Abstract
Mammalian replication protein A (RPA) undergoes DNA damage-dependent phosphorylation at numerous sites on the N terminus of the RPA2 subunit. To understand the functional significance of RPA phosphorylation, we expressed RPA2 variants in which the phosphorylation sites were converted to aspartate (RPA2D) or alanine (RPA2A). Although RPA2D was incorporated into RPA heterotrimers and supported simian virus 40 DNA replication in vitro, the RPA2D mutant was selectively unable to associate with replication centers in vivo. In cells containing greatly reduced levels of endogenous RPA2, RPA2D again did not localize to replication sites, indicating that the defect in supporting chromosomal DNA replication is not due to competition with the wild-type protein. Use of phosphospecific antibodies demonstrated that endogenous hyperphosphorylated RPA behaves similarly to RPA2D. In contrast, under DNA damage or replication stress conditions, RPA2D, like RPA2A and wild-type RPA2, was competent to associate with DNA damage foci as determined by colocalization with γ-H2AX. We conclude that RPA2 phosphorylation prevents RPA association with replication centers in vivo and potentially serves as a marker for sites of DNA damage.
DNA-damaging stress leads to the inception of a variety of cellular responses that serve to minimize mutation and prevent genomic instability. In particular, the cell cycle checkpoint apparatus is activated to block S phase entry and, in those cells in the replicative phase, to both inhibit firing of late origins of DNA replication and avert the collapse of replication forks blocked by damage (3). The DNA repair machinery is mobilized in concert to repair lesions and to allow eventual restart of stalled replication forks. One factor that plays essential roles both during DNA replication and in the repair- and recombination-mediated recovery from damage is replication protein A (RPA), the eukaryotic single-stranded (ss) DNA-binding protein (27, 52).
RPA is a heterotrimeric protein consisting, in mammalian cells, of ∼70- (RPA1), 30- (RPA2), and 14 (RPA3)-kDa subunits. During DNA replication, RPA acts at the fork, stabilizing ssDNA and facilitating nascent strand synthesis by the replicative DNA polymerases. Under DNA-damaging conditions, RPA-ssDNA complexes act to recruit and activate a key checkpoint mediator consisting of the ATR and ATRIP (ATR-interacting protein) protein-kinase complex (54). At DNA damage-dependent nuclear foci, RPA interacts with repair and recombination components to process double-strand DNA breaks and other lesions (19). RPA activity is regulated by various stress conditions. In particular, heat shock (12, 47, 48), exposure to UV radiation (9), and treatment with DNA-alkylating agents (30) each cause the generation of an RPA species that is unable to support DNA replication in vitro. In the case of heat shock, the inhibition of RPA activity is mediated by a stress-dependent association with the nucleolar protein nucleolin (12, 47).
In an area with potential regulatory significance, RPA undergoes both stress-dependent and -independent phosphorylation on the extreme N terminus of the RPA2 subunit. A basal level of RPA modification by cyclin-cdk complexes occurs at two sites (16, 35). Following stress, such as exposure to ionizing (31) or UV (9) radiation, or treatment with radiomimetic agents, such as camptothecin (CPT) (42), human RPA2 can be phosphorylated at five or more additional sites out of a possible seven by the phosphatidylinositol 3-kinase-related kinases (PIKKs) DNA-PK, ATM, and perhaps ATR (7, 10, 17, 18, 31, 33, 35, 46, 53). ATM and ATR are activated in response to DNA damage and replication stress, and they modify various effectors that facilitate the damage and cell cycle checkpoint responses (1). DNA-PK is required directly in the repair of double-strand DNA breaks and in V(D)J recombination (15). These data could suggest that the function of stress-dependent modification of RPA is to repress DNA replication or to promote recovery from DNA damage, but there are as yet no compelling data for either role. While the results of certain studies suggest that RPA modification by PIKKs may lead to the inhibition of DNA replication in vitro and in vivo (9, 37), direct testing of this possibility has not shown any appreciable effects of RPA phosphorylation on binding to ssDNA or on replication in vitro using a simian virus 40 (SV40)-based assay (7, 23).
Because previous work has primarily studied the effects of mammalian RPA phosphorylation using in vitro systems, it is possible that the modulation of RPA activity by phosphorylation might be observed only in the cellular milieu. Testing this hypothesis, we found that RPA2 phosphorylation mutants that mimic the hyperphosphorylated form were unable to localize to replication centers in normal cells. Interestingly, binding of the hyperphosphorylation mimic to DNA damage foci was unaffected, as determined by colocalization with the DNA damage marker γ-H2AX. Similar behavior was observed with endogenous hyperphosphorylated RPA. We conclude that RPA phosphorylation following damage both prevents RPA from catalyzing DNA replication and potentially serves as a marker to recruit repair factors to sites of DNA damage.
MATERIALS AND METHODS
Cell lines and stress treatments.
U2-OS and HeLa cells were maintained in McCoy's 5 M and Dulbecco's modified Eagle's media, respectively, supplemented with 10% fetal bovine serum and 50 μg of gentamicin/ml. When the effect of stress was examined, the cells were treated with either 1 μM CPT (Sigma) for 1 or 3 h, 2.5 mM hydroxyurea (HU; Sigma) for either 1 or 3 h, or 7 μM aphidicolin (Sigma) for 3 h. To inhibit the cellular checkpoint response, cells were treated with 5 mM caffeine for 30 min prior to stress. Transfection experiments were performed using Effectene (Qiagen).
Generation of RPA2 mutant constructs.
To generate the myc-RPA2wt and myc-RPA2D mammalian expression vectors, the human RPA2 genes from plasmids p11dtRPA and p11dtRPA · 32Asp8 (4, 24) were inserted into the XbaI and BstBI sites of the pEF6/Myc-HisA vector (Invitrogen), resulting in pERPA2wt and pERPA2D. Expression of the His6 tag from pEF6/Myc-HisA was prevented by mutating the ATG codon at position 1863 to a TGA codon. Vectors expressing the intermediate RPA2 phosphorylation mutants and RPA2A were constructed by a combination of site-directed mutagenesis of either pERPA2wt or pERPA2D (as appropriate) at positions 23, 29, and 33 and replacement of larger segments of the RPA2 N terminus with synthetic oligonucleotides encoding mutant phosphorylation regions. Detailed cloning procedures are available upon request.
Protein purification and in vitro replication assay.
The RPARPA2wt and RPARPA2D heterotrimers were expressed in Escherichia coli BL21 transformed with p11dtRPA and p11dtRPA · 32Asp8, respectively, and purified as described previously (24, 26). The SV40 large tumor (T) antigen used for SV40 DNA replication reactions was prepared from extracts of Sf9 cells infected with the recombinant baculovirus Ac941SVT (5) and purified using immunoaffinity chromatography (6). The AS65 fraction lacking RPA was prepared from HeLa cell extracts by ammonium sulfate fractionation according to the method of Wobbe et al. (51). SV40 DNA replication reaction mixtures (50 μl) containing 40 mM creatine phosphate (diTris salt; pH 7.8); 7 mM MgCl2; 4 mM ATP; 25 μg of creatine kinase/ml; 0.4 mM dithiothreitol; 200 μM (each) CTP, GTP, and UTP; 100 μM (each) dATP, dGTP, and dCTP; 25 μM [3H]dTTP (∼500 cpm/pmol); 0.2 μg of the ori-containing plasmid pSV01ΔEP (50); 200 μg of the AS65 fraction; 0 to 700 ng of RPARPA2wt or RPARPA2D; and 750 ng of SV40 T antigen were incubated at 37°C for 2 h. Replication activity was determined by precipitating the high-molecular-weight DNA with trichloroacetic acid and quantitating the amount of 3H in the precipitate by scintillation counting. To examine the DpnI resistance of the replication products, replication reaction mixtures containing 600 ng of either RPARPA2wt or RPARPA2D and 100 μM [α-32P]dCTP (1,000 cpm/pmol) to label the replication products were incubated at 37°C for 2.5 h. Following removal of protein by phenol extraction, the DNA products were first linearized by digestion with PstI and then either mock treated or incubated with 2.5 U of DpnI to cleave nonreplicated DNA. The digestion products were separated by electrophoresis through a 1.1% agarose gel and visualized both by ethidium bromide staining and by autoradiography.
Immunoprecipitation and immunoblotting.
Transfected U2-OS cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% [vol/vol] NP-40, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM Na3VO4, 1 mM NaF, and 1 μg each of aprotinin, leupeptin, and pepstatin per ml). The cell extracts were then incubated at 4°C for 2 h with 70A anti-RPA1 monoclonal antibody (28) conjugated to CNBr-activated Sepharose beads (Amersham Biosciences). The immunoprecipitate was washed five times with lysis buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (13% [wt/vol] polyacrylamide). To test RPA2 phosphorylation and myc-RPA2 expression, cells were directly lysed in SDS-PAGE sample buffer and proteins were separated by SDS-PAGE. For phosphatase treatment, cells were lysed in λ protein phosphatase buffer (New England Biolabs) containing 1% Triton X-100 and 1 μg each of aprotinin, leupeptin, and pepstatin per ml. Cell lysates (∼20 μg of protein) were then incubated with 400 U of λ protein phosphatase for 30 min at 30°C or mock treated in the presence of protein phosphatase inhibitors (0.1 mM Na3VO4, 1 mM NaF). The Western blots were developed with an anti-RPA2 34A mouse monoclonal antibody (28) or a rabbit polyclonal anti-pSer4/pSer8-RPA2 antibody obtained from Bethyl Laboratories, Inc. (Montgomery, Tex.). Proteins were detected using enhanced chemiluminescence (Amersham Biosciences).
Cell cycle assay.
Forty-eight hours posttransfection, cells were incubated with 10 μM bromodeoxyuridine (BrdU). After a 30-min incubation, the cells were fixed and processed according to the BrdU Flow Kit manual (BD Pharmingen). Following incubation with rat anti-BrdU (Harlan Sera-Lab) and rabbit anti-myc (Upstate Biotechnology) antibodies, the cells were stained with anti-rat fluorescein isothiocyanate-conjugated and anti-rabbit phycoerythrin-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). The DNA was stained with 7-aminoactinomycin D, and the cells were subjected to fluorescence-activated cell sorting (FACS) analysis.
Immunofluorescence microscopy.
Transfected cells were processed by two methods. To test protein expression and transfection efficiency, the cells were first washed with phosphate-buffered saline (PBS), fixed with 4% (wt/vol) formaldehyde in PBS for 15 min at room temperature, and then extracted with PBS containing 0.5% (vol/vol) Triton X-100 for 5 min. To study chromatin-bound proteins, the cells were extracted with 0.5% (vol/vol) Triton X-100 for 5 min prior to fixation as described previously (13). When required, cells were incubated in media containing 10 μM BrdU for 10 min prior to harvest. For detection of incorporated BrdU, DNA was denatured with HCl using standard procedures. RPA2 silencing was achieved using a short interfering RNA (siRNA) duplex targeted to the 5′-CCUAGUUUCACAAUCUGUU sequence found in the 3′ noncoding region of RPA2 mRNA. Prepared cells were incubated, as required, with rabbit anti-myc (Upstate Biotechnology), mouse anti-RPA1 70A and anti-RPA2 34A (28), rabbit anti-pSer4/pSer8-RPA2 (Bethyl Laboratories), rat anti-BrdU (Harlan Sera-Lab), and mouse anti-γH2AX (Upstate Biotechnology) antibodies. Following staining with the appropriate secondary antibodies (Jackson ImmunoResearch Laboratories), the cells were examined by epifluorescence microscopy using a Zeiss Axiophot microscope. To calculate the relative frequency of myc-RPA2-positive cells (see Fig. 6H and 8M), the fraction of cells transfected with myc-RPA2wt or the myc-tagged RPA mutants was first determined by processing cells without prior Triton X-100 extraction (e.g., Ftransfection:wt and Ftransfection:D4). Separately, the fraction of cells showing significant chromatin staining was also determined (e.g., Fchromatin:wt and Fchromatin:D4). The relative frequency of cells that were positive, for example, for myc-RPA2D4 chromatin staining was calculated using the following formula: relative frequency = (Fchromatin:D4/Ftransfection:D4)/(Fchromatin:wt/Ftransfection:wt) · 100%. Each value determined was the result of three independent experiments.
FIG. 6.
Increase in negative charge at the RPA2 N terminus decreases frequency of myc-positive cells. (A to F) Cells were transfected with myc-RPA2wt or myc-RPA2D41 as indicated. Representative epifluorescence images showing total myc-RPA2 staining (no extraction [A and B]), chromatin-bound myc-RPA2 (C and D), and chromatin-bound RPA1 (E and F) are shown. (G and H) Serines and threonine (boldface and underlined) at the potential phosphorylation sites in the N terminus of the RPA2 subunit were replaced with alanines (A) or aspartates (D) as indicated. Cells expressing these myc-tagged RPA2 mutants were then analyzed by immunofluorescence microscopy for the presence of myc-RPA2 bound to chromatin and, in parallel reactions, for transfection efficiency. The relative frequencies of myc-RPA2 positive cells were calculated as described in Materials and Methods.
FIG. 8.
Collapse of DNA replication forks stimulates RPA loading to damaged DNA. (A to H) U2-OS cells were transfected with plasmids expressing either myc-RPA2wt or myc-RPA2D as indicated for 48 h. The cells were then treated with 2.5 mM HU for either 1 (A and E) or 3 (B and F) h in the absence of caffeine or treated with HU for 1 (C and G) or 3 (D and H) h in the presence of 5 mM caffeine. The cells were Triton X-100 extracted and fixed and then stained for myc-RPA. (I to L) Colocalization of myc-RPA2D with sites of DNA damage. U2-OS cells were transfected with a myc-RPA2D-expressing plasmid. Forty-eight hours posttransfection, the cells were treated with 2.5 mM HU for 3 h and then extracted with 0.5% Triton X-100 and fixed. The cells were stained with γ-H2AX (I) and myc-RPA (J). The staining pattern of a representative cell and the image of the merged staining patterns (K) are provided. (L) One section (boxed) of the composite image is shown enlarged to reveal the degree of signal overlap. (M) Graph showing the fractions of myc-RPA2-transfected cells with a significant chromatin-bound myc-RPA2wt (green bars) or myc-RPA2D (brown bars) signal. The fractions of cells showing chromatin-bound RPA were quantified by visual inspection of 100 to 200 cells. The bar graph values were calculated as described in Materials and Methods. Note that although RPA2wt staining is consistently detected in a greater fraction of cells than RPA2D staining, this result is expected because RPA2wt can be observed both at replication centers and at DNA damage foci while RPA2D localizes only to damage foci. (N) Effects of stress and caffeine treatment on endogenous RPA2 phosphorylation. U2-OS cells were mock treated (mock) or treated with either 1 μM CPT for 1 or 3 h, 2.5 mM HU for 1 or 3 h, 5 mM caffeine (c) for 3 h, 2.5 mM HU for 1 or 3 h in the presence of 5 mM caffeine or with 7 μM aphidicolin (A) for 3 h and 7 μM aphidicolin for 1 or 3 h in the presence of 5 mM caffeine. The band labeled RPA2′ represents nonphosphorylated RPA, while P-RPA2 indicates the position of phosphorylated RPA.
RESULTS
The RPA2D phosphorylation mimic localizes to the nucleus but is not chromatin bound.
To understand the functional significance of RPA phosphorylation, we generated various human RPA2 constructs in which subsets of the nine potential N-terminal phosphorylation sites were mutated. Previous studies have shown that two of the RPA2 sites (S23 and S29) are phosphorylated in a cell cycle-dependent manner by cyclin-cdk2 complexes (16, 35). At least five of the other seven (S4, S8, S11, S12, S13, T21, and S33) can be phosphorylated in response to UV irradiation (53). Ionizing irradiation and treatment with the radiomimetic agent CPT cause similar RPA hyperphosphorylation and likely modification of most if not all of these same sites (31, 42). Various data strongly suggest that the PIKK members DNA-PK and ATM, and likely ATR, can independently modify the RPA stress-dependent sites (7, 10, 17, 18, 31, 33, 35, 46), although only two (T21 and S33) have canonical SQ/TQ sequences that are PIKK targets (1). Both of the cyclin-cdk2 sites and six of the stress-dependent sites (S8, S11, S12, S13, T21, and S33) were replaced by aspartate to mimic phosphate (generating the RPA2D mutant; see Fig. 6G for a schematic showing the construction of this and other mutants). Although an aspartate residue is not the same as phosphoserine or phosphothreonine, the use of aspartate residues to imitate phosphate has been shown in many cases to have identical effects on protein structure and activity (25, 49). In the RPA2A mutant, these same eight sites were converted to alanines to prevent phosphorylation (see also Fig. 6G). All of the mutants and the wild-type RPA2 control (RPA2wt) contained a C-terminal myc tag.
The RPA2wt subunit was expressed in human U2-OS cells. To detect the chromatin-bound fraction of RPA2, transfected cells were extracted with nonionic detergent prior to formaldehyde fixation (13). Under such conditions, RPA bound to chromatin in nuclear replication foci can be selectively visualized. The transfected RPA2wt subunit nearly completely colocalized with the endogenous RPA1 and exhibited a punctate distribution throughout the nucleus, consistent with its recruitment to DNA replication centers (Fig. 1A to D). To confirm this observation, transfected cells were pulse-labeled with BrdU prior to fixation, and the sites of RPA2wt localization and BrdU incorporation were examined. As expected, the RPA2wt subunit showed nearly complete colocalization with replicating chromatin (Fig. 1E to H). Taken together, these results indicate that the recombinant RPA2wt subunit can functionally replace endogenous RPA2 in supporting chromosomal DNA replication.
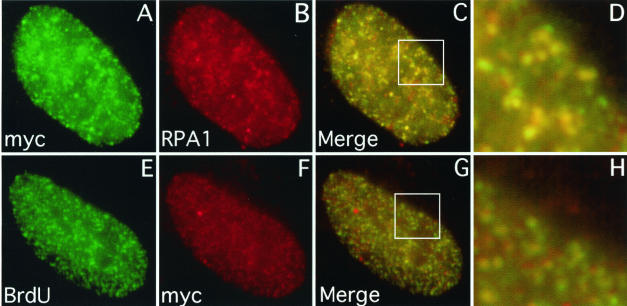
FIG. 1.
The myc-RPA2wt subunit colocalizes with endogenous RPA1 and DNA replication centers. U2-OS cells were transfected with a vector expressing myc-RPA2wt. To allow visualization of chromatin-bound myc-RPA, the cells were first extracted with 0.5% Triton X-100 for 5 min and then fixed with formaldehyde. (E to H) To detect sites of DNA replication, BrdU was added to the medium 10 min before the cells were prepared for epifluorescence microscopy. As indicated, the cells were then stained with anti-myc (A and F), anti-RPA1 (B), or anti-BrdU (E) antibody. The extent of myc-RPA2wt and endogenous RPA1 colocalization is shown (C), with enlargement of a particular nuclear region (boxed) (D). BrdU and myc-RPA2wt colocalization are similarly shown (G and H).
We next examined the localization of the RPA2A and RPA2D mutants. Transfected cells were examined both without and with prior detergent extraction to reveal transfection efficiency and to show the fraction bound to chromatin, respectively. The distribution of RPA2A on chromatin (Fig. 2L) was virtually identical to the replication pattern seen with endogenous RPA2 (data not shown) and the RPA2wt variant (Fig. 2D) and showed nearly complete colocalization with endogenous RPA1 (Fig. 2K and data not shown). RPA2 phosphorylation is therefore not required for association with replication centers.
FIG. 2.
Lack of association of the RPA2D mutant with chromatin in unstressed cells. U2-OS cells were transfected with a vector expressing myc-RPA2wt (A to D), myc-RPA2D (E to H), or myc-RPA2A (I to L). (C, D, G, H, K, and L) To allow visualization of chromatin-bound myc-RPA, cells were first extracted with 0.5% Triton X-100 for 5 min and then fixed with formaldehyde (+ extraction). (A, B, E, F, I, and J) To assay for transfection efficiency, cells were also fixed without prior extraction (− extraction). The cells were then stained with anti-myc (B, D, F, H, J, and L) or anti-RPA1 (A, C, E, G, I, and K) antibody.
In dramatic contrast, we did not observe chromatin staining for RPA2D (Fig. 2H), even though the RPA2D mutant was expressed to an equivalent similar to that of the RPA2wt construct (Fig. 2F and B, respectively). Similar experiments were performed with RPA2D and RPA2wt expressed as fusion proteins with green fluorescent protein. While a modest RPA2wt signal was detected, we did not observe an appreciable level of chromatin binding for RPA2D (data not shown). We therefore find that mutation of RPA2 to a hyperphosphorylation mimic greatly reduces its localization to DNA replication centers.
In addition to the possibility that phosphorylation of RPA inhibits its normal participation at the DNA replication fork in vivo, other explanations exist. One is that the myc-RPA2D subunit is unable to complex with the other RPA subunits. To examine this, lysates prepared from cells transfected with the RPA2wt and RPA2D expression vectors were subjected to immunoprecipitation with an anti-RPA1 antibody and immunoblotted for the presence of RPA2. The two myc-RPA2 variants, as well as the endogenous RPA2, efficiently coprecipitated with the RPA1 subunit (Fig. 3A, lanes 1 to 3). The RPA2D protein was also found in the lysate at levels comparable to those of RPA2wt, suggesting that the two proteins have similar stabilities (lanes 5 and 6). Because RPA1 and RPA2 complex formation requires the RPA3 subunit (24, 44), these data indicate that the two mutants form RPA heterotrimers with equivalent efficiencies.
FIG. 3.
RPA2D-containing RPA heterotrimers are replication competent. (A) U2-OS cells were transfected with empty vector (lanes 1 and 4), myc-RPA2wt (lanes 2 and 5), or myc-RPA2D (lanes 3 and 6). Lysates were prepared from each batch of transfected cells, and the lysates were subjected to immunoprecipitation using anti-RPA1 antibodies. Immunoprecipitates (IP) (lanes 1 to 3) and aliquots of the lysates (lanes 4 to 6) were then analyzed for the presence of RPA2 by Western blotting analysis using RPA2 antibodies (which recognize both transfected and endogenous RPA2). (B and C) RPA heterotrimers that contained either RPA2wt (lane 1) or RPA2D (lane 2) were expressed in E. coli, purified, and analyzed by SDS-PAGE and Coomassie blue staining (B). The purified RPA was then assayed for the ability to support SV40 DNA replication in combination with an AS65 fraction purified from HeLa cells (51) (C). The open triangle shows that only background levels of DNA synthesis occur in reactions containing RPA2wt but lacking T antigen. Similar results were observed using RPA2D. (D) SV40 DNA replication reactions were performed in the presence of [α-32P]dCTP to label the replication products as described in Materials and Methods. The reaction mixtures contained 600 ng of either RPARPA2wt (lanes 1, 2, 4, and 5) or RPARPA2D (lanes 3 and 6) and either lacked T antigen (lanes 1 and 4) or contained 750 ng of T antigen (lanes 2, 3, 5, and 6). After isolation, the DNA replication products were first linearized by restriction digestion and then either mock treated (lanes 1 to 3) or incubated with DpnI to cleave nonreplicated DNA (lanes 4 to 6). The digestion products were then subjected to agarose gel electrophoresis, and the images of the ethidium bromide (EtBr)-stained gel (to show the total level of DNA) and the autoradiograph of the gel (to visualize 32P-labeled reaction products) are provided. The observed bands correspond to the linearized SV40 origin-containing plasmid.
To establish if heterotrimeric RPA containing the RPA2D subunit (RPARPA2D) was inherently unable to function in DNA replication, we tested the abilities of RPARPA2D and RPARPA2wt to support SV40 DNA replication in vitro. With the exception of the viral large T antigen, SV40 replication is catalyzed by host cell components (8, 22). The SV40 system can thus provide a relatively comprehensive test of the ability of RPA to interact functionally with ssDNA and the DNA replication machinery. Previous work by J. Hurwitz and colleagues has shown that separation of human cell extracts by ammonium sulfate precipitation yields two required fractions (AS30 and AS65), with RPA found to be the only essential factor within the AS30 fraction (51). Because the AS65 fraction lacks RPA, the activities of different RPA variants can be assayed by their abilities to complement the AS65 fraction in supporting SV40 DNA replication. The RPARPA2D and RPARPA2wt variants were produced in E. coli and purified to homogeneity (Fig. 3B). Use of the AS65 fraction alone showed no significant DNA replication activity (Fig. 3C). The addition of either heterotrimeric RPA complex supported T-antigen-dependent viral DNA replication to similar extents, and the activities of the two RPA variants were similar over a range of levels (Fig. 3C). The reaction products synthesized in the presence of RPARPA2D or RPARPA2wt were equally resistant to DpnI, demonstrating that they were bona fide DNA replication products and not due to repair synthesis (Fig. 3D). RPARPA2D is therefore functionally active in supporting DNA replication in vitro. RPARPA2D was also found to bind normally to short ssDNA oligonucleotides (4). These results are not completely unexpected, as it was shown previously that the RPA phosphorylation state does not appreciably affect the ability of RPA to function in viral DNA replication or in DNA repair (2, 7, 36). In sum, mutation of the seven serines and one threonine in the N terminus of RPA2 to negatively charged aspartate residues does not have any apparent effect on the inherent activity of the heterotrimeric protein.
We next examined the possibility that expression of the RPA2D mutant generates a signal that shuts down cellular DNA synthesis and thus indirectly prevents RPA2D from associating with chromatin. To address this issue, cells were transfected with the RPA2wt or RPA2D expression construct and pulse-labeled with BrdU. The cells were then subjected to FACS based on three signals: the level of myc-RPA2, DNA content, and BrdU incorporation. In addition to confirming that the two RPA2 variants were expressed at comparable levels (Fig. 4A and C), it was found that the percentages of cells in S phase were similar regardless of whether the cells were transfected with RPA2wt (Fig. 4B), RPA2D (Fig. 4D), or empty vector (not shown). Although the percentage of cells in S phase was somewhat high compared to other experiments, perhaps because of transfection conditions, the fractions of cells in S phase were routinely found to be similar for RPA2wt and RPA2D. We conclude that expression of RPA2D does not significantly affect cell cycle progression.
FIG. 4.
Expression of RPA2D does not affect cell cycle progression. Cells transfected with myc-RPA2wt (A and B) or myc-RPA2D (C and D) were incubated with 10 μM BrdU for 30 min prior to harvest. The cells were then subjected to FACS analysis using a pairwise analysis of the levels of myc and BrdU signals. Transfected cells (with significant myc signals [boxed regions in panels A and C]) were further analyzed for the BrdU signal and the amount of DNA. (B and D) Fractions of cells in G1, S, and G2 phases. For each plot, the x and y axes indicate fluorescence intensities of the different signals.
RPA2D is unable to complement the loss of endogenous RPA2.
The data presented above suggested that the RPA2wt subunit, but not RPA2D, would be able to complement the loss of endogenous RPA2 and support chromosomal DNA replication. To test this possibility, cells were depleted of cellular RPA2 by using an siRNA molecule directed against the 3′ noncoding sequence of RPA2. The RPA2 expression cassettes do not contain the siRNA-targeted sequences, and hence the myc-RPA2 RNA produced from these vehicles is resistant to siRNA-mediated degradation. Visualization of RPA2 in these siRNA-treated cells by epifluorescence microscopy showed an apparent reduction of the RPA2 signal to nearly background level in >90% of the cells (compare Fig. 5D with A). Western blotting analysis indicated that RPA2 levels were reduced by >95% (data not shown). Upon cotransfection with myc-RPA2wt, a significant fraction of the cells demonstrated a robust myc signal bound to chromatin, with the pattern of binding similar to that seen in replicating cells (Fig. 5C). In contrast, little or no myc-RPA2D was found associated with chromatin (Fig. 5F), even though comparable levels of RPA2wt and RPA2D expression were detected in nonextracted cells (Fig. 5B and E, respectively). We therefore conclude that RPA2D, unlike RPA2wt, is unable to complement the loss of endogenous RPA2 and support DNA replication. These data also indicate that RPARPA2D is not prevented from binding ssDNA because of competition with the endogenous RPA but rather is inherently unable to productively interact with the DNA replication machinery.
