Abstract
Transforming growth factor β (TGFβ) interacts with cell surface receptors to initiate a signaling cascade critical in regulating growth, differentiation, and development of many cell types. TGFβ signaling involves activation of Smad proteins which directly regulate target gene expression. Here we show that Smad proteins also regulate gene expression by using a previously unrecognized pathway involving direct interaction with protein kinase A (PKA). PKA has numerous effects on growth, differentiation, and apoptosis, and activation of PKA is generally initiated by increased cellular cyclic AMP (cAMP). However, we found that TGFβ activates PKA independent of increased cAMP, and our observations support the conclusion that there is formation of a complex between Smad proteins and the regulatory subunit of PKA, with release of the catalytic subunit from the PKA holoenzyme. We also found that the activation of PKA was required for TGFβ activation of CREB, induction of p21Cip1, and inhibition of cell growth. Taken together, these data indicate an important and previously unrecognized interaction between the TGFβ and PKA signaling pathways.
Transforming growth factor beta (TGFβ) is one of a family of proteins that regulate a diverse array of biological functions including growth and differentiation, embryonic development, angiogenesis, and wound healing. Disruption of the ligands or components of this signaling pathway is associated with a number of human diseases, including cancer (2). The TGFβ family includes activins, inhibins, bone morphogenetic proteins, and TGFβ. Signaling begins when TGFβ binds to cell surface serine/threonine kinase receptors. TGFβ binds to the type II TGFβ receptor (RII), which then interacts with and phosphorylates the type I TGFβ receptor (RI). Phosphorylation activates the intrinsic kinase activity of RI, making it possible for the receptor to phosphorylate and, thus, activate Smad proteins (Smads). To date, at least nine Smads have been cloned, and among them, the highly related Smad2 and Smad3 are specific effectors for TGFβ signaling (24). Ligand binding to the TGFβ receptor complex results in C-terminal phosphorylation of Smad2 and Smad3. Once phosphorylated, Smad2 and Smad3 dissociate from the receptor, bind to Smad4, and enter the nucleus. In the nucleus, heteromeric complexes of Smads function as effectors of Smad signaling by binding directly to DNA and/or by interacting with other DNA-binding proteins to target genes for transcriptional regulation.
Recently, interactions of TGFβ pathway components with effectors of other signaling pathways have been described. One potentially important interaction was suggested by a report that TGFβ could activate cyclic AMP (cAMP)-dependent protein kinase (also known as protein kinase A, or PKA) through an unknown mechanism (40). PKA is a cytosolic, tetrameric holoenzyme that is composed of two regulatory subunits associated with two catalytic subunits (11, 29, 36, 39). Elevation of intracellular cAMP levels causes binding of cAMP to the regulatory subunits and leads to a dissociation of the tetrameric complex, thus allowing the free catalytic subunit to be active as a serine/threonine kinase in the cytoplasm and nucleus. The dissociated, active catalytic subunits can then affect cell physiology via phosphorylation of a wide variety of protein substrates (16, 21, 27). PKA signaling has been shown to play an important role in multiple physiological processes, including growth and differentiation, extracellular matrix production, and apoptosis (39). Since many of these cellular effects are similar to those elicited by TGFβ, we sought to understand the mechanisms involved in this interaction between the TGFβ and PKA signaling pathways.
MATERIALS AND METHODS
Cell culture and transfections.
Mv1Lu cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. The cells were maintained in a humidified incubator with 5% CO2 at 37°C. Transient transfections were performed with Lipofectamine Plus reagent (Gibco BRL, Gaithersburg, Md.) according to the manufacturer's instructions.
Preparation of pancreatic acini and adenoviral infection.
The preparation of pancreatic acini was performed as previously described (43). Briefly, pancreatic tissue was obtained from male Swiss Webster mice or Smad3−/− mice and digested with collagenase (100 U/ml) and incubated at 37°C for 45 min with shaking (120 cycles/min). Acini were then mechanically dispersed by trituration of tissue through polypropylene pipettes of decreasing orifice size (3.0, 2.4, and 1.2 mm) and filtration through a 150-μm-pore-size mesh nylon cloth. Acini were purified by centrifugation at 50 × g for 3 min through a solution containing 4% bovine serum albumin (BSA) and were resuspended in enhanced media that consisted of Dulbecco's modified Eagle's medium containing 0.5% fetal bovine serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, 0.5 mM isobutylmethylxanthine (IBMX), and 0.1 mg of soybean trypsin inhibitor per ml. Cells were maintained in a humidified atmosphere of 5% CO2 in air at 37°C during incubation times. The acinar cells were infected with adenovirus expressing either Smad3 or green fluorescent protein (106 PFU/mg of acinar protein) as described previously (43).
In vitro kinase assay for PKA activity.
PKA kinase activity was measured by a PKA kinase activity assay kit (Promega, Madison, Wis.). Mv1Lu cells or acinar cells were treated with TGFβ1 (R & D Systems, Minneapolis, Minn.), washed with phosphate-buffered saline (PBS), and harvested with cold extraction buffer containing 25 mM Tris-HCl (pH 7.4), 0.5 mM EDTA, 0.5 mM EGTA, 10 mM β-mercaptoethanol, 1 mg of leupeptin per ml, 1 mg of aprotinin per ml, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). Protein concentrations of the crude lysates were quantitated, and equal amounts of protein were added to a reaction mixture containing 40 mM Tris-HCl (pH 7.4), 20 mM MgCl2, 0.1 mg of BSA per ml, 100 mM biotinylated PKA peptide substrate (Kemptide), 3,000 Ci [γ32-P]ATP (Amersham, Arlington Heights, Ill.) per mmol, and 0.5 mM ATP per reaction. The reaction was allowed to proceed for 5 min at 30°C and then terminated by the addition of 2.5 M guanidine hydrochloride. A total of 10 μl of each sample was spotted onto streptavidin-coated disks, washed repeatedly, dried in an oven, and placed in scintillation vials for radioactive counting.
Measurement of cAMP.
Intracellular cAMP levels were measured with a Biotrak cAMP enzyme immunoassay kit (Amersham). Mv1Lu cells were treated with TGFβ or forskolin in the absence and presence of IBMX (100 mM), and the cells were collected and resuspended in PBS with 65% (vol/vol) ethanol. The cell precipitates were centrifuged, the supernatants were drawn off, and the extracts were dried in a vacuum oven. Extracts were resuspended in assay buffer, acetylated, and assayed for cAMP following the instructions supplied by the manufacturer.
Immunoblot analysis.
Whole-cell lysates were prepared by incubating cells in ice-cold lysis buffer (20 mM Tris [pH 7.8], 2 mM EDTA, 50 mM NaF, 1% Triton X-100, 5 μg of leupeptin per ml, 5 μg of pepstatin per ml, and 0.5 mM PMSF). Cells were sonicated for 8 s and then placed on ice for 15 min. The lysates were then centrifuged at 14,000 × g for 15 min at 4°C and assayed for protein with the Bio-Rad protein assay reagent. Equal amounts of protein were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Anti-p21Cip1 antibody and anti-β-actin antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.) were used. Images were visualized with an enhanced chemiluminescence (ECL) detection system (Amersham). For immunoblot analysis of phospho- and total-CREB (cAMP-response element binding protein), nuclear cellular extracts were prepared by the method of Maire as previously described (23) and anti-phospho-CREB antibody (Upstate Biotechnology, Inc., Lake Placid, N.Y.) and anti-total-CREB antibodies (Santa Cruz Biotechnology) were used.
Coimmunoprecipitation experiments.
