Abstract
The Snf1/AMP-activated protein kinase family has diverse roles in cellular responses to metabolic stress. In Saccharomyces cerevisiae, Snf1 protein kinase has three isoforms of the β subunit that confer versatility on the kinase and that exhibit distinct patterns of subcellular localization. The Sip1 β subunit resides in the cytosol in glucose-grown cells and relocalizes to the vacuolar membrane in response to carbon stress. We show that translation of Sip1 initiates at the second ATG of the open reading frame, yielding a potential site for N myristoylation, and that mutation of the critical glycine abolishes relocalization. We further show that the cyclic AMP-dependent protein kinase (protein kinase A [PKA]) pathway maintains the cytoplasmic localization of Sip1 in glucose-grown cells. The Snf1 catalytic subunit also exhibits aberrant localization to the vacuolar membrane in PKA-deficient cells, indicating that PKA regulates the localization of Snf1-Sip1 protein kinase. These findings establish a novel mechanism of regulation of Snf1 protein kinase by the PKA pathway.
The Snf1/AMP-activated protein kinase (AMPK) family is highly conserved and plays multiple roles in cellular responses to metabolic stress (for reviews see references 11 and 15). In mammals, AMPK coordinates energy homeostasis, regulating lipid and glucose metabolism. In the yeast Saccharomyces cerevisiae, the Snf1 protein kinase is required for various stress responses, most importantly the adaptation of cells to glucose limitation and the utilization of alternate carbon sources. Snf1 regulates the transcription of many genes and the activity of metabolic enzymes in response to carbon stress. Snf1 also affects meiosis and sporulation, haploid invasive growth (5), diploid pseudohyphal development (18), and aging (2).
Several factors account for the versatility of Snf1 and AMPK in contributing to diverse aspects of cellular regulation. Both Snf1 and AMPK are heterotrimeric kinases that exist in multiple forms. AMPK has different isoforms of all three subunits (α, β, and γ). Snf1 protein kinase comprises the Snf1 catalytic (α) subunit, the Snf4 stimulatory (γ) subunit, and three isoforms of the β subunit, Sip1, Sip2, and Gal83. The β subunit mediates interactions with downstream targets (38, 44) and regulates the subcellular localization of the kinase, with each β subunit displaying a unique pattern of localization (45). All three subunits are cytoplasmic during growth in glucose, but, upon a shift to a nonfermentable carbon source, Sip1 relocalizes around the vacuole, Sip2 remains cytoplasmic, and Gal83 becomes enriched in the nucleus. Presumably, localization of the kinase dictates its access to substrates.
The three β subunits exhibit considerable functional redundancy, and each alone is sufficient for growth on fermentable carbon sources, but they also play distinct roles (38, 52). Gal83 mediates the interaction of Snf1 protein kinase with specific transcription factors, such as Sip4 (38, 44), and with the transcriptional apparatus (45). Sip2 has been implicated in aging (2, 20). Finally, the three β subunits have different positive or negative roles in haploid invasive growth (46).
Another factor contributing to the functional versatility of Snf1 is its response to multiple regulatory inputs. Upstream kinases Tos3, Pak1, and Elm1 phosphorylate the activation loop threonine of the catalytic subunit and activate the kinase (12, 29, 41). Protein phosphatase 1 (Reg1-Glc7) also regulates the phosphorylation and activity of the kinase (24, 25, 37). A more modest role in regulating catalytic activity is played by Std1 (Msn3) (19), a protein that interacts with glucose sensors (39). Finally, evidence implicates glucose-6-phosphate as a signal regulating the nucleocytoplasmic distribution of Gal83 (45).
In this study, we have characterized the Sip1 β subunit with respect to the regulation of its subcellular localization. We show that Sip1 relocalizes from the cytosol to the vacuolar membrane in response to various types of carbon stress and that this relocalization requires the divergent N-terminal region of the protein and an N-myristoylation consensus site. We present evidence that the localizations of Sip1 and Gal83 are regulated by different signals. Finally, we show that the cyclic AMP (cAMP)-dependent protein kinase (protein kinase A [PKA]) controls the localization of Sip1 and Snf1-Sip1 protein kinase.
MATERIALS AND METHODS
Strains and genetic methods.
S. cerevisiae strains used in this study are listed in Table 1. MCY4908 was constructed by integration of XhoI-linearized pCN106 (3) into W303-1A, followed by selection for Ura− recombinants and identification of the desired mutants by phenotype and PCR. MCY4923, -4926, and -4931 were obtained by transformation with PCR fragments generated from genomic DNA of W303 tpk123 msn2 msn4 with primers flanking each of the tpk loci. Genotypes were confirmed by PCR or Southern blotting. The gal83Δ::natMX4 allele was constructed by transformation with a PCR fragment generated using pAG25 (7) as a template for the natMX4 cassette.
TABLE 1.
List of S. cerevisiae strains
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATaade2-1 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 | 42 |
| W303-1B | MATα ade2-1 trp1-1 his3-11,15 can1-100 ura3-1 leu2-3,112 | 42 |
| W303 tpk123 msn2 msn4 | W303-1A tpk1::URA3 tpk2::HIS3 tpk3::TRP1 msn2::HIS3 msn4::TRP1 | 8 |
| ASY62 | MATatpk1Δ::ADE8 tpk2::HIS3 tpk3::TRP1 msn2Δ::HIS3 msn4Δ::LEU2 ura3 his3 leu2 trp1 ade8 | 40 |
| MCY4039 | MCY4455 sip1Δ::kanMX6 | This study |
| MCY4040 | MATα sip1Δ::kanMX6 sip2Δ::LEU2 gal83Δ::TRP1 his3-Δ200 leu2-3,112 trp1Δ1 ura3-52 lys2-801 | 45 |
| MCY4067 | MATamsn2::HIS3 msn4::URA3 his3Δ200 leu2-3,112 lys2-801 ura3-52 trp1Δ1 | This study |
| MCY4097 | W303-1A sip2Δ::kanMX4 gal83Δ::TRP1 | This study |
| MCY4098 | W303-1A sip1Δ::kanMX6 sip2Δ::kanMX4 gal83Δ::TRP1 | This study |
| MCY4416 | MATα gal83Δ::TRP1 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 | 45 |
| MCY4455 | MATα his3Δ200 leu2-3,112 trp1Δ1 ura3-52 lys2-801 | 45 |
| MCY4456 | MATα sip2Δ3::LEU2 gal83Δ::TRP1 his3Δ200 leu2-3,112 trp1Δ1 ura3-52 lys2-801 | 45 |
| MCY4908 | W303-1A snf1Δ10 | This study |
| MCY4923 | W303-1A tpk1::URA3 | This study |
| MCY4926 | W303-1A tpk2::HIS3 | This study |
| MCY4931 | W303-1B tpk3::TRP1 | This study |
| MCY4932 | MATα tpk1::URA3 tpk2::HIS3 leu2-3,112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 | This study |
| MCY4936 | MATα tpk1::URA3 tpk3::TRP1 leu2-3,112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 | This study |
| MCY4940 | MATα tpk2::HIS3 tpk3::TRP1 leu2-3,112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 | This study |
| MCY4947 | W303 tpk123 msn2 msn4 sip2::kanMX4 gal83::natMX4 | This study |
| MCY4986 | W303-1A × W303-1B bcy1::URA3/+ | This study |
To examine the effect of bcy1 on the localization of Sip1-green fluorescent protein (GFP), an appropriate strain was constructed by a strategy designed to avoid acquiring a suppressor of bcy1. The bcy1::URA3 allele (43) was recovered by PCR of genomic DNA from strain TF1.5prFHR (13) and introduced into the diploid W303-1A × W303-1B. The diploid was transformed with plasmid pOV90 (marked by HIS3), sporulated, and subjected to tetrad analysis. A Ura+ His+ segregant was identified and examined immediately to localize Sip1-GFP. Subsequently the presence of the bcy1::URA3 allele was confirmed by PCR analysis and by its glycogen deficiency phenotype.
Selective synthetic complete (SC) media (36) contained the carbon sources indicated below at 2%, except for 3% ethanol. Cycloheximide (Sigma) was added to 10 μg/ml.
Plasmids.
Proteins were expressed from their native promoters on centromeric plasmids. pOV90, pRT12, pOV72, and pOV84 have been described (45). pRT13 and pKH32 are derivatives of pRT12 and pOV84, respectively, in pRS315. pKH20 and pKH24 are identical to pOV90, except that the vectors are pRS315 and pRS316, respectively.
pKH2 was constructed by recombination in yeast between pOV90 cut with ClaI plus AflII and a PCR fragment generated from pOV90 with primer T3 and a primer homologous to the SIP1 sequence ending at codon 564 (numbered as originally published [51]) and the beginning of the GFP coding sequence. pKH31 is a derivative of pKH2 in pRS315. pKH3 was similarly constructed by using pOV90 cut with ClaI plus BsgI and a PCR fragment generated from pOV90 with primer T7 and a primer starting at codon 565 of SIP1 with a tail homologous to sequence upstream of the first codon of SIP1. pKH9 was constructed in multiple steps. First, a PCR fragment was generated from pOV90 with primers T3 and oKH12 (AGAAGGACTGTTTGCCATGGCGGCTGC). This fragment was then used as a primer with oKH14 (CCATTTTAGAACAACTAAAGTATTTTCGG) on template pOV90 to generate a second fragment by PCR. This fragment was cotransformed into yeast with pOV90 cut with XhoI plus BsgI, and recombination yielded pKH9.
pKH15 and pKH16 carry mutations of the first and second ATG codons, respectively, to ATT. They were constructed by the same procedure described for pKH9 but with oKH33 (CGGATATATCTACGATTGTTAATTGTCAATAG) and oKH32 (GAAGGACTGTTTCCTATGGCGGCTGCAAC), respectively, instead of oKH12 as the primers. Several independent mutant plasmids were recovered, and they behaved identically. To construct pKH40, the backbone of pKH2 was replaced with that of pRS425 and the GFP coding sequence was replaced with that of the triple hemagglutinin (HA) epitope. pKH42 was derived from pKH40 by the same procedure as that used for pKH15. Mutations were verified by sequencing.
Plasmids carrying mutations in consensus PKA sites were constructed as follows. To construct pKH25, we generated a PCR fragment from pOV90 with primers T3 and oKH52 (GGCGCGTCTCTTATGAGCACTCTTCCTATTGTTAAAC). This fragment was used as a primer with oKH14 on template pOV90 to generate a second PCR fragment. The second fragment was cotransformed into yeast with pOV90 cut with XhoI plus BsgI, and recombination yielded pKH25. pKH26, pKH27, and pKH28 were constructed the same way as pKH25 except that oKH53 (CCATTATGTTCGCTAGTATGAGCGGCGCGTCTCTTATGAG), oKH54 (GGTATGCATGGATAGCTGGTTTCTTTGAATAGGG), and oKH55 (GCAAAGAAGCAAATGCGGATCTTCTCGAATTTG), respectively, were used in place of oKH52. The mutations in pKH25, pKH26, pKH27, and pKH28 result in the substitution of Ala for Thr93, Ser99, Ser282, and Ser233, respectively. To construct pKH33, which has mutations at codons 93, 99, and 282, a PCR fragment generated from pKH25 with primers T3 and oKH53 was used as a primer with oKH14 on template pKH27 to generate a second fragment by PCR. This fragment was cotransformed into yeast with pOV90 cut with XhoI plus BsgI, and recombination yielded pKH33. pKH34 carries all four mutations and was constructed in the same way as pKH28 but with pKH33 rather than pOV90. pKH36, which expresses the mutant N terminus of Sip1, was constructed by the same procedure as that used for pKH2 with pKH34 rather than pOV90 as the template. Mutations were confirmed by sequencing.
