SUMMARY
Background
The neuronal circuitry underlying sleep is poorly understood. Although dopamine (DA) is thought to play a key role in sleep/wake regulation, the identities of the individual DA neurons and their downstream targets required for this process are unknown.
Results
Here, we identify a DA neuron in each PPL1 cluster that promotes wakefulness in Drosophila. Imaging data suggest that the activity of these neurons is increased during wakefulness, consistent with a role in promoting arousal. Strikingly, these neurons project to the dorsal fan-shaped body, which has previously been shown to promote sleep. The reduced sleep caused by activation of DA neurons can be blocked by loss of DopR, and restoration of DopR expression in the fan-shaped body can rescue the wake-promoting effects of DA in a DopR mutant background.
Conclusions
These experiments define a novel arousal circuit at the single cell level. Since the dorsal fan-shaped body promotes sleep, these data provide a key link between wake and sleep circuits. Furthermore, these findings suggest that inhibition of sleep centers via monoaminergic signaling is an evolutionarily conserved mechanism to promote arousal.
INTRODUCTION
The transition from sleep to wakefulness is a key behavior, with significant implications for an animal’s survival. In mammals, studies over the past several decades have indicated the importance of specific wake-promoting nuclei [1]. However, these nuclei consist of heterogeneous groups of neurons and contain many individual cells [1, 2], making it difficult to determine the precise cellular circuits regulating sleep in mammals. In contrast, fruit flies have a simpler nervous system, and Drosophila has emerged as a powerful model system to study the molecular and cellular basis of sleep [3, 4]. Studies of sleep in Drosophila have highlighted the importance of neuronal excitability and various signaling pathways for sleep regulation [5–10]. In addition, multiple lines of evidence suggest that dopamine (DA) promotes wakefulness in Drosophila. Pharmacological manipulation of DA levels can affect sleep [11], and activation of DA neurons decreases sleep time [12]. In addition, mutations in the DA transporter DAT/fumin, which increase synaptic DA levels, cause significant reductions in sleep time and an increase in arousal [13, 14]. However, it is not known whether DA signaling is generally needed to induce wakefulness or whether specific DA neurons perform this function.
In this study, we provide evidence that, among monoaminergic transmitters, DA is the primary mediator of arousal in Drosophila. Analyses of novel Gal4 lines with expression in subsets of PPL1 and PPM3 cells suggest that a single DA neuron from each PPL1 cluster projects to the dorsal fan-shaped body to promote wakefulness. Imaging data using a surrogate marker for chronic neuronal activity suggest that these DA neurons are more active during the day and during wakefulness, consistent with a role for these neurons in promoting arousal. Because the dorsal fan-shaped body has recently been shown to promote sleep [15], our data suggest that arousal centers in Drosophila may directly regulate sleep-promoting centers. Finally, our data indicate that DopR is the DA receptor responsible for arousal in Drosophila and that it is specifically required in the fan-shaped body for this process. These studies thus define a novel arousal circuit and provide a key link between wake and sleep centers in Drosophila.
RESULTS
Dopamine signaling is the major monoaminergic pathway promoting wakefulness in Drosophila
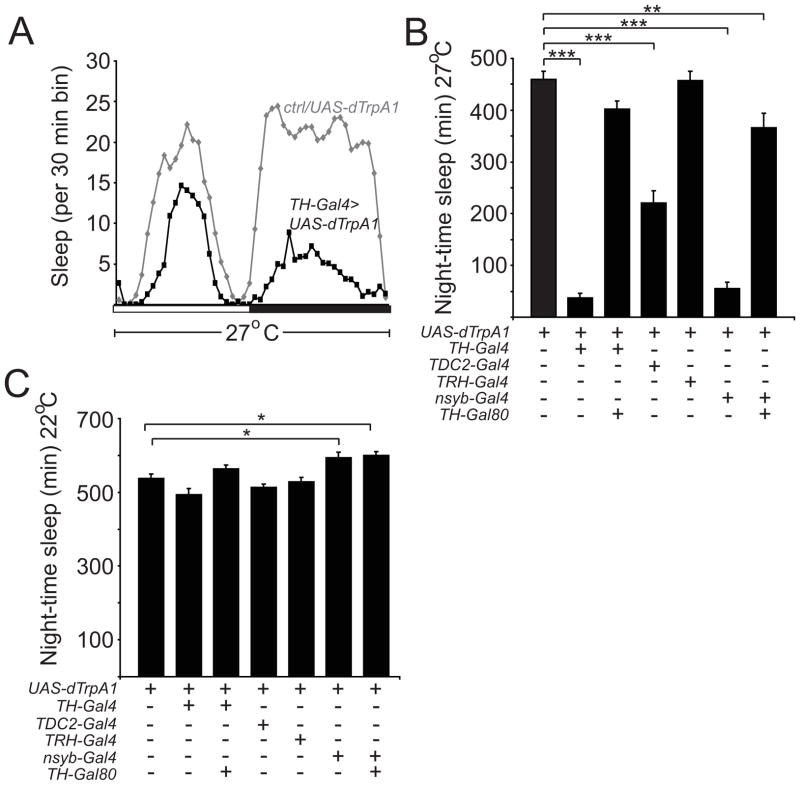
In Drosophila, there are 5 biogenic amine pathways, dopamine (DA), octopamine (OA), tyramine (TA), serotonin (5HT), and histamine (HA); DA and OA signaling have been shown to promote wakefulness, whereas 5HT signaling has been suggested to promote sleep [11, 13, 14, 16, 17]. To examine the effects of activating these monoaminergic neurons on sleep/wake, we used different Gal4 lines to drive expression of dTrpA1, a heat-inducible non-selective cation channel which can be used to depolarize neurons to trigger neurotransmitter release [18]. We used the previously characterized TH-Gal4 [19], Tdc2-Gal4 [20], and TRH-Gal4 [21] lines to drive expression in DA, OA/TA, and 5HT neurons, respectively (to our knowledge, there is no available HA-specific Gal4 driver). Using gentle activation conditions (27°C) with these different Gal4 drivers, we found that TH-Gal4/UAS-dTrpA1 flies exhibited the strongest reduction in sleep (Figures 1A and 1B). As previously described, the % decrease in sleep with activation of DA neurons was greater during the night than during the day (Figure S1A) [12]. In contrast, activation of OA/TA neurons (using Tdc2-Gal4) resulted in a moderate reduction in night-time sleep, while activation of 5HT neurons (using TRH-Gal4) had no significant effect on night-time sleep (Figure 1B). No significant decreases in night-time sleep were observed for any of the lines in the absence of heat-induction of TrpA (Figure 1C). We next examined the effects of increasing monoaminergic signaling on sleep architecture. Activation of DA neurons caused a decrease in night-time sleep bout duration and sleep bout number (Figures S1B and S1C). In contrast, activation of OA/TA neurons decreased sleep bout duration, but did not affect sleep bout number (Figures S1B and S1C). The effects of activating DA neurons on sleep amount and sleep architecture could be suppressed with TH-Gal80 [22], which inhibits Gal4 function in DA neurons (Figures 1B and S1A–S1C). To further assess the relative importance of DA signaling for wakefulness in Drosophila, we compared the effects of gentle activation of most neurons (nsyb-Gal4) vs. most neurons except DA neurons (nsyb-Gal4, TH-Gal80) using dTrpA1. Remarkably, the dramatic reduction in night-time sleep seen with activation of all neurons was largely reversed when Gal4 function is suppressed in DA cells (Figure 1B). Taken together, these data suggest that, among monoamines, DA signaling is the primary mediator of arousal.