Mv1Lu cells were treated with 100 pM TGFβ for indicated time periods. Cells were then lysed by sonicating for 5 s in 1 ml of detergent-free lysis buffer (PBS, 5 mM EDTA, 0.02% sodium azide), 10 mM iodoacetamide, 1 mM PMSF, and 2 μg of leupeptin per ml at 4°C. The lysates were cleared by microcentrifuging for 15 min at 16,000 × g at 4°C. Antibody-conjugated beads were prepared by combining 1 μg of polyclonal antibodies with 30 μl of a 50% protein A-Sepharose bead slurry in 0.5 ml of ice-cold PBS for 1 h at 4°C in a tube rotator and then were washed two times with 1 ml of lysis buffer. The antibodies used for immunoprecipitation were rabbit polyclonal anti-PKA RIβ and RIIα and anti-PKA Cα subunit antibodies (Santa Cruz Biotechnology). Cell lysate (500 μg) was incubated with the prepared beads and 10 μl of 10% BSA overnight at 4°C. The beads were washed four times with washing buffer (50 mM Tris-HCl [pH 7.4], 300 mM NaCl, 5 mM EDTA, 0.02% sodium azide, 0.1% Triton X-100) and one time with ice-cold PBS. Proteins were revealed after SDS-PAGE and Western blotting with the following antibodies: mouse anti-Flag antibody (Sigma, St. Louis, Mo.) and rabbit polyclonal antibodies to Smad4, Smad3, PKA RIβ and RIIα, and PKA Cα (Santa Cruz Biotechnology). Images were visualized by using the ECL detection system.
In vitro binding and GST pull-down assays.
Glutathione S-transferase (GST)-labeled constitutively active Smad3 (Smad3D) fusion protein and GST-Smad4 protein were produced in Escherichia coli and purified by using a bulk GST purification module (Amersham). One microgram of purified GST, GST-Smad3, or GST-Smad4 protein was immobilized on glutathione Sepharose beads and added to 1 μg of purified recombinant PKA RIIα protein in PBS supplemented with 10% BSA as a nonspecific competitor. After incubation for 1 h at 4°C, the samples were washed four times with PBS, resolved by SDS-PAGE, and blotted with anti-PKA RIIα. The same membrane was stripped and blotted with anti-Smad4 and anti-Smad3 antibodies. Images were visualized by using the ECL detection system.
PKA holoenzyme assay.
A PKA RIIα2Cα2 holoenzyme was formed and purified by sucrose gradient centrifugation as described previously (10) by using 8 μg of purified PKA Cα protein and twofold excess of purified PKA RIIα protein. Briefly, the purified proteins were incubated for 10 min at 4°C and then were loaded on the top of a 13-ml 5 to 20% sucrose (in 100 mM NaCl) gradient centrifugation column. The centrifugation was performed at 100,000 × g for 22 h. The fraction with peak cAMP-dependent kinase activity was considered as purified PKA holoenzyme. The kinase activity assay was performed as described above. The activities of RIIα2Cα2 were measured in the presence of 100 nM cAMP, a 1 μM concentration of purified Smad3D protein, a 1 μM concentration of purified Smad4 protein, or a combination of the Smad3D and Smad4 proteins, each at a concentration of 1 μM.
CREB EMSA.
Nuclear extracts were prepared and used for electrophoretic mobility shift assays (EMSAs) as previously described (34). Nuclear protein (5 μg) was incubated with gel shift binding buffer [10 mM HEPES, 10% glycerol, 1 mM dithiothreitol, 1 mg of poly(dI-dC) per 10 ml, and 5 mg of BSA per 10 ml] and a CREB oligonucleotide probe labeled with [γ-32P]ATP by T4 polynucleotide kinase. The oligonucleotide probes were provided by a gel shift assay system (E3300; Promega). The reaction was allowed to proceed for 30 min at room temperature. For cold competition experiments, the extract was preincubated for 30 min with 50-fold molar excess of unlabeled CREB oligonucleotide. For the antibody supershift assay, 1 μg of anti-CREB antibody was incubated with the nuclear extracts for 30 min at room temperature prior to the addition of labeled probe. Reactions were analyzed on a 10- by 12-cm, 0.75-mm thick, nondenaturing, 4% acrylamide gel. Gels were transferred to Whatman paper on a gel dryer, exposed to a Bio-Rad GS-250 screen overnight, and then analyzed on a Bio-Rad molecular imager.
Proliferation assay.
Cell proliferation was measured by using a CellTiter 96 AQ nonradioactive cell proliferation assay (Promega). Briefly, cells were plated in 96-well plates at a density of 2,000 cells/well in 100 μl of medium. Cells were allowed to grow up to 5 days; then combined MTS [3-(4,5-dimethylthiazol-2yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium]-phenozine methosulfate solution (20 μl/well) was added. After incubation for 2 h at 37°C in a humidified 5% CO2 atmosphere, the absorbance was measured at 490 nm by using an enzyme-linked immunosorbent assay plate reader. Data presented represent the average of three wells in one experiment which was repeated twice.
RESULTS
TGFβ activates PKA in Mv1Lu cells.
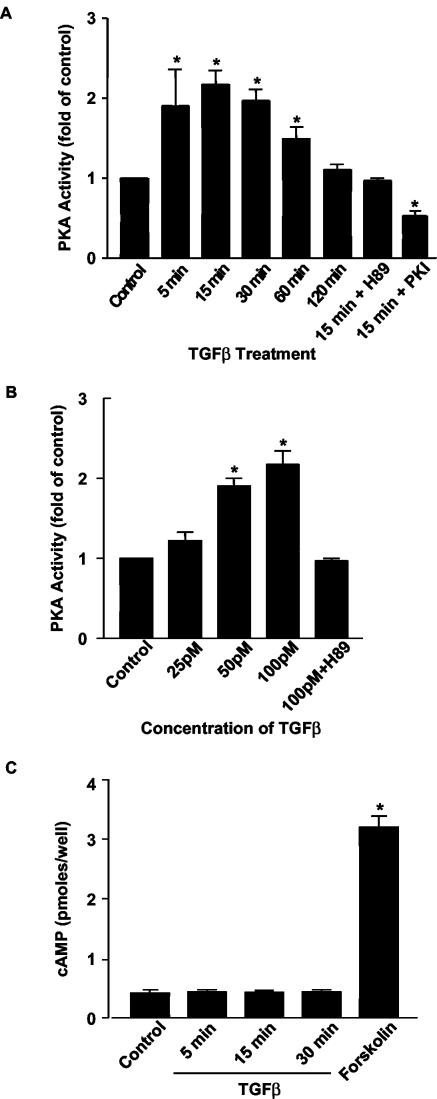
To investigate the ability of TGFβ to activate PKA, Mv1Lu cells were treated with TGFβ for specific times, and in vitro kinase assays were performed by utilizing a biotinylated substrate for PKA (Fig. 1A). It was observed that PKA activity increased more than twofold within 15 min of the administration of TGFβ and remained elevated for 60 min. The ability of TGFβ to activate PKA was completely blocked by the addition of H89, a specific PKA inhibitor. The ability of TGFβ to activate PKA was also blocked by the expression of the specific PKA inhibitory peptide, PKI, a 16-amino-acid peptide that contains a PKA pseudosubstrate sequence and specifically inhibits the catalytic subunits of PKA by binding to the substrate binding site (12, 41). Pretreatment with H89 or transfection with PKI did not affect the ability of TGFβ to phosphorylate Smad2 or to activate the TGFβ-responsive reporter 3TP-Lux (data not shown); thus, PKA inhibition does not inhibit the TGFβ receptor kinase, formation of the TGFβ receptor-Smad complex, or activation of Smads. The activation of PKA by TGFβ was also concentration dependent, with the greatest stimulation noted at the maximal tested concentration of 100 pM TGFβ (Fig. 1B). TGFβ's ability to activate PKA was independent of new protein synthesis, as it was not inhibited by pretreatment with the protein synthesis inhibitor cycloheximide (data not shown).
FIG. 1.
