified successfully in Biqi and Dongkui, and 400 (66.7%) in M. cerifera. The SSR loci (186, 31%) were identified as heterozygous loci either in Biqi or in Dongkui. Subsequently, they were used to screen 32 accessions, and detected an average of 8.25 alleles and from 3 to 15 alleles per locus (Table 2).
Table 2.
Characteristics of 158 SSR markers in this study
|
Locus |
GenBank |
Repeat motif |
Primer sequence (5'-3') |
Size range(bp)a |
Na |
Ho |
He |
PIC |
PHW |
|---|---|---|---|---|---|---|---|---|---|
| Accession | |||||||||
| ZJU001ab |
JQ318696 |
(GA)10 |
F:<NED > <Tail-1 > CCTCTCCACCCATGAGAAAC |
160-188 |
7 |
0.1667 |
0.4271 |
0.4002 |
0.0000 |
| R:CAAATCATTCCCTGCTTTCC | |||||||||
| ZJU002ac |
JQ318697 |
(TC)13 |
F:<NED > <Tail-1 > TCAAAGAGACGTTGTGGCAG |
219-229 |
4 |
0.2083 |
0.5257 |
0.4572 |
0.0005 |
| R:TCCGCTCACAGACAGAGAGA | |||||||||
| ZJU003ab |
JQ318698 |
(AG)11 |
F:<NED > <Tail-1 > GTCACCTTGCTCTTCTTGGC |
203-217 |
8 |
0.7407 |
0.8344 |
0.7949 |
0.0003 |
| R:TCCTTGTACTTGTTCTGCTGGA | |||||||||
| ZJU004ac |
JQ318699 |
(GA)10 |
F:<NED > <Tail-1 > AACAGAACCATCGTCAAGGC |
204-210 |
4 |
0.3571 |
0.7325 |
0.6704 |
0.0003 |
| R:GGTACAGTCGCTCCGGTTTA | |||||||||
| ZJU005ab |
JQ318700 |
(AG)14 |
F:<NED > <Tail-1 > CTTTGGACATGGCAACACAC |
200-228 |
11 |
0.3000 |
0.8679 |
0.8291 |
0.0000 |
| R:TCCACTTTGACAGATTCCCA | |||||||||
| ZJU006ab |
JQ318701 |
(GA)10 |
F:<NED > <Tail-1 > CTCGCCCTCTCTCTCTACCA |
193-205 |
5 |
0.2593 |
0.3305 |
0.3089 |
0.0000 |
| R:AGTTTATCCACCCGTGTCGT | |||||||||
| ZJU007ab |
JQ318702 |
(AG)13 |
F:<NED > <Tail-1 > TGATCCATTGGAACCATGTG |
193-209 |
8 |
0.5625 |
0.6617 |
0.6302 |
0.1868 |
| R:TCAGTTGATGGTGCAGAAGC | |||||||||
| ZJU008ab |
JQ318703 |
(CT)10 |
F:<NED > <Tail-1 > GGAGAAATGAACGGTGGAGA |
191-215 |
10 |
0.7931 |
0.7973 |
0.7563 |
0.0002 |
| R:TCCAAAGCTAATACCCACGC | |||||||||
| ZJU009ab |
JQ318704 |
(CT)10 |
F:<NED > <Tail-1 > AATTGTCGCAAGTAGTCGCC |
207-221 |
5 |
0.0741 |
0.3599 |
0.3371 |
0.0000 |
| R:ATATCAACCCATGGGAGCAA | |||||||||
| ZJU010ab |
JQ318705 |
(CT)11 |
F:<NED > <Tail-1 > TGCAACATCGAAATTGGAAA |
181-205 |
9 |
0.9032 |
0.8012 |
0.7614 |
0.0000 |
| R:ATGCCGGCAAGTCTTAGTGT | |||||||||
| ZJU011a |
JQ318706 |
(GA)10 |
F:<NED > <Tail-1 > GGAGGCTCGTCAGTCATCTC |
200-216 |
9 |
0.2692 |
0.7926 |
0.7554 |
0.0000 |
| R:TTAGCGTCCCTTCTCTCTCG | |||||||||
| ZJU012ab |
JQ318707 |
(CT)12 |
F:<NED > <Tail-1 > CTTCACTCACCGCCTTTCTC |
184-218 |
13 |
0.5000 |
0.8571 |
0.8251 |
0.0000 |
| R:AATGGCCTCCACATCTCAAG | |||||||||
| ZJU013ab |
JQ318708 |
(CT)10 |
F:<NED > <Tail-1 > ACTTGTCATTCCCACGTTCC |
211-221 |
6 |
0.4444 |
0.5199 |
0.4515 |
0.0094 |
| R:CACTCCATCTCAACCACCCT | |||||||||
| ZJU014ab |
JQ318709 |
(AG)15 |
F:<NED > <Tail-1 > TGGAATGTCGATCTGAAACAA |
186-212 |
13 |
0.6875 |
0.9033 |
0.8791 |
0.0251 |
| R:ACCAGCTTATACGACGGTGG | |||||||||
| ZJU015ab |
JQ318710 |
(GA)11 |
F:<NED > <Tail-1 > TTGGTGTGGTGGTAATGGTG |
199-221 |
6 |
0.6154 |
0.6614 |
0.5902 |
0.0585 |
| R:AAATAATGCAAGCAGGTGGG | |||||||||
| ZJU016ab |
JQ318711 |
(TC)10 |
F:<NED > <Tail-1 > CCGTTGACTATTGCCCAGTT |
196-216 |
11 |
0.6333 |
0.8469 |
0.8130 |
0.0179 |
| R:GGCAATTTCCAAATCGCTAA | |||||||||
| ZJU017ab |
JQ318712 |
(CT)13 |
F:<NED > <Tail-1 > ACTGAAGAACCAAACGTGGG |
180-200 |
6 |
0.6250 |
0.7093 |
0.6518 |
0.0003 |
| R:GGTGTGTTTCTCTGTGTGCG | |||||||||
| ZJU018ab |
JQ318713 |
(CT)15 |
F:<NED > <Tail-1 > ACGAAATTTGACCAATCGCT |
196-216 |
7 |
0.1429 |
0.7189 |
0.6667 |
0.0000 |
| R:AGGGTTTCTTCTGGTTCGGT | |||||||||
| ZJU019ab |
JQ318714 |
(GA)12 |
F:<NED > <Tail-1 > TTTCATAACCCGTTGGCTTC |
209-219 |
6 |
0.2800 |
0.6865 |
0.6317 |
0.0000 |
| R:AAGGTGGAAACGTGTCAAGG | |||||||||
| ZJU020b |
JQ318715 |
(AG)10 |
F:<NED > <Tail-1 > CACAGGACATGTGATGGAGG |
201-213 |
7 |
0.5172 |
0.7453 |
0.6983 |
0.0000 |
| R:CCATCCTGAGCTTTGTCGAT | |||||||||
| ZJU021a |
JQ318716 |
(TG)10 |
F:<NED > <Tail-1 > TCGCCAGCTTCCTAATGTCT |
190-212 |
8 |
0.7778 |
0.7428 |
0.7025 |
0.0663 |
| R:GAGCGCATGTTGTTGCTAAA | |||||||||
| ZJU022ab |
JQ318717 |
(GA)10 |
F:<NED > <Tail-1 > AAGCTTAAGCAAGCGTCGAG |
188-208 |
9 |
0.6923 |
0.8575 |
0.8227 |
0.0109 |
| R:TGCGAAGGGAAATTTCAGAC | |||||||||
| ZJU023ac |
JQ318718 |
(AG)15 |
F:<NED > <Tail-1 > GTGTTTGGGCAGCACCTATT |
200-226 |
14 |
0.6667 |
0.8840 |
0.8544 |
0.0251 |
| R:AAAGAGTACAACAACGCGGG | |||||||||
| ZJU024ab |
JQ318719 |
(TC)10 |
F:<NED > <Tail-1 > CCGCATGTTTGATTGATGTC |
180-196 |
6 |
0.6000 |
0.7345 |
0.6716 |
0.1624 |
| R:GCGTTGAGCGGAGAGATTAC | |||||||||
| ZJU025ab |
JQ318720 |
(TC)10 |
F:<NED > <Tail-1 > TTTGAGCGATAGTACGGAGG |
216-234 |
8 |
0.2667 |
0.7537 |
0.7044 |
0.0000 |
| R:ATATGCTACGTTGGTTGCCC | |||||||||
| ZJU026ab |
JQ318721 |
(TC)10 |
F:<NED > <Tail-1 > CCAGACAGGTTAGCCACCAT |
200-220 |
10 |
0.4545 |
0.8573 |
0.8199 |
0.0000 |
| R:GCCTCTGGATCTCGATTACG | |||||||||
| ZJU027 |
JQ318722 |
(TTC)8 |
F:<NED > <Tail-1 > GTTGCAATTTGCCTCCATTT |
203-227 |
6 |
0.3125 |
0.6250 |
0.5321 |
0.0003 |
| R:GGTGCCTATACTGCCAGCTC | |||||||||
| ZJU028ab |
JQ318723 |
(AG)10 |
F:<NED > <Tail-1 > CAACCATCCAAACCAAATCC |
164-170 |
4 |
0.1724 |
0.2789 |
0.2566 |
0.0000 |
| R:TCTACCAATCGTGGCTAGGG | |||||||||
| ZJU029ab |
JQ318724 |
(AG)10 |
F:<NED > <Tail-1 > TCTTCCGGGATGTCTACAGG |
189-205 |
6 |
0.5312 |
0.6925 |
0.6296 |
0.0480 |
| R:CAACAGCAATCGCAAAGAAA | |||||||||
| ZJU030ab |
JQ318725 |
(CA)13 |
F:<NED > <Tail-1 > AAGTGAGCTCTCCCTCCCTC |
193-205 |
7 |
0.4286 |
0.7208 |
0.6676 |
0.0000 |
