Abstract
Synthetic biology re-imagines existing biological systems by designing and constructing new biological parts, devices, and systems. In the arena of cytoskeletal-based transport, synthetic approaches are currently used in two broad ways. First, molecular motors are harnessed for non-physiological functions in cells. Second, transport systems are engineered in vitro to determine the biophysical rules that govern motility. These rules are then applied to synthetic nanotechnological systems. We review recent advances in both of these areas and conclude by discussing future directions in engineering the cytoskeleton and its motors for transport.
Keywords: dynein, kinesin, myosin, actin, microtubule, motor
Harnessing the cytoskeleton
The eukaryotic cytoskeleton is composed of fibers that exert and respond to force and serve as highways for motors. In cells this cytoskeleton performs an array of functions that includes organizing subcellular compartments, transporting diverse cargos [1], and producing unique functional structures such as the mitotic spindle [2,3] and cilia [4,5]. While synthetic approaches to harness and engineer the cytoskeleton are in the early stages, recent work utilizes the diversity of mechanochemical attributes of both the cytoskeletal motors and their tracks. In this review, we first introduce the key components of the cytoskeleton. We then discuss recent endeavors to employ these components for novel synthetic purposes. Next, we highlight how engineering molecular motors is providing a modular toolbox for synthetic in vitro transport [6–8]. Finally, we review synthetic transport systems comprised of either cytoskeletal motors that occur in nature (`natural motors'), modified natural motors, or synthetic motors. We conclude by describing promising future directions in the synthetic biology of transport.
The parts list: tracks, motors, and cargos
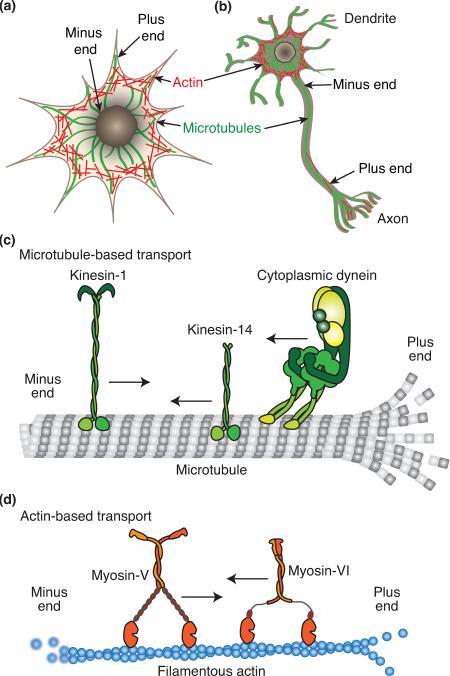
Long-distance transport within eukaryotic cells occurs on two cytoskeletal track systems: actin filaments and microtubules (Box 1, Figure I). Both tracks are polarized, with fast growing ends (called `plus' ends) and slow growing ends (called `minus' ends). Generally, dynamic actin filaments are found near the cell periphery, often with their plus ends polymerizing near the plasma membrane. Microtubules are usually nucleated from a perinuclear-organizing center and emanate from this point; thus, microtubule plus ends tend to be located in the cell periphery and minus ends near the nucleus (Box 1, Figure Ia and b). The dynamic growth and shrinkage of both actin filaments and microtubules is capable of exerting pushing and pulling forces [9]. Large, highly polarized cells, such as neurons or epithelial cells, are especially reliant on the cytoskeleton and cytoskeletal transport for maintaining spatial and temporal localization of intracellular components [10,11], as evidenced by the growing list of human diseases resulting from defects in cytoskeletal-mediated transport [12].
Box 1, Figure I.
Cellular tracks and motors.
Most naturally occurring cytoskeletal motors move unidirectionally along either actin filaments or microtubules. Dynein and kinesin motors move on microtubules, while myosin motors move on actin filaments (Box 1, Figure Ic and d). For the purposes of synthetic biology, the most useful motors may be those that are capable of moving cargo processively over long distances as these are capable of individually producing prolonged movements of cargos and/or filaments in contrast to non-processive motors, which require larger arrays to produce similar results. Of the approximately 100 genes coding for cytoskeletal motors, at least 20 of their protein products are capable of moving multiple types of intracellular cargo over long distances [1]. For microtubules, these cargo-transporting motors include the minus-end-directed cytoplasmic dyneins-1 and -2 [13,14], and the plus-end-directed kinesins-1, -2, and -3 [15]. For actin filaments, the plus-end-directed class V myosins are the best-characterized cargo-transporting motors [16]. Many organisms have expanded on the types of kinesins and myosins through gene duplication and divergent evolution. Although some of these motors do not function as long-distance cargo transporters endogenously, their range of biophysical properties could be useful for synthetic purposes. For example, there are classes of kinesins and myosins that move in reverse compared to most of their other family members: kinesin-14s are minus-end-directed kinesins [15,17] and class VI myosins are minus-end-directed myosins [18].
Cargos of motor proteins vary widely in size, shape, and function and include membranous organelles, proteinaceous signaling molecules and ribonucleoproteins. For synthetic biology uses, it is instructive to understand how motors attach to cargo such that those attachment mechanisms can be hijacked or mimicked. While the molecular connections linking natural motors to their physiological cargos are only beginning to be defined, they include both protein and lipid receptors [19,20]. The filaments themselves can also be cargos, as motors can slide filaments with respect to one another or relative to a fixed position in the cell [21]. Thus, motors and the dynamic tracks they propel are versatile building blocks for engineering synthetic systems.