TGFβ activates PKA without increasing intracellular cAMP levels. (A and B) Mv1Lu cells were serum starved (24 h) and were then treated with TGFβ (100 pM) for the indicated time periods (A) or at the indicated doses for 15 min (B). In vitro kinase assays for PKA activity were performed with a biotinylated PKA peptide substrate (Kemptide [LRRASLG]; Promega). A specific PKA inhibitor H89 (3 μM) was used to pretreat some cells for 30 min. PKA activity was also measured in cells transfected with a pcDNA3.0 plasmid which expresses the specific PKA molecular inhibitor PKI. Results are expressed as increases over the control from three separate experiments (*, P < 0.05 versus the control). (C) TGFβ does not increase cAMP. In the presence of the phosphodiesterase inhibitor IBMX (100 μM), cAMP levels were measured in Mv1Lu cells after treatment with TGFβ (100 pM) or forskolin (10 μM) for 15 min by using a Biotrak enzyme immunoassay assay kit (Amersham). Results are from three separate experiments (*, P < 0.05 versus the control).
TGFβ does not activate PKA via changes in cAMP or IκB levels.
Until now, only two mechanisms of PKA activation have been described. The predominant mechanism of PKA activation is binding of cAMP to the regulatory subunits of PKA, which promotes dissociation of the catalytic subunits (11). There has also been one report of a cAMP-independent mechanism in which the catalytic subunit of PKA was maintained in an inactive state through association with IκB and signals that caused the degradation of IκB resulted in PKA activation (45). We next sought to determine if TGFβ's ability to activate PKA was due to either of these two previously described mechanisms.
To examine if TGFβ's ability to stimulate PKA was due to increased levels of intracellular cAMP, Mv1Lu cells were treated with 100 pM TGFβ and cAMP levels were determined. TGFβ did not raise cAMP levels when it was added alone (data not shown) or in the presence of the phosphodiesterase inhibitor IBMX (Fig. 1C). In contrast, treatment with forskolin, known to directly interact with adenylate cyclase, resulted in significant increases in the levels of intracellular cAMP. Therefore, the increase in PKA activity in TGFβ-treated Mv1Lu cells was not dependent on changes in cAMP levels, in accord with what has been reported in a mesangial cell model (40). To evaluate if TGFβ-induced activation of PKA involved degradation of IκB, we analyzed protein levels of IκB after TGFβ treatment. Treatment of Mv1Lu cells with 100 pM TGFβ for 15, 30, and 60 min did not reduce IκB in whole-cell lysates (data not shown). Therefore, it is unlikely that IκB degradation is responsible for TGFβ-induced stimulation of PKA.
TGFβ's ability to activate PKA is dependent on Smad4.
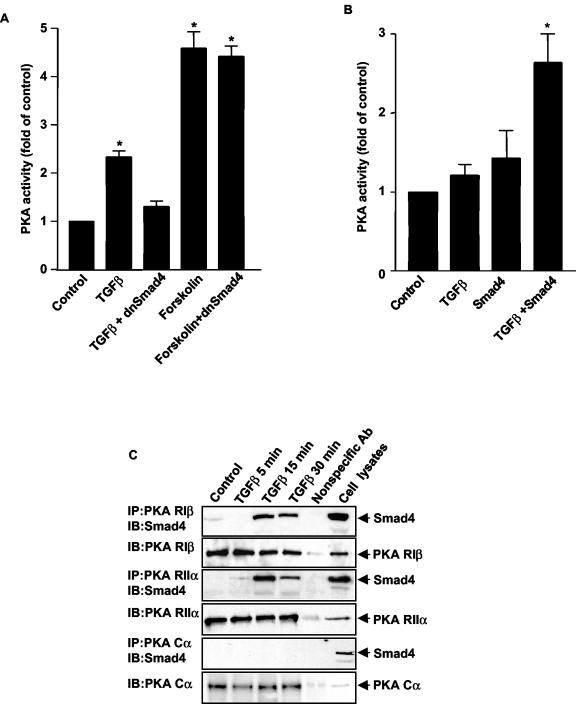
Since Smad4 is a critical component of TGFβ signaling, we evaluated whether TGFβ-induced activation of PKA was dependent on Smad4. Mv1Lu cells were transfected with a dominant negative mutant of Smad4 (dnSmad4) that has been shown to be unable to heterodimerize with other Smads (42, 44). Expression of dnSmad4 blocked TGFβ's ability to activate PKA while dnSmad4 had no effect on the ability of forskolin to activate PKA, demonstrating that PKA activation by TGFβ was Smad4 dependent (Fig. 2A). To further support the role of Smad4 in TGFβ-induced activation of PKA, we performed studies in Smad4-deficient mouse embryonic fibroblasts (35). TGFβ did not activate PKA in Smad4-deficient cells, but transfection of wild-type Smad4 was able to restore the ability of TGFβ to activate PKA (Fig. 2B). Since TGFβ did not increase cAMP levels and TGFβ's ability to activate PKA was dependent on Smad4, we hypothesized that Smad4 directly activates PKA.
FIG. 2.
TGFβ activation of PKA is dependent on TGFβ-induced interaction of a Smad3/Smad4 complex with the regulatory subunits of PKA. (A) dnSmad4 blocks TGFβ-induced PKA activation. Mv1Lu cells were transfected with the vector pCMV5dnSmad4 or empty vector for 16 h. Either 100 pM TGFβ or 10 μM forskolin was added for 15 min, and PKA assays were performed. Results are expressed as increases over the control from three separate experiments (*, P < 0.05 versus the control). (B) TGFβ does not activate PKA in Smad4 null cells. EF7(Smad4−/−) cells were transfected with the vector pCMV5Smad4 for 16 h. TGFβ (100 pM) was added to nontransfected and transfected cells for 15 min, and PKA assays were performed. Results are expressed as increases over the control from three separate experiments (*, P < 0.05 versus the control). (C) Smad4 interacts with PKA regulatory subunits but not catalytic subunits. Mv1Lu cells were treated with TGFβ (100 pM) for the indicated times. Cell lysate (500 μg) was used for immunoprecipitation (IP) with 1 μg of anti-PKA RIβ, anti-PKA RIIα, or anti-PKA Cα subunit antibodies, and then the immunoblot (IB) was detected with the indicated antibodies. One microgram of anti-His antibody from the same species and 25 μg of cell lysate served as controls. (D) Smad3, but not Smad2, interacts with PKA regulatory subunits upon TGFβ treatment. Mv1Lu cells were transfected with the vector pCMV5Flag-Smad2 or pCMV5Flag-Smad3, and cells were treated with 100 pM TGFβ for the indicated times. Coimmunoprecipitations were performed as described. (E) PKA regulatory subunits interact with endogenous Smad3. Mv1Lu cells were treated with 100 pM TGFβ for 15 min. Coimmunoprecipitations were performed with anti-PKA RIIα antibody, and the blot was detected with an anti-Smad3 antibody.
To determine if there was an interaction between Smad4 and PKA, we performed immunoprecipitation assays and Western blot analysis with Mv1Lu cell lysates. Cells untreated or treated with TGFβ were harvested and immunoprecipitated with antibodies to regulatory and catalytic subunits of PKA and blotted with an anti-Smad4 antibody. Multiple isoforms of the regulatory and catalytic subunits of PKA exist. An antibody to the Cα isoform was used to evaluate binding to the catalytic subunit because Cα is ubiquitously expressed in mammalian tissues (37). Smad4 did not bind to the catalytic subunit of PKA in either the absence or presence of TGFβ (Fig. 2C). Four regulatory subunit isoforms have been identified and shown to possess different tissue distributions (5, 13, 30, 38); therefore, we chose antibodies to two different regulatory isoforms, RIβ and RIIα, to identify an interaction with Smad4. The regulatory subunits of PKA were observed to be present in a complex with Smad4 in a TGFβ-dependent manner (Fig. 2C). This was observed with either RIβ or RIIα subunits of PKA, with peak binding at 15 min, a time course identical to that seen with TGFβ-induced PKA activation. To assess whether Smad4 and PKA regulatory subunits interact directly, GST pull-down assays were performed in vitro by using isolated, bacterially produced GST-tagged Smad4 and His-tagged RIIα proteins. GST pull-down assays did not reveal an interaction of these two proteins (data not shown), suggesting that another protein(s) was required in the complex for Smad4 to interact with the regulatory subunit of PKA.
