Abstract
Diarrhoeagenic Escherichia coli (DEC) are an important cause of diarrhoea in children and are associated with high antibiotic resistance. However, there are few studies on the molecular mechanisms of resistance in this group of bacteria. The aim of this study was to determine the mechanisms associated with antibiotic resistance in the most common phenotypes of DEC. A total of 369 E. coli strains [commensal strains and DEC from children with (‘DEC-diarrhoea’) or without (‘DEC-control’) diarrhoea] isolated from children aged <1 year in periurban districts of Lima, Peru, were analysed. In total, 154 ampicillin-resistant strains (36 commensals, 33 DEC-control and 85 DEC-diarrhoea) were studied by PCR for the most prevalent resistance mechanisms to ampicillin, trimethoprim/sulfamethoxazole (SXT), tetracycline and chloramphenicol as well as for integrase types 1 and 2. In additional, restriction fragment length polymorphism was performed for SXT-resistant strains. Commensal strains were more frequently resistant to nalidixic acid and ciprofloxacin (68% and 28%, respectively) than DEC strains (23% and 2%, respectively) (P < 0.05). DEC-diarrhoea strains were more frequently SXT-resistant (78%) compared with DEC-control strains (65%) and commensal strains (60%) (P < 0.05). The most frequent mechanisms of antibiotic resistance in DEC strains were: for β-lactams, blaTEM (31%; 37/118); for SXT, sul2 (48%; 49/103); for tetracycline, tetA (27%; 23/84); and for chloramphenicol, cat (80%; 28/35). The genes sul1 and dfrA1, related to SXT resistance, were more frequent in the DEC-diarrhoea group (41% and28%, respectively) than in the other two groups (P < 0.05). There was a high diversity of resistance genes in DEC, including symptomatic strains.
Keywords: Antibiotics, Antibiotic resistance mechanism, Children, Diarrhoeagenic E. coli, Commensal E. coli
1. Introduction
Diarrhoea is one of the leading causes of paediatric morbidity and mortality in developing countries. Every year, diarrhoea is responsible for ca. 1.8 million deaths worldwide [1]. One of the principal aetiological groups of diarrhoea is the diarrhoeagenic Escherichia coli (DEC). Based on specific virulence factors and pathogenic mechanisms, DEC are classified in six pathotypes: enterotoxigenic E. coli (ETEC); enteropathogenic E. coli (EPEC); diffusely adherent E. coli (DAEC); Shiga toxin-producing E. coli (STEC); enteroinvasive E. coli (EIEC); and enteroaggregative E. coli (EAEC) [2]. Both commensal E. coli and DEC are often resistant to antibiotics [3,4]. To facilitate appropriate empirical antibiotic selection, it is important to have a knowledge of local antibiotic susceptibility patterns [2].
In Peru, previous studies of DEC and commensal E. coli reported high antibiotic resistance to ampicillin, trimethoprim/sulfamethoxazole (SXT), tetracycline, chloramphenicol and nalidixic acid [3,4]. However, molecular mechanisms of antibiotic resistance in DEC are poorly defined in Peru and elsewhere in the developing world. Therefore, the aim of this study was to describe the molecular mechanisms of antibiotic resistance in Peruvian DEC using samples isolated from children <1 year of age.
2. Materials and methods
2.1. Samples
Commensal E. coli and DEC strains were isolated from a previous passive surveillance cohort study of diarrhoea in 1034 infants in Peru followed-up from 2 months to 12 months of age in low socioeconomic communities in the southern districts of Lima. In this study, control samples were obtained from enrolled infants when they were healthy [5].
A total of 1079 E. coli were isolated and characterised by a real-time multiplex PCR to determine DEC pathotypes [5]. This PCR uses primers designed to recognise simultaneously nine genes related to virulence factors of each DEC pathotype. A total of 592 DEC were isolated in this cohort study, comprising 326 related to diarrhoea episodes (‘DEC-diarrhoea’) and 266 related to control healthy asymptomatic children (‘DEC-control’). In addition, 487 commensal E. coli strains (strains from healthy children without either diarrhoea or virulence genes associated with DEC pathotypes) were isolated.