| R:CACCGAAATACTTGCCGTTT | |||||||||
| ZJU031ab |
JQ318726 |
(GA)16 |
F:<NED > <Tail-1 > GCACAGGAACACCAGGATCT |
179-195 |
8 |
0.8387 |
0.7948 |
0.7492 |
0.0000 |
| R:CCAAGCCCTAATTCCCTTTC | |||||||||
| ZJU032ab |
JQ318727 |
(TC)11 |
F:<NED > <Tail-1 > ATTCCCACGTTCGTTCAGAC |
204-226 |
8 |
0.6786 |
0.6442 |
0.5852 |
0.0220 |
| R:GATGCCTAACTCCGAATCCA | |||||||||
| ZJU033ab |
JQ318728 |
(TC)10 |
F:<NED > <Tail-1 > GCACAAGTTGCTGACATGCT |
195-207 |
6 |
0.0690 |
0.6655 |
0.5897 |
0.0000 |
| R:AGTTGCATTCAACCCACACA | |||||||||
| ZJU034ab |
JQ318729 |
(CT)10 |
F:<NED > <Tail-1 > ATGGGAATGTGGAGAACGAG |
191-209 |
8 |
0.4138 |
0.7762 |
0.7250 |
0.0000 |
| R:GCTTTGCTTCTTTGCTTTGG | |||||||||
| ZJU035ab |
JQ318730 |
(GA)14 |
F:<NED > <Tail-1 > TTGGATCCTGGTTACCTTCG |
201-217 |
8 |
0.1290 |
0.7425 |
0.6900 |
0.0000 |
| R:AAACTGCATGCATGGTTCCT | |||||||||
| ZJU036ab |
JQ318731 |
(GA)10 |
F:<NED > <Tail-1 > CTGCCACTCTTACTGGCCTC |
186-214 |
8 |
0.3333 |
0.5895 |
0.5516 |
0.0000 |
| R:ATGTGCCCAATCTTGACTCC | |||||||||
| ZJU037ab |
JQ318732 |
(TC)10 |
F:<NED > <Tail-1 > GTGATTTCCCTCCCATTGAC |
208-228 |
9 |
0.8125 |
0.7867 |
0.7429 |
0.0135 |
| R:ACGAAGCGGGAAGTAGGATT | |||||||||
| ZJU038b |
JQ318733 |
(AG)10 |
F:<NED > <Tail-1 > CTTATGGCCCGTTTGTAACC |
194-200 |
4 |
0.2273 |
0.5106 |
0.4646 |
0.0007 |
| R:AACGATTGCTTTAAGCGGAA | |||||||||
| ZJU039a |
JQ318734 |
(CT)10 |
F:<NED > <Tail-1 > AAACGAAAGTGGGCGTATTG |
219-229 |
6 |
0.3077 |
0.6161 |
0.5745 |
0.0004 |
| R:CACCAGTGCGTCCTATGAGA | |||||||||
| ZJU040 |
JQ318735 |
(TC)16 |
F:<NED > <Tail-1 > AAACTCCGTGCTGGAATCAA |
198-220 |
10 |
0.3182 |
0.8192 |
0.7798 |
0.0000 |
| R:GCAGACAAGCCTTCCTGTTC | |||||||||
| ZJU041ab |
JQ318736 |
(TC)11 |
F:<PET > <Tail-2 > TGATCACCTTTCAGTTGGCA |
226-244 |
5 |
0.2258 |
0.3199 |
0.3031 |
0.0000 |
| R:CACATTGGCAGAGTCCTGAA | |||||||||
| ZJU042ab |
JQ318737 |
(TC)10 |
F:<PET > <Tail-2 > AGGATTTCTCCAGAGGGACG |
220-242 |
5 |
0.3571 |
0.5331 |
0.4880 |
0.0000 |
| R:GGTTCCGCATCAAACTACAAA | |||||||||
| ZJU043b |
JQ318738 |
(CT)10 |
F:<PET > <Tail-2 > AAACCGAGCTCTCCTAAGCC |
225-245 |
4 |
0.5714 |
0.6383 |
0.5667 |
0.2655 |
| R:CTCGCAATTTCTCGGGATAC | |||||||||
| ZJU044ab |
JQ318739 |
(GA)12 |
F:<PET > <Tail-2 > GATGGTGGCTTGTCTTGGTT |
235-255 |
8 |
0.2500 |
0.5091 |
0.4853 |
0.0000 |
| R:AAGTGGGACGTCAATTCCTG | |||||||||
| ZJU045ab |
JQ318740 |
(CT)10 |
F:<PET > <Tail-2 > GAGAGAGGGAGAGAGGCCAT |
228-258 |
13 |
0.6129 |
0.8821 |
0.8544 |
0.0007 |
| R:GGAAGATTCATGGGAGAGGG | |||||||||
| ZJU046ab |
JQ318741 |
(AG)10 |
F:<PET > <Tail-2 > TTGCTGTAAGCATCGCAATC |
226-242 |
7 |
0.3871 |
0.6256 |
0.5824 |
0.0000 |
| R:AAGCTCCGGTAACACACACC | |||||||||
| ZJU047ab |
JQ318742 |
(GA)13 |
F:<PET > <Tail-2 > TTCGATCATTCATGAGGCTG |
247-259 |
7 |
0.7097 |
0.7615 |
0.7074 |
0.0019 |
| R:TTAATTGCATCCCGGATTTC | |||||||||
| ZJU048ab |
JQ318743 |
(CT)14 |
F:<PET > <Tail-2 > AGCGGACCGAGTTGTAGAGA |
230-254 |
12 |
0.2903 |
0.8493 |
0.8166 |
0.0000 |
| R:CCAACCCTACAAAGCGAGAG | |||||||||
| ZJU049ab |
JQ318744 |
(GAA)8 |
F:<PET > <Tail-2 > GTGTCTGCAGCAACTTCCAC |
234-267 |
10 |
0.8125 |
0.7262 |
0.6797 |
0.0000 |
| R:GTCGGAACCGAAGATGGTTA | |||||||||
| ZJU050ab |
JQ318745 |
(AG)11 |
F:<PET > <Tail-2 > GTCACAGCCTGGATAGCTCC |
233-245 |
7 |
0.3000 |
0.7288 |
0.6916 |
0.0000 |
| R:GTCTCTCCTGGATGAGCTGC | |||||||||
| ZJU051ab |
JQ318746 |
(AG)12 |
F:<PET > <Tail-2 > AGAGAAAGACCGGGACCAAT |
229-233 |
3 |
0.4333 |
0.4198 |
0.3594 |
0.0012 |
| R:GAGAAATAAAGCCGAGCGTG | |||||||||
| ZJU052ab |
JQ318747 |
(AG)16 |
F:<PET > <Tail-2 > CCCGAGCTGAACGAAATAGA |
230-248 |
9 |
0.4348 |
0.8628 |
0.8261 |
0.0000 |
| R:GGATCAAAGCGTTGTCGTTT | |||||||||
| ZJU053ab |
JQ318748 |
(AG)10 |
F:<PET > <Tail-2 > AAATCCGAAACACCTCTCCC |
222-240 |
8 |
0.5000 |
0.5655 |
0.5211 |
0.0001 |
| R:ATGTGGAGACTTCCACTGGG | |||||||||
| ZJU054ab |
JQ318749 |
(CT)13 |
F:<PET > <Tail-2 > TTGATTTGCTTTGTGCATTTG |
232-250 |
9 |
0.3000 |
0.8667 |
0.8268 |
0.0003 |
| R:CAAACTACCGTGCCCAACAT | |||||||||
| ZJU055ab |
JQ318750 |
(CT)10 |
F:<PET > <Tail-2 > TTATGGGTTTCATTGGGCAG |
238-254 |
6 |
0.2500 |
0.7006 |
0.6357 |
0.0000 |
| R:TCACCAGGCTACTGCATGTC | |||||||||
| ZJU056ab |
JQ318751 |
(GA)13 |
F:<PET > <Tail-2 > GACAAAGTGGGTGCCATTCT |
230-246 |
7 |
0.5714 |
0.7643 |
0.7122 |
0.0068 |
| R:TGCATGCTTCCTTTCTTCCT | |||||||||
| ZJU057ab |
JQ318752 |
(CT)10 |
F:<PET > <Tail-2 > TCATGTGGAGATTGAAGCCA |
230-244 |
6 |
0.1579 |
0.6814 |
0.6283 |
0.0000 |
| R:CGTCCCGAATGAAGATTTGT | |||||||||
| ZJU058ab |
JQ318753 |
(GT)10 |
F:<PET > <Tail-2 > TCCGGAGCTTTCAATCTCAT |
252-274 |
11 |
0.7500 |
0.8274 |
0.7900 |
0.8036 |
| R:GCCTACGAACTCAGGTTCCA | |||||||||
| ZJU059b |
JQ318754 |
(TC)14 |
F:<PET > <Tail-2 > TGTTTGTTTCTTGCTATTTCCATC |
217-235 |
7 |
0.5200 |
0.7935 |
0.7505 |
0.0016 |
| R:GACAGTTCCCACCAGCATTT | |||||||||
| ZJU060ab |
JQ318755 |
(GT)8(GA)9 |
F:<PET > <Tail-2 > TGGCCAGGAACTTTGTATCC |
223-243 |
7 |
0.6562 |
0.8110 |
0.7691 |
0.0000 |
| R:GAAAGATTGGGAATGCTGGA | |||||||||
| ZJU061ab |
JQ318756 |
(TC)11 |
F:<PET > <Tail-2 > TTTGGAGGAAGCAAACAAGC |
204-232 |
11 |
0.2812 |
0.7922 |
0.7506 |
0.0000 |
| R:TCCTGCGCCAACAATCTAAT | |||||||||
| ZJU062 |
JQ318757 |
(TC)10 |
F:<PET > <Tail-2 > GTCGAGAGAACAAAGCGACC |
240-252 |
7 |
0.2400 |
0.3282 |
0.3135 |
0.0004 |
| R:GTCCAATGCCGCACTAACTT | |||||||||
| ZJU063ab |
JQ318758 |
(TC)12 |
F:<PET > <Tail-2 > ACTCAGCAGGACCACCAACT |
232-250 |
10 |
0.7000 |
0.8593 |
0.8270 |
0.1320 |