Pathogens: model hijackers of cytoskeletal systems
Pathogenic organisms have already evolved multiple mechanisms for harnessing the power of the cytoskeleton. Here we discuss examples whereby viruses or bacteria co-opt the microtubule or actin cytoskeleton of infected cells to propagate their pathogenic lifecycle. Understanding these mechanisms can provide synthetic biologists with inspiration for reengineering the cytoskeleton.
Many viruses hijack the microtubule cytoskeleton at two major stages of their lifecycle. Invading viruses use dynein to reach the nucleus, where they replicate. Later in the infection kinesin is used to reach the cell membrane, where viral budding and exit occurs [22]. Considerable progress has been made to identify the specific viral-motor interactions required for these transport events, creating a useful resource for synthetic biologists. Interestingly, viruses have evolved a variety of mechanisms for recruiting dynein or kinesin [23]. For example, dynein is recruited by adenovirus via interactions between the dynein light intermediate and intermediate chains and a viral capsid subunit called hexon [24]. This interaction is pH-sensitive, providing an elegant mechanism for ensuring that hexon-coated adenoviruses only associate with dynein after entering the low pH environment of the lysosomal compartment. In contrast, herpes simplex virus (HSV) interacts with dynein via an interaction between the dynein light chains RP3/TcTex1 and its capsid protein, VP26 [25]. In the case of kinesin, vaccinia virus binds to kinesin-1 via an interaction with the kinesin light chain [26], whereas HSV binds directly to kinesin via a region of the kinesin heavy chain [27]. These diverse viral binding and recruiting mechanisms highlight methods by which motors could be co-opted for other non-physiological functions.
In addition to using the cytoskeletal motors, some pathogens hijack the polymerization power of the cytoskeletal filaments [28]. The best-characterized example of this is the bacteria Listeria monocytogenes, which recruits the actin polymerization process to propel itself rapidly through the cytoplasm at speeds of up to 30 nm/s. When the pathogen hits the cell cortex, actin polymerization generates enough force for the host cell to drive itself into an adjacent cell in a finger-like projection, which is then phagocytosed by that neighboring cell. This process mimics non-pathogenic lamellipodia formation in which actin networks produce membrane protrusion and ruffling at the leading edge of a motile cell [29]. Other pathogens such as Clostridium difficile use toxins to alter microtubule dynamics, causing large microtubule-filled projections to emanate out from the cell. These projections entangle the extracellular pathogen, and are thus hypothesized to help it adhere to the cell [28,30]. These distinct mechanisms demonstrate the malleability of the cytoskeletal components to be subverted for many non-physiological purposes and can provide insight for designing novel synthetic systems.
These examples highlight how understanding the mechanisms that pathogens use to co-opt the cytoskeleton may provide inspiration for synthetic designs. While purely synthetic attachment methods using standard connection technologies have been employed recently [31,32], endogenous and viral based attachments have the added benefits of being sensitive to endogenous regulation and cellular conditions. If these mechanisms are better understood, future synthetic attachment schemes could more closely mimic the dynamic and versatile connections nature has provided. Such systems would allow the integration of artificial transport more completely with endogenous processes. Additionally, synthetic in vivo cargo systems could be utilized to function in response to internal cues, a feature that current attachment systems cannot yet provide.
Harnessing transport systems to perform non-physiological functions
A major goal of synthetic biology as it relates to transport is to harness existing cellular transport systems for novel purposes, such as delivering recombinant DNA to the nucleus, targeting intracellular organelles to new locations, and mapping neuronal connectivity.
In a process similar to an invading virus navigating the cytoplasm to reach the nucleus, effective synthetic delivery of recombinant DNA to the cell`s nucleus is a critical hurdle to overcome for many applications of gene manipulation. A goal for synthetic biologists is to design a modular system capable of bypassing all barriers from cell entry through transcription of the recombinant DNA. The first experimental attempts to achieve this link plasmid DNA to the microtubule cytoskeleton [33]. While engineered viral pathogens are commonly used as vectors, they pose significant safety risks, making non-viral methods largely preferable [34]. Taking cues from viruses, recent work has sought to deliver DNA to the nucleus by coupling DNA to a dynein recruiting element [35,36]. For example, recombinant dynein light chain (LC8) linked to a DNA binding domain can interact with plasmid DNA; transfections in the presence of this reagent are more efficient compared to common transfection reagents or naked DNA [35]. Here, a likely barrier to achieving higher transfection efficiency is the ability of the plasmid DNA-dynein light chain complex to escape membrane-bound endosomes [37]. A challenge for the future will be to integrate motor recruitment with other steps required to increase the efficiency of nuclear delivery, such as the ability to escape endosomes and import across nuclear pores.
