Abstract
Cyclization of linear peptidyl precursors produced by nonribosomal peptide synthetases (NRPSs) is an important step in the biosynthesis of bioactive cyclic peptides. Whereas bacterial NRPSs use thioesterase (TE) domains to perform the cyclization, fungal NRPSs have apparently evolved to use a different enzymatic route. In verified fungal NRPSs that produce macrocyclic peptides, each megasynthetase terminates with a condensation-like (CT) domain that may perform the macrocyclization reaction. To probe the role of such a CT domain, we reconstituted the activities of the Penicillium aethiopicum trimodular NPRS TqaA in Saccharomyces cerevisiae and in vitro. Together with a reconstituted bimodular NRPS AnaPS, we dissected the cyclization steps of TqaA in transforming the linear anthranilate-D-tryptophan-L-alanyl tripeptide into fumiquinazoline F. Extensive biochemical and mutational studies confirmed the essential role of the CT domain in catalyzing cyclization in a thiolation domain-dependent fashion. Our work provided evidence of a likely universal macrocyclization strategy employed by fungal NRPSs.
Cyclic nonribosomal peptides (NRPs) from filamentous fungi represent a group of pharmaceutically important natural products, including the immunosuppressive cyclosporine A and the antifungal family of lipopeptides represented by echinocandin B1,2. Biosynthesis of the linear precursors of these molecules by their corresponding nonribosomal peptide synthetases (NRPSs) parallels that of bacterial NRPs, such as the well-studied tyrocidine A and gramicidin3,4. Following the sequential addition of each amino acid catalyzed by a NRPS module, which consists minimally of condensation (C), adenylation (A) and thiolation (T) domains, cyclization and product release take place through nucleophilic attack on the thioester carbonyl by a free amine from either the N-terminus or side chain of the peptide3,5. Given the importance of cyclization to the biological activities of these molecules, understanding the enzymatic basis of this final step in NRPs biosynthesis is a significant goal from both mechanistic and biocatalytic perspectives6,7.
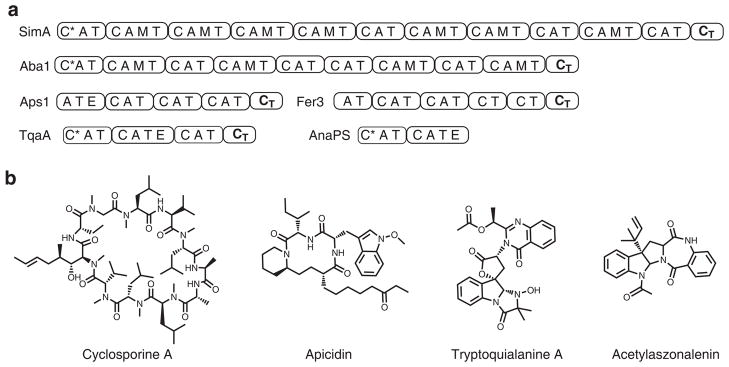
Whereas bacterial NRPSs use thioesterase (TE) domains to perform the cyclization, fungal NRPSs have apparently evolved to use a complementary strategy7–9. From the verified fungal NRPS gene clusters to date, none of the NRPSs associated with cyclic NRPs contain terminal TE domains, nor a free-standing TE-like domain encoded in the vicinity of the gene cluster10. Instead, TE domains have only been found to be associated with hydrolyzed peptide products such as the linear ACV tripeptide synthesized by the ACV synthetase5. In macrocyclic fungal NRPs such as cyclosporine A, aureobasidin A, apicidin and ferrichrome A, each corresponding NRPS terminates with a condensation-like domain (CT)11–15(Fig. 1). Similarly, in smaller polycyclic NRP alkaloids such as tryptoquialanine, the NRP core is assembled by a NRPS (TqaA) that terminates with a CT domain16 (Fig. 1). In the NRPS paradigm, C domains are canonically categorized to catalyze the formation of a peptide bond between the growing peptidyl-S-Tn and the activated aminoacyl-S-Tn+1 using an active site histidine as the general base17,18. The active site serves as a scaffold to bring together the donor and acceptor aminoacyl thiolation domains19. Therefore, it is surprising that the CT domain (a subset of C domains) can be recruited by fungal NRPS to perform the equivalent reaction of a TE domain, which relies on a serine residue for nucleophilic catalysis7. Understanding the role of the CT domain in fungal NRPSs will have broad implications, as many of the cryptic NRPSs discovered from fungal genome sequences terminate with a CT domain 9,10.
Figure 1.
Fungal nonribosomal peptide synthetases (NRPSs) that are terminated with a C-like domain. (a) Representative NRPSs: cyclosporine NRPS SimA, aureobasidin A NRPS Aba1, apicidin NRPS Aps1, ferrichrome A NRPS Fer3, tryptoquialanine NRPS TqaA, acetylaszonalenin NRPS AnaPS. Domain abbreviations: C: condensation domain; A: adenylation domain; T: thiolation domain; E: epimerization domain; C*: truncated and presumably inactive C domain; CT: Terminal C-like domain. (b) Representatives structures of final products synthesized by the clusters in part a.
