Abstract
The recently identified plant photoreceptor UVR8 triggers regulatory changes in gene expression in response to ultraviolet-B (UV-B) light via an unknown mechanism. Here, crystallographic and solution structures of the UVR8 homodimer, together with mutagenesis and far-UV circular dichroism spectroscopy, reveal its mechanisms for UV-B perception and signal transduction. β-propeller subunits form a remarkable, tryptophan-dominated, dimer interface stitched together by a complex salt-bridge network. Salt-bridging arginines flank the excitonically coupled cross-dimer tryptophan “pyramid” responsible for UV-B sensing. Photoreception reversibly disrupts salt bridges, triggering dimer dissociation and signal initiation. Mutation of a single tryptophan to phenylalanine re-tunes the photoreceptor to detect UV-C wavelengths. Our analyses establish how UVR8 functions as a photoreceptor without a prosthetic chromophore to promote plant development and survival in sunlight.
UVR8 (UV RESISTANCE LOCUS 8) orchestrates the expression of over 100 genes in Arabidopsis in response to UV-B wavelengths (280–315 nm) (1–4). The uvr8 mutant exhibits UV-B sensitivity from decreased expression of genes conferring UV-protection (1, 5). UV-B exposure promotes both rapid UVR8 accumulation in the nucleus (6), where the protein binds chromatin via histones (1, 7), and interaction with COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC 1) to initiate transcriptional responses (3, 8). In plant extracts and in heterologous systems, UV-B exposure triggers UVR8 dimer dissociation to initiate signaling (9). Tryptophan has been implicated in UV-B perception (4, 9), but the absence of detailed three-dimensional information on dimer assembly precludes understanding of the mechanisms for UVR8 photoreception and signaling.
To investigate UVR8 structure/function relationships, we made recombinant Arabidopsis UVR8 (10) for biophysical analyses (fig. S1, S2). Purified UVR8 (fig. S2A) is a homodimer that dissociates into monomers following exposure to narrowband, long wavelength UV-B (fig. S2B); the dose-response relationship (Fig. S2C) mirrors UVR8 behavior in plant extracts (9). Moreover, UV-B-induced monomerization is reversible; the active, dimeric photoreceptor spontaneously reassembles within hours in vitro, and again responds to UV-B (Fig. 1A). UVR8 absorbs strongly at 280 nm (fig. S3), as expected from its complement of aromatic residues (14 Trp, 10 Tyr and 8 Phe per 440-residue monomer). Photoactive, purified UVR8 lacks any bound cofactor, demonstrating that reversible UV-B-induced dimer dissociation is a property intrinsic to the protein.
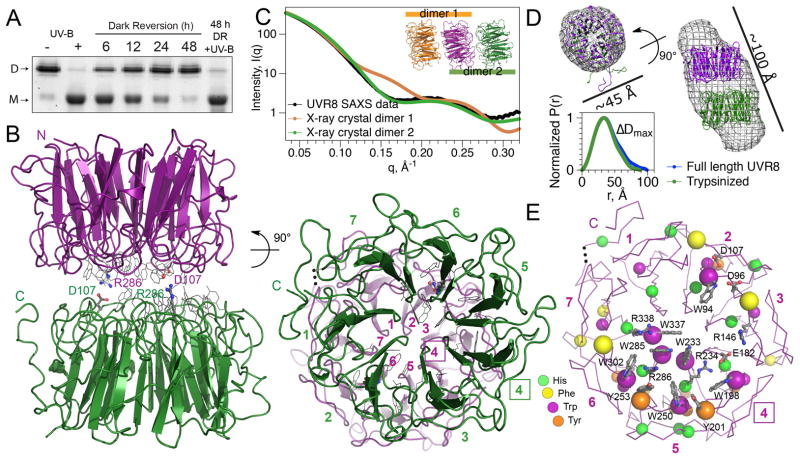
Figure 1.
Structure of Arabidopsis UVR8 dimer. (A) UV-B-induced dimer dissociation spontaneously reverses in the dark, regenerating photoactive dimers; analyzed by SDS-PAGE without sample boiling. (B) UVR8 forms a symmetric homo-dimer of 7-bladed β-propeller subunits (side-and end-views). Key salt bridges are shown as ball-and-stick. End-view is numbered to show blade pairing centered at blade 4 (box). (C) Experimental SAXS profile of UVR8 (crystallographic construct) compared with computed profiles for crystallographic dimers. (D) Crystallographic dimer docked into ab initio SAXS model of full-length UVR8 dimer (top) and pair-distance-distribution functions [P(r)] for full-length and trypsin-treated UVR8 (bottom), defining maximal diameter difference (ΔDmax). (E) Asymmetric localization of aromatic residues identifies center of photoreception. Key side chains (grey) are labeled.
The X-ray crystallographic structure of UVR8 (Fig. 1) was determined to 1.7 Å resolution (Table S1) by molecular replacement with the RCC1 (Regulator of Chromosome Condensation 1) domain of E3 ligase HERC2 as the probe (10). UVR8 has a 7-bladed β-propeller fold, like monomeric RCC1 and HERC2, but unexpectedly the topology is permuted (fig. S1, S4). In HERC2 and RCC1, the N- and C-terminal sequences are linked because each contributes two of the four β-strands to blade 1. In UVR8, however, each blade is contiguous in sequence with the N- and C-termini in the first and last blades, respectively (fig. S1, S4), potentially permitting greater conformational flexibility.
Each doughnut-shaped UVR8 monomer is 40–50 Å in diameter, by ~35 Å high with a central water-filled tunnel (Fig. 1B). Crystal packing of the trypsin-treated protein, which remains UV-B-responsive (fig. S5), suggests two potential cylindrical dimer assemblies (Fig. 1C, inset). Small angle X-ray scattering (SAXS) (Table S2) clearly distinguishes the correct dimer in solution (Fig. 1C), by agreement between calculated and observed scattering profiles (11–13). The smaller, concave surfaces of the two subunits assemble face-to-face, but offset by about 10 Å (Fig. 1B,D). The two-fold dimer symmetry juxtaposes different blades from each subunit, except self-pairing blade 4 (Fig. 1B). SAXS results (Fig. 1D) show different lengths, but the same overall shape for full-length UVR8 and the trypsin-cleaved protein used for crystallography. Molecular envelopes derived from SAXS data on full-length UVR8 match the crystallographic dimer offset and diameter, and locate the C-terminal missing ~10% at distal ends of the dimer (Fig. 1D).
The dimer interface is remarkable for a preponderance of aromatic residues (7 Trp, 3 Phe, and 2 Tyr; Fig. 1E) and charged side chains, which contribute to distinct regions of complementary electrostatic potential (Fig. 2A). The dimer offset and 2-fold symmetry align rows of arginine and carboxylate side chains (Fig. 2B, fig. S6) to form a complex network of salt bridges across the dimer interface (fig. S7). In particular, doubly hydrogen-bonded salt bridges link R286 with D107 and R146 with E182 (Fig. 2C). Additional, singly hydrogen-bonded salt bridges join R286 with D96, R338 with D44, and R354 with E43 and E53. Interestingly, the dimer interface is composed of charged, hydrophilic and aromatic residues only (except for hydrophobic A52), leading the PISA software (14) to assess dimer assembly as unstable. Yet, the solution scattering profile (Fig. 1C), size exclusion chromatography (SEC) (Table S3), multi-angle light scattering (MALS) (fig. S8A) and SDS-polyacrylamide (SDS-PAGE) gel analyses (9) (fig. S2B) demonstrate that UVR8 is a dimer.
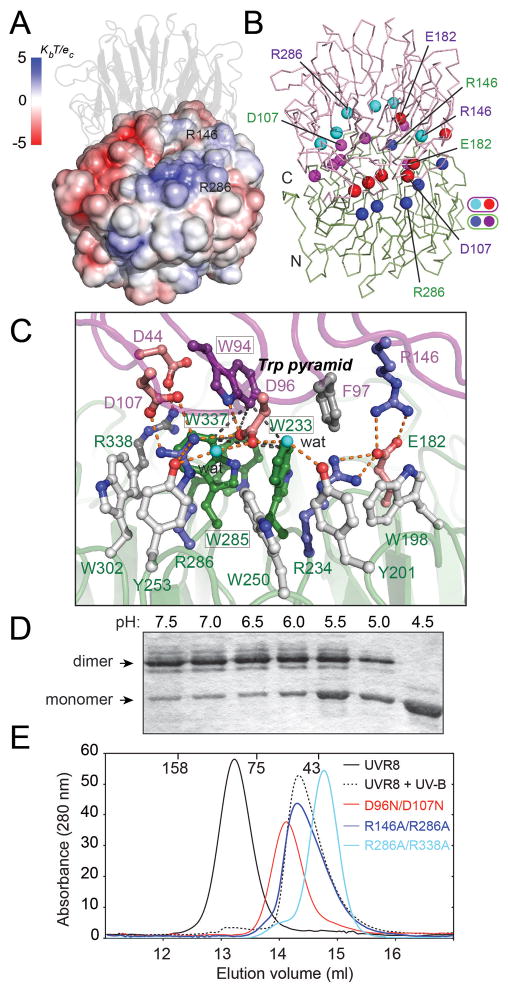
Figure 2.
Ionic interactions are key to maintaining the UVR8 dimer. (A) Electrostatic potential surface of UVR8 reveals charge complementarity at the dimer interface, (B) Arg (blue/cyan) and Glu/Asp (red/magenta) balls indicate key charged residues mediating cross-dimer salt bridges aligned across the interface. (C) Close up of key salt bridges: Arg 286 with Asp 96 and Asp 107; Arg 146 with Glu 182; Arg 338 with Asp 44, with hydrogen bonds shown as orange dots. The Trp pyramid (black dashed lines) is formed by W94 (purple) atop the Trp triad (green). (D) Acidification promotes monomerization of wild-type UVR8, as analyzed by SDS-PAGE without sample boiling (E) Size exclusion chromatography shows UVR8R146A/R286A, UVR8R286A/R338A and UVR8D96N/D107N mutants are constitutive monomers.
To test the importance of ionic interactions in maintaining the UVR8 dimer, we showed that decreasing pH in vitro promotes monomerization (Fig. 2D), and examined the effects of mutating residues that participate in cross-dimer salt bridges. The UVR8R146A and UVR8R286A proteins, each lacking one dominant salt bridge, are dimers that undergo UV-B-induced monomerization, when assayed by SEC (Table S3). However, unlike the wild-type protein, these mutants appear monomeric by SDS-PAGE without sample boiling (fig. S2D), indicating that the dimer has been destabilized. Double mutants lacking both dominant salt bridges (UVR8R146A/R286A), the salt bridges of R286 with D107 and D96 (UVR8D96N/D107N), or both the R286 and R338 salt bridges (UVR8R286A/R338A) (Fig. 2C), are constitutive monomers (Fig. 2E, fig. S8), demonstrating a key role for the R286 salt bridges in dimer formation.
In each monomer, a striking cluster of nine aromatic residues dominates the interaction surface (Fig. 1E). A conserved Gly-Trp-Arg-His-Thr sequence repeat in blades 5, 6, and 7 (fig. S1) generates a ‘triad’ of closely packed tryptophans, W233, W285 and W337, that are implicated in UVR8 photoreception (9). These pentapeptide repeats form protruding tight turns that project Trp and Arg residues outwards, and His residues (in all 7 blades) inwards to form a buried ring (Fig. 1E). Each triad Trp is flanked by the adjacent Arg guanidinium moiety, but the side-chain orientation and packing differ among the three repeats (Fig. 3A). R234 and R338 are positioned as “book ends” flanking W285 and W233. Three more pairs of aromatic residues from blades 4, 5 and 6 (W198, Y201; W250, Y253; and W302, F305 create a perimeter fence of aromatic residues that isolates the Trp triad from solvent. At the center of the aromatic cluster, W285 and R286 are π-stacked between triad W337 and perimeter Y253 (Fig. 3A). W94 of the opposing monomer forms the apex of a Trp pyramid with the Trp triad as the base (Figs. 3A, fig. S6). The close packing (less than 4.5 Å apart) of the Trp pyramid allows orbital overlap, permitting exciton coupling (15–17), which we assessed experimentally with far-UV circular dichroism spectroscopy (CD) (Fig. 3B–G)
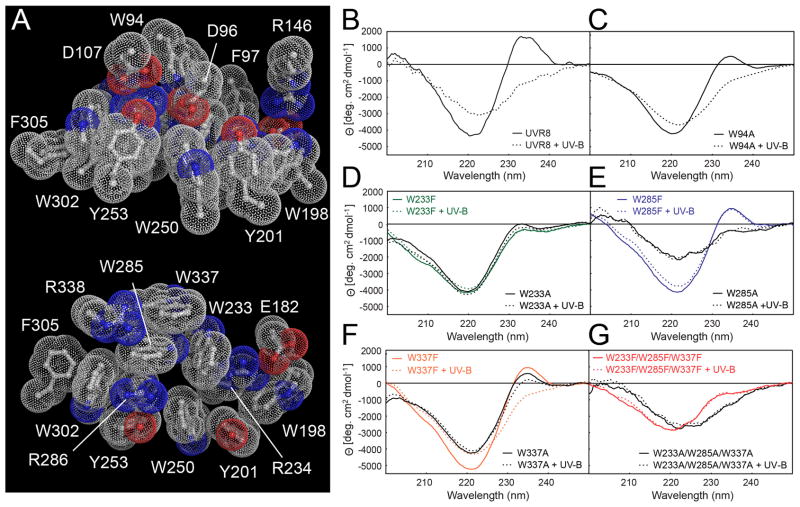
Figure 3.
Specific tryptophans mediate UV-B photoreception by UVR8. (A) Bulky aromatic residues tightly pack with key charged residues across the dimer (top) and within a monomer (bottom). (B–G) Far-UV CD spectrum change of UVR8 by UV-B. (B) Wild-type UVR8; (C) UVR8W94A; (D) UVR8W233F and UVR8W233A; (E) UVR8W285F and UVR8W285A; (F) UVR8W337F and UVR8W337A; (G) triple mutants UVR8W233F/W285F/W337F and UVR8W233A/W285A/W337A.
UV-B exposure of UVR8 strongly diminishes the large far-UV CD peak at 234 nm and trough at 221 nm that are characteristic (15) of exciton coupling between tryptophans (Fig. 3B). We mutated the pyramid Trps to investigate their contributions to exciton coupling and thus potentially to photoperception. The far-UV CD peak height is reduced in each of the single conservatively substituted W>F mutants (Fig. 3D–F), most strongly in UVR8W233F (Fig. 3D). Among four individual W>A side-chain truncation mutants (Fig. 3C–F), UVR8W285A revealed the most significant loss of exciton coupling, with its far-UV CD spectrum resembling those of triple mutant UVR8W233A/W285A/W337A (Fig. 3G) and UV-B-treated wild-type (Fig. 3B). UV-B has no effect on the altered CD signal of either the single UVR8W285A (Fig. 3E) or the triple W>A (Fig. 3G) mutants, indicating a complete loss of photoreception. Similarly, UV-B has no effect on the CD signal of the UVR8W233F and UVR8W285F mutants, whereas it substantially reduces the CD signals of UVR8W337F and UVR8W94A, although to a lesser extent than in the wild type protein (Fig. 3B, 3D–F). Consistent with these results, SEC analysis shows that UVR8W337F and UVR8W94A monomerize in response to UV-B whereas UVR8W233F and UVR8W285F are constitutive dimers (Table S3). Together, these observations indicate that the Trp pyramid is key to UVR8 photoperception, with W285 as the principal UV-B sensor. W233 is also important, not only in photoreception but particularly in maintaining exciton coupling, whereas W337 and W94 play auxilliary roles.
To examine the in vivo role of W285 in UV-B perception, we expressed GFP-UVR8W285A in mutant uvr8-1 plants and assayed UV-B induction of HY5 transcripts, mediated by UVR8 (1). GFP-UVR8W285A does not restore UV-B-mediated induction of HY5 to uvr8-1 mutants (Fig. 4A). However, loss of activity in the W285A mutant does not result from gross structural changes; SAXS analysis of UVR8W285A confirmed that the dimer protein has dimensions similar to those of the wild type (Fig. 4B). Thus, these experiments show the in vivo functional importance of W285 in UV-B photoreception. Remarkably, although UVR8W285F is unable to respond to UV-B, it does respond to UV-C, consistent with the shorter wavelength absorption of phenylalanine compared to tryptophan. Thus, UV-C exposure reduces the 234 and 221 nm CD features of UVR8W285F (Fig. 4C) and initiates monomerization (Fig. 4D). Although UV-C is clearly less efficient in initiating photoreception in UVR8W285F than UV-B in wild-type UVR8, these observations support the hypothesis that W285 has a key role in UVR8 photoreception. Furthermore, they demonstrate that the spectral sensing properties of the photoreceptor can be re-tuned by a single amino acid change, definitively establishing that UVR8 uses a tryptophan chromophore.
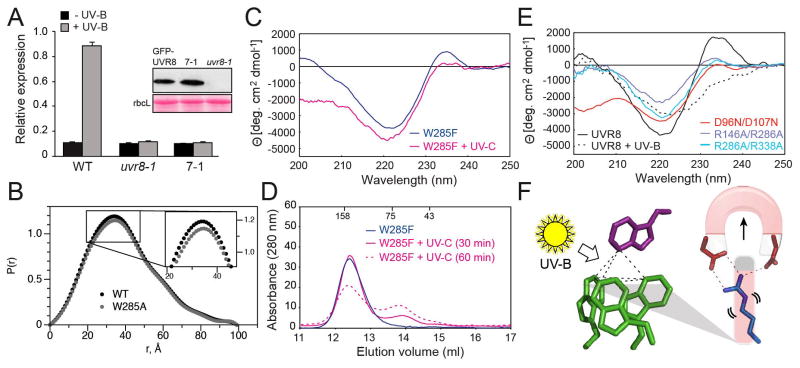
Figure 4.
Key features of UVR8 photoreceptor mechanism (A) qRT-PCR analysis of UV-B induction of HY5 transcripts in wild-type, uvr8-1, and uvr8-1 expressing GFP-UVR8W285A (line 7-1). Inset shows Western blot of GFP-UVR8 in plant lines and Ponceau S staining of rbcL protein as loading control. (B) SAXS Pair-distance-distribution functions [P(r)] show that UVR8W285A is dimeric, with only subtle conformational differences from wild-type. (C) Far-UV CD spectra of UVR8W285F before and after exposure to 40 min UV-C. (D) Size exclusion chromatography of UVR8W285F mutant protein exposed to UV-C (solid and dotted magenta lines). (E) Far-UV CD spectra of UVR8R146A/R286A, UVR8R286A/R338A and UVR8D96N/D107N mutants. (F) Model for UV-B photoreception by UVR8. UV-B sensing (by Trp “pyramid”) triggers dimer dissociation by disrupting salt bridges.
Salt-bridge mutations also influence exciton coupling as measured by far-UV CD. The constitutively monomeric, double salt-bridge mutants (Table S3, Fig. 2E) show a reduction in exciton coupling (Fig. 4E) consistent with the tight packing of arginine side chains with tryptophans within the aromatic cluster, and the role of salt bridges in maintaining the cross-dimer Trp pyramid. The impact of these salt-bridge mutations on the exciton coupling is greater than that observed for the W94A (Fig. 3C) or W337A (Fig. 3F) mutants, which remove one tryptophan from the Trp pyramid. Indeed, the CD spectrum for UVR8R146A/R286A resembles that for UVR8W285A, suggesting the importance of the π-stacking of these two residues in packing the Trp pyramid for exciton coupling.
Together our findings indicate that UV-B photoreception by the excitonically coupled Trp pyramid leads to disruption of cross-dimer salt bridges, promoting UVR8 monomerization (Fig. 4F). Notably, the proximity and coupling of arginines and tryptophans suggests a specific mechanism whereby photoreception leads to monomerization. In particular, the key chromophore W285 stacks with adjacent R286, which has a crucial role in dimerization. This closely packed W285-R286 pair is therefore positioned to link UV-B photoreception and salt-bridge status (Fig. 2C). We propose that photoreception by the Trp pyramid, predominantly W285 and W233, results in the effective transfer of an excited electron from the excitonically coupled Trp pyramid to adjacent arginine(s), leading to charge neutralization, consequent breakage of cross-dimer salt bridges and thus dimer destabilization and dissociation (Fig. 4F). The complex packing assembly of the conserved aromatic cluster surrounding the Trp pyramid, and the interconnectivity of the conserved salt bridges that zip together the dimer interface, suggest that UVR8 has evolved a robust, concerted mechanism for UV-B perception and signaling.
In conclusion, we show that UVR8 is distinct from other known photoreceptors in exploiting the UV-B-absorbance of its intrinsic tryptophans, rather than a bound chromophore. The presence of putative UVR8 orthologues in algae and mosses suggests that the photoreceptor may have evolved to promote plant survival when the earth’s early atmosphere allowed high levels of UV-B exposure. β-propeller proteins interact with partners, often at the surface that UVR8 uses for dimerization (18, 19). Hence dimer dissociation is expected to facilitate interactions of UVR8 with proteins involved in downstream signaling or photoreceptor regulation.
Supplementary Material
Summary.
UV-B photoreception by an excitonically-coupled tryptophan pyramid in the photoreceptor UVR8 disrupts cross-dimer salt-bridges leading to monomerization and signaling to promote plant development and survival in sunlight.
Acknowledgments
We thank C. Cloix, E. Kaiserli, N. Fraser, C. Hitomi, R. Rambo, J. Holton, J.A. Tainer and Structurally Integrated BiologY for Life Sciences (SIBYLS) beamline staff for discussions and assistance. This research was supported by: National Institutes of Health grant GM37684 (E.D.G.); The Skaggs Institute for Chemical Biology (K.H., A.J.P.); National Science Foundation Predoctoral Fellowship (A.J.P.); Royal Society University Research Fellowship (J.M.C.); Leverhulme Trust grant F/00179/AZ (G.I.J., J.M.C., B.O.S.); UK Biotechnology and Biological Sciences Research Council PhD studentship (A.O.). J.M.C., E.D.G., K.H. and G.I.J. designed research; A.S.A., K.B., J.M.C., K.H., M.H., S.M.K, A.O. and A.J.P. performed research; M. Hothorn provided reagents; A.A., K.B., J.M.C., E.D.G., K.H., M.H., G.I.J., S.M.K., A.O., A.J.P. and B.O.S. analyzed data; and J.M.C., E.D.G., K.H., G.I.J. and A.J.P. prepared the manuscript. Crystallography and SAXS data were collected at SIBYLS through the Integrated Diffraction Analysis Technologies program supported by the Department of Energy, Office of Biological and Environmental Research. Crystallographic coordinates and structure factors are deposited in the Protein Data Bank with accession code 4D9S.
Footnotes
Materials and Methods
References (20–38) [Note: The numbers refer to any additional references cited only within the SOM]
References and Notes
- 1.Brown BA, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci U S A. 2005;102:18225. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown BA, Jenkins GI. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 2008;146:576. doi: 10.1104/pp.107.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28:591. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenkins GI. Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol. 2009;60:407. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- 5.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130:234. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaiserli E, Jenkins GI. UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell. 2007;19:2662. doi: 10.1105/tpc.107.053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloix C, Jenkins GI. Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Mol Plant. 2008;1:118. doi: 10.1093/mp/ssm012. [DOI] [PubMed] [Google Scholar]
- 8.Oravecz A, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18:1975. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supporting material in Science online.
- 11.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura N, et al. Structural Mechanism of Abscisic Acid Binding and Signaling by Dimeric PYR1. Science. 2009;326:1373. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Grishina IB, Woody RW. Contributions of tryptophan side chains to the circular dichroism of globular proteins: exciton couplets and coupled oscillators. Faraday Discuss. 1994:245. doi: 10.1039/fd9949900245. [DOI] [PubMed] [Google Scholar]
- 16.Ohmae E, Sasaki Y, Gekko K. Effects of five-tryptophan mutations on structure, stability and function of Escherichia coli dihydrofolate reductase. J Biochem. 2001;130:439. doi: 10.1093/oxfordjournals.jbchem.a003004. [DOI] [PubMed] [Google Scholar]
- 17.Andersson D, Carlsson U, Freskgard PO. Contribution of tryptophan residues to the CD spectrum of the extracellular domain of human tissue factor: application in folding studies and prediction of secondary structure. Eur J Biochem. 2001;268:1118. doi: 10.1046/j.1432-1327.2001.01981.x. [DOI] [PubMed] [Google Scholar]
- 18.Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.