PKA activation by TGFβ requires an activated Smad3/Smad4 complex.
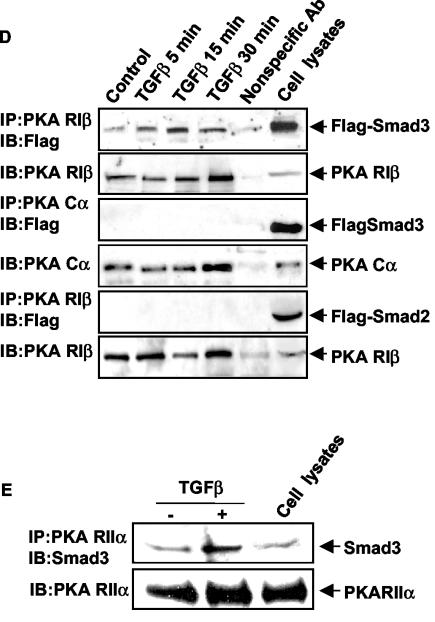
Since the ability of Smad4 to interact with the regulatory subunit of PKA was TGFβ dependent, we investigated whether Smad2 and/or Smad3 was part of the Smad4/PKA regulatory subunit complex. Mv1Lu cells were transfected with Flag-Smad2 or Flag-Smad3, and coimmunoprecipitation experiments were performed. We observed that Flag-Smad3 bound to the Smad4/PKA regulatory subunit complex in a TGFβ-dependent manner (Fig. 2D). In contrast, Flag-Smad2 was not found in this complex (Fig. 2D). The time course of the observed Smad3/Smad4/PKA regulatory subunit interaction paralleled that for PKA activation after TGFβ treatment. To examine the involvement of endogenous Smad3, we utilized an anti-Smad3 antibody. The interaction of endogenous Smad3 in the complex was demonstrated by coimmunoprecipitation with the Smad3 antibody (Fig. 2E).
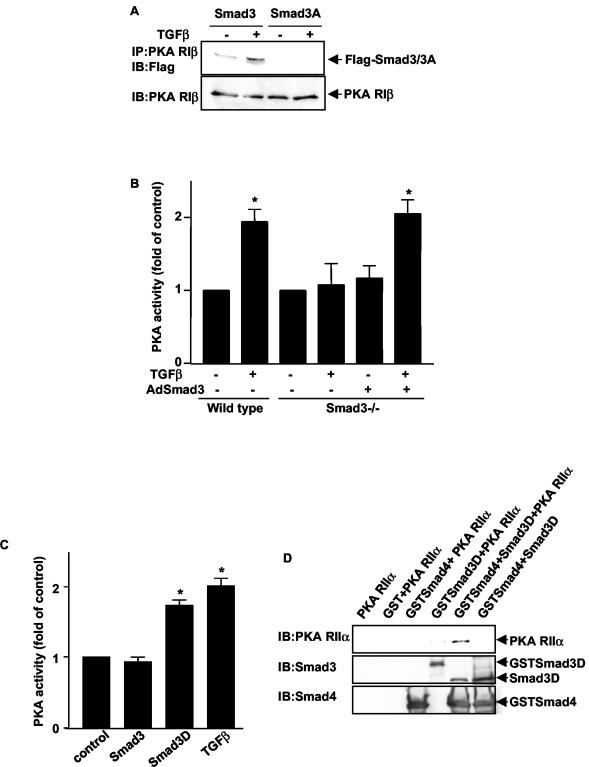
The observation that the interaction of Smad3 with the regulatory subunit of PKA was TGFβ dependent suggested that Smad3 required activation for this interaction to occur. In support of the requirement for activated Smad3 in TGFβ-induced PKA activation, a Smad3 mutant (Smad3A) in which the three C-terminal serine phosphorylation sites are mutated to alanine (which abolishes TGFβ-induced phosphorylation [17]) did not bind to the regulatory subunit of PKA (Fig. 3A). This result supports the hypothesis that the complex can form only in the presence of activated, phosphorylated Smad3. In addition, in Smad3 null cells (46), TGFβ did not activate PKA; however, transfection of wild-type Smad3 restored the ability of TGFβ to activate PKA (Fig. 3B). Further support for the hypothesis that an activated Smad3 is required for complex formation was obtained by using a constitutively active Smad3 protein. We utilized an expression vector in which the three C-terminal serines of Smad3 have been replaced by aspartic acids (Smad3D) to mimic phosphorylation of the normal serine residues by TGFβ. This construct has been shown to activate transcription of the TGFβ-inducible 3TP-Lux reporter in the absence of ligand (22). Transfection of Smad3D into Mv1Lu cells was able to induce PKA activation (Fig. 3C). Additionally, in GST pull-down assays, isolated GST-tagged Smad3D and Smad4 proteins were able to form a complex with His-tagged RIIα protein in vitro in the absence of TGFβ (Fig. 3D), suggesting that the three proteins form a trimeric complex rather than two distinct complexes. Taken together, these data support the hypothesis that the complex can only form in the presence of activated, phosphorylated Smad3.
FIG. 3.
An activated Smad3/Smad4 complex interacts with the PKA regulatory subunits to activate PKA. (A) Smad3A does not bind to the PKA regulatory subunit in TGFβ-treated cells. Mv1Lu cells were transfected with the vector pCMV5Flag-Smad3 or pCMV5Flag-Smad3A, and cells were treated with 100 pM TGFβ for 15 min. Coimmunoprecipitations (IP) were performed as described. IB, immunoblot. (B) TGFβ does not activate PKA in Smad3 null mouse pancreatic acinar cells. Pancreatic acinar cells were isolated from wild-type and Smad3 null mice and treated with TGFβ (100 pM) for 15 min. In some experiments, acinar cells from Smad3 null mice were infected with an adenovirus expressing wild-type Smad3. PKA assays were performed. Results are expressed as increases over the control from three separate experiments (*, P < 0.05 versus the control). (C) Constitutively active Smad3 (Smad3D) can activate PKA. Mv1Lu cells were either treated with TGFβ (100 pM) for 15 min or transfected with the vector pCMV5Flag-Smad3 or pCMV5Flag-Smad3D for 16 h, and PKA assays were performed. Results are expressed as increases over the control from three separate experiments (*, P < 0.05 versus the control). (D) A Smad3/Smad4 protein complex can bind with RIIα in vitro. One microgram of purified RIIα protein was incubated with 1 μg of purified GST, GSTSmad3D, Smad3D, or GSTSmad4 protein. GST pull-down assays were performed. Immunoblotting was performed by using anti-RIIα, anti-Smad3, and anti-Smad4 antibodies.
AKAPs facilitate an interaction between Smads and PKA.