Figure 1. Dopamine Signaling is a Key Arousal Pathway in Drosophila.
(A) Sleep profile of UAS-dTrpA1/+ (gray diamonds) vs TH-Gal4/UAS-dTrpA1 (black squares) flies at 27°C in 12:12 LD, plotted in 30 min bins. White and black bars denote light and dark periods, respectively. Night-time (B) sleep amount at 27°C and (C) sleep amount at 22°C plotted for UAS-dTrpA1/+ (n=54), TH-Gal4/UAS-dTrpA1 (n=36), TH-Gal80/+; TH-Gal4/UAS-dTrpA1 (n=66), TDC2-Gal4/+; UAS-dTrpA1 (n=37), TRH-Gal4/UAS-dTrpA1 (n=42), nsyb-Gal4/+; UAS-dTrpA1/+ (n=50), and nsyb-Gal4/TH-Gal80; UAS-dTrpA1/+ (n=34) flies. In this and subsequent figures, error bars represent SEM. “*”, “**”, and “***” denote P<0.05, P<0.01, and P<0.001 respectively.
The key arousal-promoting DA cells reside in PPL1 and/or PPM3
In order to study the specific DA cells that function in arousal, we set out to generate restricted DA Gal4 drivers. There are 6 major clusters of TH (tyrosine hydroxylase)-positive neurons in each hemisphere. 2 are located anteriorly (PAM and PAL), and 4 posteriorly (PPL1, PPL2, PPM1/2, and PPM3) (Figure S2B) [19, 23]. An ~11kb genomic TH fragment was previously used by Friggi-Grelin et al. (2003) to generate TH-Gal4, which drives expression in nearly all DA neurons, except the PAM subgroup. This study also suggested that the introns of the TH gene contained the key enhancers for promoting TH expression [19]. Thus, we generated 7 transgenic Gal4 driver lines containing different regions of the TH genomic locus, most of which include different combinations of introns of TH (Figure S2A).
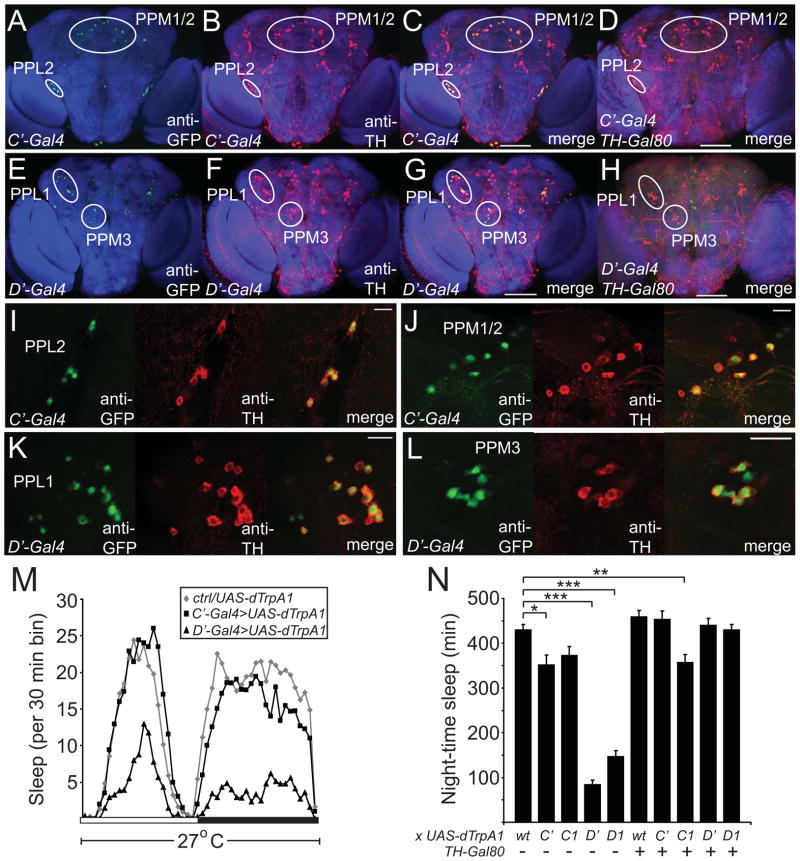
To determine which DA neurons are captured by these different restricted drivers, we used these drivers to express GFP-nls and immunostained for GFP and TH. The results of these experiments are summarized in Figures S2A and Table S1. 3 Gal4 lines (TH-A, TH-B, and TH-E) did not drive expression in any DA cells (data not shown). In contrast, 2 drivers (TH-C and TH-D) exhibited largely non-overlapping TH+ expression and together recapitulated most of the TH-Gal4 expression pattern. The TH-C lines drive expression in most of the PPM1/2 and all PPL2 cells, as well as 3 of 5 PAL cells, whereas the TH-D lines drive expression in almost all of the PPL1 and PPM3 cells, and 1–4 of the PPM1/2 neurons (Figure S2A and Table S1). With one exception discussed later, these expression patterns for TH-C and TH-D were consistently observed in multiple insertions (data not shown). The TH-F and TH-G lines demonstrate expression in the PPM1/2, PPM3, PPL1, and PPL2 subgroups, but are more variable in the number of DA cells expressed within each subgroup (Table S1). Some non-specific expression (i.e., in non-TH+ cells) was observed in the TH-C, -D, -F, and -G lines, as is often the case with promoter-Gal4 constructs (data not shown). Thus, since the TH-C and TH-D constructs seemed to be most useful in dividing the TH+ expression pattern, we made another version of those constructs by generating deletions in the original ~11kb TH-Gal4 construct (Figure S2A). TH-C′ and TH-D′ showed similar expression in TH+ subsets as TH-C and TH-D respectively, but exhibited little non-TH+ expression (Figures 2A–2L; Figures S2C–S2J). As expected, expression in TH+ cells in the TH-C′ and TH-D′ lines was suppressed by the addition of TH-Gal80 (Figures 2D, 2H and Figures S2F, S2J). Finally, all of the restricted TH drivers, like the original TH-Gal4 line [19], expressed poorly in the PAM cluster. In summary, these data confirm that the key enhancers driving expression in DA neurons (except the PAM cluster) are in the introns and also establish novel Gal4 lines driving expression in specific subsets of DA neurons.
Figure 2. Analysis of Novel DA Gal4 Drivers Suggests that the PPL1 and/or PPM3 Clusters Mediate the Arousing Effects of DA.