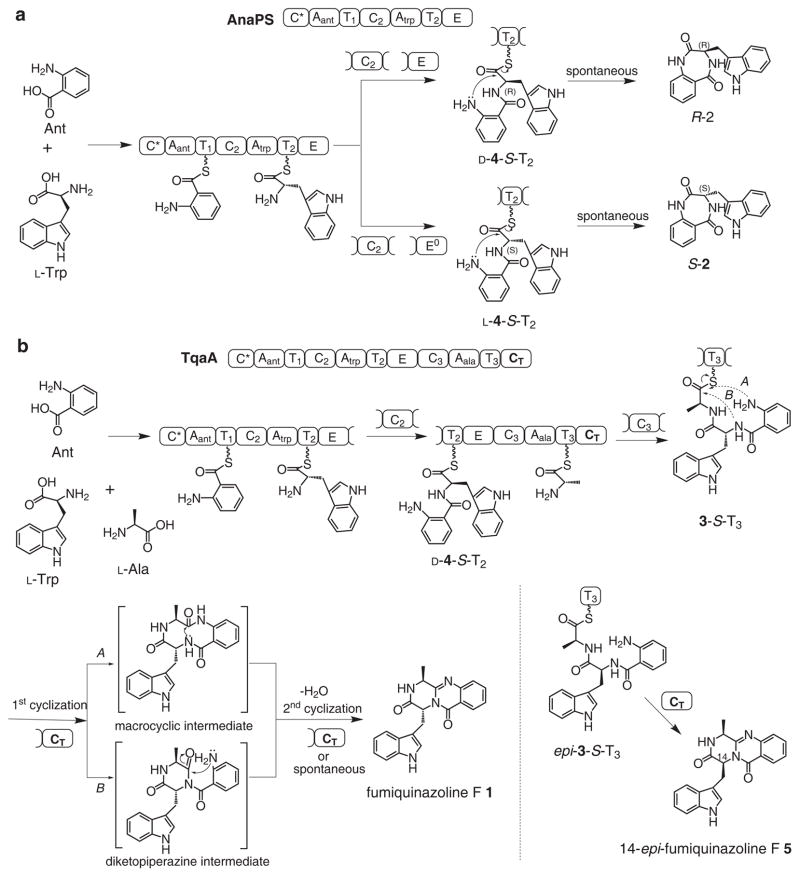
To investigate the role of the CT domain and the cyclization of fungal NRPs, we focused on the trimodular TqaA, which has been shown genetically to be involved in the biosynthesis of the tricyclic peptidyl alkaloid fumiquinazoline F 1 (Fig. 2) in Penicillium aethiopicum16. TqaA is functionally equivalent to Af12080 from the fumiquinazoline A biosynthetic pathway in Aspergillus fumigatus20. The three modules (C*AT1-CAT2E-CAT3-CT) of TqaA are proposed to activate anthranilate (Ant), L-tryptophan and L-alanine to yield the tripeptide Ant-D-Trp-L-Ala 3 attached to the last T domain (3-S-T3)16,21 (Fig. 2). Formation of the pyrazinoquinazoline ring and release of 1 require two cyclization steps, hypothesized to be catalyzed by the CT domain (Fig. 2). Interestingly, the domain organization and substrate selectivity of the first two modules of TqaA are identical to those of AnaPS (C*AT1-CAT2E), which is involved in the synthesis of the 3-indolylmethyl-3,4-dihydrobenzo-1,4-diazepine-2,5-dione precursor R-2 of acetylaszonalenin in Neosartorya fischeri22. Intriguingly, AnaPS terminates with an epimerization (E) domain rather than a CT domain, although it is known that E and C domains share sequence and structural similarities18.
Figure 2.
Proposed AnaPS (a) and TqaA (b) cyclization mechanisms. (Domain abbreviations: C: condensation domain; A: adenylation domain; T: thiolation domain; E: epimerization domain; C*: truncated and presumably inactive C domain; CT: Terminal C-like domain).
Here we present the complete in vitro reconstitution of the trimodular TqaA expressed from Saccharomyces cerevisiae, allowing biochemical investigation of the cyclization steps. The functions of AnaPS are studied in parallel to compare and contrast the cyclization steps between the two NRPSs. We show that the CT domain of TqaA is indispensable for the cyclization of 1 and requires protein-protein interactions with the T3 domain for catalysis.
RESULTS
In vitro Reconstitution of TqaA and AnaPS Activities
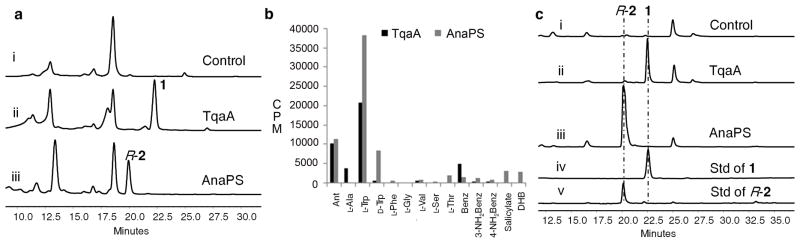
To pinpoint the domains that are involved in the cyclization of linear peptides, we first attempted to reconstitute the activities of the NRPSs in vitro. This is particularly challenging for the megasynthetases such as TqaA (450 kDa) as no NRPS with more than two modules has been successfully expressed in an active form to date. Towards this end, Saccharomyces cerevisiae 2-μm episomal vectors encoding tqaA (12 kb) and anaPS (7 kb) under the control of the ADH2 promoter were constructed using a modified yeast-based homologous recombination method23 (Supplementary Results, Supplementary Fig. 1). Both N-FLAG-tagged enzymes were expressed using the engineered S. cerevisiae BJ5464-NpgA, which contained multiple vacuolar protease knockouts and a chromosomally integrated copy of the Aspergillus nidulans npgA gene that encoded a phosphopantetheinyl transferase24. The co-expression of NpgA in BJ5464 is required for the conversion of apo thiolation domains into the phophopantetheinylated holo forms25. This strain had been successfully used as a host for heterologous reconstitution of large fungal polyketide synthases24,26. To verify functional expression of the NRPSs, small molecule metabolites in the three-day growth media were extracted with ethyl acetate and analyzed using LC-MS. Compared to the untransformed yeast strain, BJ5464-NpgA expressing TqaA and AnaPS produced 1 and R-2 at ~2 mg/L, respectively (Fig. 3a). Following this confirmation of NRPS activities, full length TqaA and AnaPS (264 kDa) were purified to near homogeneity by using anti-FLAG antibody affinity chromatography followed by gel filtration to final yields of ~ 1 mg l−1 (Supplementary Figures 2 and 3).
Figure 3.
Characterization of TqaA and AnaPS. (a) in vivo reconstitution of TqaA and AnaPS. Shown are HPLC analysis (λ = 272 nm) of metabolites extracted from 3-day cultures of i) untransformed BJ5464-NpgA; ii) BJ5464-NpgA expressing TqaA; iii) BJ5464-NpgA expressing AnaPS. (b) amino acid-dependent ATP-PPi exchange assays with AnaPS and TqaA. Abbreviations: benzoate (Benz); 1, 2-dihydroxybenzoate (DHB). (c) in vitro reconstitution of TqaA and AnaPS. Shown are HPLC analysis (λ = 272 nm) of compounds from extraction of reaction mixtures containing 2 mM of the amino acid building blocks and i) no enzyme; ii) 10μM TqaA; iii) 10 μM AnaPS; iv) standard of 1; and v) standard of R-2.
After the affinity purification step, activities of the A domains of NRPSs were assessed using amino acid-dependent γ-32P-ATP-PPi exchange assay (Supplementary Methods and Fig. 3b)27. TqaA specifically activated the three building blocks Ant, L-Trp and L-Ala over the respective D-amino acids or aryl acids. Similarly, AnaPS A domains activated Ant and L-Trp. Both NRPSs also activated aryl substrates such as benzoate (by TqaA), salicylate (by AnaPS) and 1, 2-dihydroxylbenzoate (DHB, by AnaPS), pointing to the relaxed substrate specificities of the Ant-specific A domains. The preference displayed by both NRPSs toward L-Trp over D-Trp strongly points to an active epimerization (E) domain in the second module of both NRPSs.