| R:TTAGACGGAAATTGGGCTTG | |||||||||
| ZJU064b |
JQ318759 |
(GA)10 |
F:<PET > <Tail-2 > ACCATGAAGCTGACCTGGAG |
226-244 |
6 |
0.4348 |
0.7256 |
0.6666 |
0.0001 |
| R:TTTCGTGGTCCCACCTACTC | |||||||||
| ZJU065ac |
JQ318760 |
(CA)13 |
F:<PET > <Tail-2 > TCCAGAATATCATCTCTTGCCA |
214-236 |
9 |
0.6333 |
0.7706 |
0.7219 |
0.0001 |
| R: ATATTCCTAACGTGTGCGGG | |||||||||
| ZJU066ab |
JQ318761 |
(GA)10 |
F:<PET > <Tail-2 > CTTTCCCTTGTCGCTTTCAG |
221-235 |
8 |
0.2593 |
0.6450 |
0.6075 |
0.0000 |
| R:GGTCGCAGATCAGGTCAAGT | |||||||||
| ZJU067ab |
JQ318762 |
(CT)10 |
F:<PET > <Tail-2 > CAGACAGCGAGGAGACAACA |
217-263 |
11 |
0.6923 |
0.8273 |
0.7861 |
0.0070 |
| R:GGTCTTTCGAACTTTGCTCG | |||||||||
| ZJU068ab |
JQ318763 |
(CT)10 |
F:<PET > <Tail-2 > GAAGCTAAACGCCAGAAACG |
227-239 |
6 |
0.2917 |
0.7535 |
0.6913 |
0.0000 |
| R:ACTCCTCACACGAATGGGTC | |||||||||
| ZJU069bc |
JQ318764 |
(GA)10 |
F:<PET > <Tail-2 > TGCCATAATCCTGAGAGCCT |
224-258 |
8 |
0.2609 |
0.5594 |
0.5235 |
0.0004 |
| R:TGTTCTGTAATGGCGTGGAA | |||||||||
| ZJU070ab |
JQ318765 |
(CT)11 |
F:<PET > <Tail-2 > GTGCTCGAGATGTCCTCCAT |
221-247 |
7 |
0.5200 |
0.7861 |
0.7364 |
0.0000 |
| R:ACAATCCCATCGCATACAGG | |||||||||
| ZJU071ab |
JQ318766 |
(GA)10 |
F:<PET > <Tail-2 > CTAAGGTTGGTCCCTGTCCA |
228-234 |
3 |
0.3704 |
0.6157 |
0.5305 |
0.0110 |
| R:CTTGTGTGGTGATGGTTTGG | |||||||||
| ZJU072ab |
JQ318767 |
(AG)10 |
F:<PET > <Tail-2 > AGTCAGCGTGGGAATGTACC |
223-237 |
7 |
0.5625 |
0.7604 |
0.7117 |
0.0000 |
| R:TTTCAGAACAAGTTCGTCGC | |||||||||
| ZJU073a |
JQ318768 |
(AG)12 |
F:<PET > <Tail-2 > TACGCCAAGATCCAAAGACC |
222-242 |
7 |
0.2105 |
0.7568 |
0.7087 |
0.0000 |
| R:TCTCGAGTTGAGTTTGGGCT | |||||||||
| ZJU074ab |
JQ318769 |
(CT)15 |
F:<PET > <Tail-2 > TGCAGAGGAACTGGTGACTG |
215-239 |
10 |
0.5517 |
0.8234 |
0.7831 |
0.0007 |
| R:GAGAAGGCTCAGTGGGTGAG | |||||||||
| ZJU075b |
JQ318770 |
(CT)17 |
F:<PET > <Tail-2 > AATAAACACACAGGGCGAGG |
239-255 |
9 |
0.0769 |
0.8650 |
0.8307 |
0.0000 |
| R:ATCGGGCAGACCAGAATATG | |||||||||
| ZJU076ab |
JQ318771 |
(AG)9 |
F:<FAM > <Tail-3 > ATGGTTACCGACGTCCTCTG |
131-169 |
11 |
0.8438 |
0.8353 |
0.8034 |
0.0000 |
| R:AGTGCAGAGTGCGAGATCAA | |||||||||
| ZJU077ab |
JQ318772 |
(AC)9 |
F:<FAM > <Tail-3 > TTTGGAATTCAACAACATTTAGAC |
137-153 |
8 |
0.2000 |
0.6590 |
0.6079 |
0.0000 |
| R:TGCAGCCTTGCTCCTCTTAT | |||||||||
| ZJU078ab |
JQ318773 |
(TC)10 |
F:<FAM > <Tail-3 > ACACCACGGTTCTTCGATTC |
130-146 |
6 |
0.5500 |
0.7513 |
0.6881 |
0.1339 |
| R:GTAACGAGGCTCTTGCTTGC | |||||||||
| ZJU079ab |
JQ318774 |
(TC)13 |
F:<FAM > <Tail-3 > AAGGCTAGACCGCAATCTGA |
116-134 |
9 |
0.8438 |
0.8596 |
0.8291 |
0.0008 |
| R:GGGCAACAGTTTACTTCCCA | |||||||||
| ZJU080ab |
JQ318775 |
(CT)9 |
F:<FAM > <Tail-3 > CTTGACGAAATGCAGACGAA |
124-134 |
5 |
0.2903 |
0.3411 |
0.3172 |
0.0103 |
| R:TCCGGATCAGGGTCAAATAG | |||||||||
| ZJU081ab |
JQ318776 |
(GA)8 |
F:<FAM > <Tail-3 > TGCTCTTGCAGAGAGTCGAG |
137-157 |
6 |
0.5517 |
0.5820 |
0.5379 |
0.0003 |
| R:TCATAATACCCTTGGCAAACA | |||||||||
| ZJU082ab |
JQ318777 |
(CT)10 |
F:<FAM > <Tail-3 > TATATCGAATCCCAAAGGCG |
129-141 |
5 |
0.3438 |
0.4043 |
0.3792 |
0.0169 |
| R:AAGATATTGGTCCGGCTCCT | |||||||||
| ZJU083ab |
JQ318778 |
(AG)10 |
F:<FAM > <Tail-3 > TAGCCTTGGAGATTTAGGGC |
133-157 |
11 |
0.8667 |
0.8960 |
0.8692 |
0.0000 |
| R:TTGAAATTTCGCAGCCTCTT | |||||||||
| ZJU084ab |
JQ318779 |
(AG)9 |
F:<FAM > <Tail-3 > TTTCGATTGGTGGTCTGTGA |
124-138 |
6 |
0.1379 |
0.5197 |
0.4766 |
0.0000 |
| R:TTATTAACTTCACTTTGTTTATTCGG | |||||||||
| ZJU085ab |
JQ318780 |
(AG)9 |
F:<FAM > <Tail-3 > GCTTTAACCGAGTGATGGGA |
150-184 |
8 |
0.6875 |
0.5992 |
0.5383 |
0.6352 |
| R:TAAAGGAGCGCTGGAAAGAA | |||||||||
| ZJU086ab |
JQ318781 |
(TC)10 |
F:<FAM > <Tail-3 > TCCTCTCTTTCACACTTCCGA |
118-152 |
13 |
0.9062 |
0.8720 |
0.8445 |
0.0005 |
| R:GGTCGATCATTTCTCTCCCA | |||||||||
| ZJU087ab |
JQ318782 |
(GA)9 |
F:<FAM > <Tail-3 > CGAGTGTAGCTAGGAACGGC |
135-149 |
8 |
0.4688 |
0.7748 |
0.7273 |
0.0204 |
| R:AATTGGACCTGCAAATCTCG | |||||||||
| ZJU088ab |
JQ318783 |
(CT)9 |
F:<FAM > <Tail-3 > GAGCTCCGAACTTCTTCCCT |
126-150 |
13 |
0.9677 |
0.8773 |
0.8490 |
0.0053 |
| R:CTTCTCCACAGGACTCTGCC | |||||||||
| ZJU089ab |
JQ318784 |
(GA)8 |
F:<FAM > <Tail-3 > CGTTAGGATTCGGGAACAGA |
138-152 |
7 |
0.8065 |
0.7382 |
0.6778 |
0.0000 |
| R:CAGGGCTAATGTGGACCAGT | |||||||||
| ZJU090ab |
JQ318785 |
(AG)9 |
F:<FAM > <Tail-3 > GGAAATCTCCGAATGTGATCC |
118-134 |
8 |
0.2903 |
0.6642 |
0.6089 |
0.0000 |
| R:TGGTGGATGAACCACTCAAA | |||||||||
| ZJU091bc |
JQ318786 |
(TC)15 |
F:<FAM > <Tail-3 > AAAGAGCACACAGCCCTAGC |
124-146 |
10 |
0.4615 |
0.8695 |
0.8358 |
0.0012 |
| R:GGCAGTGTCGCAGTGATAGA | |||||||||
| ZJU092ab |
JQ318787 |
(TG)10 |
F:<FAM > <Tail-3 > CTCTTGCCGACCTCATTGTT |
127-151 |
11 |
0.6875 |
0.8264 |
0.7916 |
0.0041 |
| R:CGGGACTCGCATAAATCACT | |||||||||
| ZJU093ab |
JQ318788 |
(GA)10 |
F:<FAM > <Tail-3 > ATGCCATGTTGCATGAGTGT |
130-156 |
12 |
0.9355 |
0.8662 |
0.8367 |
0.3689 |
| R:TATCCCGTAAGCAATCAGGG | |||||||||
| ZJU094ab |
JQ318789 |
(CT)10 |
F:<FAM > <Tail-3 > ATCACGGGTTCTGCTGTTCT |
124-150 |
10 |
0.9062 |
0.8646 |
0.8332 |
0.0000 |
| R:CAGAAGAAGCCATTTCTGCC | |||||||||
| ZJU095ab |
JQ318790 |
(AG)9 |
F:<FAM > <Tail-3 > TACCCACCGTACCAAAGGTC |
114-130 |
7 |
0.4839 |
0.7070 |
0.6420 |
0.0004 |
| R:GAATGAACCCAGGCGATAGA | |||||||||
| ZJU096ab |
JQ318791 |
(CT)10 |
F:<FAM > <Tail-3 > CATACTGCAATGCATCTCCC |
126-154 |
13 |
0.8000 |
0.8757 |
0.8479 |
0.0310 |