Maximum projection confocal images of the posterior half of whole mount adult brains of flies expressing GFP-nls driven by TH-C′-Gal4 (A–D) or TH-D′-Gal4 (E–H) in the absence (A–C, E–G) or presence (D, H) of TH-Gal80. Single higher magnification confocal sections showing PPL2 (I) and PPM1/2 (J) in TH-C′-Gal4 flies expressing GFP-nls, and PPL1 (K) and PPM3 (L) in TH-D′-Gal4 flies expressing GFP-nls. Brains are immunostained with nc82, anti-GFP, and anti-TH antibodies. Scale bars denote 100 μm (A–H) and 20 μm (I–L). (M) Sleep profile of UAS-dTrpA1/+ (gray diamonds), TH-C′-Gal4/+; UAS-dTrpA1/+ (black squares), and TH-D′-Gal4/+; UAS-dTrpA1/+ (black triangles) flies at 27°C in 12:12 LD, plotted in 30 min bins. White and black bars denote light and dark periods, respectively. (N) Night-time sleep plotted for UAS-dTrpA1/+ (n=63), TH-C′-Gal4/+; UAS-dTrpA1/+ (n=27), TH-C1-Gal4/+; UAS-dTrpA1/+ (n=38), TH-D′-Gal4/+; UAS-dTrpA1/+ (n=40), TH-D1-Gal4/+; UAS-dTrpA1/+ (n=46), TH-Gal80/+; UAS-dTrpA1/+ (n=44), TH-C′-Gal4/TH-Gal80; UAS-dTrpA1/+ (n=32), TH-C1-Gal4/TH-Gal80; UAS-dTrpA1/+ (n=49), TH-D′-Gal4/TH-Gal80; UAS-dTrpA1/+ (n=41), TH-D1-Gal4/TH-Gal80; UAS-dTrpA1/+ (n=51) flies. “wt” denotes background control.
To identify the groups of DA neurons regulating sleep/wake, we activated the neurons in the TH-C1, TH-C′, TH-D1, TH-D′ drivers using dTrpA1. Activation of the neurons in the TH-D1 and TH-D′ driver lines resulted in a profound decrease in night-time sleep, compared to activation of neurons in TH-C1, TH-C′ driver lines, and controls (Figures 2M and 2N). As before, these effects were more pronounced during the night compared to the day, and there was no significant decrease in night-time sleep seen in these lines when tested at 22°C (Figures S2M and S2N). As expected, TH-Gal80 reversed the decrease in sleep amount in the TH-D1 and TH-D′ lines, suggesting that the DA neurons in these drivers are responsible for this effect (Figure 2N). As with the original TH-Gal4, activation of DA cells using TH-D1 and TH-D′ caused a decrease in sleep bout duration and number at night, compared to controls (Figures S2K and S2L).
To address whether the reduction in sleep in the TH-D1>dTrpA1 flies results from changes in arousal threshold, we exposed these flies, as well as TH-C1>dTrpA1 flies, to graded light pulses at ZT16. Activation of the neurons in the TH-D1 driver resulted in a significant reduction in arousal threshold, compared to activation of neurons in the TH-C1 driver or controls (Figure S2O). Together, these data suggest that the cell groups unique to the TH-D1 and TH-D′ drivers (PPL1 and PPM3) are more important than other DA subgroups for promoting wakefulness and that this effect is mediated by an increase in arousal.
A single DA neuron in each PPL1 cluster projects to the dorsal fan-shaped body to promote wakefulness
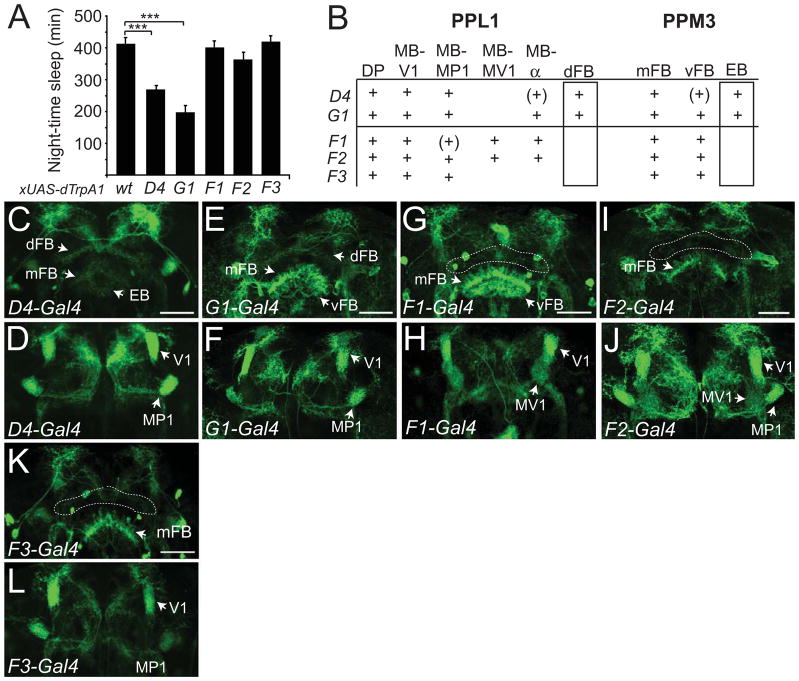
To investigate whether individual DA neurons in the PPL1 and/or PPM3 clusters promote arousal, we searched for additional restricted Gal4 drivers. We identified 3 TH-F lines, 1 TH-G line, and 1 TH-D line (TH-D4) exhibiting expression in a subset of PPL1 cells and PPM3 cells (Table S1). We next used dTrpA1 to activate the neurons in these driver lines. 2 of the lines, TH-D4 and TH-G1 exhibited a significant reduction in night-time sleep and sleep bout duration compared to controls (Figures 3A and S3A), which could be reversed with TH-Gal80 (Figures S3B and S3C). No significant differences were observed for sleep bout number or day-time sleep amount for these lines, compared to controls (Figures S3D and S3E). Therefore, these data suggest that specific neurons within PPL1 and/or PPM3, but not the entire subgroups, promote wakefulness.
Figure 3. Activation of Gal4 Drivers with Restricted Expression in PPL1 and/or PPM3 Suggests that DA cells Projecting to the Dorsal Fan-shaped Body or Ellipsoid Body Promote Wakefulness.
(A) Night-time sleep for UAS-dTrpA1/+ (n=56), TH-D4-Gal4/UAS-dTrpA1 (n=71), TH-G1-Gal4/UAS-dTrpA1 (n=40), TH-F1-Gal4/UAS-dTrpA1 (n=62), TH-F2-Gal4/UAS-dTrpA1 (n=51), and TH-F3-Gal4/UAS-dTrpA1 (n=42) flies at 27°C. (B) Identities of the projections from the PPL1 and PPM3 clusters for the TH-D4, TH-G1, TH-F1, TH-F2, TH-F3 lines, with naming as described in Table S1. “+” denotes presence of projection, and “(+)” denotes faint staining for projection. Confocal images of brains immunostained with anti-GFP of TH-D4 (C, D), TH-G1 (E, F), TH-F1 (G, H), TH-F2 (I, J), or TH-F3 (K, L) driving expression of UAS-mCD8-GFP; UAS-mCD8-GFP. Images for (C, E, G, I, K) and (D, F, H, J, L) consist of maximum projections of a ~4 μm section through the middle or anterior third of the adult brain to highlight the central complex and mushroom bodies, respectively. “dFB”, “EB”, “mFB”, “vFB”, “V1”, “MP1”, “MV1” indicate projections for the dorsal fan-shaped body, ellipsoid body, middle fan-shaped body, ventral fan-shaped body, and the V1, MP1, or MV1 regions of the mushroom bodies. Dashed lines outline the location for the dFB in (G), (I), and (K). Scale bar denotes 50μm.