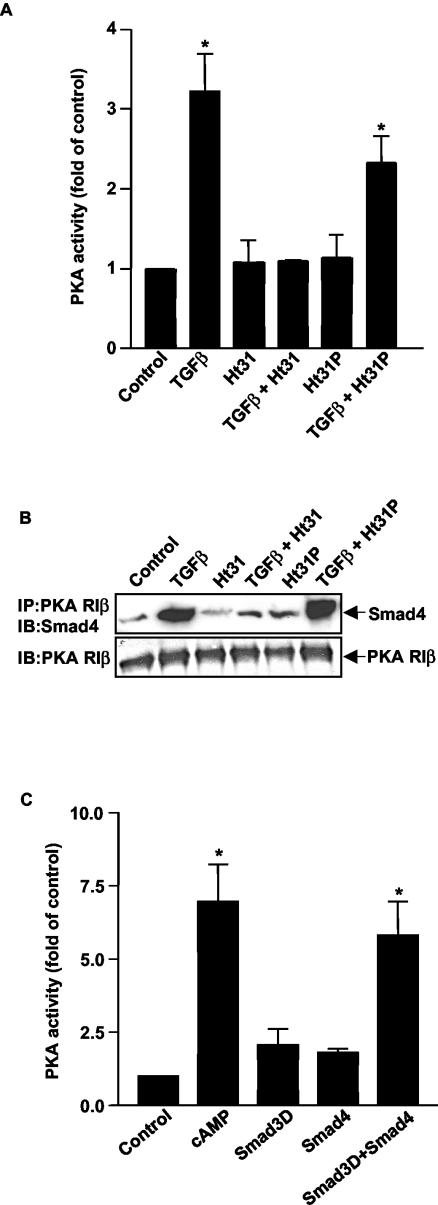
A number of studies have demonstrated that PKA may be compartmentalized in different subcellular locations through interaction with A-kinase anchoring proteins (AKAPs) (8, 26). Each AKAP has two classes of binding sites: an anchoring domain which binds the regulatory subunit of the PKA holoenzyme and a targeting domain which directs the subcellular location of the AKAP-PKA complex by interactions with structural proteins, membranes, or cellular organelles (6). Ht31 is a thyroid-anchoring peptide (3, 4) which has been shown to disrupt the interaction of AKAPs and the regulatory subunit of PKA, thus preventing PKA anchoring. To determine if AKAP-PKA interaction was necessary for TGFβ to activate PKA, cells were treated with either cell-permeant Ht31 or its control peptide Ht31P. Ht31 blocked the ability of TGFβ to activate PKA, while the control peptide had little effect (Fig. 4A). Additionally, treatment with Ht31, but not HT31P, markedly inhibited the ability of Smad4 to interact with the regulatory subunit of PKA (Fig. 4B). These data demonstrate that PKA must be in the proper subcellular location to interact with Smads. To determine if AKAPs are necessary for Smads to bind to the regulatory subunit of PKA, we performed GST pull-down assays which demonstrated that purified GST-tagged Smad3D and Smad3 proteins were able to form a complex with His-tagged RIIα protein in vitro (see Fig. 3D). Additionally, binding of purified Smad3D and Smad4 (mimicking an activated Smad heterodimer) to the regulatory subunit of PKA directly caused dissociation of the PKA holoenzyme and resultant PKA activity in vitro (Fig. 4C). These data suggest that while AKAPs are necessary to anchor PKA in the proper subcellular location to interact with Smads, Smads and the regulatory subunit are necessary and sufficient to form a complex which is functional in activating the PKA holoenzyme.
FIG.4.
TGFβ-mediated activation of PKA requires AKAP. (A) An AKAP inhibitor blocks PKA activation by TGFβ. Mv1Lu cells were pretreated with the AKAP inhibitor Ht31 or its control peptide Ht31P, each at a concentration of 25 μM for 30 min, and then 100 pM TGFβ was added for 15 min. PKA assays were performed. Results are expressed as increases over the control from three separate experiments (*, P < 0.05 versus the control). (B) An AKAP inhibitor blocks the formation of a Smad/PKA regulatory subunit complex. Mv1Lu cells were pretreated with the AKAP inhibitor Ht31 or its control peptide Ht31P at a concentration of 25 μM for 30 min, and then 100 pM TGFβ was added for 15 min. Coimmunoprecipitations (IP) were performed as described. IB, immunoblot. (C) A Smad3/Smad4 complex can dissociate PKA holoenzyme in vitro. RIIa2Ca2 PKA holoenzymes were formed and purified as described (10). The activity of RIIa2Ca2 was measured in the presence of 100 nM cAMP, Smad3D protein (1 μM), Smad4 protein (1 μM), or both Smad3D and Smad4 proteins (1 μM concentration of each protein). Results are expressed as increases over the control from three separate experiments (*, P < 0.01 versus the control).
TGFβ's ability to activate CREB is dependent on Smads and PKA.
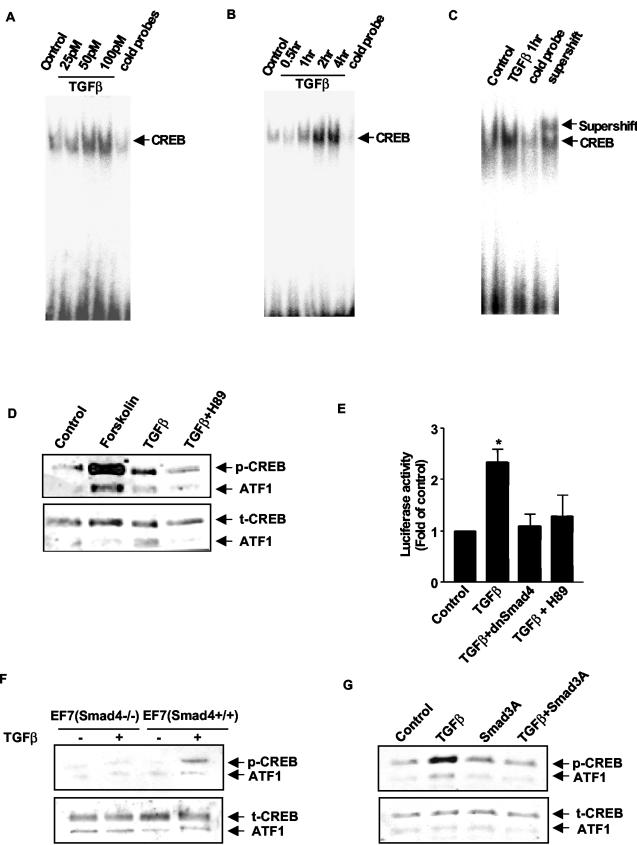
To explore the functional significance of PKA activation by TGFβ, we first tested the ability of TGFβ to activate the transcription factor CREB. CREB is a stimulus-induced transcription factor originally identified as a target of the cAMP signaling pathway and is one of the best characterized nuclear substrates of PKA. CREB is critical for a variety of cellular responses, including proliferation, differentiation, and adaptive responses (1). Although CREB phosphorylation and activation were initially characterized as mediating the response to cAMP, CREB phosphorylation and activation have subsequently been found to be stimulated by diverse extracellular signals and protein kinases in all cells (32). Signaling pathways that activate CREB lead to phosphorylation of Ser133 which is required for CREB-induced gene transcription. We found that TGFβ induced a dose- and time-dependent increase in CREB DNA binding in Mv1Lu cells, as demonstrated by EMSAs (Fig. 5A to C). Immunoblot analysis with anti-phospho-CREB antibody revealed that TGFβ stimulated phosphorylation of CREB (Fig. 5D). Immunoblotting of the same membrane with an antibody that measures total CREB revealed that TGFβ did not lead to increased levels of total CREB. Furthermore, TGFβ activated expression of a luciferase reporter construct driven by the CREB-responsive human chorionic gonadotropin promoter (25) in Mv1Lu cells (Fig. 5E). TGFβ-induced CREB activation was independent of new protein synthesis, as it was unaffected by pretreatment for 30 min with the protein synthesis inhibitor cycloheximide (10 μg/ml) (data not shown). Pretreatment of Mv1Lu cells with H89 completely blocked TGFβ-induced CREB reporter activity, as did transfection with the dominant negative Smad4 construct, demonstrating that TGFβ's ability to activate a CREB-responsive reporter was dependent on both PKA and Smad4 (Fig. 5E). The role of Smad4 in TGFβ-induced CREB activation was further strengthened by using Smad4-deficient mouse embryonic fibroblasts. TGFβ did not phosphorylate CREB in Smad4-deficient cells, but transfection of wild-type Smad4 restored the ability of TGFβ to phosphorylate CREB (Fig. 5F). In addition, the Smad3A mutant was able to block the ability of TGFβ to activate CREB (Fig. 5G), demonstrating that an activated Smad3 is also required for TGFβ to activate CREB.
FIG. 5.
