Abstract
The complexity of the human body derives from numerous modular building blocks assembled hierarchically across multiple length scales. These building blocks, spanning sizes ranging from single cells to organs, interact to regulate development and normal organismal function but become disorganized during disease. Here, we review methods for the bottom-up and directed assembly of modular, multicellular, and tissue-like constructs in vitro. These engineered tissues will help refine our understanding of the relationship between form and function in the human body, provide new models for the breakdown in tissue architecture that accompanies disease, and serve as building blocks for the field of regenerative medicine.
Keywords: Bottom-up, programmed assembly, tissue engineering, cell-cell nteractions, paracrine signaling
Investigating the relationship between tissue form and function with in vitro engineered tissues
Human beings contain trillions of cells spanning over 200 specialized subtypes. This complex cellular community grows within a web of extracellular matrix (ECM) to form the tissues and organs that perform the numerous functions of our bodies. Cells, tissues, and organs constitute a hierarchy of structures spanning tens of microns to meters, in which the arrangement of building blocks at one scale forms the building block for the next. A major goal of cell and developmental biology is to delineate how the form of the body – or the composition and physical organization of its building blocks – affects function at the level of tissues, organs, or the whole organism. However, this remains a challenging goal because direct and general methods for controlling the relative spatial position of cells in tissues and organs do not exist.
One powerful approach for elucidating fundamental principles relating form to function is an engineering strategy that builds tissue-like structures ex vivo. Like studies in model organisms, tissue-engineering strategies can incorporate genetically modified cellular building blocks. Unlike studies using model organisms, however, tissue-engineering strategies are also compatible with the use of primary or immortalized human cells, advanced imaging techniques, and techniques that control the non-cellular components of the microenvironment. Most tissue engineering approaches are “top-down” and require the use of patterned substrates, molds, or ECM scaffolds to assist cells in finding their appropriate positions and differentiation states within a tissue. In principle, a three-dimensional (3D) scaffold of ECM with the precise composition and organization can provide all the necessary structural and microenvironmental cues to direct the organization of individual cells into a functional tissue or organ, as evidenced by recent experiments using decellularized organs [1]. However, de novo construction of scaffolds with the requisite level of detail at all length scales is not currently possible. As a consequence, tissue reconstruction starting from cells or cell aggregates remains challenging because mixtures of dissociated cells do not typically reconstitute complex tissue structures or functions without pre-organization into the correct 3D geometry. Therefore, additional means of controlling the spatial organization of cells or groups of cells will facilitate tissue engineering.
Bottom-up or synthetic approaches are emerging as a valuable and alternative means to more prevalent top-down approaches for pre-organizing groups of cells into tissue-like structures. Bottom-up approaches are distinct from top-down approaches in that they link together simplified building blocks to generate objects that are structurally organized at larger length scales [2]. Directing the assembly of building blocks from the bottom-up may provide enhanced control over the relative spatial arrangement of cells in engineered tissues when used together with currently available top-down approaches. In addition to the advantages of the top-down tissue engineering strategies outlined above, bottom-up methods have several other desirable features. First, they are inherently modular, allowing for the simple replacement of specific cells or nodes in a network of interacting cells, tissues, or organs (Box 1). This feature makes bottom-up engineering attractive as a versatile method for incorporating multiple cell types into tissues as well as for building different tissue types or states (for example functional or pathological) by interchanging building blocks. Further, these methods are inherently scalable; a large number of nearly identical tissue constructs can be prepared without the need for complex or specialized scaffolds. Finally, bottom-up approaches are ideally suited for studying the direct interactions between individual building blocks. Recent research has highlighted the importance of interactions between heterogeneous cell types on tissue behaviors, whether the interactions occur within an epithelium [3–6], between the epithelium and surrounding stroma [7], or even between cells in different organ systems [8]. While spatially organizing multiple heterogeneous cellular interactions can be challenging using top-down tissue engineering approaches, a multiplicity of interacting partners can be systematically incorporated using a modular, bottom-up approach.
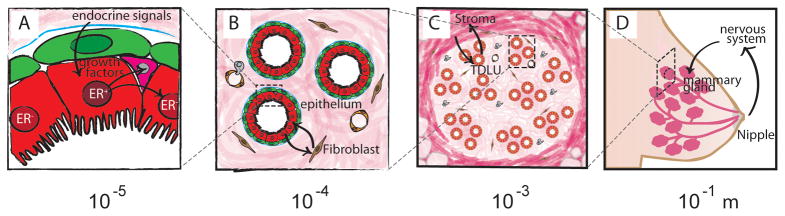
BOX. The hierarchical organization of a modular organ: the breast.
The human breast contains an organized hierarchy of structures built up from modular units, from nanometer-sized proteins of the basement membrane to micron-sized cells to millimeter-sized tissues [79]. The bilayered epithelium of the mammary gland, for example, has two principle building blocks: luminal epithelial and myoepithelial cells. Considerable heterogeneity exists even within each of these cell types. For example, sub-populations of luminal cells express estrogen and progesterone receptors. When stimulated, these cells release growth factors triggering the growth of their neighbors. In addition, luminal and myoepithelial cells play distinct functional roles, serving to secrete and pump milk, respectively (Figure Ia). These cellular building blocks are organized into ducts and acini that are further supported by fibroblasts that synthesize and reside in a collagenous ECM. Endothelial cells provide additional support for these structures through a meshwork of capillaries delivering nutrients, facilitating circulation of lymphocytes and relaying hormones from distant organs (Figure Ib). Ducts and acini are further organized into terminal ductal lobular units (TDLUs) that are surrounded by a secondary and specialized ECM containing a denser collagenous matrix and beds of adipocytes that add additional form to the organ (Figure Ic) [79]. Finally, TDLUs are organized into multiple lobes that drain into large ducts, together delivering milk to the nipple [81] (Figure Id). Though the overall architecture of the gland is drastically remodeled over the course of a woman’s lifetime, the relative position of the different cell types with respect to each other and the modular organization of the organ remain constant in healthy tissue.