In order to identify the specific DA neurons in PPL1 and PPM3 that regulate arousal in these restricted drivers, we first classified different DA cells of the PPL1 and PPM3 clusters. To do this, we studied the projection patterns of individual neurons of the TH-Gal4 driver, using images from the MARCM analysis of the FlyCircuit database [24]. Using these images, we could distinguish 9 types of PPL1 cells and 3 types of PPM3 cells with distinct projection patterns (see Table S2). We confirmed these classifications by immunostaining our Gal4 lines driving UAS-mCD8-GFP with anti-GFP and anti-TH antibodies (data not shown), and also by comparing them with analyses from previous studies [23, 25–29]. We next characterized the projection patterns of the TH+ cells in the 5 Gal4 lines with restricted expression in PPL1 and PPM3 (Figures 3C–3L and summarized in Figure 3B). In comparing the projection patterns of the 2 lines that significantly promote arousal (TH-D4 and TH-G1) to the 3 lines that do not, there were 2 classes of neurons that clearly differed: PPL1 neurons that project to the dorsal fan-shaped body (dFB) and PPM3 neurons that project to the ellipsoid body (EB) (Figure 3B). These data suggest the possibility that these 2 classes of neurons may be wake-promoting DA cells.
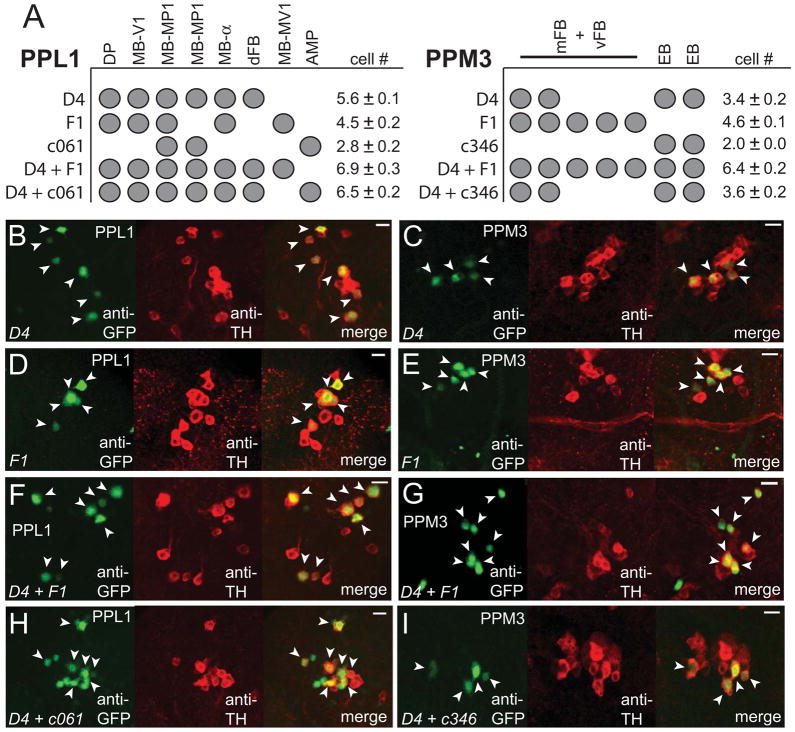
Because TH-D4 contains fewer TH+ cells than TH-G1, we focused on TH-D4 for further analysis. To further identify the individual wake-promoting DA neuron(s) in PPL1 or PPM3, we carried out cell-counting experiments (Figures 4B–4I, and summarized in Figure 4A). In TH-D4, there are 5 distinct projection patterns (Figure 3B), which are all suppressed by TH-Gal80 (Figures S4A and S4B). However, counting of TH+ cells in the PPL1 cluster in TH-D4>GFP-nls flies revealed 6 total cells (Figure 4B), suggesting that one of the classes contains 2 cells. Because there are known to be 2 MB-MP1 neurons [27, 29], we suspected that TH-D4 may contain 2 neurons of this class. We thus generated flies carrying both TH-D4 and c061-Gal4, which has previously been shown to express in 3 TH+ cells in the PPL1 cluster (2 MB-MP1 neurons and 1 MB-AMP neuron) (Figure S4C) [27, 29]. In c061-Gal4; TH-D4>GFP-nls flies, we counted a total of 7 cells in PPL1 (Figure 4H). Since TH-D4 contains 6 cells in PPL1 and c061-Gal4 contains 2 MB-MP1 cells in PPL1, these data suggest that only the MB-AMP cell is added to the TH-D4 pattern and that there are 2 MB-MP1 neurons in TH-D4. We next examined the effects of activating the 2 MB-MP1 neurons and 1 MB-AMP neuron in c061-Gal4; MB-Gal80; we used MB-Gal80 to remove mushroom body (MB) expression in this driver, as done previously [27]. We found that activation of these neurons does not significantly promote wakefulness, suggesting that the MB-MP1 neurons in TH-D4 are unlikely to be wake-promoting DA cells (Figure S4E; Figure S4F).
Figure 4. A Single Neuron from the PPL1 Subgroup, which Projects to the Dorsal Fan-shaped Body, Promotes Wakefulness.
(A) Identities of the TH+ cells in the PPL1 and PPM3 clusters in the indicated Gal4 driver lines. Mean cell counts for PPL1 and PPM3 are shown for TH-D4-Gal4 (n=13 hemispheres), TH-F1-Gal4 (n=14), c061-Gal4 (n=5), c346-Gal4 (n=3), TH-D4-Gal4/TH-F1-Gal4 (n=11), c061-Gal4; TH-D4-Gal4 (n=8), and c346-Gal4; TH-D4-Gal4 (n=9) flies expressing GFP-nls. (B–I) Single confocal sections showing the TH+ cells in the PPL1 or PPM3 clusters for the indicated driver lines expressing GFP-nls. Arrowheads indicate GFP+ TH+ cells. Scale bar denotes 10 μm.
Second, to determine which of the other DA cells in PPL1 may promote wakefulness, we next compared the cellular projections of TH-D4 with TH-F1 (which does not promote arousal). There are 5 projection patterns (Figure 3B) and 5 TH+ cells in PPL1 (Figure 4D) in TH-F1 flies, indicating that each TH+ cell can be identified (Figure 4A). We next generated flies carrying both TH-D4 and TH-F1, in combination with UAS-GFP-nls. Counting the GFP+ cells in the PPL1 cluster in these flies revealed a total of 7 cells (Figure 4F). Since there are a total of 6 TH+ PPL1 cells in TH-D4, the only cell added by the presence of TH-F1 is MB-MV1. Thus, the neurons shared between these two lines (DP, MB-V1, and MB-α) are unlikely to promote arousal. Given that the only cell in PPL1 that is unique to the TH-D4 driver, compared to the TH-F1 and c061-Gal4 drivers, is the dFB neuron, these data suggest that the PPL1-dFB neuron promotes wakefulness.