Complete reconstitution of TqaA and AnaPS activities was then performed by adding the amino acid building blocks (2 mM each) and ATP (6 mM) to each of the purified enzymes (10 μM). The reaction mixtures were incubated at room temperature for 12 hours, extracted with ethyl acetate and analyzed by LC-MS (Fig. 3c). 1 (obtained from the ΔtqaH strain of P. aethiopicum16) and chemically synthesized R-2 were used as authentic standards (Supplementary Methods). In the TqaA assay, 1 was synthesized as the single peptidyl product; no dipeptidyl or other tripeptidyl products could be detected through LC and selective ion monitoring by MS. A HPLC-based time course study revealed that the reaction proceeds with initial product formation rate of 0.40 ± 0.014 min−1 (Supplementary Fig. 4a). Similarly in the AnaPS assay, the only product detected was the 6, 7-cyclic dipeptide R-2 with an initial product formation rate of 1.47 ± 0.047 min−1 (Supplementary Fig. 4c).
Reconstitution of the synthesis of 1 and R-2 using purified TqaA and AnaPS, respectively, showed that each NRPS was sufficient to construct and cyclize the linear peptide into the multicyclic products. The proposed cyclization mechanisms are shown in Fig. 2. In the simpler case of AnaPS, attack of the Ant free amine on the thioester carbonyl of Ant-D-Trp-S-T2 (D-4-S-T2) yields R-2. However in TqaA, this reaction is apparently suppressed in favor of chain elongation by the third module to yield 3-S-T3. Two cyclization routes to 1 are possible starting from 3-S-T3 (Fig. 2b). In route A, attack of the Ant amine on the thioester carbonyl yields a 10-membered macrocyclic intermediate, which can undergo a bridging amide bond cyclization and dehydration to yield 1; in route B, the D-Trp amide serves as the initial nucleophile in attacking the thioester 3-S-T3 to yield the diketopiperazine (DKP) intermediate. Subsequent intramolecular attack by the Ant amine and dehydration completes the pyrazinoquinazoline scaffold.
Formation of R-2 can be spontaneous
To investigate the cyclization step of AnaPS, we chemically synthesized Ant-D-Trp-S-N-acetylcysteamine (D-4-SNAC). Under enzymatic assay conditions, D-4-SNAC was cyclized to yield R-2 with an initial product formation rate of 0.1 min−1 (Supplementary Fig. 5a). The significant rate of spontaneous cyclization suggested that product release from AnaPS could be uncatalyzed. The spontaneous cyclization rate, however, was at least fourteen-fold slower than that of D-4-S-T2, hinting that domains in AnaPS may promote a conformation of the dipeptide that was more favorable for formation of the seven-membered ring. Adding AnaPS to D-4-SNAC, however, did not increase the cyclization rate, suggesting that the proposed templating effect may only take place with an enzyme-tethered dipeptide (Supplementary Fig. 5b).
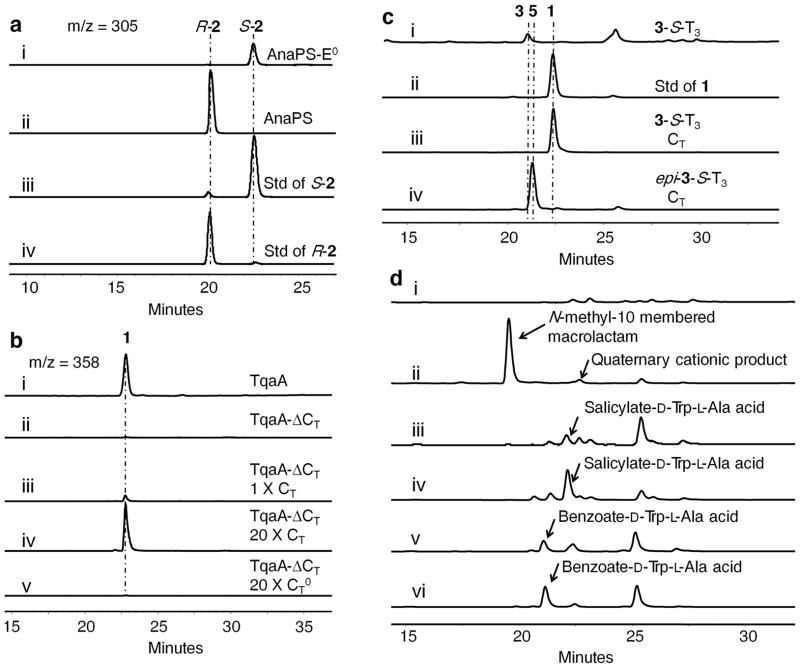
The enantiomer Ant-L-Trp-SNAC (L-4-SNAC) was also prepared synthetically, and an identical rate of cyclization to S-2 was observed (Supplementary Methods and Supplementary Fig. 5c). The availability of both R-2 and S-2 as standards allowed us to examine the enantiopurity of the product released by AnaPS using chiral HPLC (Fig. 4a). Under assay conditions, only enantiomerically pure R-2 was detected in the organic extract, suggesting AnaPS exerts strict product stereochemical control. To probe the importance of the epimerization step catalyzed by the E domain on R-2 formation, we generated a point mutant of AnaPS (AnaPS-E0) in which the second His in the H2059HxxxDx catalytic motif was mutated to Ala (Supplementary Fig. 3) 28. As expected, AnaPS-E0 no longer produces R-2 when Ant and L-Trp are added. Instead, we observed the appearance of S-2 at only ~1% of the level of R-2 produced by the wild type AnaPS (Fig. 4a, note the sizes of R-2 and S-2 peaks in traces i and ii are not drawn to scale). Based on the very low level of S-2 production by AnaPS-E0, two possible control mechanisms can be proposed. In the first model, AnaPS could suppress the rapid cyclization of L-4-S-T2 until epimerization to D-4-S-T2 takes place. The chiral environment in the E domain active site may position the Ant amine away from the thioester carbonyl. In this model, epimerization of the Trp residue took place on the dipeptidyl-S-T2, which was proposed to be favored over the epimerization of the free aminoacyl thioester due to stabilization of the carbanion intermediate through acylation of the nitrogen in the peptide bond29. In the second model, formation of L-4-S-T2 by the C2 domain was strongly disfavored in the absence of epimerization, therefore requiring the E domain to function on L-Trp-S-T2 prior to condensation with Ant-S-T1. Intriguingly, D-Trp could be activated and transferred to the T2 domain of AnaPS (Supplementary Fig. 6). However, AnaPS did not produce any product in the presence of Ant and D-Trp, suggesting that L-Trp was required for condensation to take place and thus providing evidence against the second model. Further attempts to distinguish between the two models through excision of the E domain from AnaPS were not successful because no soluble protein was recovered.