| R:TCAATTTGTGTGCCCTTACG | |||||||||
| ZJU097ab |
JQ318792 |
(AG)10 |
F:<FAM > <Tail-3 > AATTGTTAGGGAGGGCTCGT |
118-134 |
8 |
0.8438 |
0.7778 |
0.7297 |
0.0009 |
| R:TGCGTTGTGGAGACCATTTA | |||||||||
| ZJU098ab |
JQ318793 |
(CT)9 |
F:<FAM > <Tail-3 > GACGCTCCATCTCTGGTCTC |
145-167 |
10 |
0.9355 |
0.8831 |
0.8549 |
0.0483 |
| R:CCCAAACCGCACTAGAGAAA | |||||||||
| ZJU099ab |
JQ318794 |
(GA)10 |
F:<FAM > <Tail-3 > TTGTTGCACTTGTGGGTGAT |
130-150 |
9 |
0.7742 |
0.7763 |
0.7299 |
0.0000 |
| R:AACTACAAACAGCCCAACCG | |||||||||
| ZJU100ab |
JQ318795 |
(TC)9 |
F:<FAM > <Tail-3 > ACTTGTCCGGATTCCACAAC |
128-154 |
5 |
0.8333 |
0.6316 |
0.5629 |
0.2930 |
| R:TCAAGGCACACAATAATGCAA | |||||||||
| ZJU101ab |
JQ318796 |
(AG)9 |
F:<FAM > <Tail-3 > TGATTGAGCTGCCAACAAAG |
134-154 |
7 |
0.6667 |
0.7062 |
0.6527 |
0.8110 |
| R:TTTAACATTTGGCACCGACA | |||||||||
| ZJU102ab |
JQ318797 |
(GA)10 |
F:<FAM > <Tail-3 > GAACCACGAACTTCAACCGT |
118-132 |
8 |
0.4231 |
0.5890 |
0.5441 |
0.0111 |
| R:AACCACCAAACTTAGCTTCCA | |||||||||
| ZJU103ab |
JQ318798 |
(AG)9 |
F:<FAM > <Tail-3 > TGAGGAGGGAGTTGAGTTGG |
121-139 |
10 |
0.7097 |
0.7731 |
0.7359 |
0.0003 |
| R:GCGTCTTCCTCCTCCTTCTT | |||||||||
| ZJU104ab |
JQ318799 |
(TA)9 |
F:<FAM > <Tail-3 > ACGTGGCAGTTGAGTTGTTG |
114-128 |
6 |
0.3704 |
0.6296 |
0.5702 |
0.1383 |
| R:TCAGATCTCCGTTGGAGCTT | |||||||||
| ZJU105ab |
JQ318800 |
(GA)11 |
F:<FAM > <Tail-3 > TGAGAAACGCAGCAAGAGAA |
135-157 |
11 |
0.5806 |
0.8165 |
0.7801 |
0.0000 |
| R:CATCTCTCCCAAGCATCCTC | |||||||||
| ZJU106ab |
JQ318801 |
(GA)8 |
F:<FAM > <Tail-3 > GCAGTCGGATAGAGAGACGG |
134-146 |
7 |
0.3636 |
0.7717 |
0.7203 |
0.0000 |
| R:TGTTGATCAAACACACCGAGA | |||||||||
| ZJU107ab |
JQ318802 |
(TC)10 |
F:<FAM > <Tail-3 > TGGTGTCACGATTCACTGGT |
114-130 |
8 |
0.4375 |
0.5322 |
0.5012 |
0.3632 |
| R:CTGCATGTAATGGCAGTTCAA | |||||||||
| ZJU108b |
JQ318803 |
(CT)9 |
F:<FAM > <Tail-3 > TTGGTAGTGCACTGCAGGAG |
132-160 |
13 |
0.3929 |
0.8253 |
0.7909 |
0.0000 |
| R:CGAGGGTCGAGTTCAGAGAG | |||||||||
| ZJU109ab |
JQ318804 |
(TC)10 |
F:<FAM > <Tail-3 > TCCGCTCTCCTCTCTGTCTC |
138-164 |
11 |
0.8000 |
0.8441 |
0.8082 |
0.0003 |
| R:GTGAGTTGTGCTGCTGCAAT | |||||||||
| ZJU110ab |
JQ318805 |
(AG)9 |
F:<FAM > <Tail-3 > TTGCACGGTGGTAGCTGTAG |
143-159 |
5 |
0.7667 |
0.6486 |
0.5844 |
0.0000 |
| R:ACTGTGGTCCGTCGAACTCT | |||||||||
| ZJU111ab |
JQ318806 |
(TC)8 |
F:<FAM > <Tail-3 > TTTCTAATGTTGTTCGCCCA |
122-136 |
5 |
0.9000 |
0.5701 |
0.4652 |
0.0000 |
| R:TCATTCTCCTTGCAGATCCC | |||||||||
| ZJU112ac |
JQ318807 |
(GA)8 |
F:<FAM > <Tail-3 > GGAGAGTGAGAGATCGCAGC |
133-147 |
8 |
0.4839 |
0.6557 |
0.6212 |
0.0009 |
| R:GGCAACACCCTCAGTATCGT | |||||||||
| ZJU113ab |
JQ318808 |
(AG)9 |
F:<FAM > <Tail-3 > AAACGCACCAGAGAAAGACG |
138-154 |
6 |
0.6667 |
0.6588 |
0.5987 |
0.0130 |
| R:TCCATCTCTGGTCTCCATCC | |||||||||
| ZJU114a |
JQ318809 |
(GA)10 |
F:<FAM > <Tail-3 > CTAGAGCGCTCCACGATACC |
132-160 |
12 |
0.8214 |
0.8740 |
0.8448 |
0.0388 |
| R:AGAACGCTTGGAGAATCGAA | |||||||||
| ZJU115ab |
JQ318810 |
(AG)14 |
F:<FAM > <Tail-3 > GGTCTGAGGCCTTCACTCTG |
126-156 |
14 |
0.9677 |
0.9022 |
0.8775 |
0.0068 |
| R:GAGACCCAATAACCCATCCA | |||||||||
| ZJU116ab |
JQ318811 |
(AG)15 |
F:<FAM > <Tail-3 > CTTTCTCCGTCTGCTCCATC |
110-136 |
13 |
0.6875 |
0.8199 |
0.7846 |
0.0001 |
| R:GTCCAAACTTGGAGCCCATA | |||||||||
| ZJU117ab |
JQ318812 |
(AAG)9 |
F:<FAM > <Tail-3 > TCTCAGATCCCTCCACGTTC |
118-133 |
6 |
0.4688 |
0.6944 |
0.6426 |
0.0000 |
| R:CCACTGGATCAGGACAACCT | |||||||||
| ZJU118ab |
JQ318813 |
(CT)9 |
F:<FAM > <Tail-3 > CAAGCCACGTGCATACCTATT |
120-144 |
11 |
0.8750 |
0.8502 |
0.8171 |
0.0001 |
| R:CAGCTGGCTTCTAACTGCAA | |||||||||
| ZJU119a |
JQ318814 |
(AG)11 |
F:<FAM > <Tail-3 > CTTTCGACTTCAGAGGCAGC |
136-152 |
9 |
0.4828 |
0.8348 |
0.7975 |
0.0000 |
| R:TCCCTCTCAAACTTTGCCAC | |||||||||
| ZJU120ab |
JQ318815 |
(GA)8 |
F:<HEX > <Tail-4 > TTGGTTTCGTTTGCAAGTCA |
164-180 |
6 |
0.9355 |
0.7012 |
0.6354 |
0.0073 |
| R:GTCATCCATCCAATCCATCC | |||||||||
| ZJU121a |
JQ318816 |
(CT)11 |
F:<HEX > <Tail-4 > AATCACCGAAGAAATCCACG |
164-186 |
11 |
0.8621 |
0.8705 |
0.8426 |
0.0000 |
| R:ATTGCCCTCCCTTCTGTTCT | |||||||||
| ZJU122ab |
JQ318817 |
(TC)8 |
F:<HEX > <Tail-4 > TGACGGAAGGATACTGGCTC |
164-180 |
7 |
0.7742 |
0.7509 |
0.7012 |
0.0000 |
| R:CCATCAGACATGGCTTTCCT | |||||||||
| ZJU123ab |
JQ318818 |
(CT)8 |
F:<HEX > <Tail-4 > TGAATTATTCGGTTCCCTGG |
172-176 |
3 |
0.4667 |
0.6367 |
0.5499 |
0.2152 |
| R:TGCTTCAGTTCCAAACGAAA | |||||||||
| ZJU124ab |
JQ318819 |
(CT)10 |
F:<HEX > <Tail-4 > GTGGCAGCCTCTCTATCGTC |
161-187 |
12 |
0.9355 |
0.8778 |
0.8498 |
0.0001 |
| R:ATGACGTACTGCCCTTGCTT | |||||||||
| ZJU125ab |
JQ318820 |
(TC)8 |
F:<HEX > <Tail-4 > TAAGGGCAGTCAGACCAACC |
164-186 |
4 |
0.2188 |
0.3884 |
0.3453 |
0.0000 |
| R:CTGCAGCCTACAATGATCCA | |||||||||
| ZJU126ab |
JQ318821 |
(GA)10 |
F:<HEX > <Tail-4 > CCAATGTGGACAGGTGTGAG |
173-193 |
11 |
0.9677 |
0.8535 |
0.8228 |
0.0000 |
| R:GGCAGTAGTCGCTTCCCATA | |||||||||
| ZJU127 |
JQ318822 |
(GC)10 |
F:<HEX > <Tail-4 > AGGATCCTTGTCACCACCAG |
165-189 |
11 |
0.9259 |
0.8288 |
0.7900 |
0.0079 |
| R:AATTCTTCCTTCCCAGCCTC | |||||||||
| ZJU128ab |
JQ318823 |
(AG)14 |
F:<HEX > <Tail-4 > CCCAATTGACACAAATTCCC |
145-161 |
5 |
0.4194 |
0.5019 |
0.4496 |
0.1981 |
| R:TTGGCATAGCATTGTTCGTC | |||||||||
| ZJU129ab |
JQ318824 |
(CT)10 |
F:<HEX > <Tail-4 > GAGGTGCAATTACGTGGCTT |
161-189 |
10 |
0.7500 |
0.8031 |
0.7611 |
0.0234 |