For this modular and hierarchically organized tissue to function, mechanical, chemical and electrical signals must be detected by individual cells and transmitted across each hierarchy of the organ to synchronize cellular behaviors. Single epithelial cells integrate chemical and mechanical cues from the basement membrane, a specialized component of the ECM, to maintain cell polarity and secrete milk [82]. Epithelial cells are also mechanically coupled with their neighbors and distant tissues via the cytoskeleton and ECM, respectively [83, 84]. The cytosols of epithelial cells are chemically and electrically linked through gap junctions [85], while an array of secreted and membrane-localized signaling proteins coordinate tissue homeostasis and function across the epithelium. Additionally, secreted endocrine factors link the mammary gland with stromal adipocytes, the nervous system, and other reproductive organs [86]. Processes such as breastfeeding require not only coordination between multiple cell types and modules within the breast but also between sensory cells in the nervous system, which further synchronize actions in distant organs to those in the breast. While it is appreciated that overlapping signals across this hierarchy of building blocks are required for the higher order function of the breast and all other tissues, the structural organization of building blocks that serves as the foundation for the proper exchange of intercellular signaling events remains challenging to study. By spatially pre-organizing cells, tissues and organs, bottom-up and directed tissue engineering strategies aim to understand and control the exchange of signals between building blocks at all levels of structural hierarchy in the human body.
This review focuses on bottom-up and directed assembly approaches that utilize pre-defined building blocks to construct spatially defined multicellular structures containing more than one cell type. The goal of these methods is to mimic the cellular heterogeneity and physical arrangement of the modular repeating units found in mammalian tissues (Figure 1) by directing their assembly from simpler building blocks. This review will not focus on other techniques that control the spatial organization of tissues through organ printing, microscale technologies, genetic or optogenetic techniques, directed differentiation of stem cell or progenitor sources, or synthetically engineered genetic circuits. The reader is directed to several recent reviews for discussion of these topics [9–14].
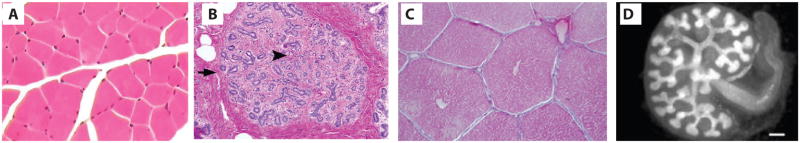
Figure 1.
Modular functional units in mammalian organs. (a) Human skeletal muscle cross section (b) Human mammary terminal ductal lobular unit (TDLU) cross section (c) Pig liver cross section showing repeating lobules (d) Mouse embryonic kidney. Reproduced from [79], Pathpedia, Werning, S., 2007 (http://calphotos.berkeley.edu/cgi/img_query?seq_num=223971&one=T), and [80] with permission.
Directing the bottom-up assembly of tissues using single cells as building blocks
Control over the relative position of single cells provides the fine resolution necessary for probing interactions between neighboring cells in a tissue. This level of spatial resolution is required for recreating stem cell niches [15] or when rebuilding cell-cell connections found in fully differentiated tissues [16]. In some cases, a group of cells has the ability to self-assemble into specific structures at these length scales. Townes and Holtfreter famously found that dissociated cells from amphibian embryos would aggregate and self-sort into germ layers without outside intervention [17]. This strategy occasionally allows a multiplicity of cell types to self-organize in wells or in hanging drops [18–20]. However, isolated mixtures of cells of multiple types do not always spontaneously organize into structures that mimic their tissue of origin without the aid of external ECM scaffolds. In the absence of these external positioning cues, bottom-up and directed assembly techniques may be used to spatially position cells in relation to each other at the microscale.
DNA-programmed assembly is a recently developed approach to direct the organization of multicellular structures in vitro with single-cell resolution [21]. Key to this approach is the covalent or non-covalent remodeling of the adhesive properties of the cell surface with single-stranded DNA (ssDNA). ssDNA is linked to the cell surface by several means. In one approach, cells are first cultured in the presence of an azide-modified monosaccharide that is incorporated into cell surface glycans. The accessible azides react with chemically modified oligonucleotides by Staudinger ligation [22] or [1,3]-dipolar cycloaddition [21] to covalently attach the DNA to the glycocalyx. In an alternative approach, N-hydroxysuccinimide-modified DNA is added to cell suspensions where it reacts covalently with free lysines on the cell surface [23]. Lastly, lipid-DNA conjugates are added to culture medium where they passively partition into the cell membrane for non-covalent cell surface modification [24–26]. Labeling or binding other interacting biomolecules to cell surfaces will also direct the programmed assembly of cells, though these are often limited to only single pairs of interacting molecules [27–30].