Figure 4.
The cyclization steps of TqaA and AnaPS. (a) chiral HPLC followed by selected mass ion monitoring (m/z 305) of R-2 or S-2 synthesized from AnaPS (trace ii) and AnaPS-E0 (trace i). Standards of R-2 (trace iv) and S-2 (trace iii) are shown for comparison. The intensities of peaks are not drawn to scale. (b) confirmation of TqaA CT domain function. All assays contained 10μM of the NRPS (TqaA or TqaA-ΔCT) and 2 mM of each amino acid. Standalone CT or CT0 was added in trans. The traces shown are selected mass ion monitoring (m/z 358) of compounds extracted from reactions containing i) wild type TqaA; ii) TqaA-ΔCT; iii) TqaA-ΔCT with 10 μM CT; iv) 10 μM TqaA-ΔCT with 200 μM CT; and v) TqaA-ΔCT with 200 μM CT0. (c) confirmation of CT cyclization activity using preloaded peptidyl thiolation domain. 3-S-T3 or epi-3-S-T3 (200 μM) was used to assay the production of 1 or 5, respectively. Traces shown are HPLC analyses (λ = 272 nm) of i) 3-S-T3 without CT; ii) standard of 1; iii) 3-S-T3 with 20 μM CT; iv) epi-3-S-T3 with 20 μM CT. (d) Analogues of 3 were used to probe the cyclization route of CT domain. 200 μM tripeptide analogueswere used to react with CT (20 μM). Traces i) and ii) are Ant-L-N-Me-Trp-L-Ala-S-T3 without and withCT, respectively; Trace iii) and iv) are salicylate-D-Trp-L-AlaS-T3 without and withCT, respectively; Trace v) and vi) are benzoate-D-Trp-L-Ala-S-T3 without and with CT, respectively.
Given that the D-4-S-T2 intermediate can rapidly cyclize spontaneously, it is intriguing how formation of R-2 can be effectively suppressed by TqaA following formation of this dipeptidyl intermediate, especially given that the first two modules of TqaA and AnaPS appear to be functionally equivalent. Efforts to express modules 1 and 2 of TqaA as a standalone enzyme, or replace them with AnaPS resulted in insoluble proteins, despite the high sequence similarity. An initial hypothesis was that the subsequent condensation with L-Ala catalyzed by C3 of TqaA was sufficiently rapid to sequester the reactive dipeptide. However, mutation of the C3 active site general base His2658 to Ala not only abolished the synthesis of 1, but also failed to yield any trace of R-2. Similarly, performing the TqaA in vitro assay with only Ant and L-Trp also did not produce R-2 (Supplementary Fig. 7). Hence, it was reasonable to propose that the C3 active site, in particular through the donor peptidyl entrance site, can structurally protect the D-4 moiety and can prevent cyclization and offloading of the intermediate as R-2.
Formation of Fumiquinazoline F requires the CT domain
Having probed the formation of R-2 by AnaPS, we next examined the cyclization of 3-S-T3 into 1 by TqaA. This was a particularly intriguing transformation as a simple linear tripeptide was morphed into the 6, 6, 6 pyrazinoquinazoline tricyclic ring system. First, to understand the stability of the tripeptidyl thioester, we synthesized 3-SNAC starting from the free acid 3 (Supplementary Methods). In contrast to 4-SNAC, 3-SNAC was remarkably stable under assay conditions, and levels of the hydrolysis product 3 only reached a noticeable level after approximately 8 hours. No cyclized products such as 1 can be observed after more than 24 hours of incubation (Supplementary Fig. 5d). Therefore, whereas formation of R-2 can be spontaneous, formation of 1 definitely required enzyme catalysis. The CT domain in TqaA was the most likely domain that could catalyze this reaction. Phylogenetic analysis showed that fungal CT domains group separately from the canonical chain-extending C domains (Supplementary Figures 8 and 9). Based on bioinformatic analysis, the percentage of fungal NRPSs incorporating a CT domain varied from 60 to 90%, depending on the genome analyzed (Supplementary Table 2).
Multiple sequence alignment of the active sites of different sequenced CT domains revealed variations in the first His residue of the highly conserved HHxxxDxxS motif of C domains (S3765HSQWDGVS3773 in TqaA)17 (Supplementary Fig. 10). The His proposed to be acting as the general base is conserved in all CT domains. To probe the role of the CT domain in pyrazinoquinazoline formation, we constructed the following mutants of TqaA: S3765A and S3765H to probe the role of the nonconserved serine, and H3766A to probe the role of the putative active site base. Among them, TqaA H3766A was expressed and purified from BJ5464-NpgA for in vitro assay (Supplementary Fig. 3). Whereas the S3765 mutants maintained production of 1 at levels comparable to that of the wild type in S. cerevisiae, the H3766A mutant failed to yield any detectable peptidyl product both in S. cerevisiae and in vitro using the purified enzyme (Supplementary Fig. 11). To exclude the possibility that an inactivated CT domain may structurally prevent the cyclization of 3-S-T3, a truncated version of TqaA without the CT domain (TqaA-ΔCT, 400 kDa) was constructed, expressed and assayed. In parallel, the standalone CT domain was expressed and purified from Escherichia coli (Supplementary Fig. 3). When assayed in vitro, whereas TqaA-ΔCT failed to produce any detectable amount of 1, complementation of TqaA-ΔCT with equimolar amounts of CT in trans restored the synthesis of 1, albeit to a lesser extent compared to full length TqaA (Fig. 4b). The lower level of 1 may be a result of inefficient in trans protein-protein interactions. Indeed, addition of twenty-fold molar excess of CT to the reaction mixture dramatically increased the production of 1, resulting in levels comparable to that observed when using intact TqaA. In contrast, in trans addition of a CT0 mutant in which the catalytic His was mutated to Ala failed to restore the synthesis of 1. With all of the inactivated mutants described above we did not detect any hydrolyzed 3 free acid in the reaction mixture, suggesting that the NRPS-tethered tripeptide was remarkably resistant to hydrolysis compared to 3-SNAC (Supplementary Fig. 5d). Collectively, these results unequivocally proved the essential role of the terminal CT domain in formation of the final product 1.
CT cyclization activity requires T domain
Having confirmed the catalytic nature of the CT domain, we next assayed the standalone CT in the cyclization reaction. Cyclizing TE domains from bacterial NRPSs can cyclize small molecule thioester mimics of the linear precursors6,30. However, when 3-SNAC or 3-S-CoA was used as a substrate for TqaA CT, no cyclized 1 was detected after prolonged incubation (Supplementary Methods and Supplementary Fig. 12). To prepare the authentic 3-S-T3 substrate for CT, the apo form of standalone T3 was expressed from E. coli (Supplementary Fig. 3) and modified in vitro in the presence of 3-S-CoA and the broadly specific phosphopantetheinyl transferase Sfp31. MALDI-TOF mass analysis confirmed the near quantitative conversion from the apo form (molecular weight 11197) to 3-S-T3 (molecular weight 11913) (Supplementary Fig. 13). In the absence of the CT domain, slow hydrolysis of this thioester to 3 was observed. When the wild type CT was combined with 3-S-T3, a new peak was observed during HPLC that corresponds to 1, and no hydrolysis was detected (Fig. 4c), confirming the importance of the T domain in facilitating CT functions. Adding the mutant CT0 did not yield any cyclized product, but partially suppressed the hydrolysis of 3-S-T3 possibly due to the templating effect of the CT active site. The kinetic parameters of the cyclizing reaction were measured as shown in Supplementary Fig. 14. The kcat of the reaction was determined to be 0.82 ± 0.053 min−1 and Km of CT for 3-S-T3 was 1.1 ± 0.12 mM. 1 can also be formed in a one-pot reaction containing apo-T3, 3-S-CoA, Sfp and CT, in which the phosphopantetheinyl modification took place in situ. When Ant-L-Trp-L-Ala-S-CoA (epi-3-S-CoA) was used to prepare epi-3-S-T3, the CT domain also efficiently cyclized the diastereomeric peptide into the corresponding 14-epi-fumiquinazoline F 5, indicating relaxed substrate specificity of the cyclization reaction towards the tripeptide (Fig. 2 and Fig. 4c).
To examine the T domain substrate specificity of CT, we also prepared noncognate T domains 3-S-TApdA and 3-S-TLovF, which correspond to the ACP domains of the fungal PKS-NPRS ApdA and PKS LovF, respectively26,32. Both TApdA and TLovF share low sequence identity/similarity (~20%/60%) with TqaA T3. TApdA naturally interacts with a downstream C domain in ApdA and the tethered polyketide intermediate is condensed with a tyrosine residue, while TlovF does not interact with any C-like domain. TqaA CT was able to recognize 3-S-TApdA to produce 1, albeit with <10% efficiency compared to 3-S-T3 (Supplementary Fig. 15) and was accompanied by hydrolysis of 3-S-TApdA. Only background hydrolysis of 3-S-TApdA was observed when TLovF was used as the peptidyl carrier. Therefore, the protein-protein interactions between CT and the T domains are selective, and may rely on T domain sequence elements that are unique for recognition by C domains. Noncognate T domains such as TLovF that do not interact with downstream C-like domains likely do not possess these elements and are not substrates of the CT domain.
Cyclization Mechanism of CT
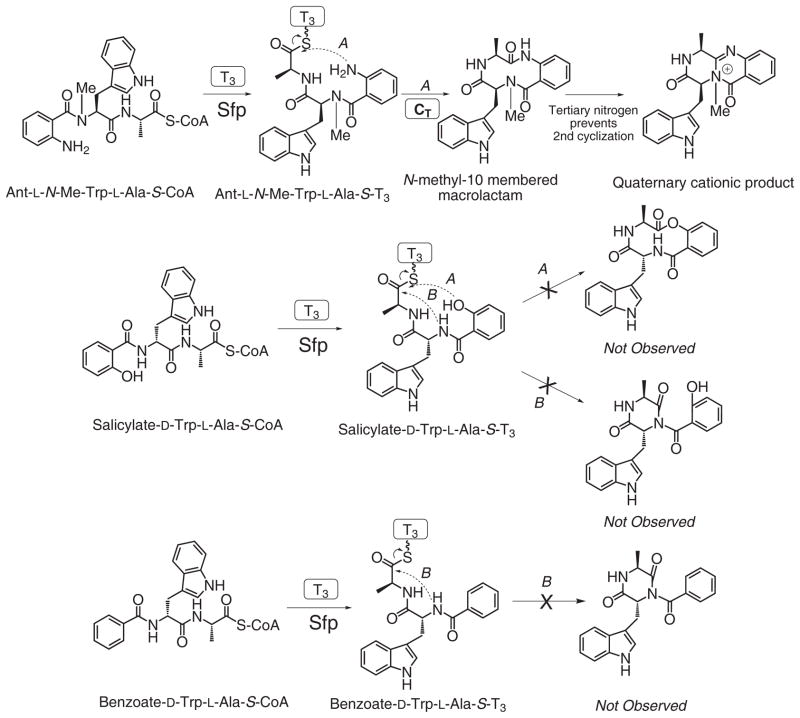
In an attempt to differentiate between the two alternate mechanisms of cyclization to the observed 6, 6, 6 tricyclic compound 1 (Fig. 2b), we examined the propensity of analogues of the natural Ant-Trp-Ala system to undergo CT mediated cyclization (Fig. 4d and Supplementary Fig. 16). Using the standalone T3 domain of TqaA and Sfp as loading catalyst we generated three tripeptidyl-T3 analogues specifically designed to disfavor either the macrocyclization (pathway A in Fig. 2b) or DKP formation (pathway B in Fig. 2b). From Ant-L-N-Me-Trp-L-Ala-S-T3, a released product with mass [M+H]+ of 391 (exact mass = 391.1765) consistent with the proposed N-methyl-10 membered macrolactam (Fig. 5 and Supplementary Fig. 17) was observed. Further cyclization of this intermediate was strongly disfavored (both sterically and by the formation of a quaternary cationic product) by the tertiary nitrogen which would be required to act as nucleophile. Nonetheless, a mass consistent with this cationic product resulting from the transannular attack of the tertiary amine actually was observed as a very minor product. The formation of this product despite the steric and electronic barriers to its formation (as stated above) lends support to the geometry of this proposed macrocycle favoring the required orientation for transannular cyclization. It is worth noting that the same mass would also be consistent with the formation of a cation N-Me-Trp derived DKP, but such a product would likely be very unstable and be subject to rapid hydrolysis to form Ant-N-Me-Trp-Ala free acid, which was not detected. Conversely salicylate-D-Trp-L-Ala-S-T3 or benzoate-D-Trp-L-Ala-S-T3, which would be expected to disfavor/prevent macrocyclization (but not interfere with DKP formation), gave no products corresponding to the mass of the dehydrated tripeptide (corresponding to either DKP or macrocycle isomers) (Fig. 5). Although the mechanism of CT mediated cyclization remains difficult to elucidate due to the reactivity of proposed intermediates, taken together these data support a route where macrolactamization precedes DKP formation in the formation of these CT mediated 6, 6, 6 tricyclic scaffolds.
Figure 5.
Probing the possible TqaA CT domain cyclization mechanisms using analogues of the tripeptide 3.