2.2. Study design
2.2.1. Bacteria
In total, 369 E. coli strains isolated in the cohort study were investigated, comprising 74 commensal, 94 DEC-control and 201 DEC-diarrhoea strains. The DEC group included strains of EPEC, ETEC, EAEC and DAEC pathotypes; STEC and EIEC strains were not included because of their very low prevalence in the same cohort of children [5]. Escherichia coli ATCC 25922 was used as a control.
2.2.2. Phenotypic characterisation of antibiotic resistance
Resistance to 11 antibiotics was determined by disk diffusion following the Clinical and Laboratory Standards Institute (CLSI) guidelines. The disks used were ampicillin (10 μg), amoxicillin/clavulanic acid (AMC) (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), trimethoprim/sulfamethoxazole (23.75/1.25 μg), ciprofloxacin (5 μg), chloramphenicol (30 μg), gentamicin (10 μg), nalidixic acid (30 μg), nitrofurantoin (30 μg) and tetracycline (30 μg).
2.2.3. Molecular mechanisms of resistance
Genes encoding common resistance mechanisms to β-lactams, tetracycline, chloramphenicol and SXT as well as integrase types 1 and type 2 were studied.
β-Lactam-related genes were studied in 154 strains with high-level resistance to ampicillin. These strains were also evaluated for genes conferring resistance to tetracycline, chloramphenicol and SXT when they were highly resistant.
DNA extraction was performed by the thermal shock lysis technique. Molecular mechanisms of antibiotic resistance and integrase types 1 and 2 detection were performed by conventional PCR using previously described primers (Table 1). PCR was performed for each gene in a 20 μL reaction mixture containing 0.25 mM of each dNTP (Promega, Madison, WI), 4 μL of 5× colourless buffer (GoTaq®; Promega), 2.4 μL of 25 mM MgCl2 (GoTaq®; Promega), 0.5 U of Taq polymerase (GoTaq®; Promega) and 2 μL of DNA template. PCR amplification was performed in a thermocycler (iCycler; Bio-Rad Laboratories, Hercules, CA) with hybridisation temperatures as specified in Table 1 for each primer. Amplified products were analysed using 1.5% agarose gel electrophoresis and were visualised by staining with ethidium bromide. A 100 bp DNA ladder (Fermentas Inc., Glen Burnie, MD) was used as a molecular marker. For all PCR amplifications, positive control E. coli strains from Centre de Recerca en Salut Internacional de Barcelona (Barcelona, Spain) were used.
Table 1.
Primers and conditions for identification of molecular mechanisms of antibiotic resistance
| Antibiotic/gene | Primer | Primer sequence (5′→3′) | Gene | Size (kb) | Hybridisation temperature (°C) | Reference |
|---|---|---|---|---|---|---|
| β-Lactams | TEM-F | ATTCTTGAAGACGAAAGGGC | blaTEM | 1150 | 60 | [6] |
| TEM-R | ACGCTCAGTGGAACGAAAAC | |||||
| SHV-F | CACTCAAGGATGTATTGTG | blaSHV | 885 | 52 | [6] | |
| SHV-R | TTAGCGTTGCCAGTTATTGTG | |||||
| carb1 | AATGGCAATCAGCGCTTC | blaCARB | 586 | 56 | [7] | |
| carb2 | GGGGCTTGATGCTCACT | |||||
| OXA-F | ACACAATACATATCAACTTCGC | blaOXA | 813 | 61 | [6] | |