| R:TCAAGCATCAGCTGCTCAGT | |||||||||
| ZJU130ab |
JQ318825 |
(GA)8 |
F:<HEX > <Tail-4 > GATTGCATGCACCAAATCAC |
160-176 |
5 |
0.3478 |
0.4599 |
0.4131 |
0.2852 |
| R:GAATGTCCACGACGTGAATG | |||||||||
| ZJU131ab |
JQ318826 |
(CT)14 |
F:<HEX > <Tail-4 > TTGAGAATCACAAACGCCTG |
153-187 |
13 |
0.8710 |
0.8990 |
0.8735 |
0.0009 |
| R:GGTGGGTGAAATGCCTAGAA | |||||||||
| ZJU132ab |
JQ318827 |
(CT)11 |
F:<HEX > <Tail-4 > AGGCACCTTTCTTTCCTCTC |
164-178 |
5 |
0.6452 |
0.6568 |
0.5834 |
0.6586 |
| R:CAAGGAAGGAGGTGACGAAG | |||||||||
| ZJU133ab |
JQ318828 |
(TC)11 |
F:<HEX > <Tail-4 > GCCCTGCAGTCTTTGTCAAT |
171-195 |
8 |
0.8710 |
0.7731 |
0.7267 |
0.0000 |
| R:CAGCTTGCAGTGTTCATTCA | |||||||||
| ZJU134ab |
JQ318829 |
(GA)11 |
F:<HEX > <Tail-4 > AGTGCCCAAGCATGACTTCT |
172-190 |
8 |
0.9688 |
0.7907 |
0.7507 |
0.0004 |
| R:AATCAGTTGTCCGAGGATGG | |||||||||
| ZJU135ab |
JQ318830 |
(AG)10 |
F:<HEX > <Tail-4 > AATTTACGGCTGTCCGTGAG |
173-191 |
10 |
0.9688 |
0.7966 |
0.7557 |
0.0000 |
| R:CCTTGGGCTTCATGAACATT | |||||||||
| ZJU136ab |
JQ318831 |
(GA)10 |
F:<HEX > <Tail-4 > TCCCACAGATCTCTAGCCGT |
173-201 |
13 |
0.7742 |
0.8953 |
0.8692 |
0.0004 |
| R:CGCTCAGTTCTTAATTTCTTACTGTC | |||||||||
| ZJU137ab |
JQ318832 |
(TC)8 |
F:<HEX > <Tail-4 > TGGATCTTGCTGCAGTTGTC |
140-168 |
12 |
0.1875 |
0.6930 |
0.6612 |
0.0000 |
| R:AGCTAGCACTGGCCTAACCA | |||||||||
| ZJU138ab |
JQ318833 |
(CT)10 |
F:<HEX > <Tail-4 > GCACAGTTGAGTTATGGGCA |
152-170 |
8 |
0.3333 |
0.7746 |
0.7261 |
0.0001 |
| R:CTCTTTCAAATCCACGCACA | |||||||||
| ZJU139ab |
JQ318834 |
(GA)12 |
F:<HEX > <Tail-4 > CCGAGCTTCGTTAGGACTTG |
138-164 |
6 |
0.3667 |
0.4418 |
0.4043 |
0.0000 |
| R:CCAACAATACCCGAACCATC | |||||||||
| ZJU140b |
JQ318835 |
(CT)14 |
F:<HEX > <Tail-4 > TGTGCTCATCTTGGATCCTG |
172-198 |
9 |
0.6538 |
0.6139 |
0.5474 |
0.0000 |
| R:ACATCAGCTTGCATCCCTCT | |||||||||
| ZJU141ab |
JQ318836 |
(CT)13 |
F:<HEX > <Tail-4 > CACAATCAGCTGCAGAATCAA |
175-201 |
11 |
0.6774 |
0.7996 |
0.7600 |
0.0002 |
| R:AATGGCCGCTTGCAATATAA | |||||||||
| ZJU142ab |
JQ318837 |
(TC)13 |
F:<HEX > <Tail-4 > CATTCACCTCCTTTCGCAAT |
166-184 |
9 |
0.6774 |
0.6912 |
0.6498 |
0.0231 |
| R:ATCCAACGGCTCAAAGAATG | |||||||||
| ZJU143ab |
JQ318838 |
(CT)12 |
F:<HEX > <Tail-4 > GTAGAGTAGATGCGCCTCGG |
181-197 |
7 |
0.6923 |
0.7044 |
0.6397 |
0.0000 |
| R:ACGTACGAGCCATACACACG | |||||||||
| ZJU144ab |
JQ318839 |
(AG)12 |
F:<HEX > <Tail-4 > GCCACTCTTCCCTCAACGTA |
148-164 |
7 |
0.5161 |
0.6864 |
0.6252 |
0.0430 |
| R:CAGGTCAGTCCTGATGGGAT | |||||||||
| ZJU145ab |
JQ318840 |
(CT)10 |
F:<HEX > <Tail-4 > TGTGGCTGTGTTCCTCCATA |
155-175 |
7 |
0.6875 |
0.7351 |
0.6912 |
0.0000 |
| R:CAATGTTGGGTGCTTTGTTG | |||||||||
| ZJU146ab |
JQ318841 |
(AG)10 |
F:<HEX > <Tail-4 > TGGAAACTTTGTCGTGTGGA |
154-168 |
6 |
0.2258 |
0.6663 |
0.6187 |
0.0000 |
| R:TTATATCGGGCAGCCAGAAC | |||||||||
| ZJU147ab |
JQ318842 |
(AG)10 |
F:<HEX > <Tail-4 > TTAGGAACCAAACTGGACGG |
173-195 |
10 |
0.8333 |
0.7169 |
0.6811 |
0.0005 |
| R:TCAAATGCCGTGCTATTGAG | |||||||||
| ZJU148ab |
JQ318843 |
(AG)18 |
F:<HEX > <Tail-4 > AAGAGCAGGAACCGAACCTT |
160-190 |
15 |
0.9375 |
0.9067 |
0.8829 |
0.4973 |
| R:ACCGAAAGACGAAGAAACGA | |||||||||
| ZJU149ab |
JQ318844 |
(TC)8 |
F:<HEX > <Tail-4 > AGCCCTCCATGTGTGCTTAT |
139-163 |
11 |
0.8333 |
0.8718 |
0.8417 |
0.0022 |
| R:AGGGAGAGAGTGGTTCTGCC | |||||||||
| ZJU150ab |
JQ318845 |
(AG)10 |
F:<HEX > <Tail-4 > ACTTAACTGAGAGGCTGCGG |
163-201 |
10 |
0.9000 |
0.8469 |
0.8123 |
0.0053 |
| R:GTGGAAACCGAACGTCCTAA | |||||||||
| ZJU151ab |
JQ318846 |
(CA)9 |
F:<HEX > <Tail-4 > GAATTGGAAATCCCTAGCCC |
156-170 |
6 |
0.3750 |
0.5511 |
0.5188 |
0.0001 |
| R:CATTTGCGCATGTCTCCTTA | |||||||||
| ZJU152ab |
JQ318847 |
(AG)11 |
F:<HEX > <Tail-4 > AAACGAAGTCGTTCAATGCC |
163-181 |
7 |
0.9355 |
0.7578 |
0.7040 |
0.0161 |
| R:CTTGATTTGGGCCTTCGATA | |||||||||
| ZJU153ab |
JQ318848 |
(AG)10 |
F:<HEX > <Tail-4 > CCAGCTCCGAATTAGCAAAC |
173-191 |
6 |
1.0000 |
0.6667 |
0.5927 |
0.0000 |
| R:GTGGCGGTTTATCTCATCGT | |||||||||
| ZJU154ab |
JQ318849 |
(AG)11 |
F:<HEX > <Tail-4 > TTGTCAATTGCCCTTCCTTC |
156-184 |
10 |
0.9333 |
0.6847 |
0.6184 |
0.0000 |
| R:TTCCTCCCTTTCCCACTTCT | |||||||||
| ZJU155ab |
JQ318850 |
(TC)9 |
F:<HEX > <Tail-4 > GAGAGCAATCAGTGAAGCCC |
160-188 |
8 |
0.8438 |
0.6731 |
0.6037 |
0.0000 |
| R:GGGAGACGGATGTCGATTTA | |||||||||
| ZJU156ab |
JQ318851 |
(TA)8 |
F:<HEX > <Tail-4 > ATACGTCGAAAGATCCACCG |
166-184 |
7 |
0.5484 |
0.6626 |
0.6063 |
0.0000 |
| R:TTCTGGAATCCTTCCCATTG | |||||||||
| ZJU157ab |
JQ318852 |
(AG)9 |
F:<HEX > <Tail-4 > CACTCACAACCAAAGCCAGA |
154-186 |
13 |
0.9677 |
0.9064 |
0.8823 |
0.0171 |
| R:GTGCATAATCACAGGCATGA | |||||||||
| ZJU158ab |
JQ318853 |
(AT)10 |
F:<HEX > <Tail-4 > CCAGATGATCACGCAGCTTA |
156-174 |
9 |
0.6452 |
0.8292 |
0.7917 |
0.0000 |
| R:CGTCCTCCAATACGTTCCTC | |||||||||
| Mean | 8.25 | 0.5636 | 0.7178 | 0.6730 | |||||
Note: a b c These SSRs are transferable for M. adenophora, M. nana and M. cerifera, respectively. SSR markers are listed according to ascending order in different fluorescent dyes. Shown for each primer pair are the repeat motif, primer sequences, size range (bp), number of alleles detected (Na), observed heterozygosity (Ho), expected heterozygosity (He), polymorphism information content (PIC) and Chi-square test for Hardy-Weinberg equilibrium (PHW). The annealing temperature was 60 °C; a, including length of tail sequences (18 bp total). PHW over 0.05 are underlined.