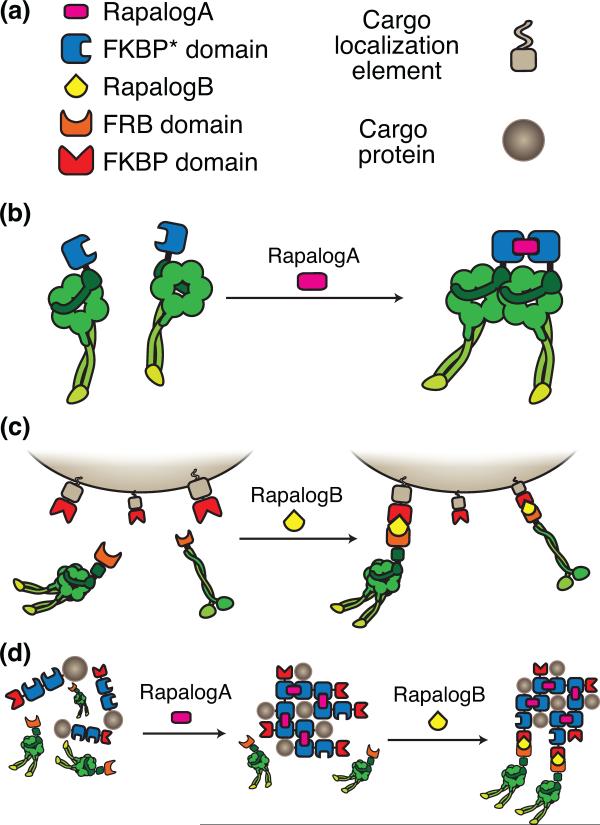
Another promising synthetic application uses actin- or microtubule-based motors to target existing intracellular cargo to new locations. This causes spatial sequestering of cargo, resulting in either blocked or modified functions. Two recent proofs of concept showed that synthetic recruitment of motors to peroxisomal organelles could drive organelle redistribution [31,32]. In the first study [32], opposite polarity microtubule-(cytoplasmic dynein and kinesin-1 or kinesin-3) or actin- (myosin-V and myosin-VI) based motors were recruited to peroxisomes in a drug-dependent manner using the FKBP-FRB system [38] (Figure 1A). The recruitment of each type of motor yielded a different peroxisomal localization pattern in fibroblasts. In the second study, similar results were obtained in hippocampal neurons [31]. Here, axonal-specific cargo was redistributed to dendrites by recruiting dynein via the FKBP-FRB system [31]. This approach is both rapid and reversible; organelle redistribution occurs on a time scale of minutes and can be reversed by drug removal. Finally, the modularity of the FKBP-FRB system can also be used to drive dynein activation in cells through motor dimerization [31], which is necessary for motor processivity [39] (Figure 1B). In future experiments, the FKBP-FRB system could also allow inducible spatial control of protein, RNA or organellar cargo through the recruitment of endogenous cytoskeletal motors (Figure 1C and 1D). Constitutive methods for co-opting motors have also been developed. By generating a myosin-kinesin chimera containing the motor domain of a kinesin and the cargo-binding domain of a myosin, fission yeast cells were rewired to use microtubules in place of actin filaments [40]. These studies highlight the versatility of motors to move non-physiological cargo and provide promise that additional innovative uses for synthetic technology will be discovered in the future.
Figure 1.
FKBP/FRB motor recruitment system.
(a) The FKBP-FRB system is a drug-induced, protein dimerization system used by several different groups to control motor properties and attachments to cargo [32,39,47]. A mutant form of FKBP, referred to as FKBP* homodimerizes in the presence of the small rapamycin analogue (AP20187) referred to as RapalogA [31]. Additionally, heterodimers can be produced using a different mutant FKBP and the protein domain FRB in the presence of the small molecule, RapalogB (AP21967) [95]. (b) Fusing FKBP* to two dynein monomers can produce an inducible dynein dimer. This construction technique can be used to activate dynein-based transport by enabling processive stepping in the presence of RapalogA. (c) Fusing FKBP to a cargo of interest through a cargo localization element (e.g. a PEX domain for peroxisome localization), allows recruitment of a recombinant FRB-motor to the cargo in an inducible fashion upon the addition of RapalogB. (d) The orthogonal nature of this system provides the ability to combine their activities for novel complex uses. For example, a protein of interest (tan circle) fused to several copies of FKBP* and a single copy of FKBP could be selectively aggregated and sequestered by addition of RapalogA and subsequently delivered to a site of interest by recruiting a recombinant FRB-motor by addition of RapalogB. These strategies (panels a–d) were successfully used to study trafficking in neurons and aspects of this figure were adapted from [31].
Recruitment of motor proteins can also be used as a tool to harness intracellular sorting mechanisms, allowing mapping of neural circuits [41]. Many viruses that infect neurons, such as rabies virus, spread uni-directionally across synapses from the peripheral nervous system to the central nervous system. This type of cell-to-cell infection requires navigation of the microtubule network within neurons. Recently, it was demonstrated that glycoproteins from different viruses were sufficient to provide directional viral particle movement [42]. This was done by engineering vesicular-stomatitis virus (VSV) with glycoproteins from viruses known to move either towards or away from the neuron`s cell body [42]. A current missing link is to determine how these viral glycoproteins differentially recruit dynein and kinesin motors, and which specific isoforms they use. These experiments demonstrate the feasibility of using synthetic approaches to recruit intracellular motors, allowing the mapping of complex neural circuits and perhaps more complex tissue organization in the future.
Interchangeable parts for molecular transport machines
Although engineering the cytoskeleton in cells for synthetic purposes is still in its infancy, significant work has been done in minimal systems in vitro. Such experiments have focused on engineering systems to gain insight into motor mechanism. These approaches have paved the way for a toolbox of interchangeable motor parts, allowing for the design of self-assembling, synthetic motile devices (termed `molecular shuttles') destined for performing programmed transport tasks for biomedical and nanomanufacturing applications.