DISCUSSION
In this work, we have reconstituted the activities of two fungal NRPSs, the trimodular TqaA and the bimodular AnaPS that make tricyclic and bicyclic heterocyclic scaffolds, respectively. The choice of the two NRPSs was based on the observation that while the first two modules of TqaA are identical to AnaPS, addition of the third module dramatically altered the structures of the resulting polycyclic NRP alkaloid products. The difference in the cyclization pattern is likely due to the precise control of the reactive dipeptide D-4 intermediate, and the use of a C-like cyclization domain that regioselectively directs the intramolecular attack of the Ant amine nucleophile. While bimodular NRPSs from both bacteria and fungi have been reconstituted previously33–35, the sheer size of NRPSs such as TqaA complicated soluble expression and functional reconstitution. The use of S. cerevisiae BJ5464-NpgA allowed high-yield heterologous expression and full manifestation of TqaA activities. The successful use of this strain in affording intact NRPSs should open the door to the functional analysis of the expanding collection of fungal NRPSs available through different sequencing efforts.
The use of the cyclic aromatic β-amino acid anthranilate as the chain initiation building block by AnaPS and TaqA is key for the generation of the 6, 7 fused bicyclic R-2 (AnaPS) and the fused 6, 6, 6 tricyclic 1 (TqaA) scaffolds. The planar 1, 3 disposition of the amino group and the carbonyl in the anthranilyl unit efficiently sets up the D-4-S-T2 intermediate for cyclization on the second module of AnaPS. This facile cyclization, demonstrable in the nonenzymatic thioester model, sets the unusual 6, 7 bicyclic ring system that is subsequently elaborated by prenylation and acetylation in the acetylaszonalenin pathway22. Notably this same 6, 7-scaffold would be an unwanted derailment product in the trimodular TqaA. Such premature release is seemingly suppressed by the action of the C-domain that results in the generation of a 3-S-T3 species. Chain release at this point mediated by CT instead creates the tricyclic scaffold of 1. The NH2 of the anthranilyl unit is again acting as an internal nucleophile on a thioester but this does not happen appreciably nonezymatically. Clearly, the use of anthranilate as a NRPS starter unit enables construction of distinct patterns of nitrogen-containing bicyclic and tricyclic scaffolds at the core of related families of fungal peptidyl alkaloids. In the case of the fumiquinazolines, these can be further elaborated into highly constrained architectures with up to seven fused rings in the mature scaffold36. Thus, understanding how anthranilate gets activated, elongated, and cyclized on fungal NRPS assembly lines offers prospects for diversification of peptidyl alkaloid scaffolds.
Fungal NRPSs have several different strategies for product release following completion of peptide elongation7,8. TE domains have been found to be responsible for the hydrolysis of full length peptidyl-NRPS adducts as in the ACV synthetase that is responsible for the production of the ACV peptide precursor to penicillin5. NADPH-dependent reductive (R) domains have been found to be associated with the family of peptaibol linear peptides, in which the C-terminal thioester is reduced to an amino alcohol37. R and R-like domains are also found at the C-termini of PKS-NRPS hybrid megasynthetases which can catalyze reductive chain release or Dieckmann cyclization, respectively38,39. However, based on recent genome sequencing efforts, a majority (60–90%) of the NRPSs found in filamentous fungi terminate with a CT domain (Supplementary Table 2). Previous sequencing of individual gene clusters has also revealed that CT domains are associated with macrocyclic and/or polycyclic NRPs. In this work, using the pyrazinoquinazoline 1 as an example, we demonstrated that the CT domain in the TqaA NRPS was responsible for the cyclization of the highly stable tripeptide 3-S-T3. The cyclization function of CT is likely to be general for other fungal NRPSs such as those shown in Fig. 1, including cyclosporine synthetase SimA11. Macrocyclization CT domains may also play a role in the biosynthesis of cyclohexadepsipeptide destruxins in the insect pathogenic fungi Metarhizium robertsii40 and in NRPSs from filamentous cyanobacteria, as illustrated in the apratoxin biosynthetic gene cluster from Lyngbya bouilloni41. CT domains have also been proposed to be involved in the cyclization via ester bond formation during the biosynthesis of bacterial polyketide-peptidyl hybrid FK520 and rapamycin42,43. However, those CT domains fall into a different group by phylogenetic analysis (Supplementary Fig. 9).
The use of a CT domain to heterocyclize or macrocyclize peptidyl products is a different strategy from the canonical TE domains found in bacterial NRPSs. The CT domains and TE domains have completely different folds and utilize different mechanisms to catalyze the same reaction. In the TE domain where a catalytic triad is present, the peptidyl intermediate is first transferred to the active site serine, followed by intramolecular nucleophilic attack on the oxyester to complete cyclization7. In contrast, as seen for canonical C domains that catalyze peptide bond formation, no covalent adduct with the enzyme is expected to be formed during CT catalyzed cyclization5. Instead, the catalytic histidine deprotonates the amine nucleophile, thus facilitating its attack on the thioester carbonyl to cyclize and release the product in one step. Another significant mechanistic difference is the requirement for a specific T domain partner in the CT domain catalyzed cyclization reaction. We showed here that only peptide substrates tethered to the natural T domain partner of the CT domain are efficiently recognized, while peptidyl-SNAC or peptidyl-CoA substrates are not recognized. This contrasts with most TE domains where small-molecule mimics of the peptidyl-thioester can be used to probe the cyclization functions6,30. One exception is the fengycin TE that involves direct interactions with the upstream T domains to mediate the cyclization44. The interactions between T and CT domains likely evolved from the interactions between upstream T and downstream C domains during peptide elongation, which facilitate the unidirectional delivery of the peptidyl substrate in a productive conformation to the donor site. This specific interaction may be advantageous to prevent spontaneous cyclization of the peptide from backbone amides or side chain amines. CT domains also appear to advantageously eliminate the competing hydrolysis reaction, which is observed in vitro with bacterial TE domains where hydrolysis of the oxyester intermediate can take place in competition with the cyclization reaction7. This is especially prevalent under in vitro conditions where a standalone TE is used. In contrast, the templating nature of the CT highly disfavors hydrolysis as illustrated in the reconstitution assay using 3-S-T3.