| OXA-R | AGTGTGTTTAGAATGGTGATC | |||||
| Tetracycline | tetA up | GTAATTCTGAGCACTGTCGC | tetA | 937 | 62 | [6] |
| tetA low | CTGCCTGGACAACATTGCTT | |||||
| tetB1 | CTCAGTATTCCAAGCCTTTG | tetB | 416 | 62 | [6] | |
| tetB2 | CTAAGCACTTGTCTCCTGTT | |||||
| Chloramphenicol | cmlA1 | TGTCATTTACGGCATACTCG | cmlA | 455 | 55 | [6] |
| cmlA2 | ATCAGGCATCCCATTCCCAT | |||||
| Cat-F | GGTGAGCTGGTGATATGG | cat | 209 | 48 | [8] | |
| Cat-R | GGGATTGGCTGAGACGA | |||||
| FloR up | CACGTTGAGCCTCTATAT | floR | 868 | 55 | [6] | |
| FloR low | ATGCAGAAGTAGAACGCG | |||||
| Sulfamethoxazole | Sul1-F | TGGTGACGGTGTTCGGCATTC | sul1 | 789 | 63 | [6] |
| Sul1-R | GCGAAGGTTTCCGAGAAGGTG | |||||
| Sul2-F | CGGCATCGTCAACATAACC | sul2 | 722 | 50 | [6] | |
| Sul2-R | GTGTGCGGATGAAGTCAG | |||||
| Trimethoprim | dfr-1 up | GTGAAACTATCACTAATGG | dfrA1A, dfrA5, dfrA15, dfrA16 a | 474 | 55 | [9] |
| dfr-1 low | TTAACCCTTTTGCCAGATTT | |||||
| dfr-7 up | TTGAAAATTTCATTGATT | dfrA7, dfrA17 a | 474 | 55 | [9] | |
| dfr-7 low | TTAGCCTTTTTTCCAAATCT | |||||
| dfr-12 up | GGTGGCGCAGAAGATTTTTCGC | dfrA12, dfrA13 a | 319 | 60 | [9] | |
| dfr-12 low | TGGGAAGAAGGCGTCACCCTC | |||||
| Integrons | Integrase1-F | GGGTCAAGGATCTGGATTTCG | intl1 | 483 | 62 | [6] |
| Integrase1-R | ACATGGGTGTAAATCATCGTC | |||||
| Integrase2-F | CACGGATATGCGACAAAAAGGT | intl2 | 788 | 62 | [6] | |
| Integrase2-R | GTAGCAAACGAGTGACGAAATG |
Restriction fragment length polymorphism was performed after PCR [9].
For trimethoprim resistance genes, PCR products were additionally analysed by restriction fragment length polymorphism (RFLP) analysis as previously described by Navia et al. [9].
3. Results
3.1. Antibiotic resistance phenotypes
Escherichia coli strains (n = 369) were commonly resistant to ampicillin (80%), SXT (71%), tetracycline (56%), chloramphenicol (21%) and nalidixic acid (32%). Quinolone (nalidixic acid and ciprofloxacin) resistance was significantly higher in commensal strains than in DEC different strains (P < 0.05). EAEC and DAEC tended to present higher resistance levels than EPEC and ETEC for most used antibiotics. Multiresistance was found in 76% of EAEC and 90% of DAEC.
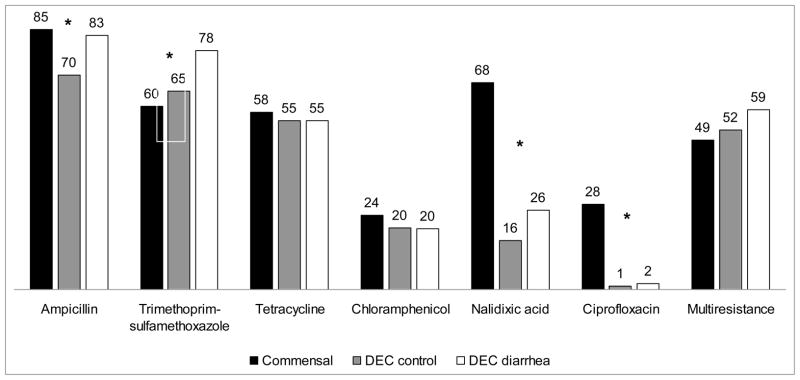
Resistance to ampicillin, SXT and nalidixic acid differed between commensals, DEC-control and DEC-diarrhoea groups (P < 0.05). Commensal strains were significantly more likely to be resistant to ampicillin, nalidixic acid and ciprofloxacin than DEC strains (Fig. 1). DEC-diarrhoea strains were significantly more commonly resistant to SXT compared with commensal strains and DEC-control (Fig. 1). Resistance rates for the other antibiotics evaluated were <5%, including resistance to third-generation cephalosporins.
Fig. 1.
Percentages of antibiotic resistance in commensal Escherichia coli (n = 74), diarrhoeagenic E. coli isolated from asymptomatic children (‘DEC-control’) (n = 94) and diarrhoeagenic E. coli isolated from children with diarrhoea (‘DEC-diarrhoea’) (n = 201). *P < 0.05 for the comparison between the three groups.