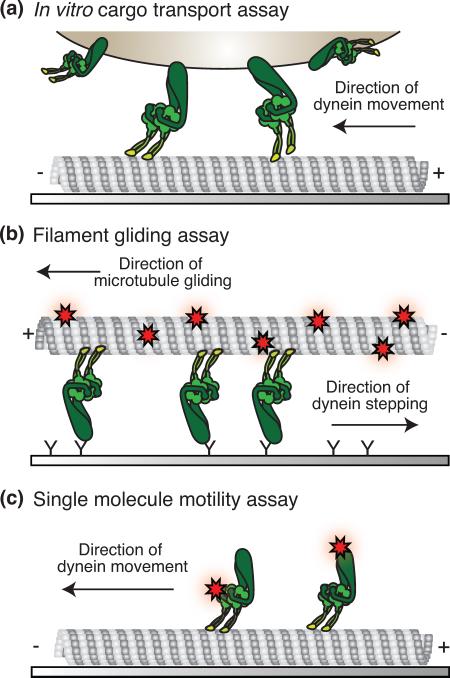
Motor proteins have been studied in vitro in both native and recombinant forms (Box 2, Figure I). Initial single-molecule experiments with kinesin, myosin, and dynein relied on genetically engineering these motors by truncating them to their smallest essential components, and adding sites for tags to aid in protein expression, purification, and labeling (see for example: [39,43,44]). These tags were originally intended for attaching fluorophores for visualization purposes, but recent advances allow them to serve as attachment sites for linking elements that provide connectivity [45–49]. This connectivity enables disparate motor and cargo components to become modular and linkable to myriad other objects. Such linking can be accomplished not only with protein, but also with DNA, arguably the most versatile connection medium [50].
Box 2, Figure I.
Motility assays.
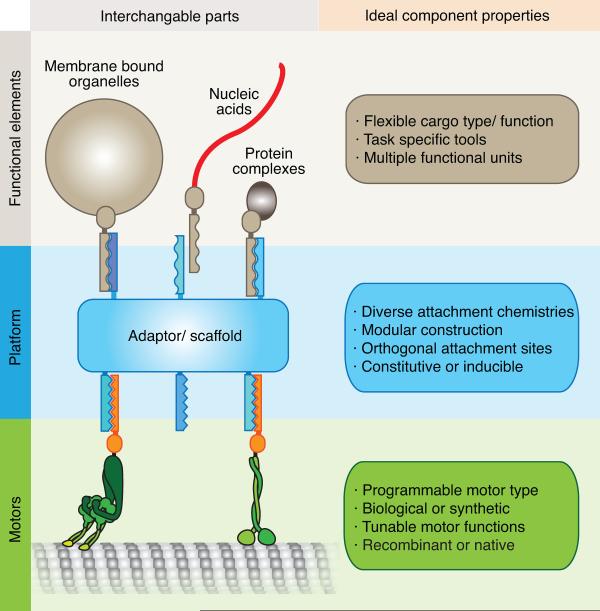
DNA connections provide not only a guaranteed linking specificity through unique sequences, but are inexpensive, easily modifiable, allow tunable affinity, and come with many enzymatic tools already provided by nature, such as restriction enzymes, polymerases and ligases. The engineering properties of DNA have been employed recently to understand the intramolecular interactions between domains in kinesin and dynein, both of which are normally homodimers. For kinesin, DNA was used to connect two motor-containing monomer subunits at non-native locations [48]. With dynein, DNA replaced the native dimerization domains such that two motor-containing monomer subunits were linked and formed a functional heterodimer [46]. This approach also enabled orthogonal fluorophore labeling of each motor subunit. Similar work using the FRB-FKBP system [38] made use of the concept of interchangeable parts to study the mechanism of wildtype and mutant dynein protomers [39,47]. Although these components have yet to be categorized into truly `standard' parts (such as has been done for nucleic acid elements [51,52]), they are interchangeable with the potential for standardization (Figure 2). Thus, a future goal for the field is to create a toolbox of mechanical components that can be applied to controlling cargo transport.
Figure 2.
Synthetic cargo systems.
An ideal synthetic motile system would have three major sets of components: task-specific functional elements (top), a scaffolding platform (middle), and motor machinery (bottom). The scaffolding platform provides connectivity for all active components of the motile system and should be modular, allowing for multiple orthogonal attachments sites to bind motors and functional elements. Depending on in vitro or in vivo applications, it can be purely synthetic in origin, or comprised of biologically produced material. Additionally, the scaffolding platform can be activated or induced as needed. The platform could be driven by motors of synthetic or biological origin. The motors are recruited to the platform through motor-specific linking chemistries, such as DNA or Rapalogs. Motors are chosen from an array of possibilities depending on need, where the potential task-specific motor attributes include directionally, velocity, track selection, and exogenous control. Finally, specific functional elements can be bound to the platform. These might include various cargo payloads or specific tools for particular applications.
Initial work demonstrating the utility, promise, and challenges associated with employing natural motors for engineering purposes has led to several devices, including molecular transporters and sorters, and has been reviewed recently [6–8,53]. Most of these efforts to produce molecular transport devices have relied on the filament-gliding assay architecture (Box 2, Figure I) with actin or microtubules serving as the cargo scaffold (see for example [54,55]). However, a modular cargo framework based on the cargo transport assay architecture (Box 2, Figure I) with multiple attachment possibilities allows construction of more complicated assemblies with programmable functions. Once a motor or motor component is connected to a chemical linker, attachments to larger scaffolding structures are possible. This has recently been done with myosin [56,57] and kinesin [58] using either quantum dots [56] or DNA [57,58] as scaffolds. Although this work with DNA-based scaffolds made use of a single DNA double helix, more complicated scaffolding could be achieved using DNA origami [59, 60] or other structure building techniques [45] in vitro or in vivo [61]. Such systems could lead to the creation of more sophisticated, varied, and modular molecular shuttles.