The PIC at each locus ranged from 0.256 to 0.883 with an average of 0.67 loci. The PCR product size ranged from 110 to 274 bp. All the primers produced amplicons in agreement with the expected sizes. Most of the SSR primers (139 primer pairs) showed significant deviation from HW equilibrium (P < 0.05). Partial correlation analysis showed that significant positive correlations existed between the repeat unit length and PIC (P < 0.01, r = 0.2747). This showed that these SSRs had high rates of transferability for M. adenophora (91.14%) and M. nana (89.87%) and low rates for M. cerifera (46.84%), indicating that these markers are suitable for genetic diversity analyses in other Myrica species.
One of the objectives of this study was to develop potential suitable SSR markers for genetic mapping using Biqi and Dongkui as crossing parents. We selected 99 heterozygous loci in Biqi and 105 in Dongkui (Table 3): 135 primer pairs can be used together in linkage mapping of the planned F1 populations between Biqi and Dongkui.
Table 3.
Distribution of the segregation types expected for the mapping population (Biqi × Dongkui)
| Segregation type | Alleles | Number | Mapping in F1 |
|---|---|---|---|
| aa × aa |
1 |
12 |
No |
| aa × bb |
2 |
11 |
No |
| aa × ab |
2 |
24 |
Yes |
| ab × aa |
2 |
18 |
Yes |
| ab × ab |
2 |
8 |
Yes |
| aa × bc |
3 |
12 |
Yes |
| ab × cc |
3 |
12 |
Yes |
| ab × ac |
3 |
41 |
Yes |
| ab × cd |
4 |
20 |
Yes |
| Total | 135 | ||
Genetic relationship analysis
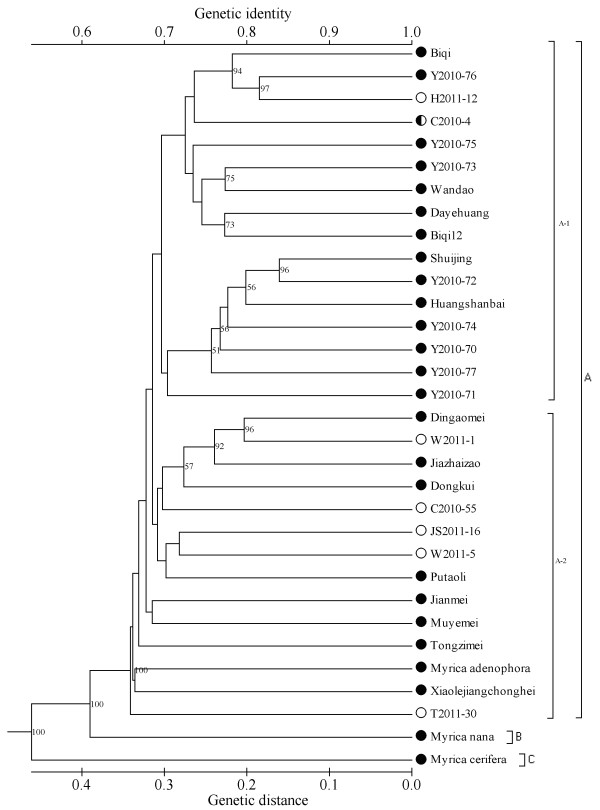
The 32 accessions were divided into three groups (A, B and C, Figure 4), based on Nei’s genetic distance coefficient [16] using UPGMA cluster analysis. The similarity among all the accessions varied from 0.54 to 0.84. At the species level, the UPGMA dendrogram produced clusters separating M. nana and M. cerifera into two distinct groups. The genetic similarity between M. cerifera and M. rubra was 0.54, lower than the 0.74 previously reported by Xie [6].
Figure 4.
Dendrogram for 32 Chinese bayberry accessions derived from UPGMA cluster analysis based on 158 SSR markers. The symbols before the accession codes indicate the sex: ○, androphyte plant, ●, common cultivars, and ◘, monoecious plant. The numbers are bootstrap values based on 1000 iterations. Only bootstrap values larger than 50 are indicated.
The main cluster ‘A’ included the subgroups A-1 and A-2. Subgroup A-1 includes 16 accessions, mainly from the cities of Ningbo (12) and Hangzhou (3), where the popular and dominant cultivar is Biqi. This demonstrates that these natural elite seedling selections are truly distinct from the local cultivars. For their genetic relationships (Figure 4), the rare monoecious individual (C2010-4) is closely related to Biqi, while the accessions ‘Shuijing’ and ‘Y2010-72’ (both white fruit type) are clearly separated in the cluster, with low genetic distance.
Subgroup A-2 includes 14 accessions, with four from Wenzhou, two from Taizhou, and one each from the cities of Hangzhou and Ningbo, and the Hunan, Guangxi, Guizhou and Jiangsu provinces. This group includes the popular cultivar Dongkui. The four accessions from Wenzhou distributed in this cluster have genetic similarity. The accession ‘Tongzimei’ from the Hunan province is on an independent branch, showing that it is genetically distinct. ‘Xiaolejiangchonghei’ and ‘M. adenophora’ grouped together, and are possibly in the same population. Six androphyte accessions, distributed in group A, are close to cultivars of the same geographic origin.
The accessions ‘Myrica nana’ from Yunnan and ‘Myrica cerifera’ from the USA were independently classified as the ‘B’ and ‘C’ group, indicating a distant relationship with cultivated Myrica rubra.
Discussion
Our major aims were to find a large set of SSR markers for Myrica rubra and understand the genetic diversity and relationship among representative cultivars, androphyate and related species. This research paves the way for constructing an SSR-based linkage map in Myrica.
The genome characteristics of genusMyrica
Shotgun sequencing is suitable for simple genomes, with no or few repeat sequences, such as Chinese bayberry. For such genomes, the genome can largely be assembled simply by merging together reads containing overlapping sequence [17]. We report the genome survey of Chinese bayberry using whole genome shotgun sequencing. The 17-nucleotide depth distribution suggests a genome size of 323 Mb, larger than peach (220 Mb, http://www.rosaceae.org/peach/genome), but close to our estimate of 250 Mb from flow cytometry using rice as the reference (date not shown). Although the highly homozygous material was selected on a limited number of SSR loci assays, the actual heterozygous rate, as revealed by 185 new SSR markers, was very high (63%). To overcome the key issue of heterozygosity and allow us to generate a high-quality genome sequence, we can use a unique homozygous form such as monoploid, derived using tissue culture or from nature and worth further study.
Marker development for under-utilised fruit crops
SSRs have been widely used for high-throughput genotyping and map construction as they have the advantage of high abundance, random distribution within the genome, high polymorphism information content and stable co-dominance [18-20]. They can be developed from either genomic or expressed sequence tag (EST) libraries. Although EST-SSRs are useful for genetic analysis, their disadvantages of relatively low polymorphism and high concentration in gene-rich regions of the genome may limit their usage, especially for the construction of linkage maps [21]. In this study, a total of 600 SSR primer pairs were designed from 28,602 SSR sites, and 581 (96.8%) primer pairs were effective. Due to the self-complementary nature to form dimers, AT/TA is not usually used to develop markers [12]. Our findings are in agreement with that published for peach, where the dinucleotide repeat motifs were also found to be the most common, and (CT)n as the most common repeat unit [22].
In the present study, the mean value of PIC was higher than the previously reported 0.62 [7], but the consistent relationship was observed between SSR polymorphism and repeat unit length. There are some reports of a positive relationship between degree of polymorphism and repeat unit length [23,24]. However, those polymorphic SSRs that are homozygous in both parents cannot be mapped in F1 populations, although they are useful for mapping in F2 or backcross populations [25], while heterozygous SSRs can be used for mapping in F1 populations (Table 2). The estimated number of SSRs that can be mapped in the F1 populations between Biqi and Dongkui was about 85%.
Recently, based on mass sequence data of Chinese bayberry obtained by RNA-Seq, 41,239 UniGenes were identified and approximately 80% of the UniGenes (32,805) were annotated, which provides an excellent platform for future EST-SSR development and functional genomic research [26].
High efficient test methods
Normally, a universal M13 primer is labelled with one of a number of fluorescent dyes. The tailed primer provides a complementary sequence to the fluorescent labelled M13 primer, leading to the amplification of fluorescent PCR products, and then the PCR products of different sizes and/or labelled with different fluorescent dyes are mixed and tested [27]. In this research, a multiplex PCR strategy was designed using the universal M13-tailed primer and three additional tail primers, designed arbitrarily, in presumed four-plex amplifications in single PCR, for a major reduction in cost and time. However, as only a few primer combinations were successful, most resulting in weak bands, we did the PCR individually and mixed the PCR products. Further optimisation of multiplex PCR is needed to evaluate its general applicability.
Evolution ofMyricaspecies
In this study, both cultivated species and wild species were analysed and their genetic diversity could easily be differentiated. M. nana and M. cerifera were clearly genetically distant to M. rubra. M. nana, also known as the dwarf or Yunnan arbutus, is indigenous to the Yunnan and Guizhou provinces, and has a plant height of < 2 m. The juvenile period of fruit tree can be shortened for breeding purposes. Studies on embryo culture in vitro of the F1 seeds of crosses between M. rubra and M. nana, [28], has shown good cross compatibility between M. rubra and M. nana, resulting in 70.5% normal seeds with intact embryo. M. adenophora and M. nana grow as wild trees, with the fruit of M. adenophora only suitable for medical purposes and not edible.
Our findings on the genetic similarity between M. adenophora and M. rubra, which are considered a progenitor-derivative species pair, are consistent with a previously published figure of 0.897 [29]. An earlier study observed little change in allelic diversity along the chronosequence and no evidence for heterosis, although there was a moderate change in genotypic diversity [30]. The markers developed in this study can be very useful in future population structure analysis.