Third, because we previously found that the restricted drivers promoting arousal also contained projections to the EB, we compared the PPM3 neurons in TH-D4 with c346-Gal4, which drives expression in a limited subset of neurons, including 2 PPM3 neurons that project to the EB [28] (Figure S4D). Activation of the 2 PPM3 neurons in the c346-Gal4 driver did not significantly promote wakefulness (Figure S4E; Figure S4F). To examine if the PPM3 EB neurons in TH-D4 are included in the c346-Gal4 driver, we combined these 2 lines to drive expression of GFP-nls. TH-D4 alone contains 4 TH+ cells in PPM3 (Figure 4C) and combined with c346-Gal4 also contains a total of 4 TH+ cells in PPM3 (Figure 4I). Thus, these data suggest that there are 2 PPM3 EB neurons in TH-D4, which are also present in c346-Gal4, and that these neurons are not critical for promoting arousal.
Finally, we investigated whether the other PPM3 neurons projecting to the mFB or vFB in TH-D4 may promote wakefulness. TH-F1 contains a total of 5 PPM3 neurons projecting to either the mFB or the vFB (Figure 4E). Combining TH-D4 with TH-F1 revealed a total of 7 PPM3 TH+ neurons (Figure 4G). These data indicate that the 2 TH+ PPM3 cells in TH-D4 are contained within the TH-F1 expression pattern and suggest that these neurons do not promote wakefulness. We next asked whether loss-of-function of the neurons in TH-D4 causes increased sleep. We electrically silenced neurons in the TH-Gal4, TH-D4, and TH-F1 drivers in an inducible fashion, using UAS-Kir2.1; tub-Gal80ts. Kir2.1 is an inward-rectifying K+ channel that hyperpolarizes the cells in which it is expressed [30]. Inhibition of the neurons in TH-Gal4 and TH-D4 resulted in increased sleep, compared to controls and TH-F1, consistent with our neuronal activation data (Figure S4G; Figure S4H). Taken together, these data suggest that the pair of PPL1 DA neurons projecting to the dFB promote wakefulness in Drosophila.
Imaging experiments using ANF-GFP suggest that the activity of PPL1-dFB neurons is increased during wakefulness
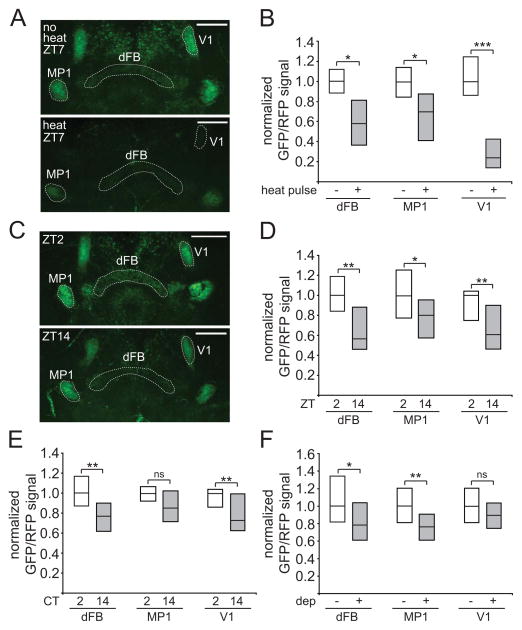
If the PPL1-dFB neurons promote wakefulness, then one would predict that these neurons would be more active during the day than the night. To address this question, it would be ideal to use a transgenic reporter of chronic neuronal activity, and so we looked for tools we could potentially adapt for this purpose. A UAS-ANF-GFP transgenic line has previously been generated and used in Drosophila to monitor neuropeptidergic vesicle trafficking [31, 32]. These flies express rat atrial natriuretic factor (ANF) fused to GFP, under UAS control. Excitation of neurons expressing ANF-GFP induces release of vesicles containing this reporter and a decrease of GFP fluorescence from the terminal; this reduction of GFP signal could potentially be used as a readout for chronic neuronal activity [31, 32]. Neuropeptides are often co-expressed with classical neurotransmitters [33], and we thus asked whether we could express ANF-GFP in DA neurons and whether this would label synaptic terminals. We used TH-D4-Gal4 to drive UAS-ANF-GFP and UAS-myr-RFP (to normalize the GFP signal) and found that GFP signal could be readily detected in the terminals of DA cells of this driver, including the PPL1-dFB, PPL1-MP1, and PPL1-V1 neurons (Figure 5A). Next, we examined whether this signal was releasable with activation of these DA neurons, by using dTrpA1 expressed in the same neurons. As shown in Figures 5A and 5B, the GFP/RFP signal decreased with activation of these neurons. Of note, the decrease in GFP/RFP signal was particularly dramatic for the V1 projections, which may reflect an increased capacity of these particular neurons for neuropeptide release.
Figure 5. Imaging experiments using ANF-GFP suggest the PPL1-dFB neurons are more active during the day and during wakefulness.
(A) Whole-mount brains from flies expressing ANF-GFP, myr-RFP, and dTrpA1, under control of TH-D4-Gal4. Images consist of maximum projections of a 12 μm section through the middle of the brain to highlight the mushroom body and central complex projections, with either no heat pulse (“no heat”) or 5 hr 32°C heat pulse (“heat”) from ZT2–ZT7. “dFB”, “MP1”, and “V1” indicate projections to the dorsal fan-shaped body or MP1 and V1 regions of the mushroom bodies, respectively. Dashed lines highlight these projections. (B) GFP/RFP signal for these flies is represented as simplified box plots, where the line inside the box indicates the median, and the top and bottom represent 75th and 25th percentiles, respectively. n=7 for no heat pulse and n=12 for heat pulse conditions. (C) Whole-mount brains from flies expressing ANF-GFP and myr-RFP under control of TH-D4-Gal4 at ZT2 or ZT14. GFP/RFP signal is shown as simplified box plots at ZT2 (n=20) and ZT14 (n=21) (D), CT2 (n=15) and CT14 (n=20) (E), or no sleep deprivation (no dep, n=22) and sleep deprivation from ZT12–24 (dep, n=31) (F) conditions. Scale bar denotes 50 μm.
Using ANF-GFP as a readout, we investigated the activity of PPL1-dFB neurons under physiological conditions. We examined GFP/RFP signal in dFB, MP1, and V1 projections near the beginning of the day (ZT2) and the night (ZT14). There was a ~43%, ~20%, and a ~39% reduction in GFP/RFP signal from ZT2 to ZT14 in the dFB, MP1, and V1 projections, respectively (Figures 5C and 5D). These data suggest that, during the day, the activity of multiple DA neurons is increased. We next asked whether these changes are dependent on the presence of light, by performing the experiment in constant darkness (DD). In the absence of light, there was a ~24% and ~27% reduction in GFP/RFP signal from CT2 to CT14 in the dFB and V1 projections, respectively, whereas there is no significant change in the MP1 projections (Figure 5E). Together, these data suggest that the PPL1-dFB and PPL1-V1 neurons are more active during the day, when flies are awake, compared to the night.