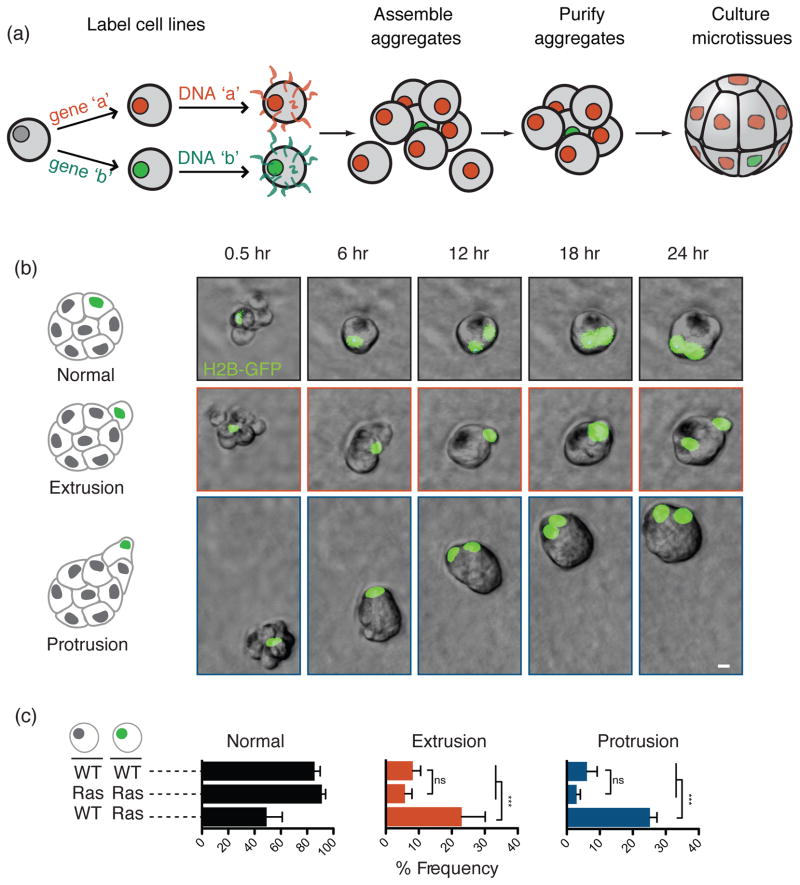
Mixing different cell populations labeled with complementary ssDNA strands or molecules directs the formation of heterogeneous microtissues, while changing the ratios of labeled populations can be used to achieve discrete multicellular arrangements (Figure 2a). This strategy has been used to recapitulate synthetic paracrine signaling networks [21] and to mimic immune cell homing to sites of inflammation [28]. More recently, a DNA-mediated programmed assembly approach was used to investigate the consequences of cell-to-cell variability among mammary epithelial cells during the dynamic process of morphogenesis [31]. Wild-type MCF10A mammary epithelial cells and derivatives with elevated Ras activation were labeled with ssDNA and combinatorially assembled to form homogeneous and mosaic microtissues of defined compositions. Mosaic epithelial aggregates assembled from single Ras-activated cells and wild-type MCF10A neighbors displayed emergent behaviors: Ras-activated cells were basally extruded or led motile multicellular protrusion that directed the motility of the surrounding WT cells across tens to hundreds of microns. Neither phenotype was observed in aggregates homogeneous for Ras-activated cells nor in aggregates assembled from single WT MCF10A cells with Ras-activated neighbors (Figure 2b and 2c). Because this method controls the initial architecture of aggregates, the emergent phenotypes could be directly attributed to the underlying cell-to-cell variability in Ras activity.
Figure 2.
DNA-programmed assembly. (a) Fluorescent epithelial cells labeled with complementary ssDNA (or other interacting molecules) are brought together through molecular recognition. Mixing cell populations 1-to-50 results in discrete multicellular aggregates that can be purified using fluorescence activated cell sorting. Assembled aggregates are then cultured in laminin-rich ECM to form polarized microtissues. Genetically distinct input cells can be incorporated to build mosaic aggregates. (b) Mosaic microtissues assembled from single H2B-GFP-expressing MCF10AT cells, which express low levels of H-RasV12, and wild-type MCF10A neighbors display emergent behaviors that are not observed in homogeneous assemblies. (scale bar, 10 μm). (c) Quantification of the emergent behaviors in homogeneous and mosaic aggregates. Mean values of great than 500 observations are displayed, with error bars representing the standard deviation of the mean. Reproduced from [21] with permission.
DNA-programmed assembly of cellular building blocks enables the study of cell-cell interactions in the context of multicellular structures with precise spatial arrangements. Because essentially any cell type can be modified with ssDNA using the various DNA-labeling methods outlined above, and a nearly unlimited set of orthogonal DNA sequences is available, DNA-programmed assembly can be used to reconstitute a variety of complex heterotypic cell-cell interactions for study. Importantly, unlike in genetic engineering, the DNA used to program cellular interactions is temporary and degrades rapidly at 37°C to leave unmodified interacting cells [23, 24]. This technique closely apposes interacting cell surfaces and is ideally suited for the study of multicellular circuits that operate over short distances. Interactions amenable to study with this approach include those mediated by electrical and chemical signals through gap junctions [32], short-range mechanical signals coupled to the cytoskeleton [33], juxtacrine signaling such as through the Notch pathway [34], and short-range paracrine signaling such as through the Wnt and Hedgehog pathways [35, 36]. However, because products of microscale cell-cell assembly have been limited to around 100 microns in diameter, cell-cell interactions that occur between neighboring or distant tissues or tissue compartments have evaded study using this strategy alone. Access to these larger structures might be achieved by combining programmed assembly with top-down techniques or by using the products of programmed assembly themselves as building blocks for further elaboration.
Directing the bottom-up assembly of tissues using cell sheets and aggregates as building blocks
Many tissues and organs contain repetitive subunits consisting of groups of cells with dimensions of hundreds of microns to a millimeter. Such structures include pancreatic islets, lymph nodes, and the lobules of the breast and liver. The majority of cells within these repeating units are fully differentiated and structurally integrated with their neighbors and ECM. Therefore, approximating these units with cell aggregates that contain fully formed cell-cell junctions may provide a means of constructing tissues at larger length scales than can be achieved from assembly of single cells alone.
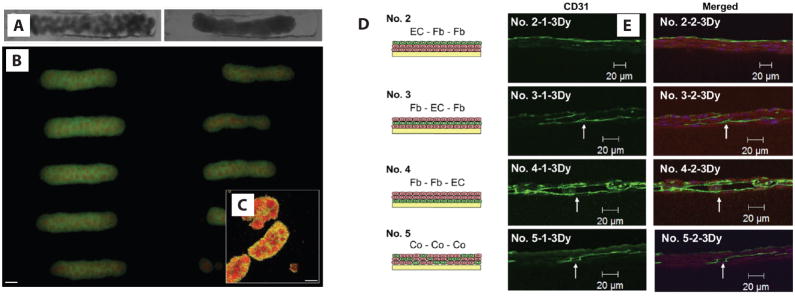
Spherical cell aggregates of the appropriate size have been engineered using top-down molding techniques and then assembled within microwells into larger structures of various shapes and sizes [37–40]. Such an approach has been used to combine aggregates of human fibroblasts and rat hepatoma cells. When pre-cultured together in spheroids, these two cell types self-sort into an inner core of fibroblasts surrounded by hepatoma cells. When directed to assemble in rectangular molds, these pre-cultured spheroidal building blocks fuse into a rod but maintain the fibroblast cores for at least 24 hours [39] (Figure 3a–c). Directed assembly with globular cell aggregates has only been demonstrated with modules of a single type, though the same basic strategy should be applicable to modules made of different cell types. However, the overall architecture in large assemblies of cell aggregates may require further structural support from the microenvironment to be stable over the long term [38].