Molecular shuttles and other sophisticated synthetic applications of cytoskeletal motors require the ability to control the basic motile properties of the motors that drive them. Methods for exogenous control over motors and motor systems will be required to achieve such applications. While progress has been made in understanding how the motile properties of biologically derived motors are modulated by the cell [14,15,62], there is a need for a greater mechanistic understanding of this process. Critical motor parameters that require innovative methods of control include regulation and control of cargo attachment, loading, and unloading; directionality; processivity; bidirectional versus unidirectional movement; and reuse or recycling of motors. Recent work to address these issues using synthetic approaches include protein-engineering to affect directionality [63,64], processivity [65–67], and dimerization [65]; chemical control over directionality and processivity [68–70]; motor copy number effects on directionality [71]; and photo-based control over motor function [72]. Of particular note is a study that engineered a switchable, bi-directional myosin motor [63].
Designing synthetic motors de novo
As a complement to the biologically evolved cytoskeletal motors, recent work has focused on the creation of synthetic motors capable of motion along a track. Although notable progress has been made in this area [73], artificial motors have yet to reach the capabilities of naturally occurring motors, particularly with regard to velocity and processivity. Yet, there are many advantages to the de novo design and fabrication of synthetic motors — attempts to create a functional motor could allow highly specific tasks to be performed and lead to a better understanding of the mechanistic details of naturally occurring motors.
Many fundamental considerations regarding how a synthetic motor might operate correlate directly with our attempts to better understand existing natural cytoskeletal motors. For example, how is directionality programmed into the structures and functions of the motors and their tracks? What enables the motors to operate processively? What is the source of fuel and how is it used? Given this fuel, what are the energy and entropic considerations that allow work to be done? Can multiple motors work together to achieve higher functions? These questions should help drive the design principles when creating synthetic motors de novo; however, purely synthetic systems also offer design possibilities beyond what is available from the biologically derived tools. For example, the structural organization of the motor could depart from cytoskeletal models and be multimeric, with many discrete track and cargo-binding domains. Track and motor design offers the potential to create many track-motor combinations or dynamically programmable routes through a network of tracks. Additionally, motor tasks could be algorithmically determined depending on the conditions and instruction sets contained within the system (see for example: [74,75]).
Given the versatility of DNA as a nanotechnological platform [50,76–78], it has become a primary construction material for synthetic molecular motors. These synthetic motors tend to be significantly smaller than cytoskeletal motors and have focused on design issues of autonomy, processivity, velocity, directionality, track selection, and visualization. Some early synthetic DNA motors are termed `clocked' because they required the sequential addition of fuel strands to propagate motility (see for example [79,80]). However, other designs allow for continuous autonomous motion [81–87] and in some cases with coordinated stepping [83,87,88]. Some of these motors consist solely of DNA [83,88], however others make use of complementary components such as DNAzymes with an RNA substrate [81,83] or ligating [82] and nicking [82,84] enzymes. In these systems, directionality can be controlled by loading the motor onto the track at a specific location [81,84] or the use of polar tracks [82,83]. Additionally, instructions contained within the motor's fuel itself [87–89] can imbue directionality, providing a directionality mechanism distinct from those of cytoskeletal motors. In contrast to cytoskeletal motor systems, in some cases the track is modified or destroyed as the synthetic motor moves along it [81,83–86].
Recently, route-based selection motifs included in the track design allowed motors to navigate particular paths by following instructions in solution or intrinsically part of the motor [89,90]. Specific construction tasks have also been demonstrated; an autonomous motor created sequential peptide bonds between building blocks analogously to a ribosome [91] and a clocked motor performed a nanomanufacturing task by assembling clusters of gold nanoparticles along an assembly line [92]. Furthermore, a DNA motor was visualized in real time taking continuous, discrete steps [84], as was recently observed for myosin-V by atomic force microscopy [93]. The visualization of single synthetic motor steps has also been achieved using single-molecule motility assays (Box 2, Figure I) [81,94]. Future designs will likely incorporate hybrid approaches that make use of both de novo and natural motor elements. By building modular components, new designs and functions could be rapidly created and tested. One promising possibility is to use biological parts as core machinery, with synthetic adapters that are able to sense and interact with their environment.
Concluding Remarks
Nature has provided excellent nanoscale transporters, and our understanding of how cells control them is rapidly increasing. The regulatory control issues cells face with molecular motors are similar to those that synthetic biologists encounter to harness and reengineer these motors for novel tasks. Similarly, these engineering problems overlap with areas of active research in cell biology. The complementary approaches of working on solutions to these design problems and understanding how the cell has solved these same issues will enrich the utility and understanding of molecular motors.
Although in the early stages of research, the ease of building in vitro transport systems shows that the core motor components are amenable to modification and inclusion within modular engineered systems. These new constructs will continue to increase in both standardization and complexity and in the future will be used to probe cellular functions and provide novel synthetic applications. Purely synthetic motors will continue to inform how endogenous motors function. The programmability of artificial motors will expand their role in synthetic nanomanufacturing, possibly outpacing the use of biological-based motors.