TGFβ's ability to activate CREB is dependent on Smads and PKA. (A to C) TGFβ induces CREB DNA binding. Mv1Lu cells were serum starved (24 h) and were treated with TGFβ at the indicated doses for 1 h (A) or with TGFβ (100 pM) for the indicated time periods (B). Nuclear extracts were prepared, and 5 μg of nuclear protein was used to perform EMSAs. Unlabeled cold probe was used as a control. Nuclear extracts were also preincubated with anti-CREB antibody for the supershift assay (C). Results are representative of three different experiments. (D) The phosphorylation of CREB by TGFβ was detected by using an antibody directed against pSer133-CREB. Ten micrograms of nuclear protein was used to perform Western blotting. TGFβ had no effect on total levels of CREB protein. (E) Mv1Lu cells were cotransfected with the CRE-luciferase and LacZ reporter genes for 8 h. The cells were also transfected with a dnSmad4 expression vector or pretreated with 3 μM H89. TGFβ (100 pM) was added for 8 h. Luciferase activity was measured and normalized to LacZ activity. The results are expressed as increases over the control and are from three separate experiments (*, P < 0.05 versus the control). (F) TGFβ does not activate CREB in Smad4 null cells. EF7(Smad4−/−) cells were serum starved for 24 h and 100 pM TGFβ was added for 60 min. In some experiments, EF7 cells were transfected with wild-type Smad4 [EF7(Smad4+/+)]. Western blotting with anti-phospho-CREB antibody was performed. Results are representative of three separate experiments. (G) Dominant negative Smad3 (Smad3A) blocks the CREB activation by TGFβ. Mv1Lu cells were transfected with vector pCMV5Flag-Smad3A for 16 h. TGFβ (100 pM) was added for 60 min. Ten micrograms of nuclear protein was used to perform Western blotting with anti-phospho-CREB antibody.
TGFβ mediates p21Cip1 induction and growth inhibition by PKA.
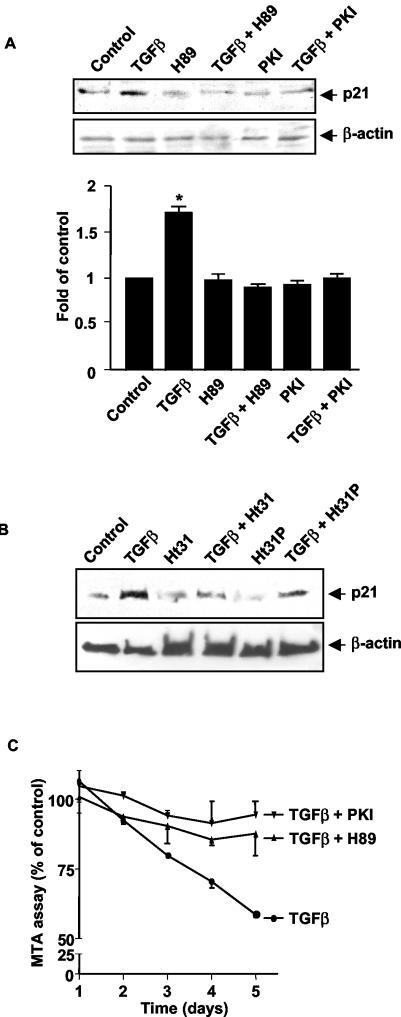
We examined the role of PKA in TGFβ's ability to regulate expression of the cyclin-dependent kinase (CDK) inhibitor p21Cip1. In many cell types, TGFβ has been shown to inhibit cell growth by increasing the expression of this molecule which inhibits the enzymatic activities of cyclin D-CDK4/6 and cyclin E-CDK2 complexes, leading to cell cycle arrest at the late phase of G1 (20). Inhibition of PKA activity either by transfection of Mv1Lu cells with an expression vector for PKI peptide or by treatment with the PKA inhibitor H89 blocked TGFβ's ability to induced p21Cip1 expression (Fig. 6A). The need for PKA to be in the proper subcellular location for TGFβ to induce p21Cip1 expression was shown by the ability of Ht31, but not Ht31P, to block induction of p21 by TGFβ (Fig. 6B). Next, we examined the role of PKA in TGFβ's ability to inhibit growth of the Mv1Lu cells. Transfection of Mv1Lu cells with a PKI expression vector or treatment with H89 blocked TGFβ-mediated growth inhibition (Fig. 6C). The addition of H89 or transfection with PKI alone had no effect on cell growth compared to growth of the controls (data not shown). Together, these results indicate that TGFβ-mediated PKA activation by a Smad3/Smad4 complex is critical for a number of TGFβ-dependent physiological responses.
FIG. 6.
TGFβ-induced p21Cip1 expression and growth inhibition is mediated through PKA activation (A) TGFβ-induced p21Cip1 expression can be blocked by H89 and PKI. Mv1Lu cells were either transfected with a PKI expression vector or pretreated with 3 μM H89 for 30 min. TGFβ (100 pM) was added for 16 h. Western blotting was performed with anti-p21Cip1 antibody. The membrane was stripped and reblotted with anti-β-actin antibody as a loading control. The lower panel represents relative density from three experiments (*, P < 0.05 versus the control). (B) TGFβ-induced p21Cip1 expression can also be blocked by an AKAP inhibitor. Mv1Lu cells were pretreated with 25 μM of either Ht31 or Ht31P for 30 min, and then 100 pM TGFβ was added for 16 h. Western blotting was performed with anti-p21Cip1 antibody. (C) TGFβ-mediated growth inhibition was determined by MTS assay. Mv1Lu cells were grown in 96-well plates at a concentration of 3,000 cells/well in the absence or presence of TGFβ (100 pM) for up to 5 days. Cells were also treated with 3 μM H89 or transfected with a plasmid expressing PKI. A total of 20 μl of MTS-phenozine methosulfate solution (Promega) was added daily, and absorbance was measured by a universal microplate spectrophotometer. Results are expressed as a percentage of the control from three separate experiments.
DISCUSSION
TGFβ activates PKA by a novel, Smad-dependent, cAMP-independent pathway.
The results presented in this paper describe a novel mechanism by which TGFβ activates PKA in a Smad-dependent and cAMP-independent manner. We demonstrated for the first time the ability of Smads to have a direct protein-protein interaction with the regulatory subunit of PKA, leading to PKA activation. We showed that during TGFβ treatment, Smad4 is present in a functional complex containing both the regulatory subunit of PKA and activated Smad3. We provided evidence that both Smad4 and an activated form of Smad3 are required for complex formation and PKA activation by use of Smad null cells and Smad dominant negative constructs. We also demonstrated that the interaction of Smad3, Smad4, and the regulatory subunit of PKA occurs in vitro in the absence of other cellular proteins and that purified Smads activate the PKA holoenzyme in vitro, verifying the functional significance of the Smad complex. Thus, TGFβ interaction with its receptor leads to the activation of both Smad and PKA signaling pathways. The interaction of these two pathways has profound implications for the regulation of cell growth, differentiation, and function.
We found that Smad2, unlike Smad3, did not participate in this complex with the regulatory subunit of PKA in TGFβ-treated Mv1Lu cells. This difference in the ability of Smad2 and Smad3 to bind to the regulatory subunit of PKA likely reflects the unique molecular characteristics of Smad2 and Smad3. For example, it has previously been reported that Smad2 and Smad3 have opposing effects on the transcriptional regulation of the mouse Goosecoid gene through the binding of FAST-2 (19). In addition, Smad2, when compared to Smad3, has a unique insert of exon 3 in the N-terminal domain, which prevents association with importin-β (18).