Figure 3.
Directed assembly of cell aggregates. (a) Pre-formed, spheroidal cell aggregates (100 μm in diameter) assembled in trough-shaped wells fused into rod-shaped structures over 24 hours. (b) Pre-cultured heterogeneous spheroids with a core of human fibroblasts (red) and surrounding rat hepatoma (green) cells cultured in wells for 24 hours. (c) Confocal image shows that the assembled spheroids retained the inner fibroblast core during spheroid fusion (scale bars, 200 μm). (d) Different combinations of three cell sheet layers are stacked to make heterogeneous tissues of endothelial (green) and fibroblast (pink) cells. (e) After 3 days, endothelial sheets formed vessels (outlined with indirect CD31 staining in green) when stacked within or under fibroblast sheets (actin staining in red in merged images; scale bars, 20 μm). Reproduced from [39] and [50] with permission.
Cell sheets have also been used as building blocks and can be manually stacked to build macroscopic tissue structures. Sheets are cultured in suspension or released using thermal- or ion-sensitive coatings [41–43]. Single sheets have been made from a variety of cell types, including fibroblasts, endothelial cells, and epithelial cells [44, 45]. Cell-cell adhesions and secreted ECM components are maintained in lifted sheets [46, 47], which can also promote tissue–like functions that are not observed with dissociated cells [48]. 3D tissue constructs comprising a variety of cell types can be prepared by manually stacking different cell sheets. For instance, myoblast cell sheets intercalated with either layers, lines, or dissociated HUVEC cells [49–51] (Figure 3d and 3e) formed vascular networks in vitro, which could integrate with host vasculature in vivo when grafted subcutaneously into rats [51].
By starting with multicellular building blocks with an established geometry, directed assembly of cell aggregates of different cell types can be used to design and spatially position tissue-sized networks of interacting cells. Higher order structures are amenable to further elaboration with layers of individual cells. For example, pancreatic islets isolated from rodents were labeled with lipid-modified DNA and surrounded with mammalian cells bearing complementary DNA to provide a barrier function to the sensitive cell aggregates [52]. In principle, such a strategy could also introduce components of the surrounding tissue stroma to provide additional trophic support between modular units. The modular organization of assembled building blocks may also be stratified with exogenous ECM-like components to mimic tissue architectures of increasing complexity.
Directing the bottom-up assembly of tissues using cell-laden hydrogels as building blocks
The minimal functional unit of many tissues consists of small groups of structurally organized cells embedded in specialized ECM. Collections of these functional units are further organized in 3D space within additional ECM that is frequently of a different composition. The terminal ductal lobular units (TDLUs) and the intervening adipocyte-rich regions of the mammary gland exemplify this organization (Box 1). These structures should be modeled with explicit inclusion of spatially segregated ECM-like materials in building blocks, particularly when matrix components are critical to cell and tissue function or when the cells cannot generate the volume and composition of ECM found in biological tissues on their own. Fortunately, many natural and synthetic polymers are available for designing ECM-like hydrogels with specific structural and mechanical properties to mimic different tissues within the human body [53, 54]. While ECM-like materials can be incorporated into tissues by mixing microspheres of hydrogel with individual cells [55], cell-laden hydrogel modules with dimensions of hundreds of microns to millimeters are closer in size to the ECM-encapsulated repeating units found in many tissues in vivo. Therefore, these may be a more tractable building block for constructing interactions between repeating units within tissues or organs.
The straightforward packing of modular cell-laden hydrogel units into a defined space can direct their assembly into larger structures. Such an approach was used with building blocks of cell-laden and collagen-based cylinders covered with an endothelial cell layer [56, 57]. The authors assembled multiple endothelial-coated cylinders containing rat myocardiocytes onto a planar nylon filter then added temporary alginate glue. The resulting modular tissue formed a sheet-like structure which contracted upon electrical stimulation [56]. Collagen hydrogels containing different cell types have also been assembled into a linear array using microfluidics to constrain hydrogel orientation [58].
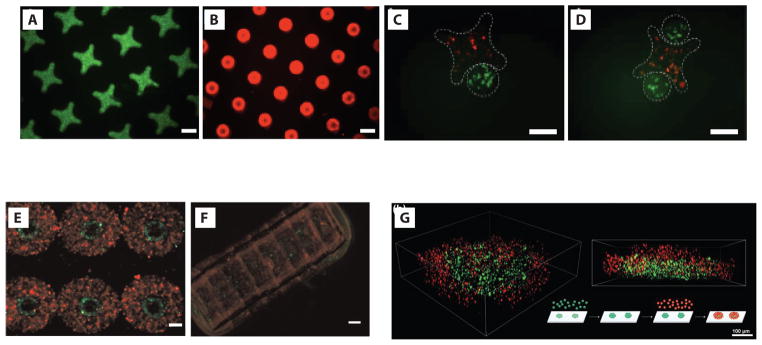
In contrast to the random but constrained assembly of subunits, directed assembly schemes can guide the formation of a multiplicity of hydrogel building blocks into more defined structures. Assembling sub-millimeter-sized polyethylene glycol (PEG)-based hydrogels in hydrophobic media, for example, promotes the self-association of the hydrophilic objects. Hydrogel building blocks containing two different cell types can be prepared by sequential photolithography steps which meld two concentric rings of cell-laden PEG hydrogel together into a single unit (Figure 4a). Additional assembly of these two-component building blocks into linear arrays generated tubes containing an inner layer of endothelial cells surrounded by smooth muscle cells that mimicked the architecture of vasculature [59] (Figure 4b). Another way to generate spatially defined heterogeneous tissues using this directed assembly method is to mix two different populations of hydrogels containing different cell types; changing the ratio of these two hydrogel monomers can bias the composition of the end products [60]. Nanofibers added to hydrogel monomers can provide additional mechanical coupling between modules, facilitating cell differentiation and tissue function [61]. An additional level of control over the interface between two populations of hydrogel-embedded cells can be added by using gels with lock-and-key designs (Figure 4c and 4d) to access precise architectures with controlled stoichiometries of building blocks [60] (Figure 4e and 4f).