Despite this exciting progress, there are a number of outstanding questions in the field and challenges for the future (Box 3). The constant feedback between synthetic and cellular biologists promises to allow this nascent field to uncover the mechanisms of cellular motility and engineer novel transport systems.
Box 1. Cellular tracks and motors.
(a) In many mammalian cells, actin filaments form a meshwork throughout the cell and near the cell membrane, while microtubules are organized radially emitting outward from a perinuclear microtubule-organizing center. (b) In mammalian neurons microtubules are similarly polarized in axons, but have mixed polarity in dendrites. (c) Microtubules are polymers of alpha and beta tubulin heterodimers. They are composed of 13–15 protofilaments (linear arrays of tubulin dimers) and can be many microns in length. In cells, they predominately polymerize and depolymerize from their dynamic plus ends near the cell cortex. The major cargo-transporting microtubule-based motors are kinesin-1, -2, and -3 (plus-end-directed) and cytoplasmic dynein (minus-end-directed). Most kinesins and dyneins are homodimers of motor containing subunits. Cytoplasmic dynein contains a number of additional subunits that play a role in cargo binding. Kinesin-14 is a minus-end-directed kinesin. (d) Filamentous actin (F-actin) is a polymer of globular actin (G-actin), with rapid polymerization in cells occurring primarily at the plus end and depolymerization at the minus end. Myosin-V moves along actin filaments toward the plus end, while myosin-VI moves toward the minus end.
Box 2. Motility assays.
In vitro motility assays provide powerful methods for studying or mimicking the behavior of cellular transport. (a) In a cargo transport assay crude or purified cellular components are observed to move on immobilized filaments by observing the cargo motion. Motors are either absorbed to beads nonspecifically such that the orientation and relative positions on the bead or cargo is not controlled, or specifically through adaptors such as antibodies or chemical linkers. (b) In gliding assays, filaments are moved by the action of motors attached to a coverslip surface. Motors are typically attached to coverslips through antibody linkages. (c) In a single-molecule motility assay the filament is immobilized to a coverslip and fluorescently labeled motors are observed to move along the filament.
Box 3. Outstanding questions.
The cytoskeleton has proved to be an amenable system for engineering novel synthetic functions. Some outstanding challenges in the field include the following questions:
Can the cytoskeleton and motor proteins be used to create additional novel functionality? This is of particular interest in large, polarized cells where cargo sorting heavily relies on transport mechanisms.
Can the cytoskeleton be reorganized for novel transport requirements? For example, can actin or microtubules be nucleated to grow from new sites?
Can motors be co-opted to enhance the efficiency of synthetic in vivo processes already developed?
Can a synthetic toolbox be built to recruit motors to any cargo of interest with standardized parts?
Can motors be exogenously controlled to produce molecular shuttles that start, stop, and change direction based on outside input?
Can the cytoskeleton be harnessed for efficient directed transport of foreign entities to the nucleus?
Can motors be designed de novo to have the efficiency, velocity, and processivity of biological motors? Can these synthetic motors be imbued with greater functionality than biological motors?
Acknowledgements
We thank J. Tytell, X. Su, and S. Wickham for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 2.Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr Biol. 2004;14:R797–805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Wittmann T, et al. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28–34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- 4.Gibbons IR. The role of dynein in microtubule-based motility. Cell Struct. Funct. 1996;21:331–342. doi: 10.1247/csf.21.331. [DOI] [PubMed] [Google Scholar]
- 5.Fisch C, Dupuis-Williams P. Ultrastructure of cilia and flagella - back to the future! Biol. Cell. 2011;103:249–270. doi: 10.1042/BC20100139. [DOI] [PubMed] [Google Scholar]
- 6.Goel A, Vogel V. Harnessing biological motors to engineer systems for nanoscale transport and assembly. Nat Nanotechnol. 2008;3:465–475. doi: 10.1038/nnano.2008.190. [DOI] [PubMed] [Google Scholar]
- 7.van den Heuvel MGL, Dekker C. Motor proteins at work for nanotechnology. Science. 2007;317:333–336. doi: 10.1126/science.1139570. [DOI] [PubMed] [Google Scholar]
- 8.Hess H. Engineering applications of biomolecular motors. Annu Rev Biomed Eng. 2011;13:429–450. doi: 10.1146/annurev-bioeng-071910-124644. [DOI] [PubMed] [Google Scholar]
- 9.McIntosh JR, et al. Tubulin depolymerization may be an ancient biological motor. Journal of Cell Science. 2010;123:3425–3434. doi: 10.1242/jcs.067611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conde C, Cáceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- 11.Müsch A. Microtubule organization and function in epithelial cells. Traffic. 2004;5:1–9. doi: 10.1111/j.1600-0854.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 12.Hirokawa N, et al. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Scholey JM. Intraflagellar transport motors in cilia: moving along the cell's antenna. J Cell Biol. 2008;180:23–29. doi: 10.1083/jcb.200709133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nature Reviews Molecular Cell Biology. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nature Reviews Molecular Cell Biology. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 16.Hammer JA, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nature Reviews Molecular Cell Biology. 2012;13:13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 17.Walker RA, et al. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature. 1990;347:780–782. doi: 10.1038/347780a0. [DOI] [PubMed] [Google Scholar]