Conclusions
In summary, the genome size of Myrica genus is small, about 320 Mb. A large set of SSRs were developed from a genome survey of Myrica rubra. The results suggest that they have high rates of transferability, making them suitable for use in other Myrica species.
Materials and methods
Plant materials and genome survey
We selected an androphyte ‘C2010-55’ for the genome survey because it was the most homozygous (10 out of 14 SSRs) individual among 230 accessions. Two DNA libraries of 250 and 500 bp insert size were constructed and sequenced by Illumina Hi-Seq 2000.
Twenty-nine accessions of the cultivated species (M. rubra) and 3 related species (M. adenophora, M. nana, M. cerifera), collected from different provinces in China (Table 4), were used to evaluate the suitability of the SSRs for genetic distance analysis. Young leaves were collected and frozen in liquid nitrogen prior to genomic DNA extraction using CTAB methods [4]. DNA concentrations were measured spectrophotometrically at 260 nm, and the extracts electrophoresed on 1% agarose to confirm the quality. The purified DNAs were standardised at 40 ng/μl and stored at -40°C.
Table 4.
The 32 bayberry accessions included in this study
| No. | Accession | Fruit/Flower coloura | Fruit maturity date | Region |
|---|---|---|---|---|
| 1 |
Biqi |
black |
Late June |
Cixi, Ningbo, Zhejiang |
| 2 |
Dongkui |
red |
Early July |
Taizhou, Zhejiang |
| 3 |
Dayehuang |
red |
Mid-June |
Hangzhou, Zhejiang |
| 4 |
Dingaomei |
black |
Mid to late June |
Wenzhou, Zhejiang |
| 5 |
Huangshanbai |
white |
Early July |
Hangzhou, Zhejiang |
| 6 |
Jiazhaizao |
black |
Mid-June |
Wenzhou, Zhejiang |
| 7 |
Jianmei |
red |
Late June |
Cixi, Ningbo, Zhejiang |
| 8 |
Muyemei |
black |
Late June |
Jinhua, Zhejiang |
| 9 |
Putaoli |
black |
Mid June |
Hangzhou, Zhejiang |
| 10 |
Shuijing |
white |
Late June/Early July |
Yuyao, Ningbo, Zhejiang |
| 11 |
Tongzimei |
black |
Mid-June |
Hunan |
| 12 |
Wandao |
black |
Early July |
Zhoushan, Zhejiang |
| 13 |
Xiaolejiangchonghei |
black |
May |
Guizhou |
| 14 |
Biqi12 |
black |
Late June |
Yuyao, Ningbo, Zhejiang |
| 15 |
Y2010-70 |
red |
Late June/Early July |
Yuyao, Ningbo, Zhejiang |
| 16 |
Y2010-71 |
black |
Mid to late June |
Yuyao, Ningbo, Zhejiang |
| 17 |
Y2010-72 |
white |
Late June/Early July |
Yuyao, Ningbo, Zhejiang |
| 18 |
Y2010-73 |
red |
Late June |
Yuyao, Ningbo, Zhejiang |
| 19 |
Y2010-74 |
red |
Late June/Early July |
Yuyao, Ningbo, Zhejiang |
| 20 |
Y2010-75 |
black |
Late June |
Yuyao, Ningbo, Zhejiang |
| 21 |
Y2010-76 |
white |
Late June/Early July |
Yuyao, Ningbo, Zhejiang |
| 22 |
Y2010-77 |
red |
Late June/Early July |
Yuyao, Ningbo, Zhejiang |
| 23 |
C2010-4 |
red |
Late June |
Cixi, Ningbo, Zhejiang |
| 24 |
*C2010-55 |
red |
- |
Cixi, Ningbo, Zhejiang |
| 25 |
*W2011-1 |
yellow- red |
- |
Wenzhou, Zhejiang |
| 26 |
*W2011-5 |
red |
- |
Wenzhou, Zhejiang |
| 27 |
*H2011-12 |
yellow-green |
- |
Hangzhou, Zhejiang |
| 28 |
*JS2011-16 |
red |
- |
Suzhou, Jiangsu |
| 29 |
*T2011-30 |
red |
- |
Taizhou, Zhejiang |
| 30 |
Myrica adenophora |
red |
February to May |
Guilin, Guangxi |
| 31 |
Myrica nana |
red |
June to July |
Yunnan |
| 32 | *Myrica cerifera | yellow-green | - | Cixi, Ningbo, Zhejiang |
Note: fruit colour for cultivar and flower colour for androphyte. * selected androphytes.
SSR identification and primer design
We used MISA scripting language ( http://pgrc.ipk-gatersleben.de/misa/misa.html) to identify microsatellite repeats in our sequence database. The SSR loci containing perfect repeat units of 2-6 nucleotides only were considered. The minimum SSR length criteria were defined as six reiterations for dinucleotide, and five reiterations for other repeat units. Mononucleotide repeats and complex SSR types were excluded from the study.
The SSR primers were designed using BatchPrimer3 interface modules ( http://pgrc.ipk-gatersleben.de/misa/primer3.html). We selected 600 primers that met the following parameters: 110–230 final product length, primer size from 18 to 22 bp with an optimum size of 20 bp, and the annealing temperature was set at 60°C. The repeat units over eight were used.
Tail-1(M13 universal sequence-TGTAAAACGACGGCCAGT), Tail-2(CGAGTCAGTATAGGGCAC), Tail-3(ATCACGAAGCAGATGCAA) and Tail-4(GAGCATCTCGTACCAGTC) were added to the 5’ end of each 150 forward primers of pairs respectively. Tail-2, Tail-3 and Tail-4 were designed so that the primer size was 18 bp and the annealing temperature was 53°C. Primers were synthesised by Invitrogen Trading (Shanghai) Co., Ltd. We primarily tested two cultivars (Biqi and Dongkui) and M. cerifera for 600 SSR loci by PAGE (polyacrylamide denaturing gel) to confirm their suitability. Tail-1, Tail-2, Tail-3 and Tail-4 labelled with one of the following dyes: NED, PET, FAM, and HEX, respectively.
Polymerase chain reaction and gel electrophoresis
Each 20 μl reaction mixture contained 10 × PCR buffer (plus Mg2+), 0.2 mM of each dNTP, 5 pmol of each reverse, 4 pmol of the tail primer, 1 pmol of the forward primer, 0.5 units of rTaq polymerase (TaKaRa, China) and 40 ng genomic DNA template. Each primer pair had an interval of 20 bp according to the expected size of amplicons.
DNA amplification was in an Eppendorf Mastercycler (Germany) programmed at 94°C for 5 min for initial denaturation, then 32 cycles at 94°C (30 s)/58°C (30 s)/72°C (30 s), followed by 8 cycles of 94°C (30 s)/53°C (30 s)/72°C (30 s). The final extension step was 10 min at 72°C. Each PCR product was run on 1% agarose gel at 110 V for a quality check.
Subsequently, PCR products were electrophoresed on 8% denaturing PAGE, according to Myers et al. [31], at 60 W in a Sequi-Gen GT Nucleic Acid electrophoresis cell (BioRad) for 4 h, depending on the fragment sizes to be separated, and visualised by silver staining [32]. Genotypes showing one and two bands were scored as homozygous and heterozygous, respectively, and the results recorded and photographed.
Multiplex PCR was designed and tested with products of different sizes and labelled with different fluorescent dyes. Each 20 μl reaction mixture contained 10 × PCR buffer (plus Mg2+), 0.8 mM of each dNTP, 1 unit of rTaq polymerase, 40 ng genomic DNA template and a total of four primer pairs with 5 pmol of each reverse primer, 4 pmol of each tail primer, and 1 pmol of each forward primer. The PCR products were diluted, mixed with the internal size standard LIZ500 (Applied Biosystems) and loaded on an ABI 3130 Genetic Analyzer. Alleles were scored using GeneMapper version 4.0 software (Applied Biosystems, Foster City, CA, USA).
Data analysis
The raw genome sequence data was first filtered to obtain high quality reads, then assembled using SOAP ( http://soap.genomics.org.cn/soapdenovo.html) denovo software to contig, scaffold and fill in gaps. In addition, we used SSPACE software to build the scaffold. K-mer analysis was to help estimate the genome size and characters, such as heterozygosis and repeats.
The number of alleles (A), observed heterozygosity (Ho) and expected heterozygosity (He) were calculated using POPGENE version 1.32 ( http://www.ualberta.ca/~fyeh/popgene_download.html). Chi-square test for Hardy-Weinberg equilibrium allele frequencies and polymorphism information content (PIC) was calculated using PowerMarker version 3.25 [33] ( http://statgen.ncsu.edu/powermarker/downloads.htm). Microsoft office Excel 2007 was used to analyse the correlation. Genetic similarity among all the accessions was calculated according to Dice’s coefficients using Nei's coefficient index [16] with the Free Tree 0.9.1.50 [34] ( http://www.natur.cuni.cz/~flegr/programs/freetree.htm) software, and the dendrogram constructed using the unweighted pair-group method with arithmetic averaging (UPGMA) option. The confidence of branch support was then evaluated by bootstrap analysis with 1,000 replicates. The dendrogram was printed using MEGA version 5.05 software [35] ( http://www.megasoftware.net/mega.php).
Authors’ contributions
ZSG: HJJ: EVW and MLC designed the experiments. YJ: CYC: GYW collected plant materials. YJ: HMJ and XWL performed the SSR experiments and analysed the data. The whole genome shotgun and sequencing assembly was performed by ZC. YJ: ZSG and EVW drafted this manuscript.
Supplementary Material
Occurrence of different SSRs in the genome survey of Chinese bayberry.
Contributor Information
Yun Jiao, Email: jydyx@163.com.
Hui-min Jia, Email: jiahuimin1988@163.com.
Xiong-wei Li, Email: lixiongweisea@yahoo.com.cn.
Ming-liang Chai, Email: mlchai@zju.edu.cn.