Figure 4.
Directed assembly of cell-laden hydrogels. (a) Donut-shaped hydrogels with an inner hydrogel ring loaded with endothelial cells (green) surrounded by an outer ring loaded with smooth muscle cells (red) were made using sequential photolithography steps. (b) Side view of tubular structure formed after sequential assembly of hydrogel units from a (scale bars, 100 μm). (c, d) Lock-and-key (cross- and rod-shaped) hydrogels stained with fluorescent dextran were made using photolithography. (e, f) Lock-and-key hydrogels loaded with fluorescent murine fibroblast cells assembled through self-association in a hydrophobic medium (scale bars, 200 μm). (g) 3D volume reconstruction of DNA-directed hydrogel assemblies. Spherical hydrogels bearing green fluorescent tracking beads and labeled with ssDNA were bound to a microarray template. A second layer of hydrogels loaded with red beads and labeled with complementary DNA was assembled onto the first population (Scale bar, 100 μm). Reproduced from [59], [60], and [62] with permission.
Similar to cellular building blocks, hydrogels can also be labeled with ssDNA or other biomolecules to program their assembly. Such an approach avoids the necessity to fabricate hydrogel units of complex shape but requires additional chemical labeling steps either before or after fabrication. A two-step approach was used to label uniform, spherical PEG-based hydrogel units with streptavidin, followed by conjugation with biotinylated ssDNA [62]. Additional control of building block positioning was demonstrated by annealing the ssDNA- hydrogels onto microarray templates or by directed assembly of two populations of hydrogels bearing complementary ssDNA on their surfaces (Figure 4g). Recent reports suggest that additional control over interfacial interactions between hydrogel building blocks using non-biological molecules could add further complexity to tissues engineered at this length scale [53, 63].
Due to advances in top-down fabrication techniques, hydrogel building blocks of various sizes, shapes, and compositions are readily designed to mimic architectures and interfaces observed in tissues in vivo. The density of cells within the hydrogel units can also be controlled, though achieving tissue-like cell densities may require extended periods of culture [58]. Because cells are physically constrained within the building blocks, cell-laden hydrogels may be especially appropriate for studying the exchange of soluble factors between different cell populations [55]. Moreover, hydrogel compositions can be adjusted using biologically derived or synthetic polymers [64, 65] designed to match the physical and chemical properties of ECM of specific tissues, or to probe the consequence of changing ECM properties on cellular behaviors. Such control would be especially useful for modeling tissues that contain multiple compartments with different matrices and cell types such as between neighboring TDLUs in the breast. Finally, cell-laden hydrogels of various levels of complexity can be implanted in vivo. In one example, gel-encapsulated cells coated with an endothelial layer show enhanced survival and differentiation, as well as a favorable host response to growth factors secreted by the encapsulated cells [66–68].
Considerations
The techniques outlined here are designed to direct the assembly of cells into a specific position relative to other cells in the context of a multicellular tissue. In order to form a functional tissue, however, the cells incorporated into building blocks must also retain the capacity to interact properly with their neighbors and to deposit and remodel their own extracellular matrix. Unfortunately, established cell lines propagated in two-dimensional culture are frequently used as building blocks in tissue engineering applications. In these cell lines, tissue-specific gene expression patterns and cell surface proteins necessary for directing heterotypic cell-cell interactions are often down-regulated or lost [69, 70]. In fact, even primary cells can begin to lose markers of differentiation after brief growth on tissue-culture plastic [71, 72]. Therefore, care must be taken to validate that the cell types used in an engineered tissue retain expression of appropriate markers of differentiation and the ability to interact with neighboring cell populations.
Optimization of other components in the microenvironment may also be required for assembled cells to transition to tissue-like organization [73]. For instance, ECM components and soluble factors in tissues can be extremely dynamic [74, 75] and may differ substantially from formulations typically used in culture. Because the chemical, mechanical, and structural organization of ECM can affect tissue and cell behaviors [76, 77], use of smart materials with adjustable and controllable properties, such as encapsulating hydrogels, could allow for dynamic control of the ECM and aid the proper morphogenesis of the engineered construct [78]. Additionally, secreted molecules can be integrated into the design of tissue modules for gradual release [21, 67, 68], or the necessary secreted factors could be delivered and controlled externally such as through microfluidics or photo-uncaging.
Concluding remarks
The applications of bottom-up strategies for directing the assembly of specific tissue architectures are still in their infancy, but they have potential to address important questions relating cellular organization to the coordination of multicellular behaviors. At the spatial resolution of single cells, directed assembly strategies will aid the study of stem cell niches from multiple tissue types, with respect to both their structure and composition. These methods will also benefit the study of cell-to-cell variability in gene expression or pathway activation in normal and diseased tissues. Finally, these methods will provide a means of studying the coordination of cellular behaviors during processes such as morphogenesis and tissue repair. At the larger spatial resolution of cell aggregates and whole tissues, directed-assembly strategies will impact the field of regenerative medicine as well as the study of mechanical and chemical coupling of cell groups, particularly between epithelial cells and the components of the stroma.
