Abstract
Analysis of cis-regulatory elements is central to understanding the genomic program for development. The scl/tal-1 transcription factor is essential for lineage commitment to blood cell formation and previous studies identified an scl enhancer (the +18/19 element) which was sufficient to target the vast majority of hematopoietic stem cells, together with hematopoietic progenitors and endothelium. Moreover, expression of scl under control of the +18/19 enhancer rescued blood progenitor formation in scl−/− embryos. However, here we demonstrate by using a knockout approach that, within the endogenous scl locus, the +18/19 enhancer is not necessary for the initiation of scl transcription or for the formation of hematopoietic cells. These results led to the identification of a bifunctional 5′ enhancer (−3.8 element), which targets expression to hematopoietic progenitors and endothelium, contains conserved critical Ets sites, and is bound by Ets family transcription factors, including Fli-1 and Elf-1. These data demonstrate that two geographically distinct but functionally related enhancers regulate scl transcription in hematopoietic progenitors and endothelial cells and suggest that enhancers with dual hematopoietic-endothelial activity may represent a general strategy for regulating blood and endothelial development.
The development of blood and endothelium is intimately linked throughout vertebrate development. The close association between the development of blood and endothelial cells in the avian and murine yolk sac gave rise to the concept of the hemangioblast, a bipotent precursor of both cell types (58, 68). This idea has been reinforced by the observation that murine embryonic stem (ES) cells can give rise to Flk-1+ cells capable of generating both blood and endothelial progeny (13, 49). Moreover, single Flk-1-positive cells from avian embryos can develop into either hematopoietic or endothelial colonies (18), Flk-1−/− mouse embryos fail to form yolk sac blood islands and vasculature (61), and the numbers of both endothelial and hematopoietic cells are severely reduced in the zebra fish mutant cloche (41, 65).
In the body of the amphibian, avian, or murine embryo, blood cells arise as clusters of cells attached to the endothelium of arteries (14-16, 26), and it has been suggested that differentiated endothelial cells may directly generate blood progenitors (39, 40, 50). Moreover, at least a proportion of the emerging hematopoietic cells in human embryos are thought to originate in vascular walls in the aorta-gonad-mesonephros (AGM) region, the fetal liver, and fetal bone marrow (52). Cells capable of giving rise to hematopoietic and endothelial progeny may also exist in the adult. Clonogenic in vitro assays have identified a population of hemangioblasts in adult human bone marrow (53) and, in the mouse, single bone marrow hematopoietic stem cells (HSCs) have been shown to generate multiple hematopoietic lineages, together with retinal endothelium (34).
The scl gene (also known as TAL-1) encodes a basic helix-loop-helix protein and is normally expressed in hematopoietic cells, in endothelium, and within specific regions of the central nervous system, a pattern of expression that is highly conserved across vertebrate species from mammals to teleost fish (reviewed in reference 7). Within the blood and endothelial system, scl is expressed in hemangioblasts, HSCs, a subset of hematopoietic lineages, and at lower levels in angioblasts and mature endothelial cells (1, 19, 57; see also reference 7 and references therein). Targeted mutation of the scl gene has shown that it is essential for the formation of HSCs during development (55, 56) and for the remodeling of primary yolk sac vascular networks (66), although scl−/− mouse embryos and ES cells both generate endothelial cells (57, 66). Conversely, dysregulated expression of scl during zebra fish development or ES cell differentiation resulted in excessive formation of hemangioblasts and endothelial and blood cells at the expense of other mesodermal lineages (22, 29). Taken together, these data indicate that scl is required for lineage commitment to blood cell formation and suggest the existence of an scl-independent route for endothelial development. Interestingly, expression of scl in adult HSCs may not be necessary for long-term repopulating activity but remains essential for normal megakaryocytic and erythroid differentiation (36, 46).
Current evidence therefore demonstrates that scl plays a pivotal role in the normal development of both blood and endothelium. This focuses attention on the mechanisms whereby transcription of scl itself is initiated and maintained, and we have therefore undertaken a systematic analysis of the transcriptional regulation of the murine scl locus. Studies of chromatin structure, long-range comparative sequence analysis, and transgenic reporter assays have identified five independent enhancers, each of which targets expression to a specific subdomain of the normal scl expression pattern (31, 32, 59, 62). Of particular note, a 3′ element (+18/19 enhancer) was shown to be active in endothelial cells and hematopoietic progenitors at multiple sites and times during ontogeny (59). This enhancer directed expression to the vast majority of long-term reopulating HSCs from adult bone marrow and fetal liver and also to hemangioblasts in frog embryos (33, 60). Biochemical and transgenic analyses defined a 640-bp core enhancer fragment and demonstrated that a novel multiprotein complex containing Ets and GATA transcription factors was essential for activity of this enhancer in blood and endothelial cells (33).
Here we demonstrate that, although sufficient to target expression to hemangioblasts and hematopoietic progenitors, the +18/19 enhancer is not necessary within the endogenous scl locus for blood cell formation or scl transcription. An scl 5′ enhancer is identified, which targets expression to hematopoietic progenitors and endothelial cells in vitro and in vivo and which is regulated by Fli-1 and Elf-1.
MATERIALS AND METHODS
Homologous recombination in ES cells.
ES cells ESF48/1 (a gift from R. Gardner, Oxford, United Kingdom) were grown on mouse embryonic fibroblast feeder cells according to standard procedures. The construct for homologous recombination was generated by inserting 5′ and 3′ homology arms (1.6-kb KpnI/HindIII fragment and 5.5-kb PCR fragment with engineered NotI and XbaI sites) on either side of a loxed pgk-promoter-hygromycin selection cassette. Homologous recombination events (1% on average) were identified by Southern blot analysis. Removal of the hygromycin selection cassette was achieved by transient transfection with a Cre recombinase expression plasmid, followed by selection for hygromycin sensitivity (on average 5% of the clones picked). Cre-mediated excision was subsequently verified by both PCR and Southern blot analysis.
In vitro differentiation and hematopoietic colony assays of ES cells.
Formation of embryoid bodies was induced after leukemia inhibitory factor withdrawal according to standard procedures. After 10 days, embryoid bodies were scored for hematopoietic activity based on the presence of hemoglobinized cells. After dissociation, cells from embryoid bodies were subjected to hematopoietic colony assays by using Methocult medium from Stem Cell Technologies according to the manufacturer's instructions. After 7 to 10 days, colonies were counted and scored based on morphology and the presence of hemoglobinized cells into erythroid, myeloid, and mixed colony types. Individual colonies were picked and cells analyzed by cytospin/May-Grunwald-Giemsa staining to further characterize hematopoietic cells.
Generation and analysis of ES cell chimeras.
ES cells were injected into C57BL/6 blastocysts and transferred into C57BL/6 recipients according to standard procedures. Chimeric mice were identified based on brown coat color. The contribution of ES cell-derived cells to hematopoietic tissues was examined by fluorescence-activated cell sorting (FACS) analysis of bone marrow, spleen, and thymus with the fluorescein isothiocyanate-conjugated Ly9.1 antibody (Pharmingen), which does not recognize C57BL/6-derived cells. The contribution to hematopoietic cells was confirmed by costaining with antibodies against phycoerythrin-conjugated B220, Mac1, CD4, and CD8 (all antibodies were from Pharmingen).
Transgenic analysis, restriction endonuclease accessibility, reporter assays, and EMSA analysis.
LacZ reporter plasmids were generated according to standard procedures (details available on request). F0 transgenic mouse embryos were prepared and analyzed as described previously (59). FACS analysis and hematopoietic colony assays were performed on embryonic day 11.5 (E11.5) fetal liver cells as described previously (59). The restriction endonuclease assay was performed as described previously (32) by using ApaI digestion and a 300-bp HindIII/ApaI fragment immediately upstream of exon 1a as the hybridization probe. Luciferase reporter constructs were generated according to standard procedures (details available on request). Mutant constructs were made as described previously (33) and verified by sequencing. 416B and MEL cells were grown as described previously (9). Transfections and luciferase assays were performed as described previously (32). Nuclear extracts for EMSA analysis were prepared as described earlier (33). Oligonucleotides used in EMSAs are shown in Fig. 6. Chromatin immunoprecipitation was also performed as described previously (33). Immunoprecipitates were analyzed by semiquantitative PCR with oligonucleotide primers flanking the −3.8 core enhancer region (FW [TGTCCCCTGCTCTTGCCTAC] and REV [ATGTCTGGGGGCAGGTTAGTTA]). PCRs were analyzed by Southern blotting with a Hewlett-Packard phosphorimager for quantification of hybridization signals.