3.2. Antibiotic resistance mechanisms
The most prevalent genes were (Table 2): for β-lactam resistance, blaTEM present in 31% of strains (47/154); for SXT resistance, dfrA1 present in 18% (23/130) and sul2 present in 49% (64/130); for tetracycline resistance, tetA present in 26% (28/106); and for chloramphenicol resistance, cat present in 78% (35/45) of strains. Results were also analysed by group, i.e. commensal E. coli, DEC-control and DEC-diarrhoea. All studied molecular mechanisms of resistance present DEC groups were also present in the commensal group, except for dfr17 conferring trimethoprim resistance (Table 2). The sul1 and dfrA1 genes related to SXT resistance were more frequent in DEC-diarrhoea strains than in commensal and DEC-control groups (P < 0.05). Integrase 1 was more frequently found in the DEC-control group than in the other two groups (P < 0.05) (Table 2). Neither DEC-diarrhoea nor DEC-control groups were found to have the cmlA gene conferring chloramphenicol resistance (Table 2). Compared with the DEC-control group, DEC-diarrhoea strains tended to have higher rates of the majority of antibiotic resistance mechanisms, although these differences were not statistically significant (Table 2).
Table 2.
Frequency of genes related to antibiotic resistance in commensal Escherichia coli, diarrhoeagenic E. coli isolated from asymptomatic children (‘DEC-control’) and diarrhoeagenic E. coli isolated from diarrhoea episodes (‘DEC-diarrhoea’)
| Genes related to antibiotic resistance |
n/N (%)
|
|||
|---|---|---|---|---|
| Commensals | DEC-control | DEC-diarrhoea | ||
| β-Lactams | blaTEM | 10/36 (28) | 7/33 (21) | 30/85 (35) |
| blaSHV | 1/36 (3) | 1/33 (3) | 5/85 (6) | |
| blaCARB | 1/36 (3) | 2/33 (6) | 0/85 (0) | |
| blaOXA | 3/36 (8) | 1/33 (3) | 5/85 (6) | |
| SXT | sul1 a | 4/27 (15) | 3/29 (10) | 30/74 (41) |
| sul2 | 15/27 (56) | 15/29 (52) | 34/74 (46) | |
| dfrA1 a | 1/27 (4) | 1/29 (3) | 21/74 (28) | |
| dfrA7 | 1/27 (4) | 1/29 (3) | 5/74 (7) | |
| dfrA17 | 0/27 (0) | 1/29 (3) | 0/74 (0) | |
| dfrA12 | 1/27 (4) | 2/29 (7) | 1/74 (1) | |
| Tetracycline | tetA | 5/22 (23) | 4/23 (17) | 19/61 (31) |
| tetB | 5/22 (23) | 1/23 (4) | 13/61 (21) | |
| Chloramphenicol | cat | 7/10 (70) | 8/12 (67) | 20/23 (87) |
| floR | 3/10 (30) | 1/12 (8) | 4/23 (17) | |
| cmlA | 1/10 (10) | 0/12 (0) | 0/23 (0) | |
| Integrons | intl1 a | 3/36 (8) | 8/33 (24) | 3/85 (4) |
| intl2 | 1/36 (3) | 4/33 (12) | 5/85 (6) | |
SXT, trimethoprim/sulfamethoxazole.
P < 0.05 for difference between three groups.
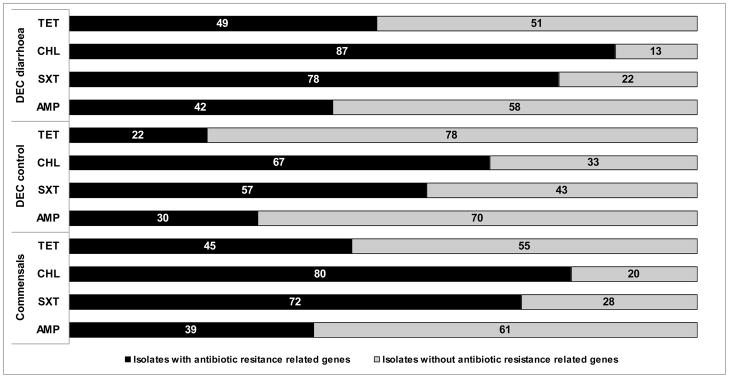
Significant proportions of antibiotic-resistant strains did not exhibit any of the resistance mechanisms tested for. For example, only 39% of ampicillin-resistant strains presented at least one of the β-lactamase genes searched for, and 42% of tetracycline-resistant strains had at least one gene related to mechanisms of tetracycline resistance. For chloramphenicol and SXT, the percentages of strains with at least one of the genes analysed were higher (80% and 72%, respectively) (Fig. 2).