Numerous opportunities exist for improving the directed assembly of tissues. One avenue of interest is in the combination of the various approaches described above to direct the hierarchical assembly of tissues with spatial precision across multiple length scales, spanning that of single cells to full organs. Some progress along these lines has been made [52] or would be a logical extension of existing work [62]. Another opportunity will be in directing the assembly of anisotropic or asymmetric tissue structures. Finally, introducing genetic circuits into modules to control interactions between neighboring cells or tissues will provide additional mechanisms for engineering the processes of development and morphogenesis. We anticipate that progress towards these challenges, better integration with top-down engineering techniques, and new applications will be forthcoming in this exciting area.
Box Figure I.
The modular and hierarchical organization of the human mammary gland. (a) Individual glandular epithelial cells exchange signals with each other and the basement membrane. (b) Epithelial cells of the ducts and acini also exchange signals with the surrounding lobular stroma. (c) Ducts and acini are organized into terminal ductal lobular units (TDLUs) that are embedded in a second type of collageneous ECM and that also contains many adipocytes. (d) The entire organ is integrated with the rest of the body to mediate its function in delivering milk during breast-feeding.
Table.
Features, advantages, and opportunities for directed assembly approaches.
| Building block | Major advantage | Size of products (relevant biological structures) | Current applications | Challenges and opportunities |
|---|---|---|---|---|
| Cell | Single cell spatial resolution | 10–100 μm (groups of cells, niches) | Study of cell-cell interactions across short length scales Study of heterotypic cell-cell interactions Drug screening |
Assembling more complex tissues Incorporation of products into multiscale tissues Control of product architecture over time |
| Cell aggregate or sheet | Mature cell-cell junctions | 100 μm - 1+ mm (functional tissue units) | Study of cell aggregation Study of interactions between cell populations Production of engineered and functional tissues |
Control of product architecture over time Nutrient delivery and/or vascularization |
| Cell-laden hydrogel | Incorporation of specialized ECM or ECM-like material Easily interfaced with microscale engineering approaches |
100 μm – 1+ mm (functional tissue units or multicomponent tissues) | Study of paracrine signaling Generation of tissue engineering constructs |
Matching matrix properties to in vivo tissue Nutrient delivery and/or vascularization Responsive or smart hydrogels |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ott HC, et al. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nature medicine. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 2.Elbert DL. Bottom-up tissue engineering. Current opinion in biotechnology. 2011;22:674–680. doi: 10.1016/j.copbio.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogan C, et al. Characterization of the interface between normal and transformed epithelial cells. Nature cell biology. 2009;11:460–467. doi: 10.1038/ncb1853. [DOI] [PubMed] [Google Scholar]
- 4.Leung CT, Brugge JS. Outgrowth of single oncogene-expressing cells from suppressive epithelial environments. Nature. 2012;482:410–413. doi: 10.1038/nature10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenhoffer GT, et al. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnston LA. Competitive interactions between cells: death, growth, and geography. Science. 2009;324:1679–1682. doi: 10.1126/science.1163862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelhardt JJ, et al. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer cell. 2012;21:402–417. doi: 10.1016/j.ccr.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeNardo DG, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mironov V, et al. Organ printing: tissue spheroids as building blocks. Biomaterials. 2009;30:2164–2174. doi: 10.1016/j.biomaterials.2008.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaji H, et al. Engineering systems for the generation of patterned co-cultures for controlling cell-cell interactions. Biochimica et biophysica acta. 2011;1810:239–250. doi: 10.1016/j.bbagen.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toettcher JE, et al. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nature methods. 2011;8:35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miesenbock G. Optogenetic control of cells and circuits. Annual review of cell and developmental biology. 2011;27:731–758. doi: 10.1146/annurev-cellbio-100109-104051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nature cell biology. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenas P, et al. Developmental engineering: a new paradigm for the design and manufacturing of cell-based products. Part I: from three-dimensional cell growth to biomimetics of in vivo development. Tissue engineering Part B, Reviews. 2009;15:381–394. doi: 10.1089/ten.TEB.2008.0575. [DOI] [PubMed] [Google Scholar]
- 15.Losick VP, et al. Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Developmental cell. 2011;21:159–171. doi: 10.1016/j.devcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai RA, et al. Cell polarity triggered by cell-cell adhesion via E-cadherin. Journal of cell science. 2009;122:905–911. doi: 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Townes PL, Holtfreter J. Directed movement and selective adhesion of embryonic amphibian cells. J Exp Zool. 1955;128:53–120. doi: 10.1002/jez.a.114. [DOI] [PubMed] [Google Scholar]
- 18.Kunz-Schughart LA, et al. Potential of fibroblasts to regulate the formation of three-dimensional vessel-like structures from endothelial cells in vitro. American journal of physiology Cell physiology. 2006;290:C1385–1398. doi: 10.1152/ajpcell.00248.2005. [DOI] [PubMed] [Google Scholar]
- 19.Wenger A, et al. Development and characterization of a spheroidal coculture model of endothelial cells and fibroblasts for improving angiogenesis in tissue engineering. Cells, tissues, organs. 2005;181:80–88. doi: 10.1159/000091097. [DOI] [PubMed] [Google Scholar]
- 20.Foty R. A simple hanging drop cell culture protocol for generation of 3D spheroids. Journal of visualized experiments: JoVE. 2011 doi: 10.3791/2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gartner ZJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandra RA, et al. Programmable cell adhesion encoded by DNA hybridization. Angew Chem Int Ed Engl. 2006;45:896–901. doi: 10.1002/anie.200502421. [DOI] [PubMed] [Google Scholar]
- 23.Hsiao SC, et al. Direct cell surface modification with DNA for the capture of primary cells and the investigation of myotube formation on defined patterns. Langmuir: the ACS journal of surfaces and colloids. 2009;25:6985–6991. doi: 10.1021/la900150n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selden NS, et al. Chemically programmed cell adhesion with membrane-anchored oligonucleotides. Journal of the American Chemical Society. 2012;134:765–768. doi: 10.1021/ja2080949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teramura Y, et al. Control of cell attachment through polyDNA hybridization. Biomaterials. 2010;31:2229–2235. doi: 10.1016/j.biomaterials.2009.11.098. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, et al. Membrane anchored immunostimulatory oligonucleotides for in vivo cell modification and localized immunotherapy. Angew Chem Int Ed Engl. 2011;50:7052–7055. doi: 10.1002/anie.201101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, et al. Targeted cell-cell interactions by DNA nanoscaffold-templated multivalent bispecific aptamers. Small. 2011;7:1673–1682. doi: 10.1002/smll.201002292. [DOI] [PubMed] [Google Scholar]
- 28.Zhao W, et al. Mimicking the inflammatory cell adhesion cascade by nucleic acid aptamer programmed cell-cell interactions. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2011;25:3045–3056. doi: 10.1096/fj.10-178384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutta D, et al. Synthetic chemoselective rewiring of cell surfaces: generation of three-dimensional tissue structures. Journal of the American Chemical Society. 2011;133:8704–8713. doi: 10.1021/ja2022569. [DOI] [PubMed] [Google Scholar]
- 30.Hamon M, et al. Avidin-biotin-based approach to forming heterotypic cell clusters and cell sheets on a gas-permeable membrane. Biofabrication. 2011;3:034111. doi: 10.1088/1758-5082/3/3/034111. [DOI] [PubMed] [Google Scholar]
- 31.Liu JS, Farlow JT, Paulson AK, LaBarge MA, Gartner ZJ. Programmed assembly of mosaic cell aggregates reveals the consequences of cell-to-cell variability in Ras activity on the collective behavior of mammary epithelial cells. Cell Reports. 2012 doi: 10.1016/j.celrep.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locke D. Gap junctions in normal and neoplastic mammary gland. The Journal of pathology. 1998;186:343–349. doi: 10.1002/(SICI)1096-9896(199812)186:4<343::AID-PATH189>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 33.Eyckmans J, et al. A hitchhiker’s guide to mechanobiology. Developmental cell. 2011;21:35–47. doi: 10.1016/j.devcel.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sprinzak D, et al. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 36.Takebe N, et al. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nature reviews. Clinical oncology. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 37.Tejavibulya N, et al. Directed self-assembly of large scaffold-free multicellular honeycomb structures. Biofabrication. 2011;3:034110. doi: 10.1088/1758-5082/3/3/034110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livoti CM, Morgan JR. Self-assembly and tissue fusion of toroid-shaped minimal building units. Tissue engineering Part A. 2010;16:2051–2061. doi: 10.1089/ten.tea.2009.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rago AP, et al. Controlling cell position in complex heterotypic 3D microtissues by tissue fusion. Biotechnology and bioengineering. 2009;102:1231–1241. doi: 10.1002/bit.22162. [DOI] [PubMed] [Google Scholar]
- 40.Rago AP, et al. Encapsulated arrays of self-assembled microtissues: an alternative to spherical microcapsules. Tissue engineering Part A. 2009;15:387–395. doi: 10.1089/ten.tea.2008.0107. [DOI] [PubMed] [Google Scholar]
- 41.Jean J, et al. Bioengineered Skin: The Self-Assembly Approach. J Tissue Sci Eng. 2011:S5. [Google Scholar]
- 42.Haraguchi Y, et al. Fabrication of functional three-dimensional tissues by stacking cell sheets in vitro. Nature protocols. 2012;7:850–858. doi: 10.1038/nprot.2012.027. [DOI] [PubMed] [Google Scholar]
- 43.Zahn R, et al. Ion-induced cell sheet detachment from standard cell culture surfaces coated with polyelectrolytes. Biomaterials. 2012;33:3421–3427. doi: 10.1016/j.biomaterials.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Yang J, et al. Reconstruction of functional tissues with cell sheet engineering. Biomaterials. 2007;28:5033–5043. doi: 10.1016/j.biomaterials.2007.07.052. [DOI] [PubMed] [Google Scholar]
- 45.Labbe B, et al. Cell sheet technology for tissue engineering: the self-assembly approach using adipose-derived stromal cells. Methods Mol Biol. 2011;702:429–441. doi: 10.1007/978-1-61737-960-4_31. [DOI] [PubMed] [Google Scholar]
- 46.Nishida K, et al. Functional bioengineered corneal epithelial sheet grafts from corneal stem cells expanded ex vivo on a temperature-responsive cell culture surface. Transplantation. 2004;77:379–385. doi: 10.1097/01.TP.0000110320.45678.30. [DOI] [PubMed] [Google Scholar]
- 47.Ohashi K, et al. Engineering functional two- and three-dimensional liver systems in vivo using hepatic tissue sheets. Nature medicine. 2007;13:880–885. doi: 10.1038/nm1576. [DOI] [PubMed] [Google Scholar]
- 48.Sekine H, et al. Cardiac cell sheet transplantation improves damaged heart function via superior cell survival in comparison with dissociated cell injection. Tissue engineering Part A. 2011;17:2973–2980. doi: 10.1089/ten.tea.2010.0659. [DOI] [PubMed] [Google Scholar]
- 49.Tsuda Y, et al. Cellular control of tissue architectures using a three-dimensional tissue fabrication technique. Biomaterials. 2007;28:4939–4946. doi: 10.1016/j.biomaterials.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 50.Asakawa N, et al. Pre-vascularization of in vitro three-dimensional tissues created by cell sheet engineering. Biomaterials. 2010;31:3903–3909. doi: 10.1016/j.biomaterials.2010.01.105. [DOI] [PubMed] [Google Scholar]
- 51.Sasagawa T, et al. Design of prevascularized three-dimensional cell-dense tissues using a cell sheet stacking manipulation technology. Biomaterials. 2010;31:1646–1654. doi: 10.1016/j.biomaterials.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 52.Teramura Y, et al. Microencapsulation of islets with living cells using polyDNA-PEG-lipid conjugate. Bioconjugate chemistry. 2010;21:792–796. doi: 10.1021/bc900494x. [DOI] [PubMed] [Google Scholar]