METHODS
Cloning, expression and purification of TqaA and AnaPS
Cloning of the intact gene into the expression vector was performed using the modified yeast-based homologous recombination method23. The tqaA gene was divided into 4 pieces (P1 ~ P4) with a maximum size of ~4.5 kb. The only intron (577 – 661 bp) of the tqaA gene within P1 was removed by RT-PCR. P2 to P4 were PCR amplified from the genomic DNA of P. aethiopicum. Each successive piece was designed to overlap (35 – 40 bp) with the two flanking ones, and the 5′-end of P1 and 3′-end of P4 overlap with the expression vector. The vector fragment was generated by digesting pXW55 (an expression vector with an engineered FLAG tag on N-terminus and 6×His-tag on C-terminus) with SpeI and PmlI26. The individual tqaA pieces (1 μg per piece) as well as the vector (~ 0.5 μg) were co-transformed using the EasyComp™ Transformation Kit (Invitrogen) into S. cerevisiae strain BJ5464-NpgA to allow assembly of the entire gene in vivo. The assembled plasmid was miniprepped from S. cerevisiae using yeast plasmid miniprep II kit (Zymo) and further verified by PCR and restriction enzyme digestion. The cloning procedure for the anaPS gene was the same as described for tqaA. The verified plasmid containing intact tqaA or anaPS gene was transformed into S. cerevisiae strain BJ5464-NpgA for protein expression. For 1 L of yeast culture, the cells were grown at 28 °C in YPD media with 1% dextrose for 72 hours. For detailed protein purification procedure, refer to Supplementary Methods. FLAG-tagged TqaA and AnaPS proteins were purified by using ANTI-FLAG® M1 Agarose Affinity Gel (Sigma-Aldrich). The cleared cell lysate was directly applied onto a column packed with ANTI-FLAG Agarose Affinity Gel. TqaA/AnaPS protein was eluted with buffer containing 0.5 mg ml−1 FLAG peptide. For the cloning and expression procedures for other proteins mentioned in this paper, see Supplementary Methods.
Synthesis of peptidyl-SNAC, peptidyl-S-CoA and peptidyl-S-T substrates
Syntheses of peptidyl-SNACs and peptidyl-S-CoAs are described in the Supplementary Methods. Loading reactions of apo-T3 or other ACP domains were carried out in 50 mM MES buffer (pH 6.8) and initiated by addition of 5 μM Sfp. After 2 hours, 3-S-T3 was buffer-exchanged into MilliQ water using an Amicon Ultra 10 MWCO 10,000 Daltons, followed by identification with MALDI-TOF MS and used for kinetics assays.
In vitro assays
To test the in vitro activities of TqaA, AnaPS or other mutant proteins, 2 mM of each amino acid building block, 6 mM ATP, 4 mM MgSO4 and 10 μM of purified proteins were combined in 50 mM Tris-HCl buffer (pH 7.5) to a total volume of 100 μl. After 12 hours incubation, reactions were quenched and extracted with 1 ml of ethyl acetate. The organic layer was dried and re-dissolved in methanol for LC-MS analyses. For kinetic characterization of the CT domain towards 3-S-T3, the concentration of the CT domain was kept constant at 2 μM. Initial velocities were determined at various 3-S-T3 concentrations ranging from 20 to 1053 μM. Samples were analyzed on a Shimadzu 2010 EV liquid chromatography mass spectrometer using positive and negative electrospray ionization and a Phenomenex Luna 5 μm, 2.0 mm × 100 mm C18 reverse-phase column. Samples were separated on a linear gradient of 5 to 95% CH3CN in water (0.1% formic acid) for 30 min at a flow rate of 0.1 ml min−1 followed by isocratic 95% CH3CN in water (0.1% formic acid) for 15 min.
Other general materials, methods and procedures. See Supplementary Methods.
Supplementary Material
Acknowledgments
This work is supported in part by National Institutes of Health Grant GM20011 (to C.T.W.), F32GM090475 (to B.D.A.) and 1R01GM092217 (to Y.T.). Wei Xu is thanked for assistance with MALDI-TOF mass analysis and providing the both ApdA and LovF ACP domains. Yen-Ting Lai is thanked for assistance with gel filtration FPLC.
Footnotes
Contributions
X.G., S.W.H., C.T.W. and Y.T. developed the hypothesis and designed the study. S.W.H. performed the synthesis of all the chemicals in this study. X.G. and P.W. performed molecular cloning. X.G., P.W. and L.P.V. performed the heterologous protein expression and purification. X.G. and B.D.A. performed in vitro and in vivo characterization of the megasynthases. All authors analyzed and discussed the results. X.G., C.T.W. and Y. T. prepared the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Bierer BE, Hollander G, Fruman D, Burakoff SJ. Cyclosporine A and FK506: molecular mechanisms of immunosuppression and probes for transplantation biology. Curr Opin Immunol. 1993;5:763–773. doi: 10.1016/0952-7915(93)90135-f. [DOI] [PubMed] [Google Scholar]
- 2.Chen SC, Slavin MA, Sorrell TC. Echinocandin antifungal drugs in fungal infections: a comparison. Drugs. 2011;71:11–41. doi: 10.2165/11585270-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 4.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 5.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 6.Kohli RM, Walsh CT, Burkart MD. Biomimetic synthesis and optimization of cyclic peptide antibiotics. Nature. 2002;418:658–661. doi: 10.1038/nature00907. [DOI] [PubMed] [Google Scholar]
- 7.Kopp F, Marahiel MA. Macrocyclization strategies in polyketide and nonribosomal peptide biosynthesis. Nat Prod Rep. 2007;24:735–749. doi: 10.1039/b613652b. [DOI] [PubMed] [Google Scholar]
- 8.Du LC, Lou LL. PKS and NRPS release mechanisms. Nat Prod Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 9.Von Doehren H. A survey of nonribosomal peptide synthetase (NRPS) genes in Aspergillus nidulans. Fungal Genet Biol. 2009;46:S45–S52. doi: 10.1016/j.fgb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Eisfeld K. Physiology and Genetics. 1. Vol. 15. Springer; Berlin Heidelberg: 2009. Non-Ribosomal Peptide Synthetases of Fungi; pp. 315–316. [Google Scholar]
- 11.Lawen A, Zocher R. Cyclosporin synthetase. The most complex peptide synthesizing multienzyme polypeptide so far described. J Biol Chem. 1990;265:11355–11360. [PubMed] [Google Scholar]
- 12.Jin JM, et al. Functional characterization and manipulation of the apicidin biosynthetic pathway in Fusarium semitectum. Mol Microbiol. 2010;76:456–466. doi: 10.1111/j.1365-2958.2010.07109.x. [DOI] [PubMed] [Google Scholar]
- 13.Slightom JL, Metzger BR, Luu HT, Elhammer AP. Cloning and molecular characterization of the gene encoding the aureobasidin A biosynthesis complex in Aureobasidium pullulans BP-1938. Gene. 2009;431:67–79. doi: 10.1016/j.gene.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 14.Winterberg B, et al. Elucidation of the complete ferrichrome A biosynthetic pathway in Ustilago maydis. Mol Microbiol. 2010;75:1260–1271. doi: 10.1111/j.1365-2958.2010.07048.x. [DOI] [PubMed] [Google Scholar]