Microscopy.
Cultures were grown to mid-log phase in selective SC media. Nuclei were stained by addition of 4′,6-diamidino-2-phenylindole (DAPI; 0.8 μg/ml) for 5 min. Cells from 1 ml of culture were harvested by brief centrifugation and resuspended in residual medium (∼20 μl), and an aliquot was placed on a microscope slide. For shift experiments, cells were grown and harvested as described above but resuspended in 1 to 5 ml of medium containing a different carbon source; washing the cells once in the new medium did not alter the results. Cells were incubated for various times, and 5 min before harvest, DAPI was added. Cells were then collected for examination as described above. Cells were viewed with a Nikon Eclipse E800 fluorescence microscope. Images were taken with an Orca100 (Hamamatsu) camera using Open Lab (Improvision) software and processed in Adobe Photoshop, version 5.5.
Immunoblot analysis.
Protein extracts were prepared essentially as described previously (44). Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 10% acrylamide and immunoblotting using a monoclonal anti-HA antibody (12CA5). Antibodies were detected by chemiluminescence using ECLPlus (Amersham Biosciences).
RESULTS
Relocalization of Sip1-GFP from the cytosol to the periphery of the vacuole in response to carbon stress.
We examined the localization of Sip1-GFP in strains with the S288C, W303, and Σ1278b genetic backgrounds. In all cases, Sip1-GFP was found in the cytosol in glucose-grown cells and relocalized to the periphery of the vacuole within 10 min of a shift to glycerol-ethanol (Fig. 1A). Fluorescence was often particularly prominent in the region between the vacuole and the nucleus. This relocalization was evident in S288C-related strains, as reported previously (45), but was more easily visualized in W303 cells, which were therefore used for subsequent experiments.
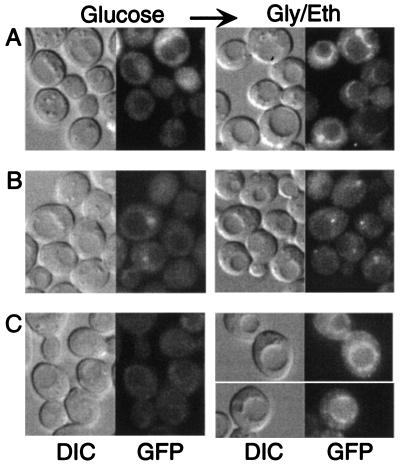
FIG. 1.
Localization of Sip1-GFP in response to the carbon source. W303-1A cells expressed Sip1-GFP from pOV90. (A) Cells were grown in glucose (Glu; 2%) and shifted to glycerol-ethanol (Gly/Eth) for 10 min. (B) Glucose-grown cells were shifted to raffinose (Raf) for 10 min. (C) Cells were grown in raffinose for 54 h with successive dilutions of the culture to maintain low cell density. (D) Glucose-grown cells were shifted to glycerol-ethanol for 30 min, and an aliquot of the culture was then shifted to glucose for 10 min. Arrows indicate that cells were shifted from one carbon source to another. GFP fluorescence and differential interference contrast (DIC) images are shown.
The response was not specific to glycerol-ethanol. When glucose-grown cells were shifted to low (0.05%) glucose, raffinose, or galactose, Sip1-GFP similarly relocalized to the vacuolar periphery within 10 min (Fig. 1B and data not shown). Relocalization was most striking for glycerol-ethanol and low glucose. As cells must adapt to utilize these carbon sources, such a shift essentially constitutes immediate carbon deprivation. These results suggest that Sip1-GFP relocalizes around the vacuole in response to carbon stress.
We also examined W303 cells during growth in glycerol-ethanol, raffinose, galactose, or sucrose after the cells had adapted to utilization of the carbon source. Although Sip1-GFP appeared exclusively cytoplasmic in cells growing on sucrose, Sip1-GFP was observed around the vacuole in 40 to 50% of the cells in the other cultures (Fig. 1C and data not shown). Some cells also had fluorescence within the vacuole, which became more pronounced if cultures were grown beyond exponential phase or were unhealthy; the vacuolar fluorescence reported previously (45) can probably be attributed to the poor growth of S288C-related cells in synthetic medium containing glycerol-ethanol.
Addition of glucose rapidly affects localization of Sip1-GFP.
To determine whether restoring glucose to the growth medium would affect the presence of Sip1 around the vacuole, we shifted glucose-grown cells to glycerol-ethanol for 30 min and verified that Sip1-GFP had relocalized (Fig. 1D). The culture was then divided into aliquots for a second shift to different carbon sources: glucose (2%), glycerol-ethanol plus glucose, or glycerol-ethanol. In the two cultures where glucose was replenished, Sip1-GFP was cytosolic within 10 min and could no longer be detected around the vacuole, whereas in the glycerol-ethanol control culture, localization was unchanged (Fig. 1D and data not shown). Addition of cycloheximide during the second shift did not noticeably affect the pattern of fluorescence (data not shown).
A subpopulation of Snf1 protein kinase localizes around the vacuole.
We next sought to determine whether Sip1 targets the Snf1 protein kinase to the vacuolar periphery. Although a subpopulation of the Snf1 catalytic subunit of the kinase, is associated with Sip1 (52), designated the Snf1-Sip1 form of the kinase, it remains possible that not all Sip1 is associated with Snf1. In glucose-grown cells, Snf1-GFP is predominantly cytoplasmic. When cells are shifted to glycerol-ethanol, Snf1 becomes enriched in the nucleus along with Gal83, but some Snf1 also remains cytoplasmic, as does Sip2; however, no ring around the vacuole was evident (45) (data not shown). Because Sip1 is not an abundant β subunit (45), we reexamined the localization of Snf1-GFP in a sip2Δ gal83Δ mutant (MCY4097), in which the only β subunit is Sip1. Upon a shift to glycerol-ethanol, Snf1 relocalized to the vacuolar membrane (Fig. 2A). No such effect was observed in a sip1Δ sip2Δ gal83Δ mutant, indicating dependence on the presence of Sip1 (Fig. 2B). These findings indicate that some, if not all, of the Sip1 around the vacuole is complexed with Snf1.
FIG. 2.
Snf1 accompanies Sip1 to the vacuolar periphery but is not required for localization. Transformants of strains MCY4097 (sip2Δ gal83Δ) (A) and MCY4098 (sip1Δ sip2Δ gal83Δ) (B) expressed Snf1-GFP from pOV84. (C) MCY4908 (snf1Δ) cells expressed Sip1-GFP from pOV90. Cells were grown in glucose and shifted to glycerol-ethanol (Gly/Eth) for 30 min. The arrow indicates a shift. GFP fluorescence and differential interference contrast (DIC) images are shown.
Localization of Sip1 is independent of Snf1.
The Snf1 protein kinase activity is important for many cellular responses to carbon stress. To determine whether the localization of Sip1 depends on Snf1, we expressed Sip1-GFP in a snf1Δ mutant. Sip1-GFP was cytosolic in glucose-grown cells and relocalized to the vacuolar periphery during a shift to glycerol-ethanol, indicating that Snf1 is not required for the localization signal (Fig. 2C).
The N terminus of Sip1 is necessary and sufficient for localization.
The N-terminal sequence of Sip1 differs substantially from those of the other β subunits, whereas the C-terminal region that interacts with Snf1 and Snf4 is conserved (Fig. 3A) (14, 52). To determine if the divergent N terminus is responsible for the unique localization pattern of Sip1, we fused the first 564 codons to the GFP coding sequence (pKH2; Fig. 3A). This polypeptide was cytoplasmic in glucose-grown cells and relocalized to the vacuolar periphery upon a shift to glycerol-ethanol (Fig. 3D). Relocalization was not dependent on the expression of native Sip1, as similar results were observed in sip1Δ cells (data not shown). In contrast, the conserved C terminus (encoded by codons 565 to 864; pKH3) did not relocalize, instead remaining cytoplasmic upon a shift to glycerol-ethanol (data not shown). The integrity of the C-terminal polypeptide was confirmed by its ability to support the growth of a sip1Δ sip2Δ gal83Δ triple mutant on raffinose (Fig. 3B); this growth assay is an assay for generic β subunit function, as any one of the β subunits suffices. Thus, the N terminus is both necessary and sufficient for the observed relocalization of Sip1 in response to carbon stress.
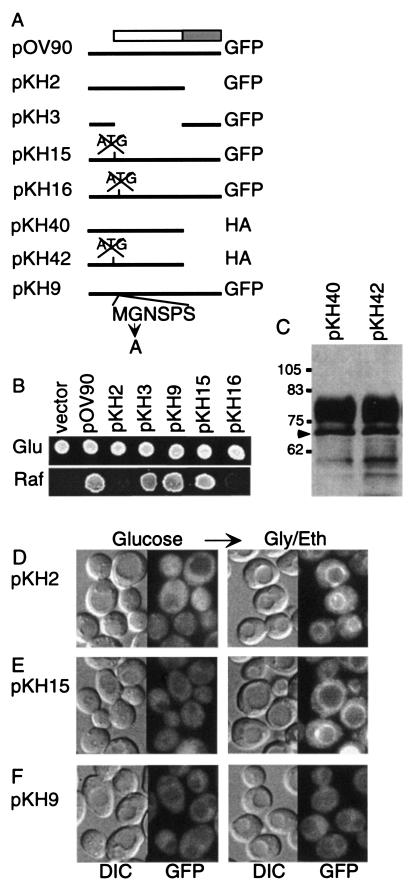
FIG. 3.
Localization and function of mutant derivatives of Sip1. (A) Structures of plasmids. Lines, DNA from the SIP1 locus, which is fused to the GFP or triple HA coding sequence, as indicated; box, SIP1 ORF, encoding a region (shaded area) that is conserved in all three β subunits and that includes sequences that interact with Snf1 and Snf4 (14). Mutation of the first and second ATG codons of the ORF is indicated by an X. For pKH9, the Gly-to-Ala substitution in the N-terminal myristoylation consensus sequence is indicated; amino acid residues are represented by single-letter code. (B) Transformants of MCY4098 (sip1Δ sip2Δ gal83Δ) carrying the indicated plasmids were tested for growth on selective SC containing glucose (Glu) or raffinose (Raf) plus antimycin (1 μg/ml). The vector is pRS313. Multiple transformants were tested (not shown). (C) Transformants of W303-1A carrying pKH40 or pKH42 were grown to mid-log phase in glucose, and proteins were analyzed by immunoblotting with an anti-HAantibody. Arrow, predicted position of the unmodified Sip1-HA polypeptide. The more slowly migrating species are presumably phosphorylated, as described previously (21). No additional bands were detected upon longer exposure, and the two samples also looked identical when loading and exposure were both reduced. Molecular size markers are indicated in kilodaltons. (D to F) Transformants of W303-1A expressing derivatives of Sip1-GFP were grown in glucose and shifted to glycerol-ethanol (Gly/Eth) for 10 to 30 min. Similar results were observed at both times. GFP fluorescence and differential interference contrast (DIC) images are shown. (D) Cells expressed the N terminus of Sip1 fused to GFP from pKH2. (E) Cells expressed Sip1-GFP from pKH15, in which the first ATG is replaced by ATT. GFP fluorescence appeared as intense as that observed for the full-length Sip1-GFP. (F) Cells expressed Sip1-GFP lacking the putative N-myristoylation site from pKH9.
A second ATG codon in the SIP1 ORF is required for expression of the protein.
The SIP1 open reading frame (ORF) is predicted to encode a protein of 864 residues (51). However, the ORF contains a second ATG at codon 49 and encodes the sequence MGNSPST, which matches the consensus sequence for sites of N myristoylation (6). There is precedent for N myristoylation of β subunits, including Sip2 and mammalian β1 (1, 26, 47). In addition, the major clusters of 5′ ends of the SIP1 RNAs map between the two ATGs (27).
To test the possibility that translation starts at the second ATG, we replaced the first ATG with ATT (pKH15; Fig. 3A). Glucose-grown cells expressing the mutant protein showed cytoplasmic GFP fluorescence that was as intense as that observed with the wild-type protein, and upon a shift to glycerol-ethanol, the mutant Sip1-GFP relocalized around the vacuole (Fig. 3E). The mutant protein also allowed growth of a sip1Δ sip2Δ gal83Δ strain on raffinose (Fig. 3B). Thus, the first ATG of the ORF is not required for expression of functional Sip1.
We next examined the products expressed from the wild-type and mutant genes. To optimize detection of polypeptides differing by only 49 amino acid residues, we expressed HA-tagged N-terminal fragments from pKH40 and pKH42 (Fig. 3A). Immunoblot analysis revealed no differences between the products of the two constructs (Fig. 3C). In both cases, a species migrated at the position predicted for the unmodified polypeptide and most of the products migrated more slowly, consistent with previous evidence that Sip1 is heavily phosphorylated (21). Although very low levels of initiation at the first ATG are not excluded, these data indicate that the second ATG serves as the major translational start site.
Finally, we replaced the second ATG with ATT (pKH16). We detected no fluorescence in wild-type cells during growth in glucose or after a shift to glycerol-ethanol, and the mutant construct did not provide Sip1 function (Fig. 3B). Similar results were obtained with three independent, sequenced mutant plasmids. Together, these findings indicate that translation of Sip1 starts at the second ATG codon.
The myristoylation consensus site is required for relocalization of Sip1 to the vacuolar membrane.
N myristoylation has been shown to facilitate the association of proteins with membranes (6). To test the role of the putative myristoylation site in the localization of Sip1-GFP, we mutated the GGA (Gly) codon for the MGNSPS sequence following the translational start site to GCA (Ala) in pKH9 (Fig. 3A). The mutant protein conferred wild-type levels of cytoplasmic fluorescence in glucose-grown cells but did not relocalize to the vacuolar periphery when cells were shifted to glycerol-ethanol (Fig. 3F). Similar results were observed with sip1Δ cells (MCY4039; data not shown). The Gly-to-Ala mutation does not abolish Sip1 function, as the mutant protein supported growth of the sip1Δ sip2Δ gal83Δ mutant on raffinose (Fig. 3B). These findings indicate that the glycine residue is required for relocalization and suggest that Sip1 is N myristoylated in vivo. However, myristoylation of a β subunit is not sufficient to confer this localization pattern because Sip2 is myristoylated (1) but remains cytoplasmic under these conditions (45).
Different signals regulate the localization of Sip1 and Gal83.
Although Sip1 and Gal83 move to different locations in response to carbon stress, it was conceivable that they respond to the same signal. In W303 cells, Gal83 exhibited the same pattern of response to a spectrum of carbon sources as did Sip1. Gal83 was cytoplasmic during growth on glucose or sucrose and relocalized to the nucleus upon a shift to glycerol-ethanol or during growth in galactose or raffinose (data not shown). However, the localization of Gal83 is apparently strain dependent; we confirmed previous findings that, during growth on galactose or raffinose, Gal83 remains cytoplasmic in the S288C-related strain MCY4416 (45) but also found that Gal83 is enriched in the nucleus in another S288C-related strain (MCY4455) (data not shown). These differences remain unexplained but may reflect differences in metabolic efficiency. Nonetheless, Gal83 and Sip1 respond similarly in W303 cells.
To address the possibility that the two β subunits respond to the same signal, we examined the effects of 2-deoxyglucose, a glucose analog that is phosphorylated but not metabolized. Addition of 2-deoxyglucose (0.02%) to glycerol-grown cells causes nuclear export of Gal83 (45), and its presence during a shift from glucose to glycerol prevents nuclear accumulation (48). These findings, together with analysis of mutants lacking hexose kinases (45), suggested that glucose-6-phosphate is a candidate for the signal controlling the localization of Gal83.
To test whether 2-deoxyglucose affects the localization of Sip1, we repeated the experiment shown in Fig. 1. We shifted glucose-grown cells to glycerol-ethanol for 30 min, confirmed that Sip1-GFP had relocalized, and divided the culture into aliquots for a second shift to different carbon sources. This time additional aliquots were taken for growth in glycerol-ethanol plus 2-deoxyglucose (0.02 and 0.04%), and Sip1-GFP remained localized around the vacuole after 10, 30, and 60 min (data not shown). Similarly, when glucose-grown cultures were shifted to glycerol-ethanol plus 2-deoxyglucose (0.02%), Sip1-GFP was visible around the vacuole at 10 and 30 min (data not shown). Thus, 2-deoxyglucose did not affect the subcellular distribution of Sip1. Controls for both experiments confirmed that 2-deoxyglucose prevented the nuclear localization of Gal83-GFP in W303 cells (data not shown). These findings indicate that Sip1 and Gal83 respond to distinct signals.
PKA regulates the localization of Sip1.
The cAMP-dependent protein kinase PKA regulates numerous cellular processes in response to glucose signals (30, 35, 49). Notably, PKA controls the nuclear localization of some transcription factors, such as Msn2 and Msn4, in response to glucose (8, 9) but does not regulate the localization of Gal83 (45). To assess the possibility that PKA regulates Sip1, we introduced Sip1-GFP into two mutant strains lacking all three catalytic subunits of PKA, strains W303 tpk123 msn2 msn4 (8) and ASY62 (40); the msn mutations allow healthy growth in the absence of PKA. In both strains, Sip1-GFP was localized around the vacuole, not only in cells that were shifted to glycerol-ethanol but also in glucose-grown cells (Fig. 4A and data not shown). The N terminus of Sip1 showed similar localization (Fig. 4B). We also noted that the intensity of fluorescence around the vacuole seemed relatively stronger in glucose-grown cells than in cells shifted to glycerol-ethanol. In control experiments, the nuclear localization of Gal83 was properly regulated in both mutant strains (Fig. 4C and data not shown). In addition, an msn2 msn4 strain (MCY4067) showed normal localization of Sip1-GFP, indicating that these mutations are not responsible for the phenotype (data not shown). These findings indicate that PKA is required to maintain the cytosolic localization of Sip1 during growth in glucose.
FIG. 4.
Localization of Sip1-GFP and Snf1-GFP in mutants with altered PKA activity. Cells were grown in glucose (Glu) and shifted to glycerol-ethanol (Gly/Eth) for 30 min, as indicated. Strain W303 tpk123 msn2 msn4 expressed Sip1-GFP from pKH20, a derivative of pOV90 in vector pRS315 (A); the N terminus of Sip1-GFP from pKH31, a derivative of pKH2 in vector pRS315 (B); or Gal83-GFP from pRT13 (C). For cells shown in panel C, DNA was stained with DAPI. (D) Strain MCY4940 (tpk2 tpk3) expressed Sip1-GFP from pKH20 and carried PDE2 in multicopy on pXY8 (13). (E) A bcy1 mutant segregant of diploid strain MCY4986 expressed Sip1-GFP from pOV90. (F) Strain MCY4947 (tpk1 tpk2 tpk3 msn2 msn4 sip2 gal83) expressed Snf1-GFP from pKH32; this strain was unhealthy. GFP fluorescence and differential interference contrast (DIC) images are shown.
Any catalytic subunit of PKA is sufficient for cytoplasmic distribution of Sip1 during growth in glucose.
The three catalytic subunits of PKA, encoded by TPK1, TPK2, and TPK3, are largely redundant but also serve some specific functions (31-33). To determine whether a particular catalytic subunit controls the localization of Sip1, we examined single and double tpk1, tpk2, and tpk3 mutants. In all six mutant strains, Sip1-GFP was cytoplasmic when cells were grown in glucose and relocalized to the vacuolar membrane when cells were shifted to glycerol-ethanol for 30 min (data not shown). Thus, any Tpk catalytic subunit suffices to confer cytoplasmic localization of Sip1-GFP during growth in glucose.
We next took advantage of the double mutants to exclude the possibility that some indirect effect resulting from the loss of PKA activity in combination with the absence of Msn2 and Msn4 was responsible for the relocalization of Sip1-GFP in the triple tpk msn2 msn4 mutant strains. We reasoned that the double mutants had reduced PKA activity and that introduction of multiple copies of the PDE2 gene, encoding cAMP phosphodiesterase, would effectively reduce PKA activity even further, perhaps enough to affect the localization of Sip1. The tpk2 tpk3 double mutant (with wild-type MSN2 and MSN4 alleles) was transformed with a multicopy plasmid carrying PDE2 (pXY8 [13]) and a plasmid expressing Sip1-GFP. In many, but not all, glucose-grown cells, Sip1-GFP was localized around the vacuole, consistent with a multicopy effect (Fig. 4D). These findings confirm that PKA activity is critical.
Absence of the Bcy1 regulatory subunit of PKA impairs relocalization of Sip1 in response to carbon stress.
To address the effect of activation of PKA, we examined a mutant lacking the Bcy1 regulatory subunit (43). Inactive PKA comprises two regulatory subunits bound to two catalytic subunits, and the binding of cAMP to Bcy1 causes a conformational change that releases the active catalytic subunits. In the bcy1 mutant (a fresh segregant from a heterozygous diploid; see Materials and Methods), Sip1-GFP was distributed in the cytosol during growth in glucose; however, Sip1-GFP failed to relocalize to the vacuole after a shift to glycerol-ethanol (Fig. 4E). Together, the analyses of this mutant and the PKA-deficient strains establish that PKA activity controls the localization of Sip1.
Aberrant localization of Snf1 in a PKA-deficient mutant.
Because it is not known whether all of the Sip1 protein in the cell is stably associated with Snf1, it remained possible that PKA controls the association of free Sip1 protein with the vacuole but has no role in regulating the localization of Snf1 protein kinase. To address this issue, we introduced sip2 and gal83 mutations into a PKA-deficient strain so that Sip1 would be the only β subunit, and we expressed Snf1-GFP in the resulting strain (MCY4947). Snf1-GFP was observed around the vacuole in glucose-grown cells (Fig. 4F); the intensity of fluorescence appeared weaker after a shift to glycerol-ethanol (data not shown), as was the case for Sip1-GFP (Fig. 4A). These findings strongly suggest that PKA regulates the localization of Snf1-Sip1 complexes.
Mutation of potential PKA phosphorylation sites in Sip1.
One mechanism by which PKA could regulate the localization of Sip1 is direct modification of the protein. Previous studies showed complex carbon-responsive patterns of Sip1 phosphorylation involving multiple sites (21). Immunoblot analysis of the N-terminal fragment of Sip1 (pKH40), which exhibits altered localization in PKA-deficient cells (Fig. 4B), revealed no differences between wild-type and PKA-deficient cells (data not shown); however, given the extent of modification (Fig. 3C), differential phosphorylation of a few sites could have escaped detection.
The N terminus of Sip1 contains four sequences that match the consensus sequence for phosphorylation by PKA (R/KR/KXS/T). We mutated these four potential PKA sites, substituting Ala for Thr93, Ser99, Ser282, and Ser233. Sip1-GFP with any single mutant site and both full-length and N-terminal Sip1-GFP carrying all four mutant sites (pKH34 and pKH36, respectively) were cytoplasmic in glucose-grown wild-type cells and relocalized around the vacuole upon a shift to glycerol-ethanol (data not shown). Thus, phosphorylation of these residues is not required for cytoplasmic localization during growth in glucose. It remains possible that PKA phosphorylates Sip1 on other sites or that PKA phosphorylates another protein that controls the localization of Sip1.
DISCUSSION
The Snf1 protein kinase has many different roles in cellular responses to stress, and the presence of three different isoforms of the β subunit confers versatility on the kinase. We have examined the unique subcellular distribution exhibited by the Sip1 β subunit of the kinase and its regulation. We show that Sip1 relocalizes from the cytoplasm to the vacuolar periphery when glucose-grown cells are subjected to acute carbon stress and that the Snf1 catalytic subunit accompanies Sip1. In addition, Sip1 is found around the vacuole in a substantial fraction of cells during growth on carbon sources such as galactose and raffinose, which are less preferred than glucose. The N terminus of Sip1, which is divergent from the N termini of the other two β subunits, is responsible for its localization to the vacuolar periphery; moreover, the glycine of the N-myristoylation consensus sequence is required, suggesting that Sip1 associates directly with the vacuolar membrane. This relocalization may target Snf1 catalytic activity to substrates present at the vacuolar membrane; however, we cannot exclude the possibility that relocalization instead serves to sequester the Sip1 form of the kinase from targets elsewhere. The observed relocalization strongly suggests a role for Sip1 during carbon source transitions. Although glucose-grown sip1Δ mutant cells appeared to adapt normally to other carbon sources on solid and liquid medium (K. Hedbacker, unpublished results), functional redundancy among the β subunits may have obscured any defect.
We further present evidence that the PKA pathway regulates the localization of Sip1. In mutants lacking PKA, Sip1 was found constitutively around the vacuole, indicating that PKA is required to maintain the cytoplasmic localization of Sip1 during growth in glucose. Similar results were obtained with a mutant lacking two of the three PKA catalytic subunits and overexpressing cAMP phosphodiesterase, confirming that PKA activity is the relevant factor. Conversely, localization of Sip1 to the vacuole in response to carbon stress was impaired in a mutant lacking the Bcy1 regulatory subunit of PKA. The Snf1 catalytic subunit was also found at the vacuolar membrane in PKA-deficient cells during growth in glucose, indicating that PKA regulates the localization of Snf1-Sip1 protein kinase complexes.
PKA is known to have broad roles in cellular regulation, controlling such processes as cell growth, metabolism, stress resistance, and filamentous invasive growth (for reviews see references 30, 35, and 49). The present findings establish a novel form of regulation by the PKA pathway, in which PKA regulates the localization of Snf1-Sip1. The possibility that PKA also regulates the catalytic activity of Snf1-Sip1 has not been excluded.
The synthesis of cAMP by adenylate cyclase is stimulated by Ras proteins and by glucose via the G protein-coupled receptor Gpr1 and the Gα protein Gpa2. It seems likely that the Gpr1-Gpa2 glucose-sensing pathway is involved in regulating the localization of Snf1-Sip1. The Gpr1 receptor is activated by glucose and interacts with Gpa2 to stimulate cAMP synthesis and to promote filamentous and invasive growth (4, 16, 17, 22, 23, 28, 50, 53). We note that Sip1 has a modest inhibitory role in haploid invasive growth (46). Glucose-induced stimulation of cAMP synthesis also requires intracellular phosphorylation of the sugar (34), and kelch repeat proteins that mimic Gβ subunits have been implicated in the signaling mechanism (10).
Previous studies showed that the catalytic activity of Snf1 protein kinase is controlled by multiple regulatory inputs, including three upstream kinases (12, 29, 41), protein phosphatase 1 (24, 25, 37), and the Std1 protein (19). It is now clear that localization of the kinase is also controlled by more than one mechanism. We present evidence that Sip1 and Gal83 respond to distinct signals: 2-deoxyglucose prevents nuclear localization of Gal83 but does not affect the localization of Sip1, and PKA regulates the localization of Sip1 but not that of Gal83. These findings are consistent with evidence that the activity of PKA toward Msn2 is insensitive to 2-deoxyglucose (8) and with genetic evidence suggesting that glucose-6-phosphate is a candidate for the signal controlling the nucleocytoplasmic distribution of Gal83 (45). Thus, these two β subunits exhibit distinct patterns of subcellular localization and their localization is regulated by different pathways. Localization affects the access of Snf1 to substrates, and regulation of localization by different pathways ensures access to different substrates in response to different signals. This complexity adds further versatility to Snf1 protein kinase function.
Acknowledgments
We thank J. Gimeno-Alcaniz for initial studies and H. Wiatrowski and V. Vyas for strains and advice.
This work was supported by Public Health Service grant GM34095 from the National Institutes of Health to M.C. Support for K.H. was provided in part by NIH training grant GM07088.
REFERENCES
- 1.Ashrafi, K., T. A. Farazi, and J. I. Gordon. 1998. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J. Biol. Chem. 273:25864-25874. [DOI] [PubMed] [Google Scholar]
- 2.Ashrafi, K., S. S. Lin, J. K. Manchester, and J. I. Gordon. 2000. Sip2p and its partner Snf1p kinase affect aging in S. cerevisiae. Genes Dev. 14:1872-1885. [PMC free article] [PubMed] [Google Scholar]
- 3.Celenza, J. L., and M. Carlson. 1989. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol. Cell. Biol. 9:5034-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombo, S., P. Ma, L. Cauwenberg, J. Winderickx, M. Crauwels, A. Teunissen, D. Nauwelaers, J. H. de Winde, M. F. Gorwa, D. Colavizza, and J. M. Thevelein. 1998. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17:3326-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farazi, T. A., G. Waksman, and J. I. Gordon. 2001. The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276:39501-39504. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 8.Gorner, W., E. Durchschlag, J. Wolf, E. L. Brown, G. Ammerer, H. Ruis, and C. Schuller. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21:135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorner, W., E. Durschschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harashima, T., and J. Heitman. 2002. The Gα protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gβ subunits. Mol. Cell 10:163-173. [DOI] [PubMed] [Google Scholar]
- 11.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67:821-855. [DOI] [PubMed] [Google Scholar]
- 12.Hong, S.-P., F. C. Leiper, A. Woods, D. Carling, and M. Carlson. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard, E. J. A., X. Yang, and M. Carlson. 1992. Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics 130:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, R., and M. Carlson. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp, B. E., D. Stapleton, D. J. Campbell, Z. P. Chen, S. Murthy, M. Walter, A. Gupta, J. J. Adams, F. Katsis, B. Van Denderen, I. G. Jennings, T. Iseli, B. J. Michell, and L. A. Witters. 2003. AMP-activated protein kinase, super metabolic regulator. Biochem. Soc. Trans. 31:162-168. [DOI] [PubMed] [Google Scholar]
- 16.Kraakman, L., K. Lemaire, P. Ma, A. W. Teunissen, M. C. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32:1002-1012. [DOI] [PubMed] [Google Scholar]
- 17.Kubler, E., H. U. Mosch, S. Rupp, and M. P. Lisanti. 1997. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272:20321-20323. [DOI] [PubMed] [Google Scholar]
- 18.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchin, S., V. K. Vyas, E. Kanter, S.-P. Hong, and M. Carlson. 2003. Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics 163:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, S. S., J. K. Manchester, and J. I. Gordon. 2001. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 276:36000-36007. [DOI] [PubMed] [Google Scholar]
- 21.Long, R. M., and J. E. Hopper. 1995. Genetic and carbon source regulation of phosphorylation of Sip1p, a Snf1-associated protein involved in carbon response in Saccharomyces cerevisiae. Yeast 11:233-246. [DOI] [PubMed] [Google Scholar]
- 22.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludin, K., R. Jiang, and M. Carlson. 1998. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:6245-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460-36466. [DOI] [PubMed] [Google Scholar]
- 26.Mitchelhill, K. I., B. Michell, C. M. House, D. Stapleton, J. Dyck, J. Gamble, C. Ullrich, L. A. Witters, and B. E. Kemp. 1997. Posttranslational modifications of the 5′-AMP-activated protein kinase β1 subunit. J. Biol. Chem. 272:24475-24479. [DOI] [PubMed] [Google Scholar]
- 27.Mylin, L. M., V. L. Bushman, R. M. Long, X. Yu, C. M. Lebo, T. E. Blank, and J. E. Hopper. 1994. SIP1 is a catabolite repression-specific negative regulator of GAL gene expression. Genetics 137:689-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakafuku, M., T. Obara, K. Kaibuchi, I. Miyajima, A. Miyajima, H. Itoh, S. Nakamura, K. Arai, K. Matsumoto, and Y. Kaziro. 1988. Isolation of a second yeast Saccharomyces cerevisiae gene (GPA2) coding for guanine nucleotide-binding regulatory protein: studies on its structure and possible functions. Proc. Natl. Acad. Sci. USA 85:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nath, N., R. R. McCartney, and M. C. Schmidt. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan, X., T. Harashima, and J. Heitman. 2000. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3:567-572. [DOI] [PubMed] [Google Scholar]
- 31.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97:5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson, L. S., and G. R. Fink. 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95:13783-13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rolland, F., J. H. De Winde, K. Lemaire, E. Boles, J. M. Thevelein, and J. Winderickx. 2000. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38:348-358. [DOI] [PubMed] [Google Scholar]
- 35.Rolland, F., J. Winderickx, and J. M. Thevelein. 2001. Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem. Sci. 26:310-317. [DOI] [PubMed] [Google Scholar]
- 36.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 37.Sanz, P., G. R. Alms, T. A. Haystead, and M. Carlson. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt, M. C., and R. R. McCartney. 2000. β-Subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 19:4936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt, M. C., R. R. McCartney, X. Zhang, T. S. Tillman, H. Solimeo, S. Wolfl, C. Almonte, and S. C. Watkins. 1999. Std1 and Mth1 proteins interact with glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4561-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutherland, C. M., S. A. Hawley, R. R. McCartney, A. Leech, M. J. Stark, M. C. Schmidt, and D. G. Hardie. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299-1305. [DOI] [PubMed] [Google Scholar]
- 42.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 43.Toda, T., S. Cameron, P. Sass, M. Zoller, J. D. Scott, B. McMullen, M. Hurwitz, E. G. Krebs, and M. Wigler. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincent, O., and M. Carlson. 1999. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 18:6672-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vyas, V. K., S. Kuchin, C. D. Berkey, and M. Carlson. 2003. Snf1 kinases with different β-subunit isoforms play distinct roles in regulating haploid invasive growth. Mol. Cell. Biol. 23:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warden, S. M., C. Richardson, J. J. O'Donnell, D. Stapleton, B. E. Kemp, and L. A. Witters. 2001. Post-translational modifications of the beta-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem. J. 354:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiatrowski, H. A., and M. Carlson. 2003. Yap1 accumulates in the nucleus in response to carbon stress in Saccharomyces cerevisiae. Euk. Cell 2:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson, W. A., and P. J. Roach. 2002. Nutrient-regulated protein kinases in budding yeast. Cell 111:155-158. [DOI] [PubMed] [Google Scholar]
- 50.Xue, Y., M. Batlle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17:1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, X., E. J. A. Hubbard, and M. Carlson. 1992. A protein kinase substrate identified by the two-hybrid system. Science 257:680-682. [DOI] [PubMed] [Google Scholar]
- 52.Yang, X., R. Jiang, and M. Carlson. 1994. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 13:5878-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yun, C. W., H. Tamaki, R. Nakayama, K. Yamamoto, and H. Kumagai. 1998. Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 252:29-33. [DOI] [PubMed] [Google Scholar]