Abstract
Suppression of protein synthesis through phosphorylation of the translation initiation factor α subunit of eukaryotic initiation factor 2 (eIF2α) is known to occur in response to many forms of cellular stress. To further study this, we have developed novel cell lines that inducibly express FLAG-tagged versions of either the phosphomimetic eIF2α variant, eIF2α-S51D, or the phosphorylation-insensitive eIF2α-S51A. These variants showed authentic subcellular localization, were incorporated into endogenous ternary complexes, and were able to modulate overall rates of protein synthesis as well as influence cell division. However, phosphorylation of eIF2α failed to induce cell death or sensitize cells to killing by proapoptotic stimuli, though it was able to inhibit viral replication, confirming the role of eIF2α in host defense. Further, although the eIF2α-S51A variant has been shown to transform NIH 3T3 cells, it was unable to transform the murine fibroblast 3T3 L1 cell line. To therefore clarify this issue, we explored the role of eIF2α in growth control and demonstrated that the eIF2α-S51A variant is capable of collaborating with hTERT and the simian virus 40 large T antigen in the transformation of primary human kidney cells. Thus, dysregulation of translation initiation is indeed sufficient to cooperate with defined oncogenic elements and participate in the tumorigenesis of human tissue.
The initiation of protein synthesis in eukaryotes is a highly complex and conserved process involving at least 13 initiation factors, many of which are themselves assembled from numerous subunits (3, 51, 56). The regulation of protein synthesis can be greatly affected following exposure to various forms of cell stress including nutrient deprivation, contact with biologic pathogens, fluctuations in temperature, or the presence of toxic compounds (18, 35). A key translation factor that is a frequent target of regulation by stress-sensitive kinases is the α subunit of the eukaryotic translation initiation factor 2 complex (eIF2α) (13, 14, 63, 72). eIF2 is a heterotrimer composed of three subunits (α, β, and γ) which functions by associating with GTP and the initiator Met-tRNAi to form a ternary complex (48, 62). The ternary complex delivers the Met-tRNAi to the 40S ribosomal subunit which, along with other translation factors including eIF3, forms the 43S preinitiation structure (8, 50, 69). Newly assembled 43S ribosome-eIF complexes associate with an mRNA transcript near the 5′ m7G cap and advance along the transcript in a 3′ direction until an AUG start codon is located within the context of an appropriate Kozak sequence (25, 44, 53). Once the AUG codon has been recognized, GTP bound by eIF2 is hydrolyzed in a reaction catalyzed, in part, by another initiation factor, eIF5 (4). The Met-tRNAi is subsequently released from the ternary complex to initiate nascent peptide chain synthesis, and eIF2 dissociates from the 43S initiation complex. The GDP associated with the free eIF2 is exchanged for GTP by the activity of the eIF2B complex, which is itself a heteropentamer comprised of α, β, γ, δ, and ɛ subunits (2, 54, 59). Following GTP exchange, eIF2 is incorporated into a new ternary complex and the next round of initiation begins (37).
Phosphorylation on serine 51 of eIF2α by stress-responsive kinases causes eIF2 to acquire an increased affinity for, and functionally sequester, the GTP exchange factor eIF2B, which is required for maintaining eIF2 activity (40). Thus, in response to stress, eIF2α kinases can depress global translation rates by inhibiting eIF2-GTP recycling and, subsequently, initiation of translation. For example, accumulation of misfolded proteins in the endoplasmic reticulum (ER) leads to activation of an ER-resident eIF2α kinase alternately named PKR-like endoplasmic reticular kinase (PERK) and pancreatic eIF2α kinase (33, 67). Similarly, the yeast eIF2α kinase GCN2 and its mammalian homologues function as cytoplasmic sensors of amino acid levels via two His-tRNA-like domains in their carboxy termini (70). GCN2 kinase activity is up-regulated under starvation conditions during which the levels of charged tRNAs fall (21). The heme-regulated inhibitor kinase, in contrast, is predominantly expressed in erythroid cells and is negatively regulated by hemin binding (11, 12). In addition, the interferon-inducible, double-stranded RNA (dsRNA)-regulated kinase PKR is activated by dsRNA produced during viral infections and functions in host defense to prevent translation of viral transcripts (6, 7, 43, 52, 71).
A number of recent reports, however, have indicated that elevated levels of phosphorylated eIF2α may actually serve to specifically enhance the translation of selected mRNAs encoding proteins that require production in response to stress in mammalian cells. The mechanism of transcript-specific translational up-regulation has been reported to be, in part, dependent upon upstream open reading frames (uORFs) in the 5′ untranslated region (UTR) of the mRNA (31, 65). Under normal physiologic conditions, these short uORFs lower the efficