Abstract
The eukaryotic genome is divided into chromosomal domains of distinct gene activities. Transcriptionally silent chromatin tends to encroach upon active chromatin. Barrier elements that can block the spread of silent chromatin have been documented, but the mechanisms of their function are not resolved. We show that the prokaryotic LexA protein can function as a barrier to the propagation of transcriptionally silent chromatin in yeast. The barrier function of LexA correlates with its ability to disrupt local chromatin structure. In accord with this, (CCGNN)n and poly(dA-dT), both of which do not favor nucleosome formation, can also act as efficient boundaries of silent chromatin. Moreover, we show that a Rap1p-binding barrier element also disrupts chromatin structure. These results demonstrate that nucleosome exclusion is one of the mechanisms for the establishment of boundaries of silent chromatin domains.
Eukaryotic DNA is compacted into chromatin. The first level of packaging is the formation of nucleosomes, each consisting of a protein core of histones H2A, H2B, H3, and H4, around which 146 bp of DNA is wrapped. Higher levels of compaction involve histone H1 and/or other proteins that associate with nucleosomes (38). Based on its cytological and molecular properties, chromatin is roughly divided into condensed heterochromatin and decondensed euchromatin, which are interspersed in the genome. In general, heterochromatin inhibits gene expression whereas euchromatin allows it, leading to a position effect on gene activity. Heterochromatin formed in one part of the genome may propagate along the chromosome, consuming euchromatin in its path. This is accomplished by the spreading of heterochromatin-specific complexes that interact with nucleosomes and condense chromatin to a higher level (20, 35). In addition, various covalent modifications of histones (e.g., acetylation and methylation) also play pivotal roles in establishing the state of chromatin at a particular locus (27). For instance, heterochromatin is associated with characteristic hypoacetylation of histones.
In Saccharomyces cerevisiae, transcriptionally silent chromatin at HMR, HML, or telomeres is the yeast equivalent of metazoan heterochromatin that is formed through coordinated actions of cis-acting elements and trans-acting factors (41). The cis-acting elements include telomeric repeats and sites flanking each HM locus that are known as silencers, and the trans-acting proteins include Sir2p-Sir4p and silencer- or telomere-binding proteins. Silencer- or telomere-binding proteins recruit the SIR complex (Sir2p/Sir3p/Sir4p), which then propagates sequentially along an array of nucleosomes. The SIR complex is an integral part of silent chromatin, and interactions between Sir3p/Sir4p and histones H3 and H4 are key to the establishment and maintenance of silenced chromatin (41). There is evidence that Sir3p has higher affinity to unacetylated histone H4 (10). Sir2p is an NAD-dependent protein deacetylase that is likely involved in reducing histone acetylation in silent chromatin (22, 23). The current model for silencing proposes that Sir2p, when recruited to a silencer or telomere, deacetylates histones in an adjacent nucleosome, which then binds another SIR complex with high affinity. The nucleosome-bound SIR complex then deacetylates the neighboring nucleosome, which in turn binds a new SIR complex. In this manner, the SIR complex promotes its own stepwise propagation along the chromatin. A similar mechanism involving a chain of events of histone H3 deacetylation followed by H3 methylation followed by binding of methylated H3 by HP1 (heterochromatin protein 1) or swi6 is believed to underlie the spread of silent chromatin in fission yeast and higher organisms (20, 35).
The fact that silent chromatin can encroach upon active chromatin poses the question of how a euchromatin region is protected from adjacent silent chromatin or how the boundaries of a distinct chromatin domain are defined. Studies of Drosophila and vertebrates have demonstrated that chromatin boundaries often coincide with nucleoprotein structures called boundary or insulator elements (52). By definition, boundary or insulator elements protect a gene in a domain from influences of adjacent domains. However, there is only a limited understanding of their mechanisms of action. Recently, sequences that can block the spread of transcriptional silencing have been discovered in S. cerevisiae (7, 16). These sequences, referred to as silent chromatin barriers, do not show sequence homology, but they all contain multiple binding sites for one or more positive and/or negative regulators of transcription. Donze and Kamakaka proposed that barriers function by recruiting chromatin-modifying or -remodeling complexes that actively promote the formation of active chromatin (15). An alternative model suggests that barriers are simply passive roadblocks to encroaching silent chromatin. Such a roadblock can be formed by tethering the barrier sequence to an “immobile” nuclear structure (24). Finally, based on the stepwise-spreading model of silent chromatin, we hypothesized that a sequence void of nucleosomes can serve as a barrier to the spread of silent chromatin (6). No definitive proof exists for any of the models, but increasing evidence indicates that different barriers employ different mechanisms in preventing the spread of silent chromatin (52).
In this work, we show that the prokaryotic LexA protein can function as a barrier to the propagation of transcriptionally silent chromatin in yeast. The barrier function of LexA correlates with its ability to alter local chromatin structure in a way indicative of nucleosome exclusion. In accord with this, we demonstrate that nucleosome-excluding sequences (CCGNN)n (where N can be G, C, A, or T) and poly(dA-dT) (43, 50) can also act as silent-chromatin barriers. In addition, we show that the previously described barrier element TEF2-UAS coincides with nuclease-hypersensitive sites. These results strongly support our hypothesis that the spread of silent chromatin can be blocked by sequences that break the regularity of nucleosomes in chromatin.
MATERIALS AND METHODS
Plasmids and strains.
Plasmid pH0 is pMB22-a containing a HindIII-BamHI fragment of chromosome III (coordinates, 14838 to 16263) with a URA3 gene inserted at its EcoRV site (5). pH1 to pH5 were derived from pH0 by inserting one to five copies, respectively, of a sequence bearing the consensus binding site of LexA, CTGTATGTACATACAG, at the SnaBI site. pH 6 and pH 7 were derived from pH0 by inserting two and three copies, respectively, of a sequence bearing the ColE1 operator, CTGTATATAAAACCAGTGGTTATATGTACAG, at the SnaBI site. pH8 was derived from pH0 by inserting four copies of the ColE1 operator at the NgoMIV site. pH10 to pH13 were derived from pH0 by inserting (CCGNN)n (see Fig. 4A) at the SnaBI site. (CCGNN)16 was inserted at the NgoMIV site of pH0 to make pH14. Note that (CCGNN)31 in pH13 is 5′-CCGTACCGATCCGAACCGGACCGCTCCGAGCCGTCCCGTACCGCACCGCGCCGTTCCGAACCGGACCGTCCCGCTCCGTACCGATCCGAACCGGACCGCTCCGAGCCGTCCCGTACCGCACCGCGCCGTTCCGAGCCGGACCTCCCGCTCCGTA-3′. (CCGNN)n in other plasmids consists of a 5′ fragment of this sequence. pH15 to pH18 were made by inserting (dT-dA)20, (dT-dA)45, (dT-dA)80, and (dA-dT)80, respectively, at the SnaBI site of pH0. pH19 was made by inserting (dT-dA)80 at the NgoMIV site of pH0. pH23 was made by inserting TEF2-UAS (coordinates, −511 to −407 relative to the TEF2 start codon) at the SnaBI sites of pH0.
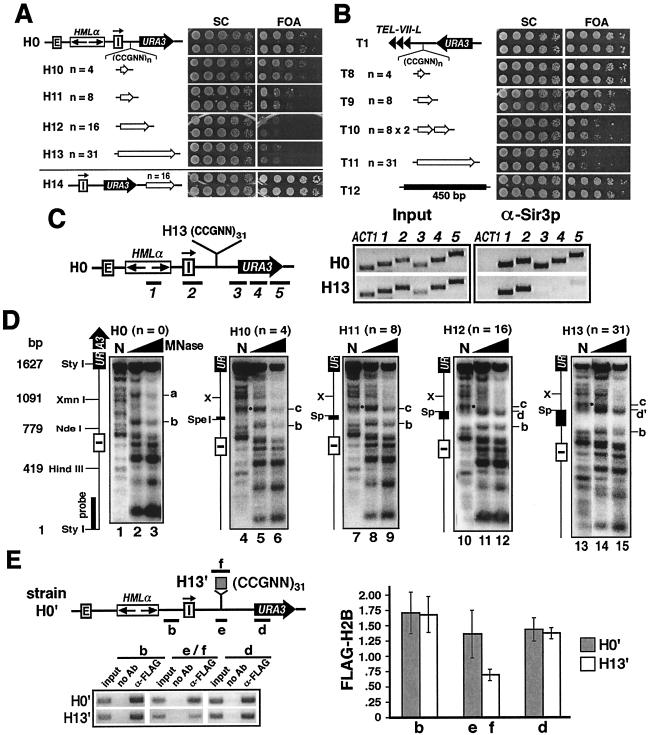
FIG. 4.
(CCGNN)n can act as a barrier to the spread of silent chromatin. (A) (CCGNN)n counteracts URA3 silencing. Left, modified HML in strains H10 to H14. Open arrows, (CCGNN)n. Right, growth phenotypes of each strain on SC and SC plus FOA (FOA) media. (B) (CCGNN)n counteracts telomeric silencing. Left, TEL-VII-L in T8 to T12. Right, growth phenotypes. (C) (CCGNN)n can block the spread of silent chromatin. ChIP was performed on strains H0 and H13 using α-Sir3p. Left, DNA fragments examined in ChIP. They were identical to those described in Fig. 2A. Right, PCR products 1 to 5 and ACT1 from H0 and H13. (D) Effect of (CCGNN)n on chromatin. Indirect end labeling was performed on strains H0 and H10 to H13. MNase-treated DNA was digested with StyI, and a 200-bp probe near the StyI site was used in Southern blotting. Sp, SpeI; X, XmnI. Sites a to d and d′ are indicated. N, naked DNA. The dots in naked DNA lanes mark bands corresponding to the gain of site c brought about by (CCGNN)n. (E) ChIP analysis of the level of histone H2B around (CCGNN)31. Left, results of ChIP using α-FLAG in strains H0′ and H13′. DNA fragments b, d, and e from strain H0′ and b, d, and f from H13′ detected by PCR were analyzed by agarose gel electrophoresis. Ab, antibody. Right, quantification and analysis of ChIP data. Each bar represents the relative abundance (α-FLAG over input) of the fragment detected. At least three independent experiments were performed, and the means (± standard deviations) of data from all of the experiments are presented.
Plasmid pT1 is pADH4UCA (18). pT2 to pT6 were derived from pT1 by inserting 1, 3, 4, 5, and 10 consensus LexA sites, respectively, at the BamHI site. pT7 was derived from pT1 by first replacing its BamHI-SalI fragment with a BamHI-ADH4-SalI fragment and subsequently inserting a BglII-TRP1-BamHI fragment, a BamHI-URA3-BglII fragment, and a BamHI-10 LexA sites-BamHI fragment at the BamHI site. pT8 to pT11 were derived from pT1 by inserting various (CCGNN)n sequences (see Fig. 4B) at the BamHI site. pT12 was made by inserting a 450-bp sequence (positions 159358 to 159682 of chromosome XV fused to 14971 to 15096 of chromosome III) at the BamHI site of pT1. pT13 and pT14 were made by inserting (dT-dA)80 at the BamHI site of pT1 in both directions. pT15 was made by inserting TEF2-UAS at the BamHI site of pT1.
pBTM116 is a 2μm-TRP1 plasmid carrying a SphI-SphI fragment consisting of the LexA gene flanked by the yeast ADH1 promoter and terminator (49). pL1 was derived from pBTM116 by deleting the SphI-LexA-SphI fragment and inserting LEU2 within TRP1. The LexA gene in pBTM116 has an extra 75 bp added to its open reading frame. Plasmid pXB319 was derived from pBTM116 by removing the 75-bp extra sequence. pL2 was derived from pXB319 by inserting LEU2 within TRP1. pL3 was derived from pL2 by fusing the sequence encoding the TADIII domain (amino acids 415 to 467) of Adr1p to the LexA gene. pG1 is pL1. pG2 is a 2μm-LEU2 plasmid carrying a sequence coding for hemagglutinin-tagged GBD (amino acids 1 to 147 of Gal4p) bracketed by the ADH1 promoter and terminator. pG3 and pG4 were derived from pG2 by fusing the LexA gene and TADIII, respectively, to hemagglutinin-GBD. pRB1840, pJK103, and pSH18-34 are 2μm-URA3 plasmids carrying a lacZ gene under the control of one, two, and eight LexA-binding sites, respectively (from Roger Brent, Molecular Sciences Institute). pRS424 is a 2μm-TRP1 plasmid.
Strain YXB76 is MATa ura3-52 leu2-3,112 ade2-1 lys1-1 his5-2 can1-100, E-HMLα-Iinverted (5). Strains H0, H1 to H8, H10 to H19, and H23 were made by transforming YXB76 to Ura+ with the HindIII-BamHI-digested plasmids pH0, pH1 to pH8, pH10 to pH19, and pH23, respectively. Strain H9 is PJ69-4α (MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::HIS3 MET2::GAL7-lacZ [26]). H20 to H22 were derived from H0, H17, and H18, respectively, by replacing DAT1 with kanMX4. T1 to T15 were made by transforming YXB76 to Ura+ with the EcoRI-SalI-digested plasmids pT1 to pT15, respectively. Strains C1, C2, and C3 were made by transforming YXB76 to Ura+ with plasmids pRB1840, pJK103, and pSH18-34, respectively. Strain YZS276 [MATa hta1-htb1Δ::LEU2 hta2-htb2Δ leu2-2,112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 ssd1 pZS145 (HTA1-FLAG-HTB1-CEN-HIS3)] was described by Sun and Allis (45). Strains H0′, H5′, and H13′ were made by transforming YZS276 to Ura+ with the HindIII-BamHI-digested plasmids pH0, pH5, and pH13, respectively. The relevant genotypes of all strains made were confirmed by Southern blotting and/or PCR.
ChIP.
Chromatin immunoprecipitation (ChIP) was carried out as described previously (12). The primers for PCR of fragments 1 to 5 (see Fig. 2A) were 5′-CCCACTTCTAAGCTGATTTCAATC-3′ and 5′-CCGCTGTGTTTCTGTATGATTTGG-3′ for fragment 1, 5′-GCTTATTGTGCTTTGTTGGGTG-3′ and 5′-GCAGCTGTTACGGAGATGC-3′ for fragment 2, 5′-CTAATAGAACGATAGATCTGGCTTTTCAATTC-3′ and 5′-AGGATGAGTAGCAGCACGTTC-3′ for fragment 3, 5′-CAAACAAACTTGTGTGCTTCATTG-3′ and 5′-TACTTCTTCTGCCGCCTGCTT-3′ for fragment 4, and 5′-GGCTCCCTATCTACTGGAGAA-3′ and 5′-GCAGTCTGTAATAAAACACACCAG-3′ for fragment 5. The primers for fragments b to f (see Fig. 3C and 4E) were 5′-GGATGTGTATACTAAGCCTTGGG-3′ and 5′-CAAGAAATTCAATAAACGTTAATGAAAGGT-3′ for fragment b, 5′-GACCATTATGCTAAAATACTGGGGTC-3′ and 5′-GCCCCTCGTTTGTATAAATACCG-3′ for fragments c, e, and f, and 5′-GGAACGTGCTGCTACTCA-3′ and 5′-TTGTACTTGGCGGATAATGCC-3′ for fragment d.
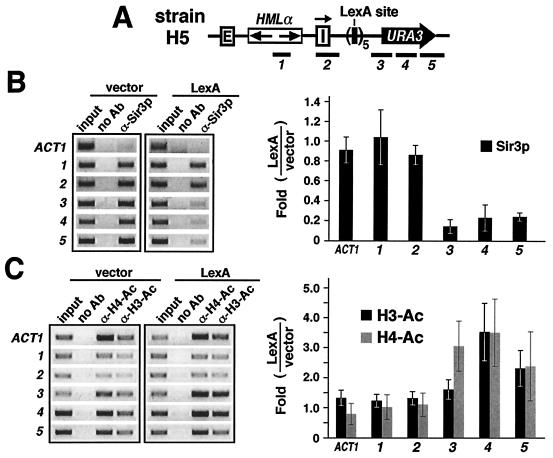
FIG. 2.
LexA blocks the spread of silent chromatin. (A) HML in strain H5 analyzed with ChIP. The positions of DNA segments 1 to 5 are shown. (B) LexA blocks the spread of Sir3p. (Left) Results of ChIP using α-Sir3p (from Danesh Moazed) in H5 bearing pL1 (vector) and pL2 (LexA). DNA fragments (ACT1 and 1 to 5) detected by PCR were analyzed by agarose gel electrophoresis. DNA from whole-cell extract (input), DNA precipitated by α-Sir3p, or DNA obtained in the same way as α-Sir3p DNA except without antibody (no Ab) was used as the PCR template. Sequence ACT1 is from the ACT1 locus. (Right) Quantification and analysis of ChIP data. Each bar represents the change (n-fold) in the intensity of a fragment detected in H5 bearing pL2 compared to that in H5 bearing pL1. At least three independent experiments were performed, and the means (± standard deviations) of data from all of the experiments are presented. Note that H5 has an endogenous ura3-52 allele resulting from a Ty insertion within the URA3 open reading frame (39). PCR primers were designed to amplify 3-5 specifically from URA3 near HML but not from ura3-52 (12). (C) LexA blocks the spread of nucleosome hypoacetylation. (Left) Results of ChIP using α-H4-Ac and α-H3-Ac (from David Allis) in H5 bearing pL1 (vector) or pL2 (LexA). The DNA sequences tested by PCR are the same as those in panel B. (Right) Quantification and analysis of ChIP data. Each solid bar represents the change (n-fold) in the level of H3 acetylation in H5 bearing pL2 compared to that in H5 bearing pL1. The shaded bars represent changes in H4 acetylation.
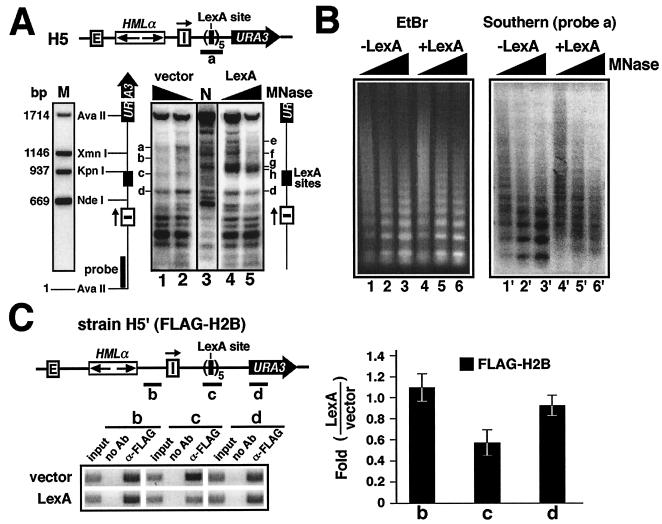
FIG. 3.
LexA binding disrupts local chromatin structure in yeast. (A) LexA induces changes in local chromatin. (Top) HML in strain H5. (Bottom) Indirect end labeling was performed on H5 harboring either pL1 (vector) or pL2 (LexA). Permeabilized cells were treated with MNase at 75 (lanes 2 and 4) or 150 (lanes 1 and 5) U/ml. DNA was isolated and digested with AvaII, which cut once in HML and once in URA3. An aliquot of permeabilized cells not treated with MNase was used to isolate naked DNA. An aliquot of naked DNA was treated with 7.5 U of MNase/ml and then digested with AvaII. This sample was designated N (lane 3). Other aliquots of naked DNA were cut directly by AvaII, AvaII-XmnI, AvaII-KpnI, or AvaII plus NdeI and then pooled as sample M (marker). Samples 1 to 5, M, and N were run on an agarose gel. After Southern blotting, the relevant DNA fragments were detected by a 200-bp probe (solid bar) near the AvaII site in HML. (B) Mononucleosome analysis of LexA-induced changes in chromatin structure. Permeabilized cells of strain H5 bearing pL1 (−LexA; lanes 1 to 3) or pL2 (+LexA; lanes 4 to 6) were treated with MNase at 75 (lanes 1 and 4), 150 (lanes 2 and 5), and 300 (lanes 3 and 6) U/ml. DNA was isolated and fractionated on an agarose gel and stained with EtBr (left). After Southern blotting, DNA was hybridized with probe a, consisting of the five LexA sites and short flanking sequences (58 bp on the left side and 98 bp on the right) (panel A). (C) ChIP analysis of the level of histone H2B around the LexA-binding sites. Left, results of ChIP using α-FLAG M2 (Sigma) in H5′ bearing pRS424 (vector) and pXB319 (LexA). DNA fragments (b to d) detected by PCR were analyzed by agarose gel electrophoresis. (Right) Quantification and analysis of ChIP data. Each bar represents the change (n-fold) in the intensity of a fragment detected in H5′ bearing pXB319 (LexA) compared to that in H5′ bearing pRS424 (vector). At least three independent experiments were performed, and the means (± standard deviations) of data from all the experiments are presented.
Chromatin analysis with MNase.
Spheroplasts made from ∼109 log-phase cells by using zymolyase were permeabilized with NP-40 as described previously (30). For indirect end labeling, micrococcal nuclease (MNase) at 75 and 150 U/ml was used to treat 2 × 108 cells at 37°C for 5 min. The reaction was stopped by 0.5% sodium dodecyl sulfate and 25 mM EDTA, andthe DNA was isolated. An aliquot of DNA was determined by electrophoresis to contain fragments representing nucleosome ladders (not shown). An aliquot of permeabilized spheroplasts not treated with MNase was used to isolate naked DNA, which was digested with MNase at 7.5 U/ml. The DNA in each sample was then digested with AvaII or StyI and run on a 1.0% agarose gel. The relevant fragments were visualized by an appropriate probe (see Fig. 3 to 6) after Southern blotting. For mononucleosome analysis, MNase at 75, 150, and 300 U/ml was used to treat 2 × 108 permeabilized cells at 37°C for 5 min. The reaction was stopped by 0.5% sodium dodecyl sulfate and 25 mM EDTA, and the DNA was isolated and run on a 1.0% agarose gel.
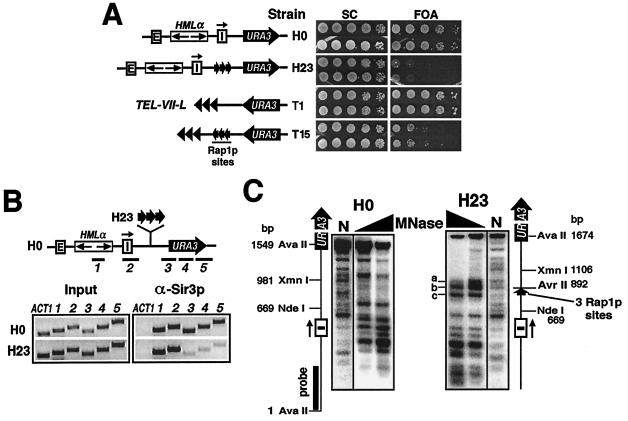
FIG. 6.
Barrier TEF2-UAS coincides with nuclease-hypersensitive chromatin. (A) TEF2-UAS counteracts HML and telomeric silencing. Strain H23 has TEF2-UAS consisting of three tandem Rap1p sites (arrows) inserted between HML and URA3. TEF2-UAS was inserted between TEL-VII-L and URA3 in T15. The growth phenotypes of H0, H23, T1, and T15 are shown. (B) TEF2-UAS can block the spread of silent chromatin. The results of ChIP performed on H0 and H23 using α-Sir3p are shown. (C) Special chromatin structure associated with TEF2-UAS. The results of indirect end labeling on strains H0 and H23 are shown. MNase-treated DNA was digested with AvaII.
RESULTS
The prokaryotic LexA protein can counteract transcriptional silencing in yeast.
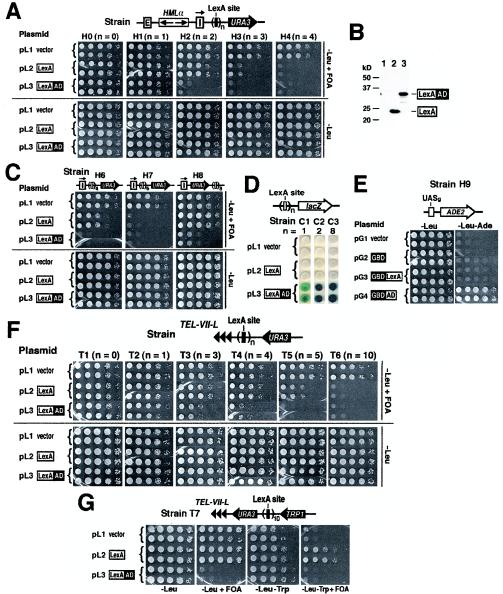
LexA is a prokaryotic sequence-specific DNA-binding protein that is widely used in eukaryotic organisms as a vehicle to target heterologous peptides to specific sites (9). LexA fusion proteins have been used to identify factors that could counteract the spread of transcriptional silencing in a silencer-blocking assay (54). In such an assay, one or two LexA-binding sites are inserted between a source of silencing (HML) and a reporter gene (URA3) (Fig. 1A, top). The direction of the HML-I silencer is inverted so that it can efficiently silence sequences to the right (centromere proximal) of HML (5). URA3 expression renders cells sensitive to 5-fluoroorotic acid (FOA), and hence, silencing of URA3 can be measured by cell growth on medium containing FOA (Fig. 1A; growth of strains H0 and H1 to H4 bearing plasmid pL1 on −Leu +FOA medium). Using this assay, it was found that the activation domains (AD) of a variety of transcriptional activators could counteract silencing (54). For example, targeting the TADIII AD of Adr1p by LexA to two LexA sites led to a robust antisilencing effect (Fig. 1A; lack of growth of H2 bearing pL3 on −Leu +FOA).
FIG. 1.
LexA can block the spread of transcriptional silencing in yeast. (A) LexA counteracts URA3 silencing at HML. Top, modified HML in strains H0 and H1 to H4. Boxes E and I, HML-E and HML-I silencers, respectively. The HML-I silencer is flipped, as indicated by the arrow above the I box, and URA3 is inserted to its right. Strains H1 to H4 have one (n = 1) to four (n = 4) consensus LexA sites (solid bar), respectively, inserted between HML and URA3. Each strain carrying pL1, pL2, or pL3 was grown to late log phase and spotted in 10-fold serial dilutions on −Leu and −Leu +FOA media. The plates were incubated for 3 days. For each strain, at least three independent clones were tested, and two representative clones are shown. −Leu medium is synthetic complete (SC) medium lacking leucine. −Leu +FOA is −Leu medium supplemented with 1 mg of FOA/ml. (B) Cellular levels of LexA and LexA-AD. Equal amounts of total protein from strain H2 bearing plasmids pL1 to pL3 (lanes 1 to 3) was analyzed by Western blotting and probed with a LexA antibody. (C) Antisilencing mediated by nonconsensus LexA sites (open bars). Growth phenotypes of strains H6 to H8 harboring plasmids pL1 to pL3, respectively, are shown. (D) LexA is not a transcriptional activator. Strains C1, C2, and C3 bearing pL1, pL2, and pL3, respectively, were grown on −Leu −Ura plates supplemented with X-Gal. (E) GBD-LexA is not an activator. Strain H9 has a genomic copy of the UASg-ADE2 construct. UASg is the UAS of GAL2 that contains Gal4p sites. H9 bearing pG1 to pG4 was tested for growth on −Leu and −Leu −Ade media. (F) LexA can block the spread of telomeric silencing. Top, modified left telomere of chromosome VII (TEL-VII-L; triple arrowhead) in strains T1 to T6. The growth phenotype of each strain bearing pL1, pL2, or pL3 is shown. (G) LexA can act as a boundary or insulator element. Top, TEL-VII-L in strain T7. The growth phenotypes of T7 bearing pL1 to pL3 on −Leu, −Leu +FOA, −Leu −Trp, and −Leu −TRP +FOA media are shown.
In experiments involving LexA fusion proteins, LexA alone ought to be used as a negative or background control. In the experiments described above, LexA did not affect URA3 silencing in strains H1 and H2 (Fig. 1A; H1 and H2 bearing pL2 on −Leu +FOA), despite the fact that cellular levels of LexA were comparable to those of LexA-AD (Fig. 1B). This validates the results concerning the antisilencing effect of TADIII of Adr1p. However, we discovered that, surprisingly, increasing the number of LexA sites to three caused a dramatic LexA-dependent decrease in URA3 silencing (Fig. 1A, H3; compare pL2 to pL1). Four or more LexA sites completely eliminated URA3 silencing as measured by the disappearance of FOA-resistant colonies (Fig. 1A, H4 bearing pL2, and data not shown). We also used reverse transcription-PCR to directly demonstrate that LexA enhanced URA3 expression by 6.5-fold in strain H5 (see supplementary Fig. 1, posted at http://www.rochester.edu/College/BIO/faculty/Bi.html). Therefore, in-triguingly, LexA by itself in sufficient amounts can also counteract transcriptional silencing.
Like many prokaryotic repressors, LexA binds as a dimer to an operator consisting of two dyad symmetric half sites. In the experiments described above, a synthetic sequence corresponding to the “perfect” consensus for LexA binding (CTGTATGTACATACAG) (8) was used. We also tested whether naturally occurring “imperfect” LexA-binding sequences could mediate antisilencing. A commonly used LexA-binding sequence is the Escherichia coli ColE1 operator consisting of two variant LexA-binding sequences with a 1-nucleotide overlap (boldface) (CTGTATATAAAACCAGTGGTTATATGTACAG) (17). Whereas one ColE1 operator had no antisilencing activity (data not shown), two operators consisting of four LexA sites caused a moderate decrease in URA3 silencing (Fig. 1C, H6; compare pL2 to pL1). Three or more operators (six or more LexA sites) abolished silencing completely (Fig. 1C, H7, and data not shown). Therefore, compared to the consensus binding sequence for LexA, more nonconsensus sites are needed to counteract silencing. This may be due to higher affinity of LexA for a consensus site than for a variant site and/or to differences in the layout of the LexA-binding sites in, e.g., H4 and H6 (Fig. 1A and C). On the other hand, LexA-AD exhibited robust antisilencing activity in H6 (Fig. 1C). In summary, the above results indicate that the strength of the antisilencing activity of LexA depends on the number of binding sites and their affinity to LexA, and it is intrinsically lower than that of a targeted activator like Adr1p.
The silencer-blocking function of LexA is not due to direct gene activation or interference with silencing of HMLα.
How does the prokaryotic LexA protein counteract silencing in yeast? One explanation is that LexA may directly activate URA3 transcription like an activator that can overcome silencing (1, 4). However, this is not likely the case for the following reasons. First, LexA sites in any of strains H1 to H7 were located ≥630 bp upstream of the translation start codon of URA3, a distance longer than what is normally allowed for an activator to function in yeast (28). Secondly, LexA has been repeatedly demonstrated to possess no activating ability in yeast (9). Here, we employed three different assays to strongly verify this notion. The first assay measures expression of a lacZ reporter under the control of one, two, or eight LexA sites (Fig. 1D, strains C1 to C3). Expression of lacZ in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) would render cells blue. As shown in Fig. 1D (pL2), LexA did not cause any expression of lacZ (as indicated by the white color of the cells). As a positive control, we showed that LexA-AD induced a high level of lacZ expression (Fig. 1D, pL3). In the second experiment, we demonstrated that tethering LexA to DNA by the DNA-binding domain of Gal4p (GBD) did not activate a downstream ADE2 gene (Fig. 1E; lack of growth of H9 on −Leu −Ade medium bearing pG3). Finally, in an experiment detailed below, we showed that LexA did not activate a telomere-linked URA3 gene with 10 LexA sites inserted adjacent to its promoter (Fig. 1G; growth of T7 bearing pL2 on −Leu +FOA). Therefore, LexA does not function as a transcriptional activator in yeast. Consequently, its antisilencing activity is not a result of direct activation of transcription.
Another explanation for the silencer-blocking function of LexA is that it affects the HML silencers directly. If this were the case, silencing of the α1 and α2 genes resident within the HML locus would be abrogated. We tested this possibility by measuring the mating ability of strain H5 bearing five LexA sites between HML and URA3 (Fig. 2A), as the mating efficiency of a MATa strain is inversely proportional to the expression state of the HMLα genes (21). As shown in supplementary Fig. 2 (posted at http://www.rochester.edu/College/BIO/faculty/Bi.html), LexA had no effect on the mating efficiency of H5 cells. Therefore, LexA does not interfere with silencing within the HML locus. This was confirmed by our later demonstration that LexA does not affect the association of Sir3p with HML (Fig. 2A). Other barrier elements described below also have no effect on HMLα silencing (supplementary Fig. 2).
LexA can block the propagation of transcriptional silencing.
An alternative explanation for the antisilencing ability of LexA is that it blocks the spread of silencing. This implies that LexA can counteract silencing only when it is targeted to sites between the source of silencing and the reporter (as is the case in strains H3 to H7) but not when it is tethered elsewhere. To test this, we inserted eight LexA sites downstream of HML and URA3 in strain H8 (Fig. 1C). It was clear that LexA then failed to alleviate URA3 silencing (Fig. 1C; H8 bearing pL2). Therefore, the antisilencing function of LexA depends on its position relative to the reporter and the source of silencing, which is consistent with it's being a barrier to the spread of silencing. On the other hand, LexA-AD significantly reduced URA3 silencing in H8 (Fig. 1C), indicating that LexA-AD is not a simple physical barrier to the propagation of silencing but can exert a long-range effect on the promoter of a gene in the path of silent chromatin.
Although the same basic mechanism is involved in silencing at both the HM loci and telomeres, telomeric silent chromatin has distinct features, such as the putative end looping mediated by the SIR proteins (44). We also examined the effect of LexA on telomeric silencing. We inserted various numbers of LexA sites between the (modified) left telomere of chromosome VII (TEL-VII-L) (18) and a URA3 gene integrated nearby (Fig. 1F, T1 to T6). URA3 was effectively silenced in these strains in the absence of LexA (Fig. 1F; growth of T1 to T6 bearing pL1 on −Leu +FOA medium). When LexA was expressed, URA3 silencing was unaffected by up to four LexA sites (Fig. 1F; T2 to T4 bearing pL2). However, 5 LexA sites induced a significant decrease in silencing (T5 bearing pL2), and 10 sites completely abolished silencing (T6). Therefore, LexA can also counteract telomeric silencing, although a greater number (≥5) of LexA sites are needed than are needed to counteract HML silencing (≥3) (Fig. 1A; H3 bearing pL2). The reduced efficiency of LexA in overcoming telomeric versus HML silencing may reflect differences between the structure and/or strength of silent chromatin at telomeres and that near HML. The strength of LexA-AD in antisilencing is also reduced at TEL-VII-L, as demonstrated by the fact that more (≥3) LexA sites are required to eliminate URA3 silencing (Fig. 1F; T3 bearing pL3).
Is LexA, as a barrier, functionally similar to boundary-insulator elements in higher eukaryotes? We addressed this question using a dual-reporter assay in which URA3 and TRP1 bracketing 10 LexA sites were integrated near TEL-VII-L (Fig. 1G; strain T7). Silencing at telomeres in yeast is reversible and is inherited in a semistable state (18). Consequently, URA3 or TRP1 can exist in either an “on” or “off” state of transcription in an individual cell. Colonies formed on −Leu +FOA medium represent cells in which URA3 is silenced (off), whereas those formed on −Leu −Trp medium represent cells in which TRP1 is on (Fig. 1G; strain T7 with pL1). However, the transcriptional states of URA3 and TRP1 are interdependent, so that in a single cell URA3 and TRP1 are cosilenced (Fig. 1G; lack of growth of strain T7 with pL1 on −Leu −Trp +FOA medium). Whereas LexA did not affect cell growth on −Leu +FOA medium, it reduced TRP1 silencing and enabled ∼10% of the cells to grow on −Leu −Trp +FOA medium (Fig. 1G; T7 bearing pL2). Therefore, LexA can protect TRP1 against silencing without affecting the silenced state of URA3. Note that in T7, LexA sites are close to the promoter of URA3, and yet LexA does not affect URA3 silencing. On the other hand, LexA-AD eliminated URA3 silencing (Fig. 1G; −Leu +FOA, T7 bearing pL3). These results reinforce the notion that the antisilencing effect of LexA is not due to direct gene activation and strongly suggest that LexA serves as a physical barrier to the spread of transcriptional silencing.
LexA blocks the propagation of silent chromatin.
The results described above suggest that LexA can act as a boundary or insulator element to delimit silent chromatin. We used the ChIP assay to test this proposal. Sir3p is an essential component of silent chromatin. We examined levels of Sir3p associated with regions on both sides of LexA sites in strain H5 (Fig. 2A). A Sir3p antibody (α-Sir3p) was used to immunoprecipitate fragments associated with Sir3p. PCR primers were designed to detect sequences 1 to 5 (200 to 450 bp in length) using immunoprecipitated template DNA from H5 carrying either pL1 (vector) or pL2 (LexA). The PCR product corresponding to each fragment was examined by gel electrophoresis. ChIP was repeated at least three times, and one representative gel picture is presented in Fig. 2B, left. The intensity of each PCR fragment was quantified and normalized against the input control. The LexA-induced change in Sir3p associated with a particular sequence was estimated as the ratio of the intensity of the PCR product from cells expressing LexA to that from cells lacking LexA. For each DNA segment, the mean of data from all the repeats together with the standard deviation was graphed in Fig. 2B, right. Also included as a control was the result for the ACT1 locus (ACT1), which is not associated with silent chromatin.
As shown in Fig. 2B, left, in the absence of LexA, Sir3p was associated with sequences 1 to 5 but not with ACT1 (compare 1 to 5 to ACT1 in the vector lanes), which is consistent with the fact that both HMLα and URA3, but not ACT1, are silenced. LexA greatly reduced the levels of Sir3p associated with sequences 3 to 5, but not 1 and 2 (Fig. 2B, left; compare LexA and vectors). As summarized in Fig. 2B, right, it is clear that LexA does not affect Sir3p associated with sequences to the left of LexA sites (Fig. 2B, bars 1 and 2) but induces an ∼5-fold reduction in Sir3p in regions to the right (Fig. 2B, bars 3 to 5). This demonstrates that LexA prevents Sir3p from spreading beyond the LexA sites.
Silent chromatin is associated with reduced histone acetylation. We also examined the levels of histone H3 and H4 acetylation on both sides of the LexA sites in strain H5. Antibodies against histone H3 with K9/K14 acetylation (α-H3-Ac) and acetyl-H4 isoforms (α-H4-Ac) were used to precipitate DNA associated with acetylated H3 and H4, respectively. As shown in Fig. 2C, right, acetylation of H4 was not affected in sequences to the left of LexA sites (shaded bars 1 and 2) but was increased 2- to 3.5-fold in sequences to the right (shaded bars 3 to 5). A similar effect of LexA on histone H3 acetylation was also observed, except that only a slight increase in H3 acetylation was detected in sequence 3 (Fig. 2C, solid bars 1 to 5). Therefore, LexA blocks the spread of histone hypoacetylation characteristic of silent chromatin. The above two lines of evidence demonstrate that LexA functions as a bona fide barrier to the propagation of silent chromatin.
LexA binding disrupts chromatin structure.
How does LexA act as a barrier to the spread of silent chromatin in yeast? To address this question, it is useful to briefly review the known properties of LexA as a DNA-binding protein. Each LexA-binding site consists of two half sites to which a pair of LexA molecules can bind in a highly cooperative manner (31). Cooperativity is achieved by dimerization of the LexA protein. Such cooperative binding, common to many prokaryotic repressors, occurs only on naked DNA and requires physical interactions between the proteins involved. When expressed in eukaryotes, LexA can access its chromosomal sites, and interestingly, it can also collaborate with heterologous DNA-binding proteins, such as yeast Gal4p and Gcn4p, in binding to their respective sites (34, 48). However, this phenomenon is fundamentally different from classic cooperative binding of prokaryotic repressors in that it can occur only on chromatin, not naked DNA, and it does not require direct contact between the two factors. Moreover, cooperation between LexA and the other factor can happen only when the two binding sites are within a nucleosome length distance. Based on these observations, it was proposed that LexA bound to a site in nucleosomal DNA will “loosen,” or destabilize, the nucleosome, thereby helping the other factor gain access to its binding site on the same nucleosome (34, 48). However, direct structural evidence for this model is lacking.
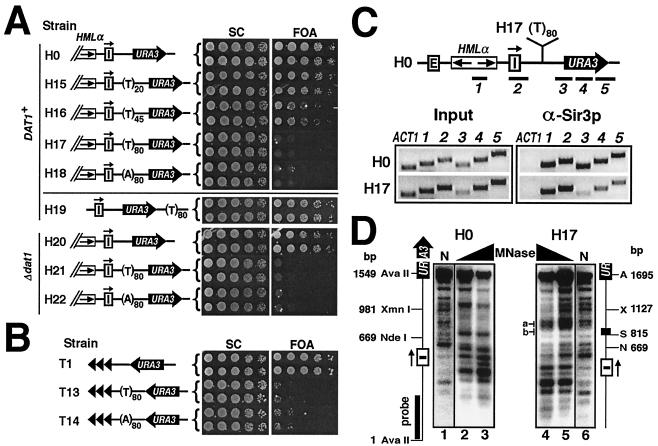
If LexA binding induces instability of the nucleosome, increased LexA occupancy should enhance this effect. In each of the strains H3 to H5, three, four, or five tandem LexA sites separated by short (13- to 20-bp) spacers exist within a sequence of <150 bp. We think that synergistic or cooperative binding of multiple LexA molecules to this sequence would disrupt or even remove a nucleosome(s). To directly examine this possibility, we analyzed the chromatin structure around the LexA-binding sites in H5 by MNase digestion, followed by indirect end labeling. MNase cleavage sites around LexA sites were mapped relative to an AvaII site within HML (Fig. 3A). LexA binding induced two prominent hypersensitive sites (g and h) <50 bp apart and immediately abutting its binding sites (Fig. 3A, lane 4), indicating that the region around g and h is not associated with a nucleosome. It is noteworthy that these hypersensitive sites persist after the elimination of silencing by deleting SIR3 (data not shown). Therefore, LexA-mediated disruption of local chromatin is independent of the silencing state of chromatin.
LexA also brought about other changes in chromatin structure in strain H5. First, the 140-bp sequence (Fig. 3A) spanning the five LexA sites appeared to be less sensitive to MNase digestion (Fig. 3A, compare lanes 2 and 4). It is possible that LexA binding renders this sequence less accessible to MNase. Given the putative nucleosome-disrupting ability of LexA, this region may be associated with a loosely structured nucleosome or no nucleosome at all. Second, in regions downstream (URA3 proximal) of LexA sites, LexA caused the disappearance of MNase sites a and b and the appearance of sites e and f (Fig. 3A, compare lanes 2 and 4). These alterations resemble those observed in an isogenic sir3− derivative of H5 lacking LexA (data not shown) and therefore likely reflect the change from silent chromatin to active chromatin as a result of the barrier activity of LexA. On the other hand, LexA caused little or no change in MNase cleavage in sequences upstream of its binding sites (Fig. 3A, lane 4; cleavage sites below site d), which is consistent with the notion that LexA blocks the spread of silent chromatin but does not actively dismantle existing silent chromatin.
We also employed a mononucleosome analysis with MNase (19) to determine whether the region spanning LexA-binding sites was wrapped into a nucleosome(s). Chromatin from strain H5 with or without LexA expression was reduced by MNase to mono-, di-, tri-, and tetranucleosome DNA fragments. These fragments, referred to as a nucleosome ladder, were resolved on an agarose gel and revealed by ethidium bromide (EtBr) staining (Fig. 3B, left). It was evident that LexA had no effect on the nucleosome ladder (Fig. 3B, compare lanes 4 to 6 to lanes 1 to 3), indicating that LexA does not alter global chromatin structure. To see whether LexA-binding sites were associated with a nucleosome, a probe spanning the LexA-binding sites (Fig. 3A) was hybridized to the nucleosome ladder after Southern blotting (Fig. 3B). In the absence of LexA, this probe revealed a clear ladder (Fig. 3B, right, lanes 1′ to 3′) similar to the EtBr-stained ladder (lanes 1 to 3), indicating that positioned nucleosomes exist at or near the LexA-binding sites. In contrast, when LexA was expressed, a smear instead of a ladder was detected (lanes 4′ to 6′), indicating the lack of positioned nucleosomes. This demonstrates that LexA prevents the positioning of nucleosomes at or near its binding sites.
Finally, to verify that LexA induces nucleosome disruption or exclusion, we used ChIP to directly show that association of histones with LexA-binding sites was reduced. For this purpose, we used strain H5′, in which a FLAG tag is fused to the N terminus of histone H2B (45) and an “HML-Iinverted-5 LexA sites-URA3” construct identical to that in strain H5 (Fig. 3C) is present. Replacement of the wild-type H2B with FLAG-H2B had no effect on URA3 silencing or the LexA barrier function (data not shown). ChIP was done with an anti-FLAG antibody, and the presence of FLAG-H2B in sequences b to d was measured (Fig. 3C). As shown in Fig. 3C, LexA induced a twofold decrease in the level of H2B associated with sequence c, which spans the five LexA-binding sites in strain H5′. On the other hand, LexA did not alter H2B binding to sequence b, which has been shown to be associated with positioned nucleosomes (51), and sequence d from the coding region of URA3. These results directly demonstrate that LexA can compete with histones at or near its binding sites and hence disrupt or remove nucleosomes.
The nucleosome-excluding (CCGNN)n sequence can stop the spread of silent chromatin.
The correlation between the ability of LexA to block the spread of silent chromatin and to disrupt chromatin structure prompted us to ask if disrupting chromatin by other means would also suffice for the barrier function. The (5′-[G/C]3NN-3′)n motif (where N can be G, C, A, or T) was predicted by Wang and Griffith to resist wrapping around a histone octamer based on its anisotropic flexibility (50). In support of this, they showed that (CCGNN)n excluded nucleosome formation in a repeat-number-dependent manner. Consistently, (CCGNN)12 or (CCGNN)48 inserted in the yeast genome induced DNase I-hypersensitive sites within and upstream of it, suggesting that (CCGNN)n represents “open,” or nucleosome-free, chromatin in vivo (32). We tested whether (CCGNN)n was able to counteract silencing. Increasing numbers of CCGNN repeats were inserted between HML and URA3 in strain H0 to make H10 to H13 (Fig. 4A). (CCGNN)4 had a barely detectable effect on URA3 silencing (Fig. 4A, strain H10). (CCGNN)8 reduced silencing by 10- to 100-fold (Fig. 4A, H11). Sixteen or more copies of CCGNN eliminated URA3 silencing (H12 and H13). Consistently, we also showed by reverse transcription-PCR that the presence of (CCGNN)31 increased the level of URA3 mRNA by 6.7-fold in strain H13 (supplementary Fig. 1). The direction of (CCGNN)n did not affect its antisilencing function (data not shown). The effect of (CCGNN)n on URA3 silencing is not a distance effect, as heterologous sequences of up to 600 bp (longer than any of the inserts in H10 to H13) do not affect silencing (54). Therefore, ≥8 CCGNN repeats can counteract silencing near HML. On the other hand, (CCGNN)16 inserted downstream of URA3 did not affect silencing (Fig. 4A, H14), indicating that (CCGNN)n functions similarly to LexA but not LexA-AD (Fig. 1C; H8 bearing pL2 and pL3, respectively). (CCGNN)n can also counteract telomeric silencing, although more (≥16) copies of it are required (Fig. 4B). The effect of (CCGNN)n on telomeric silencing was not a distance effect either, since a 450-bp heterologous sequence longer than any of the inserts in strains T8 to T11 had no effect on silencing (Fig. 4A, T12).
To test whether (CCGNN)n is a barrier to the spread of silent chromatin, we used ChIP to examine the distribution of Sir3p on both sides of (CCGNN)31 in strain H13 (Fig. 4C). As a control, Sir3p distribution in H0 was also measured. ChIP on H13 and H0 was carried out in a manner similar to ChIP on strain H5 (Fig. 2A and B). As predicted, Sir3p was associated with all of sequences 1 to 5 in strain H0 (Fig. 4C; H0, α-Sir3p, lanes 1 to 5). In H13, the level of Sir3p in regions to the left of (CCGNN)31 remained unchanged (Fig. 4C; H13, α-Sir3p, lanes 1 and 2). However, Sir3p association with sequences to the right decreased dramatically (Fig. 4C; H13, α-Sir3p, lanes 3 to 5). These results demonstrate that (CCGNN)n, like LexA, can stop the spread of silent chromatin.
We also examined whether (CCGNN)n sequences used in the experiments described above resist nucleosome formation. Indirect end-labeling experiments were performed on strains H0 and H10 to H13 bearing 0, 4, 8, 16, and 31 copies of CCGNN, respectively (Fig. 4A). As shown in Fig. 4D, (CCGNN)n induced clear changes in chromatin. (CCGNN)16 and (CCGNN)31 induced significant MNase cleavage at a site immediately downstream of it (Fig. 4C, d and d′ in lanes 11 and 14, respectively). This indicates that CCGNN repeats disrupt chromatin in their vicinity, which is consistent with data on (CCGNN)48 obtained by Kirkpatrick et al. (32). Interestingly, (CCGNN)n, especially (CCGNN)16 and (CCGNN)31, appeared to be resistant to MNase cleavage regardless of being in naked DNA in vitro or in chromatin template in vivo (e.g., Fig. 4D, compare lanes 13 and 14). The reason that CCGNN repeats resist MNase digestion may be that G+C-rich sequences are poor substrates for MNase or that a long (CCGNN)n sequence forms an unusual structure which cannot be digested by MNase. Consistent with the latter hypothesis, (CCGNN)31 also appeared to be resistant to DNase I both in vitro and in vivo (data not shown). Other changes brought about by (CCGNN)n included the loss of the MNase-sensitive site a and the gain of site c, whose intensity decreased as the number of CCGNN repeats increased (Fig. 4C, compare lanes 2, 5, 8, 11, and 14).
The results described above indicate that (CCGNN)n disrupts chromatin structure, at least in its immediate vicinity. We also used ChIP to measure the abundance of histone H2B at or near (CCGNN)31 in strain H13′ (Fig. 4E). Strain H13′ was identical to strain H5′, in which a FLAG tag was fused to histone H2B (Fig. 3C), except that instead of LexA sites (CCGNN)31 was inserted between HML and URA3 (Fig. 4E). Strain H0′ is isogenic to H13′ but lacks the (CCGNN)31 insertion. ChIP was done with an anti-FLAG antibody, and the presence of FLAG-H2B in sequences b, f, and d from strain H13′ was measured (Fig. 4E). As a control, the presence of FLAG-H2B in sequences b, e, and d from strain H0′ was also measured (Fig. 4E). Fragments b and d were described above (Fig. 3C). Fragment f consists of (CCGNN)31 and 45-bp flanking sequences from strain H13′, whereas fragment e is fragment f with (CCGNN)31 deleted. It was clear that (CCGNN)31 caused little or no change in the association of H2B with sequences b and d (Fig. 4E). However, the level of H2B present in sequence f was only about half of that in e. These results reinforced the notion that chromatin is disrupted at or near CCGNN repeats.
Poly(dA-dT) can also block the spread of silent chromatin.
Another type of DNA sequence that forms a special structure and does not favor nucleosome formation is poly(dA-dT), or T tract. Sufficiently long poly(dA-dT) adopts a straight and rigid structure with an unusually short helical repeat (10 versus 10.5 bp of typical B-DNA) and a distinctive narrow minor groove (references 2 and 37 and references therein). As a result, poly(dA-dT) does not favor the assembly or stability of nucleosomes in vitro, and long T tracts cannot be wrapped into nucleosomes (33, 43). Suter et al. showed that T tracts maintain their unusual structure in vivo, indicating that they are not folded into nucleosomes (46). Interestingly, a (dA-dT)16 sequence located within the promoter of the AMT1 gene in the yeast Candida glabrata was shown to distort, but not completely disrupt, a nucleosome, thereby providing access for an adjacent sequence in the same nucleosome to Amt1p (55). This scheme is strikingly similar to the collaborative binding of LexA and a second factor to nucleosomal DNA discussed above. In fact, in a modified HIS3 promoter, either a poly(dA-dT) sequence (25) or a LexA-binding site (in the presence of LexA) (34) can help Gcn4p bind to a nearby site to ensure gene activation. We predict that long poly(dA-dT), like LexA, can also function as a barrier to the spread of silent chromatin. To test this, we inserted (dT-dA)20, (dT-dA)45, (dT-dA)80, and (dA-dT)80 [designated (T)20, (T)45, (T)80, and (A)80] between HML and URA3 in strain H0 to make H15 to H18, respectively (Fig. 5A, left). Whereas (T)20 had little or no effect on URA3 silencing (Fig. 5A, H15), (T)45 caused a detectable decrease in silencing (H16). (T)80 or (A)80 totally eliminated URA3 silencing (Fig. 5A, H17 and H18). Consistently, we also showed that the presence of (T)80 increased the level of URA3 mRNA by 8.5-fold in strain H17 (supplementary Fig. 1). On the other hand, (T)80 inserted downstream of URA3 did not affect silencing (Fig. 5A, H19). (T)80 and (A)80 can also abolish telomeric silencing (Fig. 5B, T13 and T14). Using ChIP, we showed that (T)80 in H17 blocked the spread of Sir3p (Fig. 5C; α-Sir3p, compare 1 to 5 from H17 to those from H0). Therefore, just like LexA and (CCGNN)n, poly(dA-dT) can block the spread of silent chromatin.
FIG. 5.
Poly(dA-dT) can counteract the spread of silent chromatin and alter chromatin structure. (A) Poly(dA-dT) counteracts HML silencing in a datin-independent manner. Left, strains H0 and H15 to H22. Right, growth phenotypes of each strain. (B) Poly(dA-dT) can overcome telomeric silencing. Growth phenotypes of T1, T13, and T14 are shown. (C) Poly(dA-dT) blocks the spread of silent chromatin. ChIP was performed on H0 and H17 using α-Sir3p. (D) Effect of poly(dA-dT) on chromatin structure. The results of indirect end labeling on H0 and H17 are shown. MNase-treated DNA was digested with AvaII. A, AvaII; N, NdeI; S, SpeI; X, XmnI. Lane N, naked DNA.
In S. cerevisiae, datin encoded by DAT1 binds poly(dA-dT) longer than 10 bp (53). However, we showed that deleting DAT1 had no effect on URA3 silencing (Fig. 5A, compare H20 to H0) or the antisilencing activity of poly(dA-dT) (compare H21 and H22 to H17 and H18). Deleting DAT1 also had no effect on the ability of poly(dA-dT) to counteract telomeric silencing (data not shown). If the antisilencing effect of (T)80 were due to its special rigid structure, the above results would be in accord with the fact that the unusual T-tract structure is datin independent (46).
Correlated with its robust barrier function, (T)80 led to remarkable changes in chromatin structure (Fig. 5D). It induced strong MNase cleavage at more than one site immediately downstream of it (Fig. 5D, H17, region a), similar to a previous observation of (T)42 integrated at the HIS3 locus (25). Interestingly, similar to (CCGNN)n, (T)80 per se appeared less sensitive to MNase cleavage in both naked DNA and chromatin (Fig. 5D, compare regions b in lanes 5 and 6), consistent with the fact that the unusual structure of poly(dA-dT) persists in vivo (46). These results indicate that chromatin is disrupted, at least in the immediate vicinity of poly(T). Therefore, the barrier function of poly(dA-dT) is also linked to its ability to disrupt the chromatin structure.
The barrier function of TEF2-UAS is correlated with chromatin disruption.
It has been shown that TEF2-UAS consisting of three tandem Rap1p-binding sites could block the spread of silencing (6) (Fig. 6A, compare H23 to H0). As shown in supplementary Fig. 1, the presence of TEF2-UAS increased the level of URA3 mRNA by ninefold in strain H23. We demonstrated here that TEF2-UAS could also block telomeric silencing (Fig. 6A, compare T15 to T1). Using ChIP assays, we showed that TEF2-UAS blocked the spread of Sir3p in H23 (Fig. 6B). The three Rap1p sites are necessary and sufficient for the barrier activity of TEF2-UAS (6). We obtained evidence that binding of Rap1p to TEF2-UAS is required for the barrier function (54). Rap1p is a multifunctional protein that can act as a global positive and negative regulator of transcription and telomere maintenance (42). Interestingly, in certain yeast promoters, Rap1p, similarly to LexA and poly(dA-dT), can exclude nucleosomes, thereby facilitating binding by an activator such as Gcn4p, to a nearby site (36). It has been shown that TEF2-UAS induced the loss of one to two negative superhelical turns in local DNA, which could be explained by the elimination of one or two nucleosomes (54). Here, we directly examined the chromatin structure around TEF2-UAS in H23. As shown in Fig. 6C, there were three salient MNase-hypersensitive sites flanking TEF2-UAS (Fig. 6C, H23 lanes, bands a to c), indicative of disrupted chromatin or nucleosome exclusion. We think the barrier activity of TEF2-UAS is also linked to its ability to disrupt chromatin.
DISCUSSION
Division of the genome into distinct chromatin domains is critical to the programming of gene expression in a eukaryotic organism. Transcriptionally silent chromatin is more compact, and its formation involves the sequential spread of repressor complexes along positioned nucleosomes (35). The boundaries of silent chromatin often coincide with special nucleoprotein structures that function as barriers to its propagation. The barrier function was first ascribed to certain insulators in Drosophila and vertebrates that border specific chromosomal domains (13, 29). Recently, barriers able to block the propagation of silent chromatin have also been identified in yeast (52). Two distinct models have been proposed for their functions. One invokes a barrier that recruits chromatin-modifying or -remodeling proteins that actively prevent the silencing machinery from deacetylating and condensing active chromatin into silent chromatin (15). The other proposes that a barrier functions as a passive physical block to encroaching silent chromatin (6, 24). We have demonstrated in this report that the spread of silent chromatin can be stopped by sequences associated with disrupted chromatin characterized by nuclease hypersensitivity and reduced histone abundance.
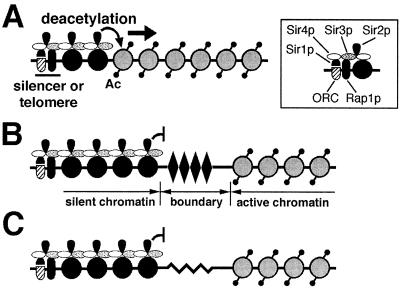
We showed that LexA bound in the path of propagation of silent chromatin acts as an efficient barrier in yeast. Data from this and other studies demonstrate that LexA is not a transcriptional activator in yeast. Thus, the barrier function of LexA is not due to direct gene activation; it requires multiple (>3) binding sites, as if a critical mass has to be reached. There is evidence that LexA binds nucleosomal DNA and destabilizes the nucleosome (34, 48). In light of this, multiple LexA molecules bound to closely positioned sites, as is the case in a group of strains used in this work, may severely destabilize or even exclude a nucleosome. Chromatin analysis by MNase digestion followed by indirect end labeling showed that LexA produced prominent MNase-hypersensitive sites immediately abutting its binding sites (Fig. 3A). Interestingly, the sequence consisting of LexA-binding sites per se appears less sensitive to MNase digestion. It is possible that LexA binding makes this region less accessible to MNase. Given that LexA binding can destabilize nucleosomes, it is likely that this region, despite being relatively resistant to MNase, is not associated with stable nucleosomes in the presence of LexA. The results of our mononucleosome analysis indicate that LexA excludes positioned nucleosomes (Fig. 3B). We also used ChIP assays to show that LexA induces a twofold reduction in histone H2B association with a sequence spanning LexA sites (Fig. 3C). Taken together, these data strongly suggest that LexA disrupts chromatin structure at and/or near its binding sites. We think that this disruption, which can be either complete removal of nucleosomes or abrogation of their stable positioning, underlies the boundary function of LexA. This is based on the proposed stepwise propagation of silent chromatin in yeast, in which an incoming SIR complex binds to the adjacent nucleosome that has been deacetylated by Sir2p (Fig. 7A) (40, 41). A region that is associated with unstable nucleosomes or is free of nucleosomes would disrupt the regularity of nucleosomes and thus not favor SIR complex binding and halt of the spread of silent chromatin (Fig. 7B).
FIG. 7.
Nucleosome-excluding sequences as barriers to the spread of silent chromatin. (A) Propagation of silent chromatin in S. cerevisiae. Silencer-binding proteins and Sir1p recruit the SIR complex. Sir2p deacetylates a nearby nucleosome (curved arrow). The hypoacetylated nucleosome (solid circle) then binds an incoming SIR complex. Repetition of this process leads to the spread of silent chromatin (indicated by the solid arrow). Shaded circles, acetylated nucleosomes; Ac, acetyl group. The inset illustrates interactions among silencer-binding proteins, SIR proteins, and the nucleosome. (B) DNA-binding proteins can form barriers to the spread of silent chromatin by excluding nucleosomes. DNA-binding proteins, such as LexA or Rap1p (solid diamond), bind to (multiple) sites in the path of the spread of silent chromatin and destabilize or remove one or more nucleosomes, thereby stalling the propagating SIR complex (curved line with bar). (C) Nucleosome-excluding sequences can form barriers to the spread of silent chromatin. Atypical DNA structures (zigzag line) formed by sequences such as (CCGNN)n or poly(dA-dT), which do not favor nucleosome formation, stop the spread of the SIR complex.
The nucleosome exclusion model implies that any sequence or structure that excludes nucleosomes has the potential to function as a barrier element. This is supported by the fact that (CCGNN)n and poly(dA-dT), both known not to favor nucleosome formation, can also block the spread of silent chromatin (Fig. 4 and 5). Interestingly, all three documented yeast barrier elements, TEF2-UAS, HMR-tRNAThr, and STARs, also have the potential to exclude nucleosomes (7, 36). We showed here that TEF2-UAS coincided with MNase-hypersensitive sites, indicative of nucleosome exclusion, while functioning as a barrier (Fig. 6). In addition, we also found that the naturally occurring nucleosome-free sequences from the ILV1, GCY1, AKY2, and PFY1 loci (reference 3 and references therein) can all counteract silencing (Y. Zou and X. Bi, unpublished results). We think, but have not tested, that other nucleosome-excluding sequences or structures, including Z-DNA and cruciform DNA, can also act as barriers to silent chromatin.