- 15.Zocher R, et al. Biosynthesis of cyclosporin A: partial purification and properties of a multifunctional enzyme from Tolypocladium inflatum. Biochemistry. 1986;25:550–553. doi: 10.1021/bi00351a005. [DOI] [PubMed] [Google Scholar]
- 16.Gao X, et al. Fungal indole alkaloid biosynthesis: genetic and biochemical investigation of the tryptoquialanine pathway in Penicillium aethiopicum. J Am Chem Soc. 2011;133:2729–2741. doi: 10.1021/ja1101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergendahl V, Linne U, Marahiel MA. Mutational analysis of the C-domain in nonribosomal peptide synthesis. Eur J Biochem. 2002;269:620–629. doi: 10.1046/j.0014-2956.2001.02691.x. [DOI] [PubMed] [Google Scholar]
- 18.Keating TA, Marshall CG, Walsh CT, Keating AE. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat Struct Biol. 2002;9:522–526. doi: 10.1038/nsb810. [DOI] [PubMed] [Google Scholar]
- 19.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure. 2007;15:781–792. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Ames BD, Walsh CT. Anthranilate-activating modules from fungal nonribosomal peptide assembly lines. Biochemistry. 2010;49:3351–3365. doi: 10.1021/bi100198y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ames BD, Liu X, Walsh CT. Enzymatic processing of fumiquinazoline F: a tandem oxidative-acylation strategy for the generation of multicyclic scaffolds in fungal indole alkaloid biosynthesis. Biochemistry. 2010;49:8564–8576. doi: 10.1021/bi1012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin WB, Grundmann A, Cheng J, Li SM. Acetylaszonalenin biosynthesis in Neosartorya fischeri Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J Biol Chem. 2009;284:100–109. doi: 10.1074/jbc.M807606200. [DOI] [PubMed] [Google Scholar]
- 23.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma SM, et al. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mootz HD, Schorgendorfer K, Marahiel MA. Functional characterization of 4′ phosphopantetheinyl transferase genes of bacterial and fungal origin by complementation of Saccharomyces cerevisiae lys5. Fems Microbiol Lett. 2002;213:51–57. doi: 10.1111/j.1574-6968.2002.tb11285.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu W, Cai XL, Jung ME, Tang Y. Analysis of intact and dissected fungal polyketide synthase-nonribosomal peptide synthetase in vitro and in Saccharomyces cerevisiae. J Am Chem Soc. 2010;132:13604–13607. doi: 10.1021/ja107084d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusnak F, Sakaitani M, Drueckhammer D, Reichert J, Walsh CT. Biosynthesis of the Escherichia-coli siderophore enterobactin: sequence of the entf gene, expression and purification of entf, and analysis of covalent phosphopantetheine. Biochemistry. 1991;30:2916–2927. 28. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 28.Stachelhaus T, Walsh CT. Mutational analysis of the epimerization domain in the initiation module PheATE of gramicidin S synthetase. Biochemistry. 2000;39:5775–5787. doi: 10.1021/bi9929002. [DOI] [PubMed] [Google Scholar]
- 29.Linne U, Marahiel MA. Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization. Biochemistry. 2000;39:10439–10447. doi: 10.1021/bi000768w. [DOI] [PubMed] [Google Scholar]
- 30.Sieber SA, Tao JH, Walsh CT, Marahiel MA. Peptidyl thiophenols as substrates for nonribosomal peptide cyclases. Angew Chem Int Ed Engl. 2004;43:493–498. doi: 10.1002/anie.200352787. [DOI] [PubMed] [Google Scholar]
- 31.Quadri LEN, et al. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 32.Xie X, Meehan MJ, Xu W, Dorrestein PC, Tang Y. Acyltransferase mediated polyketide release from a fungal megasynthase. J Am Chem Soc. 2009;131:8388–8389. doi: 10.1021/ja903203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balibar CJ, Walsh CT. GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry. 2006;45:15029–15038. doi: 10.1021/bi061845b. [DOI] [PubMed] [Google Scholar]
- 34.Keating TA, Miller DA, Walsh CT. Expression, purification, and characterization of HMWP2, a 229 kDa, six domain protein subunit of yersiniabactin synthetase. Biochemistry. 2000;39:4729–4739. doi: 10.1021/bi992923g. [DOI] [PubMed] [Google Scholar]
- 35.Lee JH, Evans BS, Li GY, Kelleher NL, van der Donk WA. In vitro characterization of a heterologously expressed nonribosomal peptide synthetase involved in phosphinothricin tripeptide biosynthesis. Biochemistry. 2009;48:5054–5056. doi: 10.1021/bi900164d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ames BD, et al. Complexity generation in fungal peptidyl alkaloid biosynthesis: oxidation of fumiquinazoline A to the heptacyclic hemiaminal fumiquinazoline C by the flavoenzyme Af12070 from Aspergillus fumigatus. Biochemistry. 2011;50:8756–8769. doi: 10.1021/bi201302w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel JFD, Rodrigues E. Peptaibols of Trichoderma. Nat Prod Rep. 2007;24:1128–1141. doi: 10.1039/b618086h. [DOI] [PubMed] [Google Scholar]
- 38.Liu XY, Walsh CT. Cyclopiazonic acid biosynthesis in Aspergillus sp.: characterization of a reductase-like R* domain in cyclopiazonate synthetase that forms and releases cyclo-acetoacetyl-L-tryptophan. Biochemistry. 2009;48:8746–8757. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sims JW, Schmidt EW. Thioesterase-like role for fungal PKS-NRPS hybrid reductive domains. J Am Chem Soc. 2008;130:11149–11155. doi: 10.1021/ja803078z. [DOI] [PubMed] [Google Scholar]
- 40.Wang B, Kang Q, Lu Y, Bai L, Wang C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc Natl Acad Sci USA. 2012;109:1287–1292. doi: 10.1073/pnas.1115983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grindberg RV, et al. Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS One. 2011;6(4):e18565. doi: 10.1371/journal.pone.0018565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gatto GJ, McLoughlin SM, Kelleher NL, Walsh CT. Elucidating the substrate specificity and condensation domain activity of FkbP, the FK520 pipecolate-incorporating enzyme. Biochemistry. 2005;44:5993–6002. doi: 10.1021/bi050230w. [DOI] [PubMed] [Google Scholar]
- 43.Schwecke T, et al. The biosynthetic gene-cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sieber SA, Walsh CT, Marahiel MA. Loading peptidyl-coenzyme A onto peptidyl carrier proteins: a novel approach in characterizing macrocyclization by thioesterase domains. J Am Chem Soc. 2003;125:10862–10866. doi: 10.1021/ja0361852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.