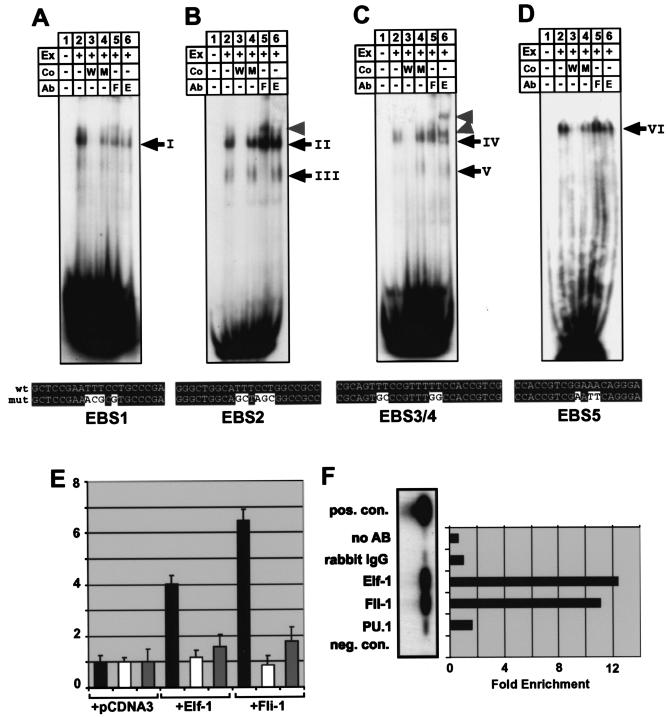
FIG. 6.
Fli-1 and Elf-1 can activate the −3.8 core enhancer and bind to it in vitro and in vivo. (A to D) Electrophoretic mobility shift assays for oligonucleotides EBS1 (A), EBS2 (B), EBS3/4 (C), and EBS5 (D) containing the Ets consensus-binding sites indicated in Fig. 5. Ex, nuclear extract; Co, competitor oligo; Ab, supershift antibody; W, wild type; M, mutant; F, Fli-1; E, Elf-1. The sequences of the wild-type oligonucleotides and the respective mutant oligonucleotides are shown underneath each panel. wt, wild-type oligonucleotide; mut, mutant oligonucleotide. (A) One major complex (complex I) was bound by the EBS1 oligonucleotide (compare lanes 1 and 2). Complex I binding was competed for by excess wild-type oligonucleotide and, to a lesser extent, by mutant oligonucleotide (compare lanes 3 and 4). Complex I was not supershifted with antibodies against Fli-1 or Elf-1 (see lanes 5 and 6). (B) Two major complexes (complexes II and III) were bound by the EBS2 oligonucleotide (compare lanes 1 and 2). Both complexes were competed by excess wild-type, but not mutant oligonucleotide (compare lanes 3 and 4). Complex II was not supershifted with antibodies against Fli-1 or Elf-1, whereas complex III was supershifted with antibody to Fli-1 but not Elf-1 (see lanes 5 and 6 and arrowhead). (C) One major complex (complex IV) was bound by the EBS3/4 oligonucleotide (compare lanes 1 and 2). Complex IV binding was competed by excess wild-type but not mutant oligonucleotide (compare lanes 3 and 4). Complex IV was supershifted with Elf-1 antibody (see lanes 5 and 6 and arrowheads). An additional complex (V) of higher mobility could be seen after prolonged exposure (see lanes 4 and 6) and was supershifted with antibodies against Fli-1 but not Elf-1 (compare lanes 5 and 6). (D) One major complex (complex VI) was bound by the EBS5 oligonucleotide (compare lanes 1 and 2). Complex VI was competed for by excess wild-type but not mutant oligonucleotide (compare lanes 3 and 4) and was not supershifted with antibodies against Fli-1 or Elf-1. (E) Transactivation of the −3.8 core enhancer in MEL cells. Black bars represent the fold activation of the core enhancer when coelectroporated with expression plasmids for Elf-1 and Fli-1 relative to the empty expression vector (pcDNA3). No transactivation was seen with the enhancerless TK minimal promoter luciferase control plasmid (white bars), and only marginal transactivation was seen with a −3.8 enhancer construct with the five Ets sites EBS1 to -5 mutated (gray bars). (F) Chromatin immunoprecipitation of the −3.8 enhancer in 416B cells. Semiquantitative PCR of immunoprecipitates, followed by Southern blotting, showed enrichment for Elf-1 and Fli-1 but not for PU.1. The positive control consists of cross-linked and sheared genomic DNA removed before immunoprecipitation. Immunoprecipitates obtained by using rabbit IgG or no antibody served as negative controls. The enrichment was calculated as the ratio of band intensities to the IgG control band.
RESULTS
The scl 3′ stem cell enhancer is dispensable for blood cell formation.
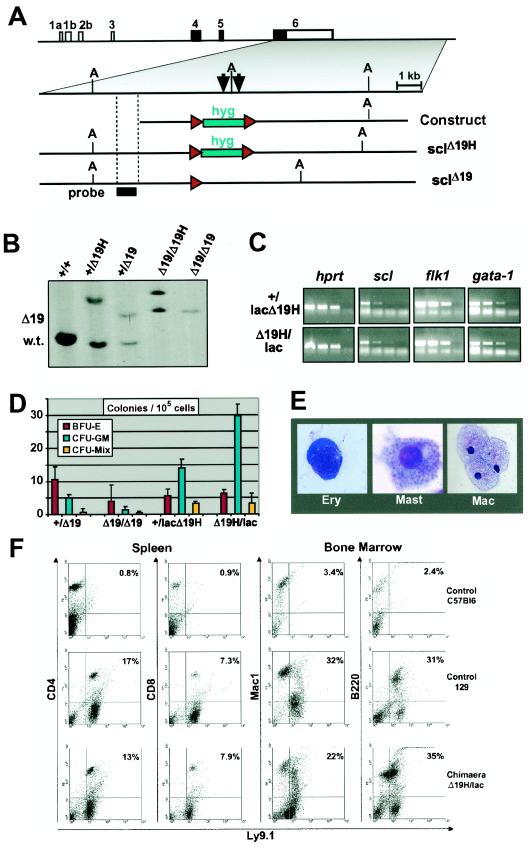
We have previously identified an scl 3′ stem cell enhancer with striking properties. A 5.5-kb fragment, containing two conserved regions of open chromatin (+18 and +19 elements), was sufficient to direct reporter gene expression to hemangioblasts, hematopoietic stem and progenitor cells, and endothelium (33, 59, 60). When used to drive expression of an scl cDNA, the same fragment was also sufficient to rescue hematopoietic progenitor formation in scl−/− mice (60). In order to assess the function of the stem cell enhancer within the context of the endogenous scl locus, homologous recombination was used to delete the enhancer in ES cells (Fig. 1A and B). A 2.5-kb region containing both the +18 and the +19 elements was replaced by a loxed hygromycin cassette to generate scl+/HΔ19 ES cells. Removal of the hygromycin cassette by using Cre recombinase produced scl+/Δ19 ES cells. ES cells lacking the enhancer in both alleles were generated by a second round of homologous recombination and removal of the hygromycin cassette.
FIG.1.
The scl 3′ enhancer is not required for blood formation from ES cells. (A) Strategy for deleting the mouse scl 3′ stem cell enhancer by homologous recombination in ES cells. Shown are the structure of the mouse scl locus, together with the enhancer knockout construct and the two knockout alleles (sclHΔ19 and sclΔ19) generated in the present study. Arrows indicate previously mapped DNase I-hypersensitive sites +18 and +19. (A) ApaI restriction site; hyg, hygromycin selection cassette; red arrowheads, loxP sites; probe, position of probe used for Southern analysis shown in panel B. (B) Southern blot analysis of targeted ES cell lines. DNA was digested with ApaI and analyzed by using the probe shown in panel A. Bands distinguishing the three alleles are indicated as wt (5.2 kb), HΔ19 (9.2 kb), or Δ19 (6.9 kb). (C) Levels of expression of scl, FLK1, and GATA-1 are not significantly different between embryoid bodies derived from scl+/lacHΔ19 and sclHΔ19/lac ES cells. Total RNA extracted from day 7 embryoid bodies with the indicated genotypes was reverse transcribed, and the resultant cDNA was analyzed by semiquantitative PCR. Shown are three fivefold serial dilutions and a negative control for each primer pair. (D) ES cells lacking the 18/19 enhancer can give rise to hematopoietic colonies in vitro. Cells derived from day 10 embryoid bodies of the indicated genotypes were used to perform hematopoietic colony assays. Red bars, BFU-E; turquoise bars, CFU-GM; yellow bars, CFU-Mix. The data shown are from a representative experiment. Similar results were obtained in three independent experiments. (E) Morphology of hematopoietic cells in colonies derived from sclΔ19/Δ19 ES cells (May-Grunwald-Giemsa stain). (F) sclHΔ19/lac ES cells can give rise to blood cells in vivo. FACS analysis of spleen and bone marrow cells from chimeras generated by using sclHΔ19/lac ES cells, together with control C57BL/6 and 129 mice (negative and positive controls, respectively). Ly9.1 is expressed by ES cell-derived progeny but not by host C57BL/6 cells.
Previous studies have demonstrated that scl−/− ES cells are unable to produce hematopoietic cells after in vitro differentiation (21, 55, 57). In contrast, scl+/Δ19 and sclΔ19/Δ19 ES cells both generated hemoglobinized embryoid bodies, which expressed scl mRNA (data not shown). In order to confirm the above results in a second independent ES cell line, we took advantage of scl+/lac ES cells in which one scl allele has been disrupted by insertion of lacZ (20). Using the strategy described above, the +18/19 enhancer was deleted from both alleles to generate scl+/lacHΔ19 and sclHΔ19/lac ES cells. We then performed semiquantitative reverse transcription-PCR (RT-PCR) on RNA isolated from day 7 embryoid bodies, which demonstrated that scl expression levels were not significantly different for those two genotypes (Fig. 1C). In addition to the positive hprt control, we included flk-1 and gata-1 in this analysis. During ES cell differentiation, flk-1 expression was previously shown not to be affected by deleting scl, whereas gata-1 levels were drastically reduced (21). Deletion of the scl +19 enhancer did not change expression of either flk-1 or gata-1, a finding consistent with our observation that there was no significant change in scl expression levels.
Hematopoietic progenitor assays demonstrated that scl+/Δ19 and sclΔ19/Δ19 embryoid bodies contained erythroid, myeloid, and multipotent progenitors, as assessed by colony assays (Fig. 1D) and morphology of cells from individual colonies (Fig. 1E). The numbers of colonies generated from sclΔ19/Δ19 embryoid bodies were slightly reduced compared to scl+/Δ19 embryoid bodies, even though we failed to notice qualitative differences in colonies generated from these two genotypes or indeed scl+/+ ES cells. Since the sclΔ19/Δ19 ES cells had undergone additional rounds of targeting and selection, which may have resulted in a reduced capacity to differentiate, we analyzed colony-forming ability in independently targeted scl+/lacHΔ19 and sclHΔ19/lac ES cells. The latter did not produce fewer BFU-E, CFU-GM, or CFU-Mix (Fig. 1D), suggesting that, at least under the assay conditions (i.e., saturating levels of cytokines), there appears to be no quantitative difference in colony-forming activity. These results demonstrate that the scl stem cell enhancer is not required for blood cell formation in vitro.
The hematopoietic potential of ES cells lacking the 3′ stem cell enhancer was then assessed in vivo. Wild-type and mutant (sclHΔ19/lac or sclHΔ19/Δ19) ES cells were injected into C57BL/6 blastocysts. Given that ES cell derived cells express the marker Ly9.1 which is not expressed by C57BL/6 cells, the resultant chimeras were analyzed for the presence of ES cell derived Ly9.1+ hematopoietic cells (Fig. 1F). Four chimeras were generated (three sclHΔ19/lac and one sclHΔ19/Δ19), with degrees of chimerism ranging from 50 to 100%, as estimated by coat color. As exemplified in Fig. 1F, spleen and bone marrow from all four chimeras contained readily identifiable Ly9.1+ hematopoietic cells, which included B cells (B220+), myeloid cells (Mac1+), and T cells (CD4+ or CD8+).
Taken together, these results demonstrate that the scl +18/19 enhancer is not required for blood cell formation in vitro or in vivo and suggested that the scl locus contained an additional regulatory element capable of driving scl expression during hematopoietic development.
The mouse scl 5′ flanking region directs expression to hematopoietic progenitor cells.
Previous analysis of transgenic mouse embryos had not identified any enhancers with significant hematopoietic activity apart from the 3′ stem cell enhancer. However, a 6.3-kb fragment from the 5′ region of the mouse scl gene directed expression to the vast majority of embryonic endothelial cells, together with rare round cells in the fetal liver (62). Moreover, the same fragment had also been shown to contain three regions of open chromatin in murine myeloid cell lines (24, 32), suggesting that the rare round fetal liver cells may be of hematopoietic origin. We therefore investigated whether the scl 5′ region contained an enhancer that might ensure scl expression during hematopoietic differentiation of sclΔ19/Δ19 ES cells.
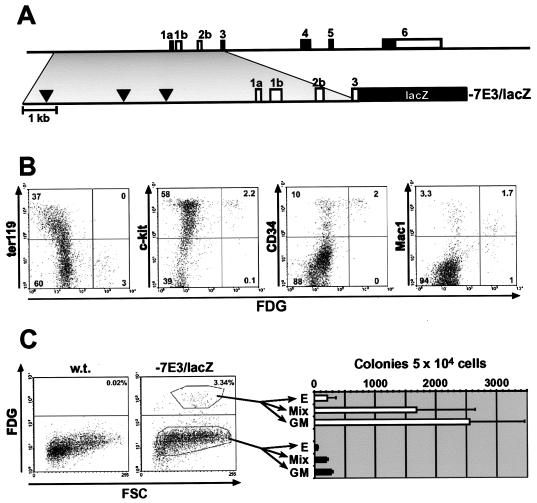
To this end, we utilized transgenic mice containing the −7E3/lacZ transgene (62), which consists of lacZ driven by a genomic fragment stretching from 7 kb upstream of exon 1a to exon 3 and includes the three regions of open chromatin identified in myeloid cell lines (Fig. 2A). In the two mouse lines analyzed, a small minority (1 to 4%) of E11.5 fetal liver cells expressed the lacZ transgene. These cells did not express the erythroid marker Ter119 but were positive for the hematopoietic progenitor markers c-kit and CD34 (Fig. 2B). A proportion of the enhancer positive cells was also positive for Mac1, which in fetal liver marks hematopoietic progenitors, as well as more mature myeloid cells. Colony assays confirmed that the lacZ-expressing fraction was indeed highly enriched for hematopoietic progenitors (Fig. 2C). These results demonstrate that the −7E3/lacZ construct was capable of directing expression to hematopoietic progenitors, as well as to endothelial cells. The endothelial enhancer activity was previously shown to be present within a 6.3-kb fragment extending from −1 to −7 kb upstream of exon 1a (62). Since the scl promoter region (from 1 kb upstream of exon 1a to exon 3) has been shown to have neither hematopoietic nor endothelial activity in transgenic mice (59, 62), our data suggested that both hematopoietic and endothelial activity were located between 1 and 7 kb upstream of exon 1a.
FIG. 2.
The mouse scl 5′ flanking region directs expression to hematopoietic progenitor cells. (A) Diagram of mouse scl locus indicating the position of the 5′ fragment present in the −7E3/lacZ transgene. Black arrowheads indicate the positions of DNase I-hypersensitive sites previously mapped in myeloid cell lines. (B) Characterization of hematopoietic cells targeted by the 5′ enhancer. FACS analysis of E11.5 fetal liver demonstrates enhancer activity in a subset of c-kit+ cells, CD34+ cells, and Mac1+ cells, markers previously shown to be expressed by fetal liver hematopoietic stem/progenitor cells. (C) E11.5 fetal liver cells expressing the −7E3/lacZ transgene are enriched for hematopoietic progenitors. lacZ-positive and lacZ-negative cells from mice carrying the −7E3/lacZ transgene were sorted by FACS and assessed for hematopoietic colony-forming activity. FDG, fluorescein di-β-d-galactopyranoside fluorescent β-galactosidase/lacZ substrate; FSC, forward scatter; E, burst-forming units; GM, granulocyte/macrophage colonies; Mix, multipotent colonies.
A core enhancer located at −3.8 kb is necessary and sufficient for hematopoietic and endothelial expression.
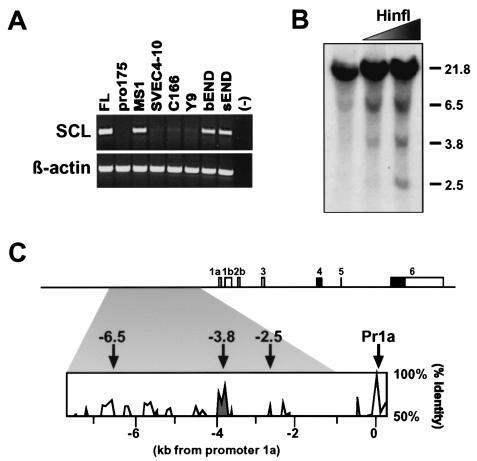
In order to identify the likely location of scl 5′ endothelial and/or hematopoietic core enhancers, the chromatin structure of the scl 5′ flanking region was investigated. Previous analysis of hematopoietic cell lines had revealed three regions of open chromatin at kb −2.5, −3.8, and −6.5 upstream of exon 1a (24, 32), but endothelial cells had not been studied. A survey of seven endothelial cell lines identified three (MS1, bEND, and sEND), which expressed readily detectable levels of scl mRNA by reverse transcription-PCR (RT-PCR) (Fig. 3A). Regions of open chromatin were mapped in the MS1 and sEND endothelial cell lines by using a restriction endonuclease accessibility assay. Three regions of accessible chromatin were identified (Fig. 3B; data not shown), all of which coincided with the regions previously mapped in murine hematopoietic cell lines (24). The positions of the accessible restriction endonuclease sites were identified in the mouse sequence and compared to a human/mouse homology profile. As shown in Fig. 3C, the region at −3.8 displayed the highest level of sequence conservation and thus stood out as a prime candidate for the location of an endothelial and/or hematopoietic enhancer.
FIG. 3.
Chromatin accessibility and sequence homology identify a candidate scl 5′ enhancer in endothelial cell lines. (A) RT-PCR analysis demonstrates that three of seven mouse endothelial cell lines (MS1, bEND, and sEND) express readily detectable levels of scl. FL, E14 fetal liver-positive control; (−), water-negative control. (B) Restriction endonuclease accessibility assay identifies three regions of accessible chromatin at kb −2.5, −3.8, and −6.5 upstream of mouse scl exon 1a. (C) Diagram of murine scl locus indicating 5′ regions of open chromatin (arrows) and the 6.3-kb fragment with endothelial activity in transgenic mice (shaded area). The homology profile of a mouse/human sequence alignment demonstrates that the kb −3.8 region exhibits the highest level of sequence conservation within the 6.3-kb fragment. Pr1a, promoter 1a.