Fig. 2.
Presence of antimicrobial resistance-related genes. Percentages of resistant Escherichia coli isolates (commensals, DEC-diarrhoea and DEC-control) that present at least one of the analysed genes related to mechanisms of antibiotic resistance for each antibiotic family. ‘DEC-control’, diarrhoeagenic E. coli isolated from asymptomatic children; ‘DEC-diarrhoea’, diarrhoeagenic E. coli isolated from children with diarrhoea; AMP, ampicillin; SXT, trimethoprim/sulfamethoxazole; CHL, chloramphenicol; TET, tetracycline.
4. Discussion
In this study, we found a high percentage of multiple antibiotic resistance in commensal strains that was probably due to frequent antibiotic use in the subjects. Antibiotics are often used in patients with severe enteritis. For dysenteric and persistent diarrhoea, antibiotic therapy is usually recommended [10]. However, in paediatric diarrhoea the risk benefit of antibiotic use is not fully defined. Previous studies suggest that commensal strains could be acting as antibiotic resistance reservoirs in the community [11]. In this report, commensal E. coli were more resistant to quinolones (nalidixic acid and ciprofloxacin) than DEC, although the use of this family of antibiotics in children in Peru is limited.
The DEC isolates were separated into DEC-control and DEC-diarrhoea. It is likely that these groups would have had different histories of antibiotic exposure, and previous data have shown that DEC-diarrhoea strains are associated with higher resistance rates [5]. However, few significant differences in antibiotic bacterial resistance rates were found in the three groups (including commensal isolates) for all the tested antibiotics, with the exceptions being nalidixic acid, ampicillin and SXT. Resistance to third-generation cephalosporins in commensal E. coli and DEC as well as resistance to ciprofloxacin in the DEC group were found to be low in Peru.
A total of 19 mechanisms of resistance and two integrases were searched in this study. The principal mechanisms of resistance found in commensal strains were cat (70%), sul2 (56%) floR (30%) and blaTEM (28%). The majority of these mechanisms are related to mobile elements of antibiotic resistance, which would be consistent with antibiotic exposure explaining their high prevalence in the study population [11]. A previous study in E. coli strains from healthy children in Spain reported the presence of a variety of β-lactamases in 24 ampicillin-resistant strains, such as blaTEM (83%), blaSHV (2%) and blaOXA-30 (2%). For tetracycline, tetA (57%), tetB (24%) and tetD (2%) were reported from 21 resistant isolates [12].
The high percentage of resistance to SXT correlates with the high prevalence of genes of antibiotic resistance to both antibiotics in the DEC-diarrhoea group. High levels of resistance to this combined antibiotic in commensal E. coli and DEC have been reported previously [13]. One given explanation was their widespread use in the treatment of diseases associated with Gram-negative bacteria, especially in children under 2 years of age with acute infectious diarrhoea [13].
In conclusion, the present study describes for the first time a comprehensive assessment of molecular mechanisms of antibiotic resistance in DEC isolated from children with diarrhoea and from healthy controls in a large number of strains. We were unable to detect the antibiotic resistance mechanisms in all of the strains analysed, especially for ampicillin- and tetracycline-resistant strains; among the principal mechanisms of resistance reported in E. coli that we did not search for are the different families of Gram-negative efflux pumps directly related to high multidrug resistance [14]. However, the fact that a large number of antibiotic resistance genes was demonstrated highlights the importance of continued surveillance studies, especially in developing areas such as Peru where the most commonly used antibiotics in children and adults [15] are available without prescription.