- 53.Seliktar D. Designing cell-compatible hydrogels for biomedical applications. Science. 2012;336:1124–1128. doi: 10.1126/science.1214804. [DOI] [PubMed] [Google Scholar]
- 54.Correia AL, Bissell MJ. The tumor microenvironment is a dominant force in multidrug resistance. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy. 2012;15:39–49. doi: 10.1016/j.drup.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott EA, et al. Modular scaffolds assembled around living cells using poly(ethylene glycol) microspheres with macroporation via a non-cytotoxic porogen. Acta biomaterialia. 2010;6:29–38. doi: 10.1016/j.actbio.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leung BM, Sefton MV. A modular approach to cardiac tissue engineering. Tissue engineering Part A. 2010;16:3207–3218. doi: 10.1089/ten.tea.2009.0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGuigan AP, Sefton MV. Vascularized organoid engineered by modular assembly enables blood perfusion. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11461–11466. doi: 10.1073/pnas.0602740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bruzewicz DA, et al. Fabrication of a modular tissue construct in a microfluidic chip. Lab on a chip. 2008;8:663–671. doi: 10.1039/b719806j. [DOI] [PubMed] [Google Scholar]
- 59.Du Y, et al. Sequential assembly of cell-laden hydrogel constructs to engineer vascular-like microchannels. Biotechnology and bioengineering. 2011;108:1693–1703. doi: 10.1002/bit.23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du Y, et al. Directed assembly of cell-laden microgels for fabrication of 3D tissue constructs. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9522–9527. doi: 10.1073/pnas.0801866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dvir T, et al. Nanowired three-dimensional cardiac patches. Nature nanotechnology. 2011;6:720–725. doi: 10.1038/nnano.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li CY, et al. DNA-templated assembly of droplet-derived PEG microtissues. Lab on a chip. 2011;11:2967–2975. doi: 10.1039/c1lc20318e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Y, et al. Switching of macroscopic molecular recognition selectivity using a mixed solvent system. Nat Commun. 2012:3. doi: 10.1038/ncomms1841. [DOI] [PubMed] [Google Scholar]
- 64.Van Vlierberghe S, et al. Biopolymer-based hydrogels as scaffolds for tissue engineering applications: a review. Biomacromolecules. 2011;12:1387–1408. doi: 10.1021/bm200083n. [DOI] [PubMed] [Google Scholar]
- 65.Jabbari E. Bioconjugation of hydrogels for tissue engineering. Current opinion in biotechnology. 2011;22:655–660. doi: 10.1016/j.copbio.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta R, Sefton MV. Application of an endothelialized modular construct for islet transplantation in syngeneic and allogeneic immunosuppressed rat models. Tissue engineering Part A. 2011;17:2005–2015. doi: 10.1089/ten.tea.2010.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Butler MJ, Sefton MV. Cotransplantation of Adipose-derived Mesenchymal Stromal Cells and Endothelial Cells in a Modular Construct Drives Vascularization in SCID/bg Mice. Tissue engineering Part A. 2012 doi: 10.1089/ten.tea.2011.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vallbacka JJ, Sefton MV. Vascularization and improved in vivo survival of VEGF-secreting cells microencapsulated in HEMA-MMA. Tissue engineering. 2007;13:2259–2269. doi: 10.1089/ten.2006.0284. [DOI] [PubMed] [Google Scholar]
- 69.Birgersdotter A, et al. Gene expression perturbation in vitro--a growing case for three-dimensional (3D) culture systems. Seminars in cancer biology. 2005;15:405–412. doi: 10.1016/j.semcancer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 70.Petersen OW, et al. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lacorre DA, et al. Plasticity of endothelial cells: rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood. 2004;103:4164–4172. doi: 10.1182/blood-2003-10-3537. [DOI] [PubMed] [Google Scholar]
- 73.Rivron NC, et al. Tissue assembly and organization: developmental mechanisms in microfabricated tissues. Biomaterials. 2009;30:4851–4858. doi: 10.1016/j.biomaterials.2009.06.037. [DOI] [PubMed] [Google Scholar]
- 74.Maller O, et al. Extracellular matrix composition reveals complex and dynamic stromal-epithelial interactions in the mammary gland. Journal of mammary gland biology and neoplasia. 2010;15:301–318. doi: 10.1007/s10911-010-9189-6. [DOI] [PubMed] [Google Scholar]
- 75.Muller P, Schier AF. Extracellular movement of signaling molecules. Developmental cell. 2011;21:145–158. doi: 10.1016/j.devcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gudjonsson T, et al. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. Journal of cell science. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu P, et al. The extracellular matrix: a dynamic niche in cancer progression. The Journal of cell biology. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Samchenko Y, et al. Multipurpose smart hydrogel systems. Advances in colloid and interface science. 2011;168:247–262. doi: 10.1016/j.cis.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 79.Damassa DA, et al. Purification and characterization of the sex hormone-binding globulin in serum from Djungarian hamsters. Comparative biochemistry and physiology. Part B, Biochemistry & molecular biology. 1996;113:593–599. doi: 10.1016/0305-0491(95)02084-5. [DOI] [PubMed] [Google Scholar]
- 80.Davies JA. A method for cold storage and transport of viable embryonic kidney rudiments. Kidney international. 2006;70:2031–2034. doi: 10.1038/sj.ki.5001884. [DOI] [PubMed] [Google Scholar]
- 81.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annual review of cell and developmental biology. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Streuli CH, et al. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. The Journal of cell biology. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 84.Guo CL, et al. Long-range mechanical force enables self-assembly of epithelial tubular patterns. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5576–5582. doi: 10.1073/pnas.1114781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McLachlan E, et al. Connexins and gap junctions in mammary gland development and breast cancer progression. The Journal of membrane biology. 2007;218:107–121. doi: 10.1007/s00232-007-9052-x. [DOI] [PubMed] [Google Scholar]
- 86.Brisken C, O’Malley B. Hormone action in the mammary gland. Cold Spring Harbor perspectives in biology. 2010;2:a003178. doi: 10.1101/cshperspect.a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]