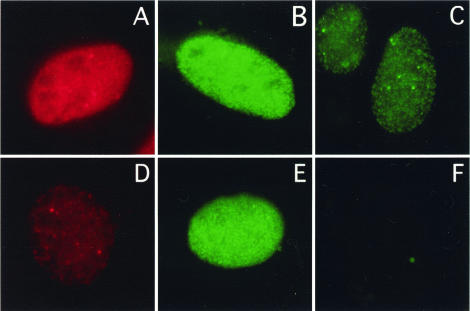
FIG. 5.
Lack of RPA2D chromatin association in cells lacking endogenous RPA2. U2-OS cells were incubated with a control (i.e., scrambled) siRNA (A) or an siRNA specific for the 3′ noncoding region of the RPA2 mRNA (B to F). The cells were simultaneously cotransfected with an empty control vector (D), myc-RPA2wt (B and C), or myc-RPA2 D (E and F). Forty-eight hours posttransfection, the cells were extracted with 0.5% (vol/vol) Triton X-100 for 5 min prior to formaldehyde fixation to reveal RPA associated with chromatin (C and F) or were fixed to show total endogenous or transfected RPA2 (A, B, D, and E). The cells were then stained with anti-RPA2 (A and D) or anti-myc (panels B, C, E, and F) antibody and then visualized by epifluorescence microscopy. Cells with representative signals were chosen.
RPA association with replication centers is dependent on the RPA2 N terminus negative charge.
We next examined whether mutation of particular serine or threonine residues to aspartate was responsible for the lack of RPA2D association with replication centers or whether it was a consequence of the heightened negative charge at the RPA2 N terminus. We first constructed serine-to-aspartate substitutions at the cyclin-cdk2 sites S23 and S29 (RPA2D2) (Fig. 6G). S29 is invariably modified in each form of phosphorylated RPA (53), and thus, the RPA2D2 mutant resembles the form found in the initial steps of the RPA phosphorylation pathway. Further intermediate RPA2 mutants were designed to roughly follow the phosphorylation pathway, as suggested by the data of Zernik-Kobak and colleagues (53). However, it must be mentioned that the exact pathway of RPA2 modification from the hypophosphorylated to the hyperphosphorylated form is not known, and it is unlikely that a strict order of modification occurs in vivo. Additional serine-to-aspartate changes were generated in the background of the RPA2D2 mutant, with a total of three (RPA2D3 and RPA2D31), four (RPA2D4 and RPA2D41), or five (RPA2D5) positions mutated. The sites mutated in these RPA2 variants are also found to be modified in RPA with an intermediate phosphorylation state in vivo.
Transfection of U2-OS cells indicated that all of the intermediate RPA2 mutants were expressed at similar levels (Fig. 6A and B and data not shown). Relative to RPA2wt, the RPA2 mutants with two or three Ser→Asp changes had two notable effects: (i) a modestly reduced fraction of transfected cells showing mutant RPA2 bound to chromatin (Fig. 6H) and (ii) a reduction in the intensity of RPA2 bound to chromatin (see below). More dramatic effects were observed when four or five serines were converted. For RPA2D41, the fraction of cells with significant chromatin binding was threefold less than for RPA2wt, and this fraction was reduced to 8% for the RPA2D5 mutant (Fig. 6H). The intensities of chromatin staining for the intermediate RPA2 mutants were also greatly reduced in individual cells, as demonstrated by comparing the average staining patterns of cells transfected with RPA2wt and RPAD41 (Fig. 6C and D, respectively [taken with identical exposure times]).
The decrease in association of RPA2 with replication centers was most strongly correlated with the number of aspartate residues rather than with changes at any particular positions. The notion that the mutation of serines to aspartates per se (i.e., irrespective of the changes in the RPA2 negative charge) causes decreased RPA binding to replication centers is argued against because the N terminus of RPA2 is not critical for DNA replication in vitro for mammalian RPA (23) or in vivo for yeast RPA (38). These data therefore suggest that the increase in net negative charge afforded by the increased number of aspartate residues is the primary factor regulating RPA binding to chromatin. Although the location of the aspartate residues did not appear to have major effects on RPA2 activity, we did note that mutation of the S33 site, known as a consensus sequence for PIKKs, appeared to have a somewhat more deleterious effect.
RPA2D is recruited to DNA damage foci following genotoxic stress.
Under DNA damage conditions, a significant change occurs in the nuclear distribution of RPA, with the more diffuse punctate pattern seen during DNA replication transforming to bright, well-distinguished foci. In this state, RPA colocalizes with a number of repair and checkpoint proteins (e.g., ATR and Rad51) and is thought to demarcate the sites of DNA repair and/or unrepairable lesions (19, 20, 39, 54). Such stress conditions cause a subset of the endogenous RPA pool to become hyperphosphorylated (see below). We therefore reexamined the behavior of RPA2D and RPA2A in cells undergoing genotoxic stress.
Cells were transfected with the RPA2wt, RPA2A, or RPA2D expression construct and then treated with CPT. CPT inhibits topoisomerase I, indirectly causing DNA double-strand breaks, and leads to rapid and massive RPA phosphorylation (42). Similar to RPA2wt (Fig. 7A to C), the RPA2A variant colocalized with RPA1 in bright foci following CPT treatment (Fig. 7G to I). Very similar foci were observed for endogenous RPA2 (not shown). Thus, the phosphorylation-defective RPA2A variant is apparently competent to bind chromatin both in normal (above [Fig. 2L]) and in stressed cells.
FIG. 7.
RPA2D binds chromatin and colocalizes with RPA1 after CPT treatment. U2-OS cells were transfected with myc-RPA2wt (A to C), myc-RPA2D (D to F), or myc-RPA2A (G to I) vector. Forty-eight hours posttransfection, the cells were incubated with 1 μM CPT for 3 h, extracted with 0.5% (vol/vol) Triton X-100 for 5 min, fixed, and stained with anti-RPA1 (A, D, and G) and anti-myc (B, E, and H) antibodies. (C, F, and I) Colocalization of the two stains, enlarged from the boxed regions.
In sharp contrast to the inability of RPA2D to stably associate with replication centers, CPT treatment caused the RPA2D variant to colocalize with RPA1 in DNA damage foci (Fig. 7D to F). The number and distribution of these foci, as well as the intensity of staining, were indistinguishable from those observed with the RPA2wt (and RPA2A) construct. Thus, although the RPA2D mutant is unable to localize to replication centers, this defect does not extend to the involvement of RPA2D in the DNA damage response.
We determined if the CPT-dependent recruitment of RPA2D to DNA damage foci was applicable to other stresses. We tested HU and aphidicolin, agents that do not directly cause DNA damage but rather result in stalling of the DNA replication fork. As cells were incubated with HU from 1 to 3 h (Fig. 8E and F), a progressive increase in RPA2D association with chromatin was observed, with most cells demonstrating a dispersed staining pattern. A fraction of cells exhibited distinctive foci, and these showed significant colocalization with γ-H2AX, the phosphorylated form of histone variant H2AX that is a marker for sites of DNA damage (Fig. 8I to L) (40). Similar effects of HU were noted on cells transfected with RPA2wt (Fig. 8A and B). In contrast, treatment with aphidicolin for 3 h did not stimulate RPA2D association with chromatin (Fig. 8M; data not shown), demonstrating reduced toxicity of aphidicolin relative to HU under these conditions. Exposure to ionizing radiation (10 Gy) gave rise to staining patterns of RPA2wt and RPA2D similar to that found with CPT (data not shown).
In the functional absence of the budding yeast homologs of ATR and its downstream effector Chk1 (Mec1p and Rad53, respectively), replication forks have a greater propensity to collapse when encountering DNA damage, yielding unregulated production of long ssDNA regions (32, 43, 45). We therefore hypothesized that addition of caffeine, an inhibitor of the ATR-ATM-dependent checkpoint response (21, 41), to HU-treated cells would similarly lead to replication fork degradation. This in turn would cause faster induction of DNA damage foci and of RPA2D localization. To test this hypothesis, RPA2wt- or RPA2D-transfected cells were treated with HU for 1 or 3 h in the presence of caffeine. Particularly for RPA2D, addition of caffeine dramatically increased the number and intensity of RPA2 foci at both the 1- and 3-h time points (Fig. 8G and H). Quantification of the effects on myc-RPA2 localization demonstrated that caffeine greatly increased the fraction of HU-treated cells with significant RPA2D and RPA2wt signals (Fig. 8M).
The effects of these various stress conditions on endogenous RPA phosphorylation were also examined (Fig. 8N). Those stress conditions that resulted in significant RPA2D chromatin binding also caused increased phosphorylation of endogenous RPA2, although CPT caused modification of a greater fraction of the RPA pool, as well as phosphorylation of more RPA2 sites, than HU. Enhanced phosphorylation of RPA following a 1-h treatment with HU and caffeine was occasionally seen. Consistent with our results showing the inability of aphidicolin to stimulate the chromatin binding of RPA2D, aphidicolin also did not induce RPA2 phosphorylation. Because caffeine has been demonstrated to be an inhibitor of ATM and ATR kinase activities (21, 41), the observed hyperphosphorylation of RPA probably results from the caffeine-insensitive activity of DNA-PK that is stimulated by collapsed replication forks. However, we note that a recent study found that caffeine can inhibit the checkpoint response without inhibiting ATR-ATM kinase activity in vivo (11), leaving open the possibility that these kinases may still be responsible. In any case, these data indicate that the rate and extent of RPA2D (and RPA2wt) localization to sites of DNA damage correlate with the degree of DNA damage sustained during stress.
Localization of endogenous hyperphosphorylated RPA.
The properties of endogenous hyperphosphorylated RPA were examined using an antibody generated against an RPA2 peptide doubly phosphorylated on serine residues 4 and 8. Lysates prepared from untreated or CPT-treated U2-OS cells were probed with either a general RPA2 antibody or the pSer4/pSer8-RPA antibody (Fig. 9J). The phosphospecific antibody selectively recognized a species from CPT-treated cells that comigrated with hyperphosphorylated RPA2 by Western blotting analysis. Prior incubation of the CPT-treated lysates with phosphatase resulted in the loss of both the hyperphosphorylated RPA2 form and reactivity by the phosphospecific antibody. We conclude that the phosphospecific antibody recognizes a hyperphosphorylated RPA2 species that is modified on Ser4 and Ser8.
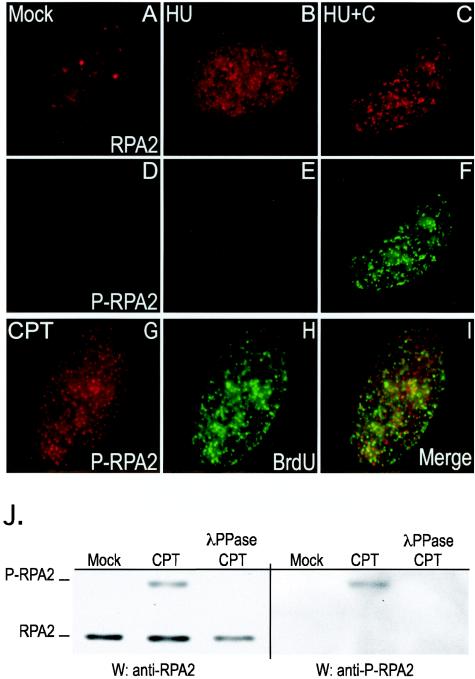
FIG. 9.
Endogenous phosphorylated RPA (P-RPA2) does not localize to sites of DNA synthesis. (A to F) U2-OS cells were either mock treated (A and D) or treated with 2.5 mM HU for 3 h (B and E) or with 2.5 mM HU and 5 mM caffeine for 3 h (C and F). The cells were extracted to visualize the chromatin-bound form of RPA, fixed, and then stained either with anti-RPA2 (A to C) or anti-pSer4/pSer8-RPA2 (D to F) antibody. (G to I) U2-OS cells were either mock treated or treated with 1 μM CPT for 30 min, followed by an additional 2.5-h incubation in medium lacking CPT. The cells were incubated with 10 μM BrdU for 15 min prior to being processed. The cells were then extracted to visualize the chromatin-bound form of RPA, fixed, and stained either with anti-pSer4/pSer8-RPA2 (G) or anti-BrdU (H) antibody. (I) Merged staining pattern. (J) Extracts prepared from mock-treated or CPT-treated (1 μM for 3 h) cells were subjected to Western blotting (W) analyses using either anti-RPA2 (34A) monoclonal antibody (anti-RPA2) or a rabbit anti-pSer4/pSer8-RPA2 antibody (anti-P-RPA2). CPT-treated extracts were also incubated with λ protein phosphatase (λPPase), as indicated.
The phosphospecific antibodies were used to examine the localization of the pSer4/pSer8 form of RPA in untreated U2-OS cells and in cells treated with HU alone or with HU and caffeine. In control cells or in cells treated only with HU (Fig. 9D and E), little if any pSer4/pSer8-RPA staining was detected. Following treatment with both HU and caffeine, the cells showed a dramatic increase in pSer4/pSer8-RPA staining (Fig. 9F). The staining pattern was nearly identical to that of total RPA2 (compare Fig. 9C and F), and also showed good overlap with γ-H2AX staining (data not shown). The colocalization of pSer4/pSer8-RPA with sites of DNA synthesis was also examined. Cells were treated with CPT and then incubated with BrdU. The areas of pSer4/pSer8-RPA staining did not colocalize with sites of remaining DNA synthesis to any significant degree (Fig. 9L). A majority of the RPA pool is hyperphosphorylated under these conditions (Fig. 8N), rendering similar experiments using general RPA2 antibodies uninformative. We conclude that the hyperphosphorylated form of RPA localizes only to chromatin following DNA damage and is not significantly associated with sites of chromosomal DNA synthesis.
DISCUSSION
We find that the RPA2D mutant that mimics the hyperphosphorylated state is prevented from stable association with replication centers in vivo. The lack of association with sites of DNA synthesis is also observed for endogenous hyperphosphorylated RPA and is not a result of competition with the nonphosphorylated protein. Importantly, the RPARPA2D protein has activity equivalent to the wild-type protein both in ssDNA binding (4) and in SV40 DNA replication in vitro. The inherent activity of hyperphosphorylated RPA or RPARPA2D in vivo also appears normal because genotoxic stress causes these RPA species to localize to DNA damage foci similarly to endogenous RPA2 and RPA2wt. Our data therefore indicate that the chromosomal DNA replication machine has the ability to discriminate between RPA species with different phosphorylation states. In addition to providing a means to regulate RPA loading and hence DNA replication, RPA phosphorylation also has the potential to mark sites of DNA damage or replication stress for recruitment of repair factors.
Our data suggest a novel feature of eukaryotic DNA replication, namely, that RPA is actively loaded onto the ssDNA by the chromosomal replication machinery. This model arises from the fact that RPARPA2D, and by inference hyperphosphorylated RPA, is inherently active in binding the ssDNA at a DNA replication fork but is unable to do so in vivo. The most logical explanation is that, as the duplex DNA is unwound by the advancing DNA helicase, the hypophosphorylated RPA is loaded onto the ssDNA by protein components of the replication fork machinery. One could easily envision, for example, that the minichromosome maintenance (MCM) complex, suggested to be the eukaryotic replicative helicase (29) and known to interact with RPA (55), would load RPA molecules in a step-by-step fashion as the ssDNA is generated. Selective binding of nonphosphorylated RPA (i.e., endogenous RPA, RPARPA2wt, or RPARPA2A) to MCM would therefore allow this RPA species to bind only to unwound DNA. (The MCM complex is not involved in SV40 DNA replication.) However, RPA interacts with various proteins, including the DNA polymerase α-DNA primase complex (14), and RPA phosphorylation has been found to inhibit the association with DNA polymerase α (34). Thus, discrimination of the RPA phosphorylation state can be achieved by these or other replication factors. One alternative model that does not require concerted RPA loading would involve a discrimination filter that prevents access of the phosphorylated RPA to the replication fork. The nature of such a filter would be difficult to envisage.
DNA-damaging stress relieves the inhibition of RPARPA2D chromatin binding and causes RPARPA2D association with DNA damage foci, as evident by colocalization with γ-H2AX. That HU causes RPARPA2D foci to form and increases the level of RPA2 phosphorylation while aphidicolin does neither indicates that replication fork blockage is not sufficient for RPARPA2D chromatin binding but that the presence of DNA damage or aberrant replication fork structures is also required. This conclusion is strengthened by our observation that inhibition of ATR- or ATM-mediated checkpoint response by caffeine stimulates the rate of RPA association with DNA damage foci. Mutation of MEC1, the Saccharomyces cerevisiae ATR homolog, is known to cause the collapse of DNA replication forks that have been stalled by treatment with HU or methyl methanesulfonate (32, 45), and such treatment leads to the production of long ssDNA regions (43). Because of the high affinity of RPA for ssDNA (27, 52), we propose that the increased availability of ssDNA releases the constraints on RPA loading seen during normal S-phase progression. Thus, under damage conditions, the RPA phosphorylation state no longer regulates the association of RPA with chromatin.
Our data indicate that hyperphosphorylated RPA is preferentially associated with sites of DNA damage. The specific association of repair factors with this modified form of RPA would therefore provide a mechanism to recruit repair factors to sites of DNA damage. Interestingly, the ATRIP-ATR complex has been found to sense damaged DNA by recognition of RPA-ssDNA complexes. Clearly, RPA phosphorylation has the potential to regulate the binding of ATRIP-ATR and thereby modify the cellular checkpoint response. Although our examination of RPA2D expression did not detect any notable effects on cell cycle progression, it will be interesting to examine whether RPA2D and RPA2A expression in cells lacking endogenous RPA alters cellular proliferative capacity or response to DNA damage.
Finally, our data indicate that hyperphosphorylation of RPA can limit its ability to support chromosomal DNA replication. It is unlikely that this mechanism alone could cause significant reductions in the level of DNA synthesis during genotoxic stress. Under severe stress conditions, such as 1-h exposure to 1 μM CPT (Fig. 8N) or irradiation with 30 J of UV light/m2 (53), the hyperphosphorylated form of RPA contributes ∼50% of the total RPA pool prepared from asynchronous cells. Even though the fraction of hyperphosphorylated RPA may be higher in S-phase cells, these data suggest that enough hypophosphorylated RPA would be available to sustain DNA replication. That being said, we and others have found that stress conditions also lead to the inhibition of RPA activity by other processes (9, 30, 48), including sequestration of RPA by nucleolin (12, 47). Combined, these data suggest that inhibition of RPA activity by multiple mechanisms can serve to repress chromosomal DNA replication following stress.
Acknowledgments
We thank Kyung Kim and Diana Dimitrova for helpful discussions during the course of these experiments, Kristine Carta for expert technical assistance, and John Hirsch for assistance with FACS analysis.
J.A.B. was supported by NIH grant AI29963, DOD Breast Cancer Research Program DAMD17-03-1-0299, Philip Morris grant 15-B0001-42171, and the NYU Cancer Institute and the Rita J. and Stanley Kaplan Comprehensive Cancer Center (NCI P30CA16087). M.S.W. was supported by NIH grant GM44721.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Ariza, R. R., S. M. Keyse, J. G. Moggs, and R. D. Wood. 1996. Reversible protein phosphorylation modulates nucleotide excision repair of damaged DNA by human cell extracts. Nucleic Acids Res. 24:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartek, J., and J. Lukas. 2001. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13:738-747. [DOI] [PubMed] [Google Scholar]
- 4.Binz, S. K., Y. Lao, D. F. Lowry, and M. S. Wold. 2003. The phosphorylation domain of the 32-kDa subunit of replication protein A modulates RPA-DNA interactions: evidence for an intersubunit interaction. J. Biol. Chem. 278:35584-35591. [DOI] [PubMed] [Google Scholar]
- 5.Borowiec, J. 1992. Inhibition of structural changes in the simian virus 40 core origin of replication by mutation of essential origin sequences. J. Virol. 66:5248-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowiec, J. A., F. B. Dean, and J. Hurwitz. 1991. Differential induction of structural changes in the simian virus 40 origin of replication by T antigen. J. Virol. 65:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brush, G. S., C. W. Anderson, and T. J. Kelly. 1994. The DNA-activated protein kinase is required for the phosphorylation of replication protein A during simian virus 40 DNA replication. Proc. Natl. Acad. Sci. USA 91:12520-12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 9.Carty, M. P., M. Zernik-Kobak, S. McGrath, and K. Dixon. 1994. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J. 13:2114-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, D. W., S. C. Son, W. Block, R. Ye, K. K. Khanna, M. S. Wold, P. Douglas, A. A. Goodarzi, J. Pelley, Y. Taya, M. F. Lavin, and S. P. Lees-Miller. 2000. Purification and characterization of ATM from human placenta. A manganese-dependent, wortmannin-sensitive serine/threonine protein kinase. J. Biol. Chem. 275:7803-7810. [DOI] [PubMed] [Google Scholar]
- 11.Cortez, D. 2003. Caffeine inhibits checkpoint responses without inhibiting the ataxia-telangiectasia-mutated (ATM) and ATM- and Rad3-related (ATR) protein kinases. J. Biol. Chem. 278:37139-37145. [DOI] [PubMed] [Google Scholar]
- 12.Daniely, Y., and J. A. Borowiec. 2000. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J. Cell Biol. 149:799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimitrova, D. S., and D. M. Gilbert. 2000. Stability and nuclear distribution of mammalian replication protein A heterotrimeric complex. Exp. Cell Res. 254:321-327. [DOI] [PubMed] [Google Scholar]
- 14.Dornreiter, I., L. F. Erdile, I. U. Gilbert, D. von Winkler, T. J. Kelly, and E. Fanning. 1992. Interaction of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 11:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durocher, D., and S. P. Jackson. 2001. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol. 13:225-231. [DOI] [PubMed] [Google Scholar]
- 16.Dutta, A., and B. Stillman. 1992. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. EMBO J. 11:2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fotedar, R., and J. M. Roberts. 1992. Cell cycle regulated phosphorylation of RPA-32 occurs within the replication initiation complex. EMBO J. 11:2177-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gately, D. P., J. C. Hittle, G. K. T. Chan, and T. J. Yen. 1998. Characterization of ATM expression, localization, and associated DNA-dependent protein kinase activity. Mol. Biol. Cell 9:2361-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golub, E. I., R. C. Gupta, T. Haaf, M. S. Wold, and C. M. Radding. 1998. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 26:5388-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haaf, T., E. Raderschall, G. Reddy, D. C. Ward, C. M. Radding, and E. I. Golub. 1999. Sequestration of mammalian Rad51-recombination protein into micronuclei. J. Cell Biol. 144:11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall-Jackson, C. A., D. A. Cross, N. Morrice, and C. Smythe. 1999. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene 18:6707-6713. [DOI] [PubMed] [Google Scholar]
- 22.Hassell, J. A., and B. T. Brinton. 1996. SV40 and polyomavirus DNA replication, p. 639-677. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Henricksen, L. A., T. Carter, A. Dutta, and M. S. Wold. 1996. Phosphorylation of human replication protein A by the DNA-dependent protein kinase is involved in the modulation of DNA replication. Nucleic Acids Res. 24:3107-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. [PubMed] [Google Scholar]
- 25.Huang, W., and R. L. Erikson. 1994. Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc. Natl. Acad. Sci. USA 91:8960-8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iftode, C., and J. A. Borowiec. 1998. Unwinding of origin-specific structures by human replication protein A occurs in a two-step process. Nucleic Acids Res. 26:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iftode, C., Y. Daniely, and J. A. Borowiec. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34:141-180. [DOI] [PubMed] [Google Scholar]
- 28.Kenny, M. K., U. Schlegel, H. Furneaux, and J. Hurwitz. 1990. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem. 265:7693-7700. [PubMed] [Google Scholar]
- 29.Lei, M., and B. K. Tye. 2001. Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci. 114:1447-1454. [DOI] [PubMed] [Google Scholar]
- 30.Liu, J. S., S. R. Kuo, M. M. McHugh, T. A. Beerman, and T. Melendy. 2000. Adozelesin triggers DNA damage response pathways and arrests SV40 DNA replication through replication protein A inactivation. J. Biol. Chem. 275:1391-1397. [DOI] [PubMed] [Google Scholar]
- 31.Liu, V. F., and D. T. Weaver. 1993. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol. Cell. Biol. 13:7222-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 33.Niu, H., H. Erdjument-Bromage, Z. Q. Pan, S. H. Lee, P. Tempst, and J. Hurwitz. 1997. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J. Biol. Chem. 272:12634-12641. [DOI] [PubMed] [Google Scholar]
- 34.Oakley, G. G., S. M. Patrick, J. Yao, M. P. Carty, J. J. Turchi, and K. Dixon. 2003. RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry 42:3255-3264. [DOI] [PubMed] [Google Scholar]
- 35.Pan, Z.-Q., A. A. Amin, E. Gibbs, H. Niu, and J. Hurwitz. 1994. Phosphorylation of the p34 subunit of human single-stranded-DNA-binding protein in cyclin A-activated G1 extracts is catalyzed by cdk-cyclin A complex and DNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 91:8343-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan, Z.-Q., C. H. Park, A. A. Amin, J. Hurwitz, and A. Sancar. 1995. Phosphorylated and unphosphorylated forms of human single-stranded DNA-binding protein are equally active in simian virus 40 DNA replication and in nucleotide excision repair. Proc. Natl. Acad. Sci. USA 92:4636-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, J. S., S. J. Park, X. Peng, M. Wang, M. A. Yu, and S. H. Lee. 1999. Involvement of DNA-dependent protein kinase in UV-induced replication arrest. J. Biol. Chem. 274:32520-32527. [DOI] [PubMed] [Google Scholar]
- 38.Philipova, D., J. R. Mullen, H. S. Maniar, J. Lu, C. Gu, and S. J. Brill. 1996. A hierarchy of SSB protomers in replication protein A. Genes Dev. 10:2222-2233. [DOI] [PubMed] [Google Scholar]
- 39.Raderschall, E., E. I. Golub, and T. Haaf. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA 96:1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 41.Sarkaria, J. N., E. C. Busby, R. S. Tibbetts, P. Roos, Y. Taya, L. M. Karnitz, and R. T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375-4382. [PubMed] [Google Scholar]
- 42.Shao, R. G., C. X. Cao, H. Zhang, K. W. Kohn, M. S. Wold, and Y. Pommier. 1999. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 18:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 44.Stigger, E., F. B. Dean, J. Hurwitz, and S.-H. Lee. 1994. Reconstitution of functional human single-stranded DNA-binding protein from individual subunits expressed by recombinant baculoviruses. Proc. Natl. Acad. Sci. USA 91:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 46.Wang, H., J. Guan, A. R. Perrault, Y. Wang, and G. Iliakis. 2001. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res. 61:8554-8563. [PubMed] [Google Scholar]
- 47.Wang, Y., J. Guan, H. Wang, D. Leeper, and G. Iliakis. 2001. Regulation of DNA replication after heat shock by replication protein A-nucleolin interactions. J. Biol. Chem. 276:20579-20588. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Y., A. R. Perrault, and G. Iliakis. 1998. Replication protein A as a potential regulator of DNA replication in cells exposed to hyperthermia. Radiat. Res. 149:284-293. [PubMed] [Google Scholar]
- 49.Wittekind, M., J. Reizer, J. Deutscher, M. H. Saier, and R. E. Klevit. 1989. Common structural changes accompany the functional inactivation of HPr by seryl phosphorylation or by serine to aspartate substitution. Biochemistry 28:9908-9912. [DOI] [PubMed] [Google Scholar]
- 50.Wobbe, C. R., F. Dean, L. Weissbach, and J. Hurwitz. 1985. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc. Natl. Acad. Sci. USA 82:5710-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wobbe, C. R., L. Weissbach, J. A. Borowiec, F. B. Dean, Y. Murakami, P. Bullock, and J. Hurwitz. 1987. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc. Natl. Acad. Sci. USA 84:1834-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wold, M. S. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 53.Zernik-Kobak, M., K. Vasunia, M. Connelly, C. W. Anderson, and K. Dixon. 1997. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 272:23896-23904. [DOI] [PubMed] [Google Scholar]
- 54.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]
- 55.Zou, L., and B. Stillman. 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20:3086-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]