Although our data indicate that the complex formation with Smad3, Smad4, and the regulatory subunit of PKA is sufficient for the observed activation of PKA, there may be other proteins in the physiological complex. One potential participant would be AKAPs. There is considerable literature on the role of AKAPs and their interaction with the regulatory subunit of PKA (8, 26). To examine the possibility that Smad3/Smad4 may bind to an AKAP, thereby bringing PKA into proximity with the Smad complex, we utilized the bioactive peptide Ht31 that is capable of disrupting PKA location within cells. We demonstrated that Ht31 blocked the ability of Smads to interact with the regulatory subunit of PKA and activate PKA. This suggests that AKAPs are important in placing the regulatory subunit of PKA in the correct subcellular location to interact with Smads. However, the in vitro studies demonstrated that Smad3, Smad4, and the regulatory subunit of PKA can form a complex in the absence of other proteins, including AKAPs. Furthermore, a purified, activated Smad3/Smad4 complex can activate the PKA holoenzyme. Thus, while AKAPs are necessary to properly localize PKA in the cell to interact with Smads, Smads do not need to physically interact with AKAPs to form a complex with and directly activate PKA.
The role of PKA in TGFβ-mediated cellular responses.
The observed interaction between TGFβ and PKA signaling pathways has many implications for the regulation of cell function. For example, in the present study, we demonstrated that PKA activation by Smads is critical in mediating the TGFβ-induced responses of CREB activation, p21Cip1 induction, and growth regulation. TGFβ-regulated activation of PKA leads to increased DNA binding and phosphorylation of CREB. We demonstrated that the TGFβ-regulated transcriptional activation of CREB occurs by the ability of Smads to activate PKA. Active, phosphorylated CREB affects transcription of CRE (cAMP response element)-dependent genes via interaction with the coactivator CREB-binding protein CBP, which bridges the CRE/CREB complex to components of the basal transcriptional apparatus (7, 14). Previous studies have demonstrated that Smads can also regulate CREB activity by interacting with the coactivator CBP (9, 15, 28, 33). Thus, the ability of Smads to regulate the CREB signaling pathway appears to occur at several levels and may help cells more finely control the expression of genes regulated by TGFβ.
The involvement of PKA in TGFβ-mediated cell cycle and growth regulation has not been previously demonstrated. Because TGFβ signaling is often disrupted in cancer, these aspects of TGFβ regulation are of particular interest. A role for PKA in mediating some TGFβ-induced responses has been suggested in two studies. Sharma and colleagues recently demonstrated that TGFβ-induced phosphorylation of the type I inositol 1,4,5-trisphosphate receptor in mesangial cells is mediated by PKA (31). Also, inhibition of PKA has been found to attenuate TGFβ-induced stimulation of CREB phosphorylation and fibronectin gene expression (40), supporting the hypothesis that activation of PKA by TGFβ participates in TGFβ-mediated cell regulation.
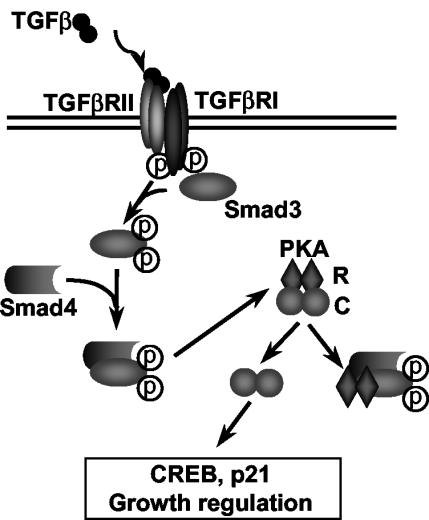
Model and conclusions.
The data in the present study support a model for the mechanism by which Smads function to regulate cellular gene expression through both direct and indirect mechanisms. In this model (Fig. 7), TGFβ treatment initiates a kinase cascade that results in the phosphorylation of Smad3, followed by its heteromerization with Smad4. This complex can directly influence gene transcription. Smad3/Smad4 complexes can also bind the regulatory subunit of PKA, releasing the catalytic subunit and resulting in the activation of downstream target genes. In this model, TGFβ signaling activates PKA without an increase of intracellular cAMP and with no effect on IκB. This study demonstrates that in addition to the traditional role of Smads as transcription factors, Smads also possess a DNA binding-independent role by mediating activation of PKA signaling.
FIG. 7.
Model of how a TGFβ-induced Smad3/Smad4 complex directly activates PKA. TGFβ directly binds to TGFβ RII, which leads to the phosphorylation of TGFβ RI. This phosphorylation activates the RI protein kinase, which then phosphorylates Smad3. Phosphorylated Smad3 binds to Smad4, and this complex binds to the regulatory subunits of PKA (R), leading to the release of catalytic subunits (C) and resulting in the activation of downstream target genes.
In summary, Smad3 and Smad4, two essential Smad proteins involved in mediating TGFβ transcriptional responses, were shown to interact with the regulatory subunits of PKA. This interaction was specific, ligand dependent, and occurred via formation of an activated Smad3/Smad4 complex. We have also demonstrated that PKA activation by TGFβ was important in mediating several physiological responses elicited by TGFβ, including CREB activation, p21Cip1 induction, and growth inhibition. This report reveals new insight on how Smad-PKA interactions may be an important locus of signal integration in the cell.
Acknowledgments
We thank J. Massague for the Smad constructs, H. Lodish for the Smad3D construct, J. Graff for the Smad3 null mice, and C. Sirard for the Smad4 null fibroblasts.
This work was supported by NIH grants DK02137-1 (to D.S.), 1R03DK60486-01 (to D.S.), and DK 41225 (to C.L.), by University of Michigan Peptide Center grant 5P30 DK34933 (to D.S.), and by an American College of Surgeons Faculty Research Fellowship (to D.S.).
REFERENCES
- 1.Andrisani, O. M. 1999. CREB-mediated transcriptional control. Crit. Rev. Eukaryot. Gene Expr. 9:19-32. [PubMed] [Google Scholar]
- 2.Blobe, G. C., W. P. Schiemann, and H. F. Lodish. 2000. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 342:1350-1358. [DOI] [PubMed] [Google Scholar]
- 3.Carr, D. W., Z. E. Hausen, I. D. Fraser, R. E. Stofko-Hahn, and J. D. Scott. 1992. Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein. Cloning and characterization of the RII-binding domain. J. Biol. Chem. 267:13376-13382. [PubMed] [Google Scholar]
- 4.Carr, D. W., R. E. Stofko-Hahn, I. D. Fraser, S. M. Bishop, T. S. Acott, R. G. Brennan, and J. D. Scott. 1991. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 266:14188-14192. [PubMed] [Google Scholar]
- 5.Clegg, C. H., G. G. Cadd, and G. S. McKnight. 1988. Genetic characterization of a brain-specific form of the type I regulatory subunit of cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 85:3703-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dell'Acqua, M. L., and J. D. Scott. 1997. Protein kinase A anchoring. J. Biol. Chem. 272:12881-12884. [DOI] [PubMed] [Google Scholar]
- 7.Eckner, R. 1996. p300 and CBP as transcriptional regulators and targets of oncogenic events. Biol. Chem. 377:685-688. [PubMed] [Google Scholar]
- 8.Edwards, A. S., and J. D. Scott. 2000. A-kinase anchoring proteins: protein kinase A and beyond. Curr. Opin. Cell Biol. 12:217-221. [DOI] [PubMed] [Google Scholar]
- 9.Feng, X. H., Y. Zhang, R. Y. Wu, and R. Derynck. 1998. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 12:2153-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flockhart, D. A., and J. D. Corbin. 1982. Regulatory mechanisms in the control of protein kinases. Crit. Rev. Biochem. 12:133-186. [DOI] [PubMed] [Google Scholar]