- 18.Wells AL, et al. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 19.Akhmanova A, Hammer JA. Linking molecular motors to membrane cargo. Curr Opin Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamal A, Goldstein LSB. Principles of cargo attachment to cytoplasmic motor proteins. Curr Opin Cell Biol. 2002;14:63–68. doi: 10.1016/s0955-0674(01)00295-2. [DOI] [PubMed] [Google Scholar]
- 21.Kulic IM, et al. The role of microtubule movement in bidirectional organelle transport. Proceedings of the National Academy of Sciences. 2008;105:10011–10016. doi: 10.1073/pnas.0800031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Döhner K, et al. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 2005;13:320–327. doi: 10.1016/j.tim.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bremner KH, et al. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe. 2009;6:523–535. doi: 10.1016/j.chom.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas MW, et al. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J Biol Chem. 2004;279:28522–28530. doi: 10.1074/jbc.M311671200. [DOI] [PubMed] [Google Scholar]
- 26.Ward BM, Moss B. Vaccinia virus A36R membrane protein provides a direct link between intracellular enveloped virions and the microtubule motor kinesin. J. Virol. 2004;78:2486–2493. doi: 10.1128/JVI.78.5.2486-2493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diefenbach RJ, et al. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 2002;76:3282–3291. doi: 10.1128/JVI.76.7.3282-3291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haglund CM, Welch MD. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J Cell Biol. 2011;195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambrechts A, et al. Listeria comet tails: the actin-based motility machinery at work. Trends Cell Biol. 2008;18:220–227. doi: 10.1016/j.tcb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Schwan C, et al. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 2009;5:e1000626. doi: 10.1371/journal.ppat.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapitein LC, et al. Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr Biol. 2010;20:290–299. doi: 10.1016/j.cub.2009.12.052. [DOI] [PubMed] [Google Scholar]
- 32.Kapitein LC, et al. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys J. 2010;99:2143–2152. doi: 10.1016/j.bpj.2010.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lechardeur D, et al. Intracellular routing of plasmid DNA during non-viral gene transfer. Adv. Drug Deliv. Rev. 2005;57:755–767. doi: 10.1016/j.addr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Edelstein ML, et al. Gene therapy clinical trials worldwide to 2007--an update. J Gene Med. 2007;9:833–842. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- 35.Toledo MAS, et al. Development of a recombinant fusion protein based on the dynein light chain LC8 for non-viral gene delivery. J Control Release. 2012;159:222–231. doi: 10.1016/j.jconrel.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Bergen JM, Pun SH. Evaluation of an LC8-binding peptide for the attachment of artificial cargo to dynein. Mol. Pharm. 2007;4:119–128. doi: 10.1021/mp060086o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller AM, Dean DA. Tissue-specific and transcription factor-mediated nuclear entry of DNA. Adv. Drug Deliv. Rev. 2009;61:603–613. doi: 10.1016/j.addr.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Banaszynski L, et al. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 39.Reck-Peterson SL, et al. Single-Molecule Analysis of Dynein Processivity and Stepping Behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Presti, Lo L, Martin SG. Shaping fission yeast cells by rerouting actin-based transport on microtubules. Curr Biol. 2011;21:2064–2069. doi: 10.1016/j.cub.2011.10.033. [DOI] [PubMed] [Google Scholar]
- 41.Enquist LW, Card JP. Recent advances in the use of neurotropic viruses for circuit analysis. Curr. Opin. Neurobiol. 2003;13:603–606. doi: 10.1016/j.conb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Beier KT, et al. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proceedings of the National Academy of Sciences. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warshaw DM, et al. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys J. 2005;88:L30–2. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 45.Diehl MR, et al. Engineering cooperativity in biomotor-protein assemblies. Science. 2006;311:1468–1471. doi: 10.1126/science.1122125. [DOI] [PubMed] [Google Scholar]
- 46.Qiu W, et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat Struct Mol Biol. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeWitt MA, et al. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyazono Y, et al. Strain through the neck linker ensures processive runs: a DNA-kinesin hybrid nanomachine study. EMBO J. 2010;29:93–106. doi: 10.1038/emboj.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guydosh NR, Block SM. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michelotti N, et al. Beyond DNA origami: the unfolding prospects of nucleic acid nanotechnology. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4:139–152. doi: 10.1002/wnan.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shetty RP, et al. Engineering BioBrick vectors from BioBrick parts. J Biol Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Canton B, et al. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 53.Korten T, et al. Towards the application of cytoskeletal motor proteins in molecular detection and diagnostic devices. Current Opinion in Biotechnology. 2010;21:477–488. doi: 10.1016/j.copbio.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt C, et al. Tuning the “roadblock” effect in Kinesin-based transport. Nano Lett. 2012;12:3466–3471. doi: 10.1021/nl300936j. [DOI] [PubMed] [Google Scholar]
- 55.Malcos JL, Hancock WO. Engineering tubulin: microtubule functionalization approaches for nanoscale device applications. Appl Microbiol Biotechnol. 2011;90:1–10. doi: 10.1007/s00253-011-3140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali MY, et al. Myosin Va and myosin VI coordinate their steps while engaged in an in vitro tug of war during cargo transport. Proceedings of the National Academy of Sciences. 2011;108:E535–41. doi: 10.1073/pnas.1104298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu H, et al. Collective dynamics of elastically-coupled myosinV motors. Journal of Biological Chemistry. 2012 doi: 10.1074/jbc.M112.371393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers AR, et al. Negative interference dominates collective transport of kinesin motors in the absence of load. Physical chemistry chemical physics : PCCP. 2009;11:4882–4889. doi: 10.1039/b900964g. [DOI] [PubMed] [Google Scholar]
- 59.Shih WM, Lin C. Knitting complex weaves with DNA origami. Current Opinion in Structural Biology. 2010;20:276–282. doi: 10.1016/j.sbi.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krishnan Y, Bathe M. Designer Nucleic Acids to Probe and Program the Cell. Trends Cell Biol. 2012;22 doi: 10.1016/j.tcb.2012.10.001. in press. [DOI] [PubMed] [Google Scholar]
- 61.Delebecque CJ, et al. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 62.Hartman MA, et al. Principles of unconventional myosin function and targeting. Annu Rev Cell Dev Biol. 2011;27:133–155. doi: 10.1146/annurev-cellbio-100809-151502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen L, et al. Engineering controllable bidirectional molecular motors based on myosin. Nat Nanotechnol. 2012;7:252–256. doi: 10.1038/nnano.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carter AP, et al. Structure and functional role of dynein's microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shishido H, Maruta S. Engineering of a novel Ca(2+)-regulated kinesin molecular motor using a calmodulin dimer linker. Biochem Biophys Res Commun. 2012;423:386–391. doi: 10.1016/j.bbrc.2012.05.135. [DOI] [PubMed] [Google Scholar]
- 66.Clancy BE, et al. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hodges AR, et al. Engineering the processive run length of Myosin V. J Biol Chem. 2007;282:27192–27197. doi: 10.1074/jbc.M703968200. [DOI] [PubMed] [Google Scholar]
- 68.Gerson-Gurwitz A, et al. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011;30:4942–4954. doi: 10.1038/emboj.2011.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cochran JC, et al. A metal switch for controlling the activity of molecular motor proteins. Nat Struct Mol Biol. 2012;19:122–127. doi: 10.1038/nsmb.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walter WJ, et al. Two independent switches regulate cytoplasmic dynein's processivity and directionality. Proceedings of the National Academy of Sciences. 2012;109:5289–5293. doi: 10.1073/pnas.1116315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roostalu J, et al. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- 72.Yamada MD, et al. Photocontrol of kinesin ATPase activity using an azobenzene derivative. Journal of Biochemistry. 2007;142:691–698. doi: 10.1093/jb/mvm183. [DOI] [PubMed] [Google Scholar]
- 73.Delius, von M, Leigh DA. Walking molecules. Chem Soc Rev. 2011;40:3656–3676. doi: 10.1039/c1cs15005g. [DOI] [PubMed] [Google Scholar]
- 74.Qian L, et al. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 75.Soloveichik D, et al. DNA as a universal substrate for chemical kinetics. Proceedings of the National Academy of Sciences. 2010;107:5393–5398. doi: 10.1073/pnas.0909380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seeman NC. Nanomaterials based on DNA. Annu. Rev. Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bath J, Turberfield AJ. DNA nanomachines. Nat Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 78.Aldaye FA, et al. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 79.Sherman WB, Seeman NC. A precisely controlled DNA biped walking device. Nano Lett. 2004;4:1203–1207. [Google Scholar]
- 80.Shin J-S, Pierce NA. A synthetic DNA walker for molecular transport. J Am Chem Soc. 2004;126:10834–10835. doi: 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- 81.Lund K, et al. Molecular robots guided by prescriptive landscapes. Nature. 2010;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng Y, et al. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed. Engl. 2004;43:4906–4911. doi: 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- 83.Omabegho T, et al. A bipedal DNA Brownian motor with coordinated legs. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wickham SFJ, et al. Direct observation of stepwise movement of a synthetic molecular transporter. Nat Nanotechnol. 2011;6:166–169. doi: 10.1038/nnano.2010.284. [DOI] [PubMed] [Google Scholar]
- 85.Pei R, et al. Behavior of polycatalytic assemblies in a substrate-displaying matrix. J Am Chem Soc. 2006;128:12693–12699. doi: 10.1021/ja058394n. [DOI] [PubMed] [Google Scholar]
- 86.Bath J, et al. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. Engl. 2005;44:4358–4361. doi: 10.1002/anie.200501262. [DOI] [PubMed] [Google Scholar]
- 87.Bath J, et al. Mechanism for a directional, processive, and reversible DNA motor. Small. 2009;5:1513–1516. doi: 10.1002/smll.200900078. [DOI] [PubMed] [Google Scholar]
- 88.Green SJ, et al. Coordinated chemomechanical cycles: a mechanism for autonomous molecular motion. Phys. Rev. Lett. 2008;101:238101. doi: 10.1103/PhysRevLett.101.238101. [DOI] [PubMed] [Google Scholar]
- 89.Wickham SFJ, et al. A DNA-based molecular motor that can navigate a network of tracks. Nat Nanotechnol. 2012;7:169–173. doi: 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- 90.Muscat RA, et al. A programmable molecular robot. Nano Lett. 2011;11:982–987. doi: 10.1021/nl1037165. [DOI] [PubMed] [Google Scholar]
- 91.He Y, Liu DR. Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker. Nat Nanotechnol. 2010;5:778–782. doi: 10.1038/nnano.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gu H, et al. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kodera N, et al. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 94.Masoud R, et al. Studying the Structural Dynamics of Bipedal DNA Motors with Single-Molecule Fluorescence Spectroscopy. ACS Nano. 2012 doi: 10.1021/nn301709n. [DOI] [PubMed] [Google Scholar]
- 95.Clackson T, et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci USA. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]