To address whether these changes are circadian- or sleep state-dependent, we deprived flies of sleep during the night (ZT12–ZT24) and measured GFP/RFP signal at ZT0. As compared to control non-deprived flies, sleep deprived flies exhibited a ~21% and a ~24% reduction in GFP/RFP signal in the dFB and MP1 projections respectively, whereas there was no significant change in the V1 projections (Figure 5F). These data suggest that PPL1-dFB and PPL1-MP1 neurons, but not PPL1-V1 neurons, are active when flies are forced to stay awake during the night. Together, these data suggest that among these 3 groups of PPL1 neurons, only the PPL1-dFB neurons are more active both during the day and during wakefulness, consistent with a role in promoting arousal.
DopR in the fan-shaped body is required for DA-mediated arousal
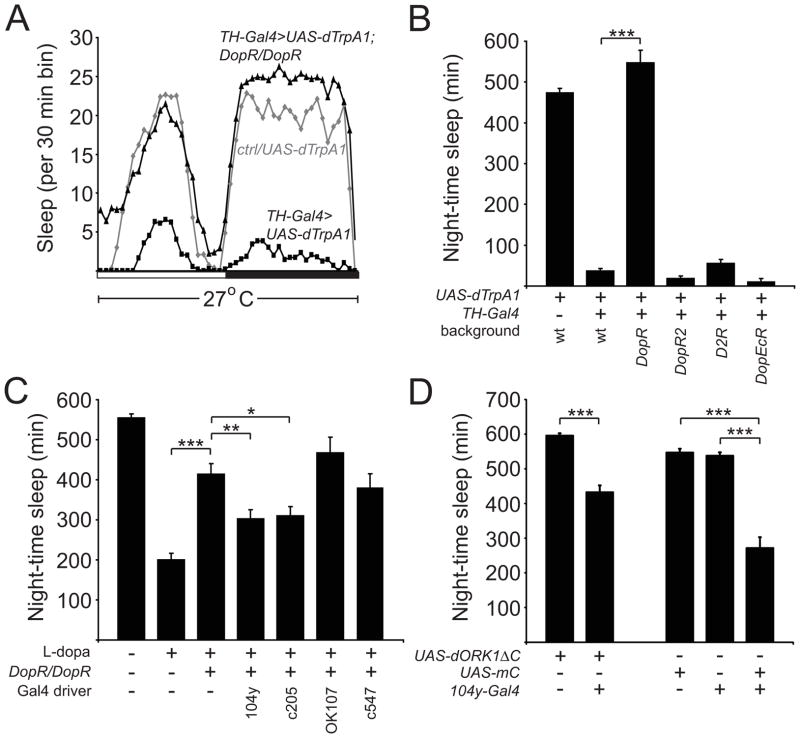
We next set out to determine the downstream target of the PPL1-dFB neurons that promote wakefulness. There are 4 DA receptors in Drosophila: DopR, DopR2, D2R, and DopEcR [34–37]. To examine which DA receptor is required for DA-mediated arousal, we activated DA neurons using TH-Gal4 and UAS-dTrpA1 in DopRf02676, DopR2MB05108, D2Rf06521, and DopEcRc02142 mutant backgrounds, respectively. DopRf02676 and DopEcRc02142 have been previously shown to severely reduce DopR and DopEcR levels, respectively [38, 39]. For the DopR2MB05108 and D2Rf06521 mutant lines, we performed quantitative PCR analysis and found that these mutants exhibited a ~14% and ~74% reduction in transcript levels, respectively (Figure S5). As shown in Figures 6A and 6B, only loss of DopR fully suppressed the marked reduction in sleep observed when DA neurons are activated. Similarly, DopRf02676 markedly suppressed the increased wakefulness seen when flies are fed L-Dopa to enhance DA signaling (Figure 6C). These data suggest that DopR is the main DA receptor required for promoting wakefulness.
Figure 6. DopR Mediates the Arousing Effects of DA and is Required in the Fan-shaped Body to Promote Wakefulness.
(A) Sleep profile of UAS-dTrpA1/+ (gray diamonds), TH-Gal4/UAS-dTrpA1 (black squares), and TH-Gal4, DopR/UAS-dTrpA1, DopR flies (black triangles) at 27°C in 12:12 LD, plotted in 30 min bins. White and black bars denote light and dark periods, respectively. (B) Night-time sleep for UAS-dTrpA1/+ (n=62), TH-Gal4/UAS-dTrpA1 (n=81), TH-Gal4, DopR/UAS-dTrpA1, DopR (n=31), TH-Gal4, DopR2/UAS-dTrpA1, DopR2 (n=31), D2R;;TH-Gal4/UAS-dTrpA1 (n=41), and TH-Gal4, DopEcR/UAS-dTrpA1, DopEcR (n=32) flies at 27°C. (C) Night-time sleep for control flies not fed L-dopa (n=64) and control (n=145), DopR/DopR (n=69), 104y-Gal4/+; DopR/DopR (n=82), c205-Gal4, DopR/DopR or c205-Gal4, DopR (n=67), DopR/DopR; OK107/+ (n=26), and c547-Gal4, DopR/DopR (n=32) flies fed 4 mg/ml L-dopa at 25°C. (D) Night-time sleep for UAS-dORK1ΔC/+ (n=59), 104y-Gal4/+; UAS-dORK1ΔC/+ (n=35), UAS-mC*/+ (n=62), 104y-Gal4/+ (n=31), and 104y-Gal4/UAS-mC* (n=37) flies at 25°C.
The 104y-Gal4 driver was recently used to show that the dFB neurons in this driver regulate sleep [15]. Thus, we next examined if DopR expression in the FB using 104y-Gal4 is sufficient to rescue the wake-promoting effects of DA in DopRf02676 flies. Because this transposon itself contains UAS sequences and is inserted in the 5′ non-essential portion of the gene, it can directly be used, in combination with a Gal4 driver, for tissue-specific rescue of DopR [28, 38]. When DopR expression was driven in the FB by 104y-Gal4 in the DopRf02676 mutant background, the reduced sleep caused by DA signaling was significantly restored, suggesting that DA signaling to the FB is necessary for promoting wakefulness (Figure 6C). Because there are non-FB neurons in the 104y-Gal4 expression pattern, we repeated these experiments with an additional driver (c205-Gal4) that also expresses in the dFB and obtained similar results. This rescue was specific, because these effects were not observed when DopR is expressed in MB using OK107-Gal4 (which expresses in all lobes of the MB) or EB using c547-Gal4 (which expresses in R2/R4m ring neurons, where PPM3 neurons project) [28] (Figure 6C).
It was previously shown that activation of the FB neurons in the 104y-Gal4 driver results in an increase in sleep time [15]. Because our data suggest that the PPL1-dFB neurons promote arousal by signaling to the dFB, DA signaling to the dFB would be predicted to inhibit the activity of the neurons in 104y-Gal4. In order to address whether inhibition of the FB neurons of the 104y-Gal4 driver would decrease sleep time, we used 104y-Gal4 to drive expression of UAS-dORK1ΔC [40]. dORK1ΔC is a “leak” K+ channel, which electrically silences neurons [40]. 104y-Gal4>UAS-dORK1ΔC flies exhibited reduced sleep, compared to controls (Figure 6D). DopR is a D1-type DA receptor [41], which, when activated, stimulates adenyl cyclase activity and increases cAMP levels. Thus, to mimic the effects of activating DopR in the dFB, we expressed mC*, a constitutively active catalytic subunit of cAMP-dependent kinase (PKA), in the dFB using 104y-Gal4. Increasing PKA activity in the neurons in the 104y-Gal4 driver resulted in a significant decrease in sleep time (Figure 6D), consistent with a model whereby increasing DopR activity and increasing cAMP signaling in the dFB inhibits the function of these neurons, resulting in reduced sleep time.
DISCUSSION
In Drosophila, DA has been suggested to play an important role not only in sleep and arousal, but also in courtship behaviors, addiction, learning and memory, and appetite/taste [11, 13, 14, 26, 27, 38, 39, 42, 43]. Recent work has identified specific DA neurons in the PPL1 cluster (which are distinct from the dFB-projecting neurons identified in this study) and in the PAM cluster that play an important role in appetitive and olfactory memory [26, 27, 29, 44, 45]. These studies, along with our data, support the notion that individual DA cells, with their distinct targets, play important and qualitatively different roles in animal behavior. Our data suggests that a single DA neuron in each PPL1 subgroup, projecting to the dFB, significantly promotes wakefulness. However, because we did not directly activate single DA neurons, it is possible that these PPL1-dFB cells act in concert with other DA neurons to promote arousal. Indeed, there may be additional wake-promoting DA neurons, since activation of the neurons in TH-Gal4 causes a stronger phenotype than that seen with activation of the TH-D4 driver.
Next, to address the physiological regulation of these PPL1-dFB neurons, we used ANF-GFP as a reporter to monitor chronic neuronal activity. The sensitivity of using ANF-GFP to monitor chronic neuronal activity may be somewhat limited, as the ANF-GFP signal cannot be fully depleted even with supraphysiologic neuronal excitation. In addition, because neuropeptides and classical neurotransmitters are processed differently and have different kinetics of release during synaptic transmission, the ANF-GFP decrease may not completely correlate with DA release. Nonetheless, our data suggest that only the PPL1-dFB neurons are consistently active during the day under LD and DD conditions and during forced wakefulness at night, suggesting a specific role for these neurons in arousal. Although UAS-ANF-GFP has previously been used to study neuropeptide trafficking and release [31, 32], this work represents, to our knowledge, the first use of this tool to monitor chronic neuronal activity. Given the lack of a transgenic chronic neuronal activity reporter in Drosophila, this approach could be potentially useful for measuring neuronal activity on longer timescales (e.g., hours) in other types of neuronal circuits.
We also show that DopR is likely the sole DA receptor responsible for promoting wakefulness and that it is specifically required in the FB for this function. Since the dFB has recently been shown to promote sleep [15], our study provides an important link between wake and sleep circuits in Drosophila. The notion that a wake-promoting neuron may act on and inhibit a sleep-promoting center is reminiscent of the “flip-flop switch” model described in mammals, where wake-promoting nuclei (e.g., acetylcholinergic and monoaminergic nuclei) and sleep-promoting nuclei (e.g., ventrolateral/median preoptic nuclei, VLPO/MnPO) inhibit each other. In this mutually inhibitory relationship, when one side of the switch becomes more active, it suppresses the other side of the switch, facilitating a rapid and complete transition between wake and sleep [1]. In this model, the dFB in flies may be the analog of VLPO/MnPO in mammals. Because our data suggests that DopR is specifically required in the FB to promote arousal, our model predicts that the PPL1-dFB DA neurons signal to DopR in the FB and inhibit the function of downstream neurons. DopR is a D1-type DA receptor, which has adenyl cyclase activity [46]. In flies, it is unclear whether DopR signaling depolarizes or hyperpolarizes target neurons, but evidence from primary cultured Drosophila neurons suggests that activation of DopR can inhibit neurotransmission [47]. Finally, we find that expressing UAS-mC* to simulate potential downstream effects of elevated cAMP in the FB mimics the effects of electrically inhibiting the FB and is consistent with a model whereby increased DopR activation and subsequent cAMP signaling inhibits the dFB in order to promote wakefulness.
As the circuit map for sleep/wake regulation becomes more delineated, important questions regarding the regulation of the DA circuitry can be addressed. As in mammals, arousal circuits in flies are likely to be regulated by influences such as light, circadian, and homeostatic inputs [48]. For example, a recent study implicated light as modulating the arousing effects of DA specifically in PDF (Pigment Dispersing Factor) neurons [12]. In addition, DA is required for circadian entrainment to dim light, and cycling of DA expression may play a role in circadian rhythm strength [49]. Ultimately, unraveling the sleep/wake circuitry in flies should lead to a better understanding of how external stimuli, circadian, and homeostatic influences regulate arousal at the cellular and molecular level.
Experimental Procedures
Behavioral assays and immunostaining were generally performed as previously described [7]. Details of behavioral assays, imaging experiments, fly strains used, and molecular biology constructs and assays are available in the online Supplemental Information.
Supplementary Material
Highlights.
DA signaling is the primary mediator of arousal in Drosophila
Two DA neurons in PPL1 project to the dorsal fan-shaped body to promote wakefulness
Imaging experiments suggest these PPL1 neurons are more active during wakefulness
DopR is specifically required in the fan-shaped body to promote wakefulness
Acknowledgments
We thank Fred Wolf, Serge Birman, Hugo Bellen, Paul Garrity, Amita Sehgal, Chris Potter, Tim Lebestky, Ed Kravitz, and the Bloomington Stock Center for fly stocks. We also thank T. Ueno and K. Kume for communication of unpublished results. This work was funded by NINDS K08NS059671 and a Burroughs-Wellcome Fund Career Award for Medical Scientists (M.N.W.).
Footnotes
Supplemental Information includes 5 figures, 2 tables, and Supplemental Experimental Procedures and can be found with this article online at XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE. Sleep state switching. Neuron. 2010;68:1023–1042. doi: 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka C, Ishikawa M, Shimada S. Histochemical mapping of catecholaminergic neurons and their ascending fiber pathways in the rhesus monkey brain. Brain Res Bull. 1982;9:255–270. doi: 10.1016/0361-9230(82)90139-3. [DOI] [PubMed] [Google Scholar]
- 3.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 4.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 5.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 6.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, Hoshi T, Sehgal A, Koh K. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010;13:69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18:R670–R679. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 10.Stavropoulos N, Young MW. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron. 2011;72:964–976. doi: 10.1016/j.neuron.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 12.Shang Y, Haynes P, Pirez N, Harrington KI, Guo F, Pollack J, Hong P, Griffith LC, Rosbash M. Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci. 2011;14:889–895. doi: 10.1038/nn.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu MN, Koh K, Yue Z, Joiner WJ, Sehgal A. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep. 2008;31:465–472. doi: 10.1093/sleep/31.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332:1571–1576. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crocker A, Sehgal A. Octopamine regulates sleep in Drosophila through protein kinase A-dependent mechanisms. J Neurosci. 2008;28:9377–9385. doi: 10.1523/JNEUROSCI.3072-08a.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 18.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- 20.Cole SH, Carney GE, McClung CA, Willard SS, Taylor BJ, Hirsh J. Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. J Biol Chem. 2005;280:14948–14955. doi: 10.1074/jbc.M414197200. [DOI] [PubMed] [Google Scholar]
- 21.Alekseyenko OV, Lee C, Kravitz EA. Targeted manipulation of serotonergic neurotransmission affects the escalation of aggression in adult male Drosophila melanogaster. PLoS One. 2010;5:e10806. doi: 10.1371/journal.pone.0010806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sitaraman D, Zars M, Laferriere H, Chen YC, Sable-Smith A, Kitamoto T, Rottinghaus GE, Zars T. Serotonin is necessary for place memory in Drosophila. Proc Natl Acad Sci U S A. 2008;105:5579–5584. doi: 10.1073/pnas.0710168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, Wu CC, Chen GY, Ching YT, Lee PC, Lin HH, Hsu HW, Huang YA, Chen JY, Chiang HJ, Lu CF, Ni RF, Yeh CY, Hwang JK. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2010;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508:711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- 26.Claridge-Chang A, Roorda RD, Vrontou E, Sjulson L, Li H, Hirsh J, Miesenbock G. Writing memories with light-addressable reinforcement circuitry. Cell. 2009;139:405–415. doi: 10.1016/j.cell.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, Waddell S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139:416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong EC, Woo K, Li H, Lebestky T, Mayer N, Sniffen MR, Heberlein U, Bainton RJ, Hirsh J, Wolf FW. A pair of dopamine neurons target the D1-like dopamine receptor DopR in the central complex to promote ethanol-stimulated locomotion in Drosophila. PLoS One. 2010;5:e9954. doi: 10.1371/journal.pone.0009954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aso Y, Siwanowicz I, Bracker L, Ito K, Kitamoto T, Tanimoto H. Specific dopaminergic neurons for the formation of labile aversive memory. Curr Biol. 2010;20:1445–1451. doi: 10.1016/j.cub.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21:1523–1531. doi: 10.1523/JNEUROSCI.21-05-01523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao S, Lang C, Levitan ES, Deitcher DL. Visualization of neuropeptide expression, transport, and exocytosis in Drosophila melanogaster. J Neurobiol. 2001;49:159–172. doi: 10.1002/neu.1072. [DOI] [PubMed] [Google Scholar]
- 32.Shakiryanova D, Tully A, Hewes RS, Deitcher DL, Levitan ES. Activity-dependent liberation of synaptic neuropeptide vesicles. Nat Neurosci. 2005;8:173–178. doi: 10.1038/nn1377. [DOI] [PubMed] [Google Scholar]
- 33.Hokfelt T, Broberger C, Xu ZQ, Sergeyev V, Ubink R, Diez M. Neuropeptides--an overview. Neuropharmacology. 2000;39:1337–1356. doi: 10.1016/s0028-3908(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 34.Feng G, Hannan F, Reale V, Hon YY, Kousky CT, Evans PD, Hall LM. Cloning and functional characterization of a novel dopamine receptor from Drosophila melanogaster. J Neurosci. 1996;16:3925–3933. doi: 10.1523/JNEUROSCI.16-12-03925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han KA, Millar NS, Grotewiel MS, Davis RL. DAMB, a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron. 1996;16:1127–1135. doi: 10.1016/s0896-6273(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 36.Hearn MG, Ren Y, McBride EW, Reveillaud I, Beinborn M, Kopin AS. A Drosophila dopamine 2-like receptor: Molecular characterization and identification of multiple alternatively spliced variants. Proc Natl Acad Sci U S A. 2002;99:14554–14559. doi: 10.1073/pnas.202498299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srivastava DP, Yu EJ, Kennedy K, Chatwin H, Reale V, Hamon M, Smith T, Evans PD. Rapid, nongenomic responses to ecdysteroids and catecholamines mediated by a novel Drosophila G-protein-coupled receptor. J Neurosci. 2005;25:6145–6155. doi: 10.1523/JNEUROSCI.1005-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lebestky T, Chang JS, Dankert H, Zelnik L, Kim YC, Han KA, Wolf FW, Perona P, Anderson DJ. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron. 2009;64:522–536. doi: 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inagaki HK, Ben-Tabou de-Leon S, Wong AM, Jagadish S, Ishimoto H, Barnea G, Kitamoto T, Axel R, Anderson DJ. Visualizing Neuromodulation In Vivo: TANGO-Mapping of Dopamine Signaling Reveals Appetite Control of Sugar Sensing. Cell. 2012;148:583–595. doi: 10.1016/j.cell.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 41.Gotzes F, Balfanz S, Baumann A. Primary structure and functional characterization of a Drosophila dopamine receptor with high homology to human D1/5 receptors. Receptors Channels. 1994;2:131–141. [PubMed] [Google Scholar]
- 42.Kaun KR, Azanchi R, Maung Z, Hirsh J, Heberlein U. A Drosophila model for alcohol reward. Nat Neurosci. 2011;14:612–619. doi: 10.1038/nn.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marella S, Mann K, Scott K. Dopaminergic modulation of sucrose acceptance behavior in Drosophila. Neuron. 2012;73:941–950. doi: 10.1016/j.neuron.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Placais PY, Trannoy S, Isabel G, Aso Y, Siwanowicz I, Belliart-Guerin G, Vernier P, Birman S, Tanimoto H, Preat T. Slow oscillations in two pairs of dopaminergic neurons gate long-term memory formation in Drosophila. Nat Neurosci. 2012;15:592–599. doi: 10.1038/nn.3055. [DOI] [PubMed] [Google Scholar]
- 45.Berry JA, Cervantes-Sandoval I, Nicholas EP, Davis RL. Dopamine is required for learning and forgetting in Drosophila. Neuron. 2012;74:530–542. doi: 10.1016/j.neuron.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang YY, Kandel ER. D1/D5 receptor agonists induce a protein synthesis-dependent late potentiation in the CA1 region of the hippocampus. Proc Natl Acad Sci U S A. 1995;92:2446–2450. doi: 10.1073/pnas.92.7.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan N, Lee D. Suppression of excitatory cholinergic synaptic transmission by Drosophila dopamine D1-like receptors. Eur J Neurosci. 2007;26:2417–2427. doi: 10.1111/j.1460-9568.2007.05870.x. [DOI] [PubMed] [Google Scholar]
- 48.Saper CB, Cano G, Scammell TE. Homeostatic, circadian, and emotional regulation of sleep. J Comp Neurol. 2005;493:92–98. doi: 10.1002/cne.20770. [DOI] [PubMed] [Google Scholar]
- 49.Hirsh J, Riemensperger T, Coulom H, Iche M, Coupar J, Birman S. Roles of dopamine in circadian rhythmicity and extreme light sensitivity of circadian entrainment. Curr Biol. 2010;20:209–214. doi: 10.1016/j.cub.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.