Many well-studied insulators in higher organisms are associated with nuclease-hypersensitive sites. The Drosophila scs or scs′ insulator consists of a 250- to 350-bp nuclease-resistant core flanked on both sides by hypersensitive sites (47). The gypsy insulator from Drosophila bearing 12 Su(Hw) binding sites contains several DNase I-hypersensitive sites (11). Nuclease-hypersensitive sites are also associated with other known Drosophila insulator elements (14). In addition, DNase I-hypersensitive sites exist in the 5′ HS4 insulator at the chicken β-globin locus (13). Therefore, most, if not all, known insulators have features indicative of open, or nucleosome-free, chromatin. Given the similarities between the mechanisms of the spread of silent chromatin in yeast and higher organisms, it will be interesting to investigate whether nucleosome exclusion also plays a role in the function of insulators as barriers to the spread of heterochromatin in higher cells.
Acknowledgments
We thank Danesh Moazed for the gift of α-Sir3p antibody, David Allis for α-H3-Ac and α-H4-Ac, and Zu-Wen Sun, David Allis, Kevin Struhl, Virginia Zakian, Roger Brent, and Philip James for gifts of plasmids and strains. We thank Hengping Xu, Ya-Hui Chiu, Travis Foland, Dan Schellhorn, and John Bishay for assistance. We also thank the reviewers for suggestions on improving this work.
This work was supported by National Institutes of Health grant GM 62484 to X.B.
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2001. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104:839-847. [DOI] [PubMed] [Google Scholar]
- 2.Alexeev, D. G., A. A. Lipanov, and I. Y. Skuratovskii. 1987. Poly(dA) · poly(dT) is a B-type double helix with a distinctively narrow minor groove. Nature 325:821-823. [DOI] [PubMed] [Google Scholar]
- 3.Angermayr, M., U. Oechsner, and W. Bandlow. 2003. Reb1p-dependent DNA bending affects nucleosome positioning and constitutive transcription at the yeast profilin promoter. J. Biol. Chem. 278:17918-17926. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio, O. M., and D. E. Gottschling. 1994. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 8:1133-1146. [DOI] [PubMed] [Google Scholar]
- 5.Bi, X., M. Braunstein, G. J. Shei, and J. R. Broach. 1999. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc. Natl. Acad. Sci. USA 96:11934-11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi, X., and J. R. Broach. 1999. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 13:1089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi, X., and J. R. Broach. 2001. Chromosomal boundaries in S. cerevisiae. Curr. Opin. Genet. Dev. 11:199-204. [DOI] [PubMed] [Google Scholar]
- 8.Brent, R., and M. Ptashne. 1984. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature 312:612-615. [DOI] [PubMed] [Google Scholar]
- 9.Brent, R., and M. Ptashne. 1985. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43:729-736. [DOI] [PubMed] [Google Scholar]
- 10.Carmen, A. A., L. Milne, and M. Grunstein. 2001. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277:4778-4781. [DOI] [PubMed] [Google Scholar]
- 11.Chen, S., and V. G. Corces. 2001. The gypsy insulator of Drosophila affects chromatin structure in a directional manner. Genetics 159:1649-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu, Y. H., Q. Yu, J. J. Sandmeier, and X. Bi. 2003. A targeted histone acetyltransferase can create a sizable region of hyperacetylated chromatin and counteract the propagation of transcriptionally silent chromatin. Genetics 165:115-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung, J. H., M. Whiteley, and G. Felsenfeld. 1993. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74:505-514. [DOI] [PubMed] [Google Scholar]
- 14.Corces, V. G., and G. Felsenfeld. 2000. Chromatin boundaries, p. 278-299. In S. C. R. Elgin, and J. L. Workman (ed.), Chromatin structure and gene expression, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 15.Donze, D., and R. T. Kamakaka. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomycese cerevisiae. EMBO J. 20:520-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donze, D., and R. T. Kamakaka. 2002. Breaking the silence: how heterochromatic gene repression is stopped in its tracks. BioEssays 24:344-349. [DOI] [PubMed] [Google Scholar]
- 17.Ebina, Y., Y. Takahara, F. Kishi, A. Nakazawa, and R. Brent. 1983. LexA protein is a repressor of the colicin E1 gene. J. Biol. Chem. 258:13258-13261. [PubMed] [Google Scholar]
- 18.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 19.Gregory, P. D., and W. Hörz. 1999. Mapping chromatin structure in yeast. Methods Enzymol. 304:365-376. [DOI] [PubMed] [Google Scholar]
- 20.Grewal, S. I., and S. C. Elgin. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12:178-187. [DOI] [PubMed] [Google Scholar]
- 21.Herskowitz, I. 1988. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52:536-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie, S. P. Gygi, and D. Moazed. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 24.Ishii, K., G. Arib, C. Lin, G. Van Houwe, and U. K. Laemmli. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109:551-562. [DOI] [PubMed] [Google Scholar]
- 25.Iyer, V., and K. Struhl. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14:2570-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 28.Keegan, L., G. Gill, and M. Ptashne. 1986. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231:699-704. [DOI] [PubMed] [Google Scholar]
- 29.Kellum, R., and P. Schedl. 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64:941-950. [DOI] [PubMed] [Google Scholar]
- 30.Kent, N. A., L. E. Bird, and J. Mellor. 1993. Chromatin analysis in yeast using NP-40 permeabilised sphaeroplasts. Nucleic Acids Res. 21:4653-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, B., and J. W. Little. 1992. Dimerization of a specific DNA-binding protein on the DNA. Science 255:203-206. [DOI] [PubMed] [Google Scholar]
- 32.Kirkpatrick, D. T., Y. H. Wang, M. Dominska, J. D. Griffith, and T. D. Petes. 1999. Control of meiotic recombination and gene expression in yeast by a simple repetitive DNA sequence that excludes nucleosomes. Mol. Cell. Biol. 19:7661-7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkel, G. R., and H. G. Martinson. 1981. Nucleosomes will not form on double-stranded RNA or over poly(dA) · poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 9:6869-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. A., and J. Widom. 2003. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 23:1623-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 36.Morse, R. H. 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16:51-53. [DOI] [PubMed] [Google Scholar]
- 37.Nelson, H. C., J. T. Finch, B. F. Luisi, and A. Klug. 1987. The structure of an oligo(dA) · oligo(dT) tract and its biological implications. Nature 330:221-226. [DOI] [PubMed] [Google Scholar]
- 38.Richmond, T. J., and J. Widom. 2000. Nucleosome and chromatin structure, p. 1-23. In S. C. R. Elgin, and J. L. Workman (ed.), Chromatin structure and gene expression, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 39.Rose, M., and F. Winston. 1984. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol. Gen. Genet. 193:557-560. [DOI] [PubMed] [Google Scholar]
- 40.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell. 13:2207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [Online.] [DOI] [PubMed] [Google Scholar]
- 42.Shore, D. 1994. RAP1: a protean regulator in yeast. Trends Genet. 10:408-412. [DOI] [PubMed] [Google Scholar]
- 43.Simpson, R. T., and P. Kunzler. 1979. Chromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 6:1387-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 45.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-1088. [Online.] [DOI] [PubMed] [Google Scholar]
- 46.Suter, B., G. Schnappauf, and F. Thoma. 2000. Poly(dA · dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Res. 28:4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Udvardy, A., E. Maine, and P. Schedl. 1985. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 185:341-358. [DOI] [PubMed] [Google Scholar]
- 48.Vashee, S., K. Melcher, W. V. Ding, S. A. Johnston, and T. Kodadek. 1998. Evidence for two modes of cooperative DNA binding in vivo that do not involve direct protein-protein interactions. Curr. Biol. 8:452-458. [DOI] [PubMed] [Google Scholar]
- 49.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Y. H., and J. D. Griffith. 1996. The [(G/C)3NN]n motif: a common DNA repeat that excludes nucleosomes. Proc. Natl. Acad. Sci. USA 93:8863-8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss, K., and R. T. Simpson. 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol. Cell. Biol. 18:5392-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 53.Winter, E., and A. Varshavsky. 1989. A DNA binding protein that recognizes oligo(dA) · oligo(dT) tracts. EMBO J. 8:1867-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, Q., R. Qiu, T. B. Foland, D. Griesen, C. S. Galloway, Y. H. Chiu, J. Sandmeier, J. R. Broach, and X. Bi. 2003. Rap1p and other transcriptional regulators can function in defining distinct domains of gene expression. Nucleic Acids Res. 31:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, Z., and D. J. Thiele. 1996. A specialized nucleosome modulates transcription factor access to a C. glabrata metal responsive promoter. Cell 87:459-470. [DOI] [PubMed] [Google Scholar]