Analysis of the scl 5′ flanking region by using bioinformatic tools, including cpgplot (http://www.emboss.org/) and promoter prediction software such as TSSG (63) showed that the −3.8 region was G/C-rich and raised the possibility that it represented an additional scl promoter. To address this issue, the 6.3-kb fragment was inverted to generate the −1-7/SV/lac construct. All three transgenic F0 embryos generated with this construct showed strong endothelial lacZ expression (Fig. 4A and B and Table 1). These results demonstrate that the activity of the 6.3-kb fragment was orientation independent and that endothelial expression did not reflect the presence of a previously unrecognized scl promoter.
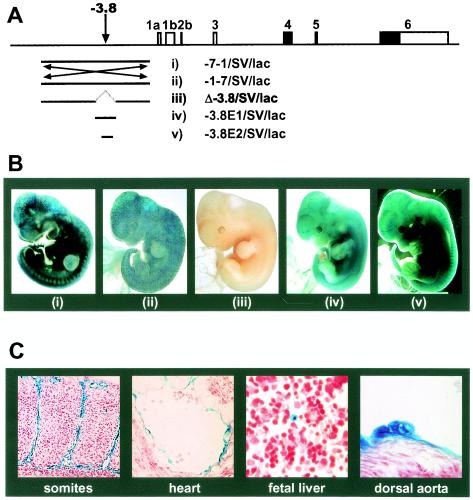
FIG. 4.
The −3.8 region is both necessary and sufficient for endothelial and hematopoietic activity in transgenic mouse embryos. (A) Transgenic reporter constructs in relation to the mouse scl locus. (B) Panels i to v show representative E11.5 embryos transgenic for the constructs indicated in panel A. (C) Sections of embryo shown in panel Bv demonstrating specific expression in endothelial cells, endocardium, round cells in the fetal liver, and presumed hematopoietic clusters on the ventral wall of the dorsal aorta.
TABLE 1.
Activity of scl 5′ enhancer constructs in transgenic mouse embryosa
| Construct | No. of F0 transgenic embryos
|
|||
|---|---|---|---|---|
| Total | Staining in endothelium | Ectopic staining only | Not staining | |
| −7-1/SV/lac | 6 | 5 | 1 | 0 |
| −1-7/SV/lac | 3 | 3 | 0 | 0 |
| Δ-3.8/SV/lac | 7 | 0 | 6b | 1 |
| −3.8E1/SV/lac | 7 | 4 | 1 | 2 |
| −3.8E2/SV/lac | 3 | 3 | 0 | 0 |
The lacZ reporter constructs described in Fig. 2 were used to generate transgenic embryos. F0 embryos were collected at E10.5 to E11.5 and stained overnight with X-Gal to determine lacZ activity.
Staining was weak, punctate, and mostly confined to the head. Analysis of multiple histological sections identified no β-galactosidase-positive endothelial cells.
We next deleted a 797-bp fragment encompassing the −3.8 region from the original 6.3-kb fragment and assessed the activity of the resultant construct (Δ-3.8/SV/lac) in F0 transgenic embryos (Fig. 4A and B). Of seven transgenic embryos, none showed endothelial expression by whole-mount analysis (Fig. iii) or by histological analysis of multiple tissues (Table 1). Reporter constructs containing 797- and 394-bp core enhancer fragments (−3.8E1/SV/luc and −3.8E2/SV/luc) were then used to generate additional transgenic mice. Four of seven and three of three F0 embryos showed endothelial expression with the 797- and 394-bp constructs, respectively (Fig. 4A and B; Table 1).
Analysis of histological sections from E11.5 F0 transgenic embryos carrying the 394-bp core enhancer construct confirmed that the extensive staining seen in whole-mount analyses reflected widespread endothelial expression in multiple tissues, including skin, brain, somites, heart, fetal liver, dorsal aorta, and yolk sac (Fig. 4C; data not shown). Two populations of nonendothelial cells were also found to express lacZ reproducibly in multiple embryos: first, rare round cells (presumably hematopoietic) in the fetal liver and, second, clusters of round cells attached to the ventral floor of the dorsal aorta (Fig. 4C). Similar clusters of cells in the dorsal aorta are thought to represent emerging HSCs (15, 52, 70). Taken together, our data therefore demonstrate that a 394-bp region is both necessary and sufficient for enhancer activity within the 6.3-kb fragment. Moreover, our results suggest that the 394-bp region contains an enhancer capable of targeting expression to both endothelium and hematopoietic progenitors during embryonic development.
Conserved Ets sites are critical for the activity of the −3.8 core enhancer in endothelial and hematopoietic cells.
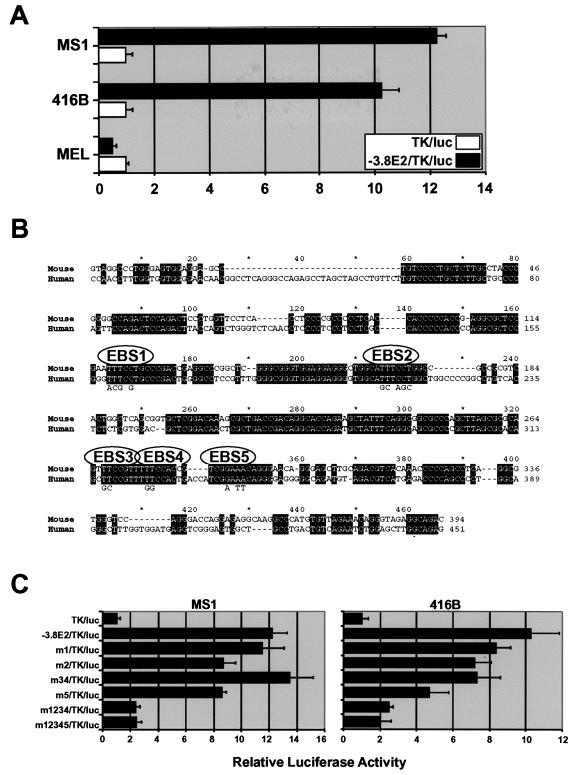
In order to perform biochemical analysis of the 5′ core enhancer, it was important to develop cellular assays for enhancer function. Luciferase reporter constructs were therefore introduced into the MS1 (endothelial), 416B (hematopoietic progenitor), and MEL (erythroid) cell lines. All three cell lines express scl, but the chromatin structure of the −3.8 region is nuclease sensitive only in MS1 and 416B cells (Fig. 3; data not shown) and not in MEL cells (32). The simian virus 40 minimal promoter itself was highly active in MS1 cells, possibly due to the fact that MS1 cells were generated by transformation of mouse pancreatic islet endothelial cells using simian virus 40 large T antigen (3). We therefore generated alternative luciferase reporter constructs with the thymidine kinase minimal promoter, which had low baseline activity in MS1, and used these in transient reporter assays. The 394-bp core enhancer was strongly active in MS1 and 416B but not in MEL cells (Fig. 5A). These results show that the 394-bp core enhancer exhibited lineage-restricted activity, which correlated with the chromatin structure of the endogenous enhancer. The data are also consistent with the in vivo activity of the core enhancer in endothelial and hematopoietic progenitor cells but not in fetal liver erythroid cells.
FIG. 5.
Activity of the −3.8 core enhancer requires conserved Ets transcription factor-binding sites. (A) Lineage-specific activity of the −3.8 core enhancer in endothelial and hematopoietic cell lines. Transient-transfection assays with luciferase reporter constructs show that the −3.8 core region has strong enhancer activity in MS1 (endothelial) and 416B (hematopoietic progenitor) but not MEL cells (erythroid). (B) Mouse/human sequence alignment of the −3.8 core enhancer. EBS1 to -5 indicate the positions of the four conserved TTCC/GGAA Ets binding sites. (C) Reporter assays of wild-type and mutant core enhancer constructs in MS1 and 416B cells. m1 to m5 indicate mutations in sites EBS1 to -5.
Human/mouse sequence comparisons revealed several blocks of homology within the 400-bp core enhancer, which contained five conserved Ets family binding sites (EBS1 to -5, Fig. 5B). All five Ets sites (TTCC or GGAA, respectively) conformed precisely to the consensus binding site recognized by most members of the Ets family. Mutations were generated in each region, and their effect on enhancer activity was quantified by using transfection assays in MS1 and 416B cells (Fig. 5C). In both cell types, mutation of EBS1 alone (m1) or EBS3 and EBS4 together (m34) had no effect on activity of the core enhancer, and mutation of EBS2 and EBS5 alone (m2, m5) produced a 30 to 50% reduction of activity (Fig. 5C). In contrast, mutation of the first four Ets sites (m1234) or all five Ets sites (m12345) resulted in marked impairment of enhancer function in both MS1 and 416B cells. Given that transient assays do not always give a true representation of enhancer activity after chromosomal integration, we also performed stable transfection assays. These assays confirmed that the wild-type enhancer was highly active and that this activity was greatly diminished when all Ets sites were mutated (data not shown). Our data demonstrate a degree of redundancy between the five Ets motifs and identify a critical role for Ets factors in regulating the activity of the −3.8 core enhancer in both endothelial and hematopoietic cells.
Fli-1 and Elf-1 bind to Ets sites in the −3.8 core enhancer.
An RT-PCR expression survey of 20 Ets family members was performed to assess which were expressed in MS1 and 416B cells. MS1 and 416B expressed 15 and 10 Ets family members, respectively (data not shown). Interestingly, both cell lines coexpressed Fli-1 and Elf-1, the two Ets family members previously shown to be part of a protein complex regulating the scl +19 enhancer in hematopoietic progenitors cells (33). Moreover, the pattern of expression of Fli-1 and Elf-1 during embryogenesis is consistent with a role for both in the regulation of scl transcription within endothelial, as well as hematopoietic progenitor cells (5, 10, 17, 37).
To investigate whether Fli-1 and Elf-1 were capable of binding to the core enhancer, gel shift assays were performed with nuclear extracts from both MS1 and 416B cells. Four oligonucleotides containing the conserved sequence blocks surrounding the EBS1, EBS2, EBS3/4, and EBS5 motifs were synthesized, together with corresponding competitor oligonucleotides containing the same mutations that were used for the functional assays described in the previous section. The same pattern of band shifts was obtained with nuclear extracts prepared from MS1 and 416B cells (Fig. 6A to D; data not shown). The EBS1 oligonucleotide was bound by one complex (complex I, Fig. 6A; data not shown), which could be competed by a wild type and to a lesser extent also by a mutant oligonucleotide (Fig. 6A, compare lanes 2, 3, and 4). Using specific antibodies against Ets-1, Fli-1, and Elf-1, we assessed whether complex I contained any of these factors. However, these three antibodies neither competed for binding of complex I to the EBS1 oligonucleotide, nor did they generate a supershift (Fig. 6A, lanes 5 and 6; data not shown). These results suggest that complex I does not contain either Fli-1 or Elf-1 and, in view of the competition data, may not bind with high specificity to the EBS1 motif.
The EBS2 oligonucleotide was bound by two specific complexes (complexes II and III, Fig. 6B), which could be competed for by wild-type but not by mutant oligonucleotide (Fig. 6B, compare lanes 2, 3, and 4). Complex III was supershifted by using anti-Fli-1 antibody (Fig. 6B, lane 5), whereas neither anti-Ets-1 nor anti-Elf-1 antibodies affected binding of complex II (Fig. 6B, lane 6, and data not shown). These results demonstrate that Fli-1 and an as-yet-unidentified Ets factor can bind to the EBS2 site.
The EBS3/4 oligonucleotide was bound by one major specific complex (complex IV, Fig. 6C), which could be competed by wild-type but not mutant oligonucleotide (Fig. 6C, compare lanes 2, 3, and 4). Complex IV was supershifted by using anti-Elf-1 antibody giving rise to two complexes with slower mobility, possibly due to the fact that the presence of two Ets motifs allows for different stoichiometries of Elf-1/antibody complexes (Fig. 6C, lane 6). Prolonged exposure revealed an additional weaker band of faster mobility (complex V), which was supershifted by using anti-Fli-1 antibody (Fig. 6C, compare lanes 4, 5, and 6). These results demonstrate that Elf-1 and, to a lesser degree, Fli-1 are capable of binding to the EBS3/4 site in vitro.
The EBS5 oligonucleotide was bound by one major specific complex (complex VI, Fig. 6D), which could be competed for by wild-type but not by mutant oligonucleotide (Fig. 6D, compare lanes 2, 3, and 4). Using specific antibodies against Fli-1 and Elf-1, we assessed whether complex I contained any of these factors. However, these antibodies neither competed for binding of complex VI to the EBS5 oligonucleotide, nor did they generate a supershift (Fig. 6D, lanes 5 and 6). These results demonstrate that an as yet unidentified Ets factor can bind to the EBS5 site in vitro.
The scl −3.8 core enhancer is activated and bound in vivo by Fli-1 and Elf-1.
Two further experimental approaches were then adopted to confirm the importance of Fli-1 and Elf-1 for the activity of the −3.8 core enhancer. In order to assess the ability of Fli-1 and Elf-1 to activate the −3.8 enhancer, transactivation experiments were performed in MEL cells in which the core enhancer is inactive (Fig. 5C). Both Elf-1 and Fli-1 were able to increase the transcriptional activity of the −3.8 core enhancer by 4- and 6.5-fold, respectively, relative to the expression vector control (Fig. 6E). Importantly, neither Elf-1 nor Fli-1 were able to transactivate the enhancerless TK/luc control plasmid and only showed marginal transactivation of a −3.8 enhancer construct in which all five Ets sites had been mutated (Fig. 6E). Marginal levels of transactivation with the mutant enhancer may be mediated by binding of excess Elf-1/Fli-1 to nonconserved Ets motifs still present after mutation of the five conserved Ets sites. These data are consistent with direct transactivation of the −3.8 enhancer by Fli-1 and Elf-1 but do not demonstrate that these factors act directly on the core enhancer in cells where the enhancer is active.
To address this issue, chromatin immunoprecipitation studies were performed to investigate whether Fli-1 and Elf-1 are bound to the −3.8 core enhancer in 416B cells. Briefly, proteins bound to DNA were cross-linked by using formaldehyde and, after shearing by sonication, protein-DNA complexes were immunoprecipitated by using control antibodies and antibodies specific to Elf-1, Fli-1, and PU.1. The DNA content of immunoprecipitates was then analyzed by PCR. The region of the mouse scl 5′ core enhancer was clearly enriched in immunoprecipitates obtained with antibodies to Fli-1 and Elf-1 relative to immunoprecipitates obtained by using rabbit immunoglobulin G (IgG) or the no antibody controls (Fig. 6E). In contrast, no significant enrichment was observed after chromatin immunoprecipitation with PU.1 antibody, which thus served as an additional negative control. Moreover, scl exon 6, a region not involved in transcriptional regulation, was not enriched in immunoprecipitates obtained by using antibodies to Fli-1 or Elf-1 (33). These data therefore demonstrate that the −3.8 enhancer is bound by Fli-1 and Elf-1 in vivo.
DISCUSSION
The scl gene is essential for formation of all hematopoietic lineages (55, 56), and several lines of evidence suggest that its transcriptional activation is required for commitment of mesodermal progenitors to hematopoietic fates, including the generation of HSCs (23, 29, 57). Systematic analysis of the scl locus has identified a panel of cis-acting regulatory elements, including enhancers that target expression to endothelium, mid-brain, hind-brain, and spinal cord, all subdomains of the normal scl expression pattern (31, 32, 62). However, only a single element targeting hematopoietic cells had been identified. This element contains two prominent regions of open chromatin (+18 and +19 regions) and directs reporter gene expression in transgenic mice to endothelium, as well as to the vast majority of hematopoietic progenitors and long-term repopulating HSCs (59, 60). A 640-bp fragment containing the +19 region is necessary and sufficient for this pattern of expression (33). The +18 element is not necessary for the pattern of expression in vivo but functions to enhance activity of the +19 element in transfection experiments (B. Göttgens, unpublished observations). Here we demonstrate that within the endogenous stem cell leukemia (SCL) locus the +18 and +19 regions are not required for the formation of hematopoietic cells either in vitro or in vivo. A search for additional hematopoietic elements resulted in the identification and molecular characterization of a second bifunctional regulatory element (−3.8 enhancer) capable of targeting expression to hematopoietic progenitors and endothelium.
A second scl enhancer targets hematopoietic progenitors.
The −3.8 and +19 scl enhancers share several attributes. Both direct reporter gene expression to embryonic endothelium and to hematopoietic progenitors in transgenic mice (59, 60; the present study). Our results also demonstrate that the functional similarities exhibited by the two enhancers are reflected in their molecular control. Both enhancers contain functionally important Ets binding sites, which are bound by Fli-1 and Elf-1 in myeloid progenitor cell lines. The expression patterns and functions of Fli-1 and Elf-1, where known, are consistent with a role for these proteins in the regulation of scl expression during blood and endothelial development (17, 67). Chicken Fli-1 is expressed in endothelial cells and splanchnopleural mesoderm, including hematopoietic clusters attached to the wall of the dorsal aorta (43), and a lacZ knockin into the murine Fli-1 gene resulted in widespread endothelial lacZ expression, although expression in dorsal aorta clusters was not examined (37). Elf-1 was found to be expressed in developing chicken blood vessels (17) and was present in a subtracted cDNA library from highly purified murine fetal liver HSCs (54). Interestingly, the complete absence of primitive and definitive hematopoiesis that typifies the scl−/− phenotype is not reproduced by targeted mutation of either Fli-1 or Elf-1 (27, 37, 64). This is likely to reflect redundancy within the Ets family of transcription factors; in particular, Erg and Mef/Elf-4 are closely related in both sequence and expression pattern to Fli-1 and Elf-1, respectively (4, 47).
However, the −3.8 and +19 elements do display biological and biochemical differences. In transgenic mice, a 5.5-kb fragment containing the +18/19 enhancer targeted expression to 5 to 17% of E11.5 fetal liver cells (59) and was active in adult endothelium (L. Gambardella, unpublished observation). In contrast, a 6.3-kb fragment containing the −3.8 element directed expression to fewer (1 to 4%) E11.5 fetal liver cells (the present study) and a 10-kb fragment of the SCL locus (−7E3/lacZ construct) also containing the −3.8 element was inactive in adult endothelium (62). At a molecular level, the +19 enhancer contains a functionally important GATA-binding site (33), whereas no GATA sites are present in the −3.8 enhancer. Our data do not exclude the possibility that other transcription factors may serve to recruit GATA proteins to the 5′ enhancer without the need for a GATA binding site. A protein complex between the Ets factor PU.1 and GATA-1 was postulated to bind DNA via a single Ets binding site (72). Although this complex was inhibitory, recent data suggest that interactions between Fli-1 and GATA-1 are activating, although these latter studies did not investigate whether interaction could occur in the absence of a GATA-1 binding site (38, 69).
Mechanistically the scl −3.8 and +19 enhancers also exhibit some differences. The −3.8 enhancer functions in transient and stable transfections (the present study and data not shown), whereas the +19 enhancer only functions in stable transfections and thus displays a requirement for integration into chromatin (33). Moreover, several lines of evidence suggest that the +19 enhancer is activated by a multiprotein complex, which contains Ets and GATA proteins (33). Consistent with this model, mutation or altered spacing of individual Ets and GATA binding sites abolished enhancer activity. In contrast, mutations of individual Ets binding sites did not significantly affect activity of the −3.8 enhancer, thus demonstrating that individual Ets sites were dispensable for enhancer activity. Nonetheless, existing data are consistent with the concept that activity of both the −3.8 and +19 enhancers requires occupancy of a minimum of two Ets sites.
Notably, a construct carrying both the 5′ and 3′ scl enhancers showed similar activity to the 3′ enhancer alone when assayed in transgenic mice (30). Together with our current data, which demonstrate that the 5′ and 3′ enhancers have overlapping activities and that the 3′ enhancer is not required for SCL expression in blood progenitors, these results are consistent with partial redundancy between the 5′ and 3′ scl enhancers. Partial redundancy of regulatory elements is an emerging theme in the context of mammalian gene regulation. It has been noted less frequently in lower organisms such as Drosophila melanogaster, an observation that may reflect the fact that Drosophila mutagenesis relies largely on screening for abnormal phenotypes. There are a number of potential advantages to having partially redundant regulatory elements. The existence of redundant enhancers controlling a key physiological process means that loss or inactivation of a single element will not necessarily be detrimental. Furthermore, although such elements may be redundant in some aspects of their function, they may also exhibit unique attributes. Duplication of regulatory elements, and their subsequent partial specialization, are likely to represent important mechanisms by which organisms evolve additional levels of transcriptional control.
The β-globin locus contains several enhancer elements (collectively referred to as the β-globin LCR), which are distributed over a 20-kb region. The individual elements (HS 1 to 5) display functional differences in transfection and transgenic assays and together form a powerful composite erythroid enhancer. However, individual elements are dispensable for nearly normal expression and contribute additively to overall expression levels (reference 8 and references therein). In the mouse α-globin locus, deletion of the dominant erythroid enhancer (+26 element) also gives rise to a surprisingly mild phenotype, raising the possibility that additional α-globin enhancers remain to be characterized (2). Other examples of regulatory elements with partially redundant functions have been described at the TCRγ (71), MyoD (12), GATA-1 (45), and HoxD loci (6). Interestingly, whereas the individual β-globin enhancers contain overlapping sets of transcription factor binding sites, this is not the case for the MyoD and TCRγ genes. In both of these loci, two enhancers have been identified that lack overt sequence similarities and that display both redundant and unique functions during development. Of further note is the observation that some enhancers such as the β-globin HS2 and HS3 function in transgenic mice when assayed in conjunction with heterologous promoters (44), while others such as the GATA-1 erythroid enhancer require interaction with the endogenous GATA-1 promoter (51).
Our data show that the scl −3.8 and +19 enhancers share a subset of transcription factor binding sites, and both function in transgenic assays in conjunction with a heterologous promoter. Transfection and transgenic assays demonstrate functional differences between the two scl enhancers, but it is not yet clear whether the two elements perform unique functions within the endogenous scl locus.
Transcriptional regulation of blood and endothelial development.
Blood and endothelium have long been postulated to share a common origin and the concept of a common progenitor, the hemangioblast (48), has received support from experiments with avian embryos (18) and murine ES cells (13). In addition, there is mounting evidence for the existence of hematopoietic endothelium, differentiated endothelial cells which can give rise to hematopoietic cells, in the AGM region of avian (40), murine (50), and human (52) embryos. The close relationship between blood and endothelium may also continue in later stages of development and in the adult. The BCR-ABL fusion gene associated with chronic myeloid leukemia is reported to be present in endothelial cells, as well as hematopoietic cells (35), suggesting that this leukemia results from transformation of an adult cell capable of giving rise to both lineages. Moreover, endothelial cells from human fetal liver and bone marrow can generate hematopoietic progeny (52), and single cells from adult bone marrow can generate both hematopoietic and endothelial progeny in vivo (34) and in vitro (53).
We have now identified two scl enhancers (the −3.8 and +18/19 elements) that each target expression to endothelium and hematopoietic progenitors. In both cases endothelial and hematopoietic activities are inseparable and are contained within small core enhancer fragments. These data demonstrate that the close developmental and phenotypic relationship between blood progenitors and endothelium is reflected at a molecular level within the scl locus by the existence of individual bifunctional regulatory elements. Our data do not exclude the possibility that the enhancers are activated independently in endothelial and hematopoietic progenitors. However, several arguments suggest that that this is unlikely and that both enhancers are probably activated at the stage of a common precursor. First, both −3.8 and +18/19 enhancers are active in the extraembryonic mesoderm of E7.5 mouse embryos (59, 62). Second, in transgenic frog embryos, the +18/19 enhancer is active within dorsolateral plate mesoderm in progenitors, which are thought to give rise to both blood and endothelium (33). Third, targeted mutation of the scl gene results in enhanced formation of smooth muscle cells (22), an observation which would be consistent with scl function in a mesodermal progenitor, although a non-cell-intrinsic effect of SCL on smooth muscle differentiation has not been excluded. Additional circumstantial evidence that scl functions relatively early during mesoderm differentiation comes from studies which show that the rescue of hematopoiesis in scl−/− ES cells requires scl expression before the development of differentiated endothelial cells (23).
Endothelial cells and hematopoietic progenitors coexpress a large number of genes, and several genes originally thought to be endothelium specific (for example, see reference 25) have been found to be expressed in blood progenitors, with some emerging as useful markers for HSC purification protocols (11). Our results lead us to speculate that enhancers with dual hematopoietic-endothelial activity may represent a general strategy for regulating blood and endothelial development. Consistent with this concept, colocalization of hematopoietic and endothelial activity to the same enhancer region is a recurrent theme, as illustrated by the recent transgenic analysis of enhancers from the c-mpl, vav, and Ly-6 genes (15, 28, 42, 73). However, core enhancer regions have not been defined thus far for these genes (functional cassettes range from 2 to 14 kb). Consequently, bifunctional activity has not yet been narrowed down to a single small fragment, and molecular analysis has not been performed in both hematopoietic and endothelial lineages. Nonetheless, existing data lead us to hypothesize that dual hematopoietic-endothelial enhancers may be widespread and mark genes expressed in hemangioblasts and hematogenic endothelium, as well as in their respective progeny. This arrangement may represent an economical strategy for regulating the transcriptional programming necessary for transitions between hemangioblasts, endothelium, and hematopoietic progenitors.
Acknowledgments
This study was supported by the Leukemia Research Fund, the Wellcome Trust, the Kay Kendall Leukemia Fund, the Cambridge MIT Institute, the Medical Research Council (United Kingdom), the National Health and Medical Research Council (Canberra, Australia), and the British Heart Foundation.
We gratefully acknowledge the expert assistance of Andy Riddell with FACS and F. Köntgen (Ozgene, Perth, Australia) for generating the sclHΔ19/lac ES cells.
REFERENCES
- 1.Akashi, K., X. He, J. Chen, H. Iwasaki, C. Niu, B. Steenhard, J. Zhang, J. Haug, and L. Li. 2003. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood 101:383-389. [DOI] [PubMed] [Google Scholar]
- 2.Anguita, E., J. A. Sharpe, J. A. Sloane-Stanley, C. Tufarelli, D. R. Higgs, and W. G. Wood. 2002. Deletion of the mouse alpha-globin regulatory element (HS-26) has an unexpectedly mild phenotype. Blood 100:3450-3456. [DOI] [PubMed] [Google Scholar]
- 3.Arbiser, J. L., M. A. Moses, C. A. Fernandez, N. Ghiso, Y. Cao, N. Klauber, D. Frank, M. Brownlee, E. Flynn, S. Parangi, H. R. Byers, and J. Folkman. 1997. Oncogenic H-ras stimulates tumor angiogenesis by two distinct pathways. Proc. Natl. Acad. Sci. USA 94:861-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltzinger, M., A. M. Mager-Heckel, and P. Remy. 1999. Xl erg: expression pattern and overexpression during development plead for a role in endothelial cell differentiation. Dev. Dyn. 216:420-433. [DOI] [PubMed] [Google Scholar]
- 5.Bassuk, A. G., K. P. Barton, R. T. Anandappa, M. M. Lu, and J. M. Leiden. 1998. Expression pattern of the Ets-related transcription factor Elf-1. Mol. Med. 4:392-401. [PMC free article] [PubMed] [Google Scholar]
- 6.Beckers, J., and D. Duboule. 1998. Genetic analysis of a conserved sequence in the HoxD complex: regulatory redundancy or limitations of the transgenic approach? Dev. Dyn. 213:1-11. [DOI] [PubMed] [Google Scholar]
- 7.Begley, C. G., and A. R. Green. 1999. The SCL gene: from case report to critical hematopoietic regulator. Blood 93:2760-2770. [PubMed] [Google Scholar]
- 8.Bender, M. A., J. N. Roach, J. Halow, J. Close, R. Alami, E. E. Bouhassira, M. Groudine, and S. N. Fiering. 2001. Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNase I hypersensitive sites. Blood 98:2022-2027. [DOI] [PubMed] [Google Scholar]
- 9.Bockamp, E. O., F. McLaughlin, B. Gottgens, A. M. Murrell, A. G. Elefanty, and A. R. Green. 1997. Distinct mechanisms direct SCL/tal-1 expression in erythroid cells and CD34-positive primitive myeloid cells. J. Biol. Chem. 272:8781-8790. [DOI] [PubMed] [Google Scholar]
- 10.Brown, L. A., A. R. Rodaway, T. F. Schilling, T. Jowett, P. W. Ingham, R. K. Patient, and A. D. Sharrocks. 2000. Insights into early vasculogenesis revealed by expression of the ETS- domain transcription factor Fli-1 in wild-type and mutant zebrafish embryos. Mech. Dev. 90:237-252. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. Z., M. Li, D. de Graaf, S. Monti, B. Gottgens, M. J. Sanchez, E. S. Lander, T. R. Golub, A. R. Green, and H. F. Lodish. 2002. Identification of endoglin as a functional marker that defines long-term repopulating hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 99:15468-15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J. C., R. Ramachandran, and D. J. Goldhamer. 2002. Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Dev. Biol. 245:213-223. [DOI] [PubMed] [Google Scholar]
- 13.Choi, K., M. Kennedy, A. Kazarov, J. C. Papadimitriou, and G. Keller. 1998. A common precursor for hematopoietic and endothelial cells. Development 125:725-732. [DOI] [PubMed] [Google Scholar]
- 14.Ciau-Uitz, A., M. Walmsley, and R. Patient. 2000. Distinct origins of adult and embryonic blood in Xenopus. Cell 102:787-796. [DOI] [PubMed] [Google Scholar]
- 15.de Bruijn, M. F., X. Ma, C. Robin, K. Ottersbach, M. J. Sanchez, and E. Dzierzak. 2002. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity 16:673-683. [DOI] [PubMed] [Google Scholar]
- 16.Dieterlen-Lievre, F. 1975. On the origin of haemopoietic stem cells in the avian embryo: an experimental approach. J. Embryol. Exp. Morph. 33:607-619. [PubMed] [Google Scholar]
- 17.Dube, A., S. Thai, J. Gaspar, S. Rudders, T. A. Libermann, L. Iruela-Arispe, and P. Oettgen. 2001. Elf-1 is a transcriptional regulator of the Tie2 gene during vascular development. Circ. Res. 88:237-244. [DOI] [PubMed] [Google Scholar]
- 18.Eichmann, A., C. Corbel, V. Nataf, P. Vaigot, C. Bréant, and N. M. Le Douarin. 1997. Ligand-dependent development of the endothelial and hemopoietic lineages from embryonic mesodermal cells expressing vascular endothelial growth factor receptor 2. Proc. Natl. Acad. Sci. USA 94:5141-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elefanty, A. G., C. G. Begley, L. Hartley, B. Papaevangeliou, and L. Robb. 1999. SCL expression in the mouse embryo detected with a targeted lacZ reporter gene demonstrates its localization to hematopoietic, vascular, and neural tissues. Blood 94:3754-3763. [PubMed] [Google Scholar]
- 20.Elefanty, A. G., C. G. Begley, D. Metcalf, L. Barnett, F. Kontgen, and L. Robb. 1998. Characterization of hematopoietic progenitor cells that express the transcription factor SCL, using a lacZ “knock-in” strategy. Proc. Natl. Acad. Sci. USA 95:11897-11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elefanty, A. G., L. Robb, R. Birner, and C. G. Begley. 1997. Hematopoietic-specific genes are not induced during in vitro differentiation of scl-null embryonic stem cells. Blood 90:1435-1447. [PubMed] [Google Scholar]
- 22.Ema, M., P. Faloon, W. J. Zhang, M. Hirashima, T. Reid, W. L. Stanford, S. Orkin, K. Choi, and J. Rossant. 2003. Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 17:380-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endoh, M., M. Ogawa, S. Orkin, and S. Nishikawa. 2002. SCL/tal-1-dependent process determines a competence to select the definitive hematopoietic lineage prior to endothelial differentiation. EMBO J. 21:6700-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fordham, J. L., B. Gottgens, F. McLaughlin, and A. R. Green. 1999. Chromatin structure and transcriptional regulation of the stem cell leukaemia (SCL) gene in mast cells. Leukemia 13:750-759. [DOI] [PubMed] [Google Scholar]
- 25.Fraser, S. T., M. Ogawa, T. Yokomizo, Y. Ito, and S. Nishikawa. 2003. Putative intermediate precursor between hematogenic endothelial cells and blood cells in the developing embryo. Dev. Growth Differ. 45:63-75. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Porrero, J. A., I. E. Godin, and F. Dieterlen-Lievre. 1995. Potential intraembryonic hemogenic sites at pre-liver stages in the mouse. Anat. Embryol. 192:425-435. [DOI] [PubMed] [Google Scholar]
- 27.Garrett-Sinha, L. A., R. Dahl, S. Rao, K. P. Barton, and M. C. Simon. 2001. PU. 1 exhibits partial functional redundancy with Spi-B, but not with Ets-1 or Elf-1. Blood 97:2908-2912. [DOI] [PubMed] [Google Scholar]
- 28.Georgiades, P., S. Ogilvy, H. Duval, D. R. Licence, D. S. Charnock-Jones, S. K. Smith, and C. G. Print. 2002. vavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 34:251-256. [DOI] [PubMed] [Google Scholar]
- 29.Gering, M., A. R. Rodaway, B. Gottgens, R. K. Patient, and A. R. Green. 1998. The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 17:4029-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottgens, B., L. M. Barton, M. A. Chapman, A. M. Sinclair, B. Knudsen, D. Grafham, J. G. Gilbert, J. Rogers, D. R. Bentley, and A. R. Green. 2002. Transcriptional regulation of the stem cell leukemia gene (SCL): comparative analysis of five vertebrate SCL loci. Genome Res. 12:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottgens, B., L. M. Barton, J. G. Gilbert, A. J. Bench, M. J. Sanchez, S. Bahn, S. Mistry, D. Grafham, A. McMurray, M. Vaudin, E. Amaya, D. R. Bentley, A. R. Green, and A. M. Sinclair. 2000. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 18:181-186. [DOI] [PubMed] [Google Scholar]
- 32.Gottgens, B., F. McLaughlin, E. O. Bockamp, J. L. Fordham, C. G. Begley, K. Kosmopoulos, A. G. Elefanty, and A. R. Green. 1997. Transcription of the SCL gene in erythroid and CD34-positive primitive myeloid cells is controlled by a complex network of lineage-restricted chromatin-dependent and chromatin-independent regulatory elements. Oncogene 15:2419-2428. [DOI] [PubMed] [Google Scholar]
- 33.Gottgens, B., A. Nastos, S. Kinston, S. Piltz, E. C. Delabesse, M. Stanley, M. J. Sanchez, A. Ciau-Uitz, R. Patient, and A. R. Green. 2002. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21:3039-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grant, M. B., W. S. May, S. Caballero, G. A. Brown, S. M. Guthrie, R. N. Mames, B. J. Byrne, T. Vaught, P. E. Spoerri, A. B. Peck, and E. W. Scott. 2002. Adult hematopoietic stem cells provide functional hemangioblast activity during retinal neovascularization. Nat. Med. 8:607-612. [DOI] [PubMed] [Google Scholar]
- 35.Gunsilius, E., H. C. Duba, A. L. Petzer, C. M. Kahler, K. Grunewald, G. Stockhammer, C. Gabl, S. Dirnhofer, J. Clausen, and G. Gastl. 2000. Evidence from a leukaemia model for maintenance of vascular endothelium by bone-marrow-derived endothelial cells. Lancet 355:1688-1691. [DOI] [PubMed] [Google Scholar]
- 36.Hall, M. A., D. J. Curtis, D. Metcalf, A. G. Elefanty, K. Sourris, L. Robb, J. R. Gothert, S. M. Jane, and C. G. Begley. 2003. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc. Natl. Acad. Sci. USA 100:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart, A., F. Melet, P. Grossfeld, K. Chien, C. Jones, A. Tunnacliffe, R. Favier, and A. Bernstein. 2000. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity 13:167-177. [DOI] [PubMed] [Google Scholar]
- 38.Holmes, M. L., N. Bartle, M. Eisbacher, and B. H. Chong. 2002. Cloning and analysis of the thrombopoietin-induced megakaryocyte-specific glycoprotein VI promoter and its regulation by GATA-1, Fli-1, and Sp1. J. Biol. Chem. 277:48333-48341. [DOI] [PubMed] [Google Scholar]
- 39.Jaffredo, T., R. Gautier, V. Brajeul, and F. Dieterlen-Lievre. 2000. Tracing the progeny of the aortic hemangioblast in the avian embryo. Dev. Biol. 224:204-214. [DOI] [PubMed] [Google Scholar]
- 40.Jaffredo, T., R. Gautier, A. Eichmann, and F. Dieterlen-Lievre. 1998. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 125:4575-4583. [DOI] [PubMed] [Google Scholar]
- 41.Liao, W., B. W. Bisgrove, H. Sawyer, B. Hug, B. Bell, K. Peters, D. J. Grunwald, and D. Y. R. Stainier. 1997. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development 124:381-389. [DOI] [PubMed] [Google Scholar]
- 42.Ma, X., C. Robin, K. Ottersbach, and E. Dzierzak. 2002. The Ly-6A (Sca-1) GFP transgene is expressed in all adult mouse hematopoietic stem cells. Stem Cells 20:514-521. [DOI] [PubMed] [Google Scholar]
- 43.Mager, A. M., A. Grapin-Botton, K. Ladjali, D. Meyer, C. M. Wolff, P. Stiegler, M. A. Bonnin, and P. Remy. 1998. The avian fli gene is specifically expressed during embryogenesis in a subset of neural crest cells giving rise to mesenchyme. Int. J. Dev. Biol. 42:561-572. [PubMed] [Google Scholar]
- 44.Magram, J., K. Niederreither, and F. Costantini. 1989. Beta-globin enhancers target expression of a heterologous gene to erythroid tissues of transgenic mice. Mol. Cell. Biol. 9:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDevitt, M. A., R. A. Shivdasani, Y. Fujiwara, H. Yang, and S. H. Orkin. 1997. A “knockdown” mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 94:6781-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikkola, H. K., J. Klintman, H. Yang, H. Hock, T. M. Schlaeger, Y. Fujiwara, and S. H. Orkin. 2003. Hematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421:547-551. [DOI] [PubMed] [Google Scholar]
- 47.Miyazaki, Y., X. Sun, H. Uchida, J. Zhang, and S. Nimer. 1996. MEF, a novel transcription factor with an Elf-1 like DNA binding domain but distinct transcriptional activating properties. Oncogene 13:1721-1729. [PubMed] [Google Scholar]
- 48.Murray, P. D. F. 1932. The development in vitro of the blood of the early chick embryo. Proc. Roy Soc. B 111:497-521. [Google Scholar]
- 49.Nishikawa, S. I., S. Nishikawa, M. Hirashima, N. Matsuyoshi, and H. Kodama. 1998. Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Development 125:1747-1757. [DOI] [PubMed] [Google Scholar]
- 50.Nishikawa, S. I., S. Nishikawa, H. Kawamoto, H. Yoshida, M. Kizumoto, H. Kataoka, and Y. Katsura. 1998. In vitro generation of lymphohematopoietic cells from endothelial cells purified from murine embryos. Immunity 8:761-769. [DOI] [PubMed] [Google Scholar]
- 51.Nishimura, S., S. Takahashi, T. Kuroha, N. Suwabe, T. Nagasawa, C. Trainor, and M. Yamamoto. 2000. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol. Cell. Biol. 20:713-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oberlin, E., M. Tavian, I. Blazsek, and B. Peault. 2002. Blood-forming potential of vascular endothelium in the human embryo. Development 129:4147-4157. [DOI] [PubMed] [Google Scholar]
- 53.Pelosi, E., M. Valtieri, S. Coppola, R. Botta, M. Gabbianelli, V. Lulli, G. Marziali, B. Masella, R. Muller, C. Sgadari, U. Testa, G. Bonanno, and C. Peschle. 2002. Identification of the hemangioblast in postnatal life. Blood 100:3203-3208. [DOI] [PubMed] [Google Scholar]
- 54.Phillips, R. L., R. E. Ernst, B. Brunk, N. Ivanova, M. A. Mahan, J. K. Deanehan, K. A. Moore, G. C. Overton, and I. R. Lemischka. 2000. The genetic program of hematopoietic stem cells. Science 288:1635-1640. [DOI] [PubMed] [Google Scholar]
- 55.Porcher, C., W. Swat, K. Rockwell, Y. Fujiwara, F. W. Alt, and S. H. Orkin. 1996. The T-cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86:47-57. [DOI] [PubMed] [Google Scholar]
- 56.Robb, L., N. J. Elwood, A. G. Elefanty, F. Kontgen, R. Li, L. D. Barnett, and C. G. Begley. 1996. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 15:4123-4129. [PMC free article] [PubMed] [Google Scholar]
- 57.Robertson, S. M., M. Kennedy, J. M. Shannon, and G. Keller. 2000. A transitional stage in the commitment of mesoderm to hematopoiesis requiring the transcription factor SCL/tal-1. Development 127:2447-2459. [DOI] [PubMed] [Google Scholar]
- 58.Sabin, F. R. 1920. Studies on the origin of blood vessels and of red blood corpuscles as seen in the living blastoderm of chicks during the second day of incubation. Contrib. Embryol. 36:213-262. [Google Scholar]
- 59.Sanchez, M., B. Gottgens, A. M. Sinclair, M. Stanley, C. G. Begley, S. Hunter, and A. R. Green. 1999. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult hematopoietic progenitors. Development 126:3891-3904. [DOI] [PubMed] [Google Scholar]
- 60.Sanchez, M. J., E. O. Bockamp, J. Miller, L. Gambardella, and A. R. Green. 2001. Selective rescue of early hematopoietic progenitors in Scl−/− mice by expressing Scl under the control of a stem cell enhancer. Development 128:4815-4827. [DOI] [PubMed] [Google Scholar]
- 61.Shalaby, F., J. Rossant, T. P. Yamaguchi, M. Gertsenstein, X. F. Wu, M. L. Breitman, and A. C. Schuh. 1995. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376:62-66. [DOI] [PubMed] [Google Scholar]
- 62.Sinclair, A. M., B. Gottgens, L. M. Barton, M. L. Stanley, L. Pardanaud, M. Klaine, M. Gering, S. Bahn, M. Sanchez, A. J. Bench, J. L. Fordham, E. Bockamp, and A. R. Green. 1999. Distinct 5′ SCL enhancers direct transcription to developing brain, spinal cord, and endothelium: neural expression is mediated by GATA factor binding sites. Dev. Biol. 209:128-142. [DOI] [PubMed] [Google Scholar]
- 63.Solovyev, V., and A. Salamov. 1997. The Gene-Finder computer tools for analysis of human and model organisms genome sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5:294-302. [PubMed] [Google Scholar]
- 64.Spyropoulos, D. D., P. N. Pharr, K. R. Lavenburg, P. Jackers, T. S. Papas, M. Ogawa, and D. K. Watson. 2000. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 20:5643-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stainier, D. Y. R., B. M. Weinstein, B. M. Detrich, H. W. Detrich, L. I. Zon, and C. F. Mark. 1995. cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development 121:3141-3150. [DOI] [PubMed] [Google Scholar]
- 66.Visvader, J. E., Y. Fujiwara, and S. H. Orkin. 1998. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 12:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vlaeminck-Guillem, V., S. Carrere, F. Dewitte, D. Stehelin, X. Desbiens, and M. Duterque-Coquillaud. 2000. The Ets family member erg gene is expressed in mesodermal tissues and neural crests at fundamental steps during mouse embryogenesis. Mech. Dev. 91:331-335. [DOI] [PubMed] [Google Scholar]
- 68.Wagner, R. C. 1980. Endothelial cell embryology and growth. Adv. Microcirc. 9:45-75. [Google Scholar]
- 69.Wang, X., J. D. Crispino, D. L. Letting, M. Nakazawa, M. Poncz, and G. A. Blobel. 2002. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 21:5225-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wood, H. B., G. May, L. Healy, T. Enver, and G. M. Morriss-Kay. 1997. CD34 expression patterns during early mouse development are related to modes of blood vessel formation and reveal additional sites of hematopoiesis. Blood 90:2300-2311. [PubMed] [Google Scholar]
- 71.Xiong, N., C. Kang, and D. H. Raulet. 2002. Redundant and unique roles of two enhancer elements in the TCRγ locus in gene regulation and gammadelta T-cell development. Immunity 16:453-463. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, P., X. Zhang, A. Iwama, C. Yu, K. A. Smith, B. U. Mueller, S. Narravula, B. E. Torbett, S. H. Orkin, and D. G. Tenen. 2000. PU. 1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 96:2641-2648. [PubMed] [Google Scholar]
- 73.Ziegler, S., K. Burki, and R. C. Skoda. 2002. A 2-kb c-mpl promoter fragment is sufficient to direct expression to the megakaryocytic lineage and sites of embryonic hematopoiesis in transgenic mice. Blood 100:1072-1074. [DOI] [PubMed] [Google Scholar]