- 11.Francis, S. H., and J. D. Corbin. 1994. Structure and function of cyclic nucleotide-dependent protein kinases. Annu. Rev. Physiol. 56:237-272. [DOI] [PubMed] [Google Scholar]
- 12.Glass, D. B., H. C. Cheng, B. E. Kemp, and D. A. Walsh. 1986. Differential and common recognition of the catalytic sites of the cGMP-dependent and cAMP-dependent protein kinases by inhibitory peptides derived from the heat-stable inhibitor protein. J. Biol. Chem. 261:12166-12171. [PubMed] [Google Scholar]
- 13.Jahnsen, T., L. Hedin, V. J. Kidd, W. G. Beattie, S. M. Lohmann, U. Walter, J. Durica, T. Z. Schultz, E. Schiltz, M. Browner, C. B. Lawrence, D. Goldman, S. L. Ratoosh, and J. S. Richards. 1986. Molecular cloning, cDNA structure, and regulation of the regulatory subunit of type II cAMP-dependent protein kinase from rat ovarian granulosa cells. J. Biol. Chem. 261:12352-12361. [PubMed] [Google Scholar]
- 14.Janknecht, R., and T. Hunter. 1996. Versatile molecular glue: transcriptional control. Curr. Biol. 6:951-954. [DOI] [PubMed] [Google Scholar]
- 15.Janknecht, R., N. J. Wells, and T. Hunter. 1998. TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev. 12:2114-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krebs, E. G., and J. A. Beavo. 1979. Phosphorylation-dephosphorylation of enzymes. Annu. Rev. Biochem. 48:923-959. [DOI] [PubMed] [Google Scholar]
- 17.Kretzschmar, M., F. Liu, A. Hata, J. Doody, and J. Massague. 1997. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 11:984-995. [DOI] [PubMed] [Google Scholar]
- 18.Kurisaki, A., S. Kose, Y. Yoneda, C. H. Heldin, and A. Moustakas. 2001. Transforming growth factor-β induces nuclear import of Smad3 in an importin-β1 and Ran-dependent manner. Mol. Biol. Cell 12:1079-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labbe, E., C. Silvestri, P. A. Hoodless, J. L. Wrana, and L. Attisano. 1998. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell 2:109-120. [DOI] [PubMed] [Google Scholar]
- 20.Laiho, M., J. A. Decaprio, J. W. Ludlow, D. M. Livingston, and J. Massague. 1990. Growth inhibition by TGF-beta linked to suppression of retinoblastoma protein phosphorylation. Cell 62:175-185. [DOI] [PubMed] [Google Scholar]
- 21.Levitan, I. B. 1994. Modulation of ion channels by protein phosphorylation and dephosphorylation. Annu. Rev. Physiol. 56:193-212. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X., Y. Sun, S. N. Constantinescu, E. Karam, R. A. Weinberg, and H. F. Lodish. 1997. Transforming growth factor β-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc. Natl. Acad. Sci. USA 94:10669-10674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maire, P., J. Wuarin, and U. Schibler. 1989. The role of cis-acting promoter elements in tissue-specific albumin gene expression. Science 244:343-346. [DOI] [PubMed] [Google Scholar]
- 24.Massague, J., and Y.-G. Chen. 2000. Controlling TGF-β signaling. Genes Dev. 14:627-644. [PubMed] [Google Scholar]
- 25.Mellon, P. L., C. H. Clegg, L. A. Correll, and G. S. McKnight. 1989. Regulation of transcription by cyclic AMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 86:48874891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel, J. J., and J. D. Scott. 2002. AKAP mediated signal transduction. Annu. Rev. Pharmacol. Toxicol. 42:235-257. [DOI] [PubMed] [Google Scholar]
- 27.Montminy, M. R., G. A. Gonzalez, and K. K. Yamamoto. 1990. Regulation of cAMP-inducible genes by CREB. Trends Neurosci. 13:184-188. [DOI] [PubMed] [Google Scholar]
- 28.Pouponnet, C., L. Jayaraman, and J. Massague. 1998. Physical and functional interaction of SMADs and p300/CBP. J. Biol. Chem. 273:22865-22868. [DOI] [PubMed] [Google Scholar]
- 29.Scott, J. D. 1991. Cyclic nucleotide-dependent protein kinases. Pharmacol. Ther. 50:123-145. [DOI] [PubMed] [Google Scholar]
- 30.Scott, J. D., M. B. Glaccum, M. J. Zoller, M. D. Uhler, D. M. Helfman, G. S. McKnight, and E. G. Krebs. 1987. The molecular cloning of a type II regulatory subunit of the cAMP-dependent protein kinase from rat skeletal muscle and mouse brain. Proc. Natl. Acad. Sci. USA 84:5192-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma, K., L. Wang, Y. Zhu, S. Bokkala, and S. K. Joseph. 1997. Transforming growth factor-beta1 inhibits type I inositol 1, 4, 5-trisphosphate receptor expression and enhances its phosphorylation in mesangial cells. J. Biol. Chem. 272:14617-14623. [DOI] [PubMed] [Google Scholar]
- 32.Shaywitz, A. J., and M. E. Greenberg. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 33.Shen, X., P. P. Hu, N. T. Liberati, M. B. Datto, J. P. Frederick, and X.-F. Wang. 1998. TGF-beta-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol. Biol. Cell 9:3309-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simeone, D. M., L. Zhang, K. Graziano, B. Nicke, T. Pham, C. Schaefer, and C. D. Logsdon. 2001. Smad4 mediates activation of mitogen-activated protein kinases by TGFβ in pancreatic acinar cells. Am. J. Physiol. Cell Physiol. 281:C311-C319. [DOI] [PubMed] [Google Scholar]
- 35.Sirard, C., S. Kim, C. Mirtso, P. Tadich, P. A. Hoodless, A. Itie, R. Maxson, J. L. Wrana, and T. W. Mak. 2000. Targeted disruption in murine cells reveals variable requirement for Smad4 in transforming growth factor β-related signaling. J. Biol. Chem. 275:2063-2070. [DOI] [PubMed] [Google Scholar]
- 36.Taylor, S. S., J. A. Buechler, and W. Yonemoto. 1990. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 59:971-1005. [DOI] [PubMed] [Google Scholar]
- 37.Uhler, M. D., J. C. Chrivia, and G. S. McKnight. 1986. Evidence for a second isoform of the catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 261:15360-15363. [PubMed] [Google Scholar]
- 38.Ventra, C., A. Porcellini, A. Feliciello, A. Gallo, M. Paolillo, E. Mele, V. E. Avvedimento, and G. Schettini. 1996. The differential response of protein kinase A to cyclic AMP in discrete brain areas correlates with the abundance of regulatory subunit II. J. Neurochem. 66:1752-1761. [DOI] [PubMed] [Google Scholar]
- 39.Walsh, D. A., and S. M. Van Patten. 1994. Multiple pathway signal transduction by the cAMP-dependent protein kinase. FASEB J. 8:1227-1236. [DOI] [PubMed] [Google Scholar]
- 40.Wang, L., Y. Zhu, and K. Sharma. 1998. Transforming growth factor-β1 stimulates protein kinase A in mesangial cells. J. Biol. Chem. 273:8522-8527. [DOI] [PubMed] [Google Scholar]
- 41.Whitehouse, S., and D. A. Walsh. 1983. Mg X ATP2-dependent interaction of the inhibitor protein of the cAMP-dependent protein kinase with the catalytic subunit. J. Biol. Chem. 258:3682-3692. [PubMed] [Google Scholar]
- 42.Wu, R. Y., Y. Zhang, X. H. Feng, and R. Derynck. 1997. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol. Cell. Biol. 17:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, L., G. Graziano, T. Pham, C. D. Logsdon, and D. M. Simeone. 2001. Adenovirus-mediated gene transfer of dominant-negative Smad4 blocks TGF-beta signaling in pancreatic acinar cells. Am. J. Physiol. Gastrointest. Liver Physiol. 280:G1247-G1253. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Y., X. H. Feng, R. Y. Wu, and R. Derynck. 1996. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature 383:168-172. [DOI] [PubMed] [Google Scholar]
- 45.Zhong, H., H. SuYang, H. Erdjument-Bromage, P. Tempst, and S. Ghosh. 1997. The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89:413-424. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, Y., J. A. Richardson, L. F. Parada, and J. M. Graff. 1998. Smad3 mutant mice develop metastatic colon cancer. Cell 94:703-714. [DOI] [PubMed] [Google Scholar]