Acknowledgments
Funding: This study was partially supported by the Agencia Española de Cooperación Internacional (AECID), Spain, Programa de Cooperación Interuniversitaria e Investigación Científica con Iberoamérica (D/019499/08, D/024648/09 and D/030509/10) (TJO and JR). TJO is supported by the US National Institutes of Health Public Health Service award RO1-HDO67694-01A1. JR is supported by ‘Miguel Servet’ ISCIII (CP05/0130).
Footnotes
Competing interests: None declared.
Ethical approval: Ethical approval was given by the Institutional Ethics Committee of Universidad Peruana Cayetano Heredia (Lima, Peru) (project no. 57758).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boschi-Pinto C, Velebit L, Shibuya K. Estimating child mortality due to diarrhoea in developing countries. Bull World Health Organ. 2008;86:710–7. doi: 10.2471/BLT.07.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ochoa TJ, Ruiz J, Molina M, Del Valle LJ, Vargas M, Gil AI, et al. High frequency of antimicrobial drug resistance of diarrheagenic Escherichia coli in infants in Peru. Am J Trop Med Hyg. 2009;81:296–301. [PMC free article] [PubMed] [Google Scholar]
- 4.Bartoloni A, Pallecchi L, Fiorelli C, Di Maggio T, Fernandez C, Villagran AL, et al. Increasing resistance in commensal Escherichia coli, Bolivia and Peru. Emerg Infect Dis. 2008;14:338–40. doi: 10.3201/eid1402.070138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochoa TJ, Ecker L, Barletta F, Mispireta ML, Gil AI, Contreras C, et al. Age-related susceptibility to infection with diarrheagenic Escherichia coli among infants from periurban areas in Lima, Peru. Clin Infect Dis. 2009;49:1694–702. doi: 10.1086/648069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sáenz Y, Briñas L, Domínguez E, Ruiz J, Zarazaga M, Vila J, et al. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother. 2004;48:3996–4001. doi: 10.1128/AAC.48.10.3996-4001.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arlet G, Philippon A. Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB) [corrected] FEMS Microbiol Lett. 1991;66:19–25. doi: 10.1016/0378-1097(91)90414-6. [DOI] [PubMed] [Google Scholar]
- 8.Sunde M, Norstrom M. The prevalence of, associations between and conjugal transfer of antibiotic resistance genes in Escherichia coli isolated from Norwegian meat and meat products. J Antimicrob Chemother. 2006;58:741–7. doi: 10.1093/jac/dkl294. [DOI] [PubMed] [Google Scholar]
- 9.Navia MM, Ruiz J, Sanchez-Cespedes J, Vila J. Detection of dihydrofolate reductase genes by PCR and RFLP. Diagn Microbiol Infect Dis. 2003;46:295–8. doi: 10.1016/s0732-8893(03)00062-2. [DOI] [PubMed] [Google Scholar]
- 10.Pickering LK. Antimicrobial resistance among enteric pathogens. Semin Pediatr Infect Dis. 2004;15:71–7. doi: 10.1053/j.spid.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Bailey JK, Pinyon JL, Anantham S, Hall RM. Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J Med Microbiol. 2010;59:1331–9. doi: 10.1099/jmm.0.022475-0. [DOI] [PubMed] [Google Scholar]
- 12.Domínguez E, Zarazaga M, Sáenz Y, Briñas L, Torres C. Mechanisms of antibiotic resistance in Escherichia coli isolates obtained from healthy children in Spain. Microb Drug Resist. 2002;8:321–7. doi: 10.1089/10766290260469589. [DOI] [PubMed] [Google Scholar]
- 13.Garcia PG, Silva VL, Diniz CG. Occurrence and antimicrobial drug susceptibility patterns of commensal and diarrheagenic Escherichia coli in fecal microbiota from children with and without acute diarrhea. J Microbiol. 2011;49:46–52. doi: 10.1007/s12275-011-0172-8. [DOI] [PubMed] [Google Scholar]
- 14.Nikaido H. Antibiotic resistance caused by Gram-negative multidrug efflux pumps. Clin Infect Dis. 1998;27(Suppl 1):S32–41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 15.Wirtz VJ, Dreser A, Gonzales R. Trends in antibiotic utilization in eight Latin American countries, 1997–2007. Rev Panam Salud Publica. 2007;27:219–25. doi: 10.1590/s1020-49892010000300009. [DOI] [PubMed] [Google Scholar]