Hui-juan Jia, Email: jiahuijuan@zju.edu.cn.
Zhe Chen, Email: chenzhe2@genomics.org.cn.
Guo-yun Wang, Email: yywgy@sina.com.
Chun-yan Chai, Email: cxccy@sina.com.
Eric van de Weg, Email: eric.vandeweg@wur.nl.
Zhong-shan Gao, Email: gaozhongshan@zju.edu.cn.
Acknowledgements
This work was supported by grants from the Zhejiang Province (2006 C14016) and Special Research Fund for International Cooperation with European Union (1114) and Public Welfare in Chinese Agriculture (contract no. 200903044) We thank Dr Rangjin Xie for technical assistance in the PAGE experiment, and Dr. Shirley Burgess for correcting the English.
Author details
1Department of Horticulture, The State Agriculture Ministry Laboratory of Horticultural Plant Growth, Development and Quality Improvement, Zhejiang University, Hangzhou 310058, China. 2BGI-Shenzhen, Beishan Industrial Zone, Yantian District, Shenzhen 518083, China. 3Fruit Research Institute, Yuyao, Ningbo 315400, China. 4Forestry Technology Extension Center, Cixi Ningbo 315300, China. 5Plant Breeding-Wageningen University and Research Centre, P.O. Box 166700 AA, Wageningen, The Netherlands.
References
- Chen K, Xu C, Zhang B, Ferguson IB. In: Red bayberry: botany and horticulture, in Horticultural Reviews. Jules Janick, editor. John Wiley & Sons, Inc, Oxford; 2004. pp. 83–114. 30. [Google Scholar]
- Zhang B, Kang MX, Xie QP, Xu B, Sun CD, Chen KS, Wu YL. Anthocyanins from Chinese bayberry extract protect beta cells from oxidative stress-mediated injury via HO-1 upregulation. J Agr Food Chem. 2011;59(2):537–545. doi: 10.1021/jf1035405. [DOI] [PubMed] [Google Scholar]
- Terakawa M, Kikuchi S, Kanetani S, Matsui K, Yumoto T, Yoshimaru H. Characterization of 13 polymorphic microsatellite loci for an evergreen tree, Myrica rubra. Mol Ecol Notes. 2006;6(3):709–711. doi: 10.1111/j.1471-8286.2006.01318.x. [DOI] [Google Scholar]
- Zhang SM, Xu CJ, Gao ZS, Chen KS, Wang GY. Development and characterization of microsatellite markers for Chinese bayberry (Myrica rubra Sieb. & Zucc.) Conserv Genet. 2009;10(5):1605–1607. doi: 10.1007/s10592-008-9804-x. [DOI] [Google Scholar]
- Xie XB, Qiu YY, Ke LP, Zheng XL, Wu GT, Chen JQ, Qi XJ, Ahn S. Microsatellite primers in red bayberry, myrica rubra (Myricaceae) Am J Bot. 2011;98(4):E93–E95. doi: 10.3732/ajb.1000271. [DOI] [PubMed] [Google Scholar]
- Xie R-J, Zhou J, Wang G-Y, Zhang S-M, Chen L, Gao Z-S. Cultivar identification and genetic diversity of chinese bayberry (Myrica rubra) accessions based on fluorescent SSR markers. Plant Mol Biol Rep. 2011;29(3):554–562. doi: 10.1007/s11105-010-0261-6. [DOI] [Google Scholar]
- Zhang SM, Gao ZS, Xu CJ, Chen KS. Genetic diversity of Chinese bayberry (Myrica rubra Sieb. et Zucc.) accessions revealed by Amplified Fragment Length Polymorphism. HortSci. 2009;44(2):487–491. [Google Scholar]
- Cheng YJ, Zhang R, Zhu AD, Wang XJ, Yu J, Zhang HR, Gao JS, Deng XX. Development of Juglans regia SSR Markers by data mining of the EST database. Plant Mol Biol Rep. 2010;28(4):646–653. doi: 10.1007/s11105-010-0192-2. [DOI] [Google Scholar]
- Aravanopoulos FA, Ganopoulos IV, Avramidou E, Fasoula DA, Diamantidis G. Assessing inter- and intra-cultivar variation in Greek Prunus avium by SSR markers. Plant Genet Resour-C. 2010;8(3):242–248. doi: 10.1017/S1479262110000298. [DOI] [Google Scholar]
- Audergon JM, Bourguiba H, Khadari B, Krichen L, Trifi-Farah N, Santoni S. Grafting versus seed propagated apricot populations: two main gene pools in Tunisia evidenced by SSR markers and model-based Bayesian clustering. Genetica. 2010;138(9–10):1023–1032. doi: 10.1007/s10709-010-9488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin L, Martinez RT, Berger A, Dollet M. Characterization of the genetic diversity of the Tall coconut (Cocos nucifera L.) in the Dominican Republic using microsatellite (SSR) markers. Tree Genet Genomes. 2010;6(1):73–81. doi: 10.1007/s11295-009-0229-6. [DOI] [Google Scholar]
- Wu YQ, Wang YW, Samuels TD. Development of 1,030 genomic SSR markers in switchgrass. Theor Appl Genet. 2011;122(4):677–686. doi: 10.1007/s00122-010-1477-4. [DOI] [PubMed] [Google Scholar]
- Goff SA. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) (April, pg 92, 2002) Science. 2005;309(5736):879–879. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen C, Na J-K, Yu Q, Hou S, Paull R, Moore P, Alam M, Ming R. Genome-wide comparative analyses of microsatellites in papaya. Tropical Plant Biology. 2008;1(3):278–292. doi: 10.1007/s12042-008-9024-z. [DOI] [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 2000;18(2):233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- Nei M. Genetic Distance between Populations. Am Nat. 1972;106(949):283–292. doi: 10.1086/282771. [DOI] [Google Scholar]
- Batzoglou S, Jaffe DB, Stanley K, Butler J, Gnerre S, Mauceli E, Berger B, Mesirov JP, Lander ES. ARACHNE: A whole-genome shotgun assembler. Genome Res. 2002;12(1):177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe KN, Hirano R, Ishii H, Oo TH, Gilani SA, Kikuchi A. Propagation management methods have altered the genetic variability of two traditional mango varieties in Myanmar, as revealed by SSR. Plant Genet Resour-C. 2011;9(3):404–410. doi: 10.1017/S1479262111000049. [DOI] [Google Scholar]
- Chen C, Zhou P, Choi Y, Huang S, Gmitter F. Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet. 2006;112(7):1248–1257. doi: 10.1007/s00122-006-0226-1. [DOI] [PubMed] [Google Scholar]
- Fraser LG, Harvey CF, Crowhurst RN, De Silva HN. EST-derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet. 2004;108(6):1010–1016. doi: 10.1007/s00122-003-1517-4. [DOI] [PubMed] [Google Scholar]
- McCouch SR, Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, Park WD, Ayres N, Cartinhour S. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.) Theor Appl Genet. 2000;100(5):713–722. doi: 10.1007/s001220051343. [DOI] [Google Scholar]
- Sosinski B, Gannavarapu M, Hager LD, Beck LE, King GJ, Ryder CD, Rajapakse S, Baird WV, Ballard RE, Abbott AG. Characterization of microsatellite markers in peach [Prunus persica (L.) Batsch] Theor Appl Genet. 2000;101(3):421–428. doi: 10.1007/s001220051499. [DOI] [Google Scholar]
- Cuc LM, Mace ES, Crouch JH, Quang VD, Long TD, Varshney RK. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea) BMC Plant Biol. 2008;8 doi: 10.1186/1471-2229-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapila RK, Yadav RS, Plaha P, Rai KN, Yadav OP, Hash CT, Howarth CJ. Genetic diversity among pearl millet maintainers using microsatellite markers. Plant Breeding. 2008;127(1):33–37. [Google Scholar]
- Lu J, Huang H, Ren ZB, Hunter W, Dowd SE, Dang P. Mining and validating grape (Vitis L.) ESTs to develop EST-SSR markers for genotyping and mapping. Mol Breeding. 2011;28(2):241–254. doi: 10.1007/s11032-010-9477-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Chen M, Xu CJ, Bai L, Yin XR, Li X, Allan AC, Ferguson IB, Chen KS. Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-Seq. BMC Genomics. 2012;13(1):19. doi: 10.1186/1471-2164-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zheng D, Vimolmangkang S, Khan MA, Beever JE, Korban SS. Integration of physical and genetic maps in apple confirms whole-genome and segmental duplications in the apple genome. J Exp Bot. 2011;62(14):5117–5130. doi: 10.1093/jxb/err215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XB, Qiu YY, Zheng XL, Qi XJ, Qiu LJ, Huang ZP, Wang T, Liang SM. Studies on the crossing between Myrica rubra and M. nana and embryo culture in vitro of hybrid F1. Journal of Fruit Science. 2009;26(4):507–510. [Google Scholar]
- Cheng YP, Chien CT, Lin TP. Population genetics of geographically restricted and widespread species of Myrica (Myricaceae) J Hered. 2000;91(1):61–66. doi: 10.1093/jhered/91.1.61. [DOI] [PubMed] [Google Scholar]
- Erickson DL, Hamrick JL. Genetic and clonal diversity for Myrica cerifera along a spatiotemporal island chronosequence. Heredity. 2003;90(1):25–32. doi: 10.1038/sj.hdy.6800172. [DOI] [PubMed] [Google Scholar]
- Myers RM, Maniatis T, Lerman LS. Detection and localization of single base changes by denaturing gradient gel-electrophoresis. Method Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- Bassam BJ, Caetanoanolles G. Silver staining of DNA in polyacrylamide gels. Appl Biochem Biotech. 1993;42(2–3):181–188. [Google Scholar]
- Liu KJ, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Hampl V, Pavlicek A, Flegr J. Construction and bootstrap analysis of DNA fingerprinting-based phylogenetic trees with the freeware program FreeTree: application to trichomonad parasites. Int J Syst Evol Micr. 2001;51:731–735. doi: 10.1099/00207713-51-3-731. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Occurrence of different SSRs in the genome survey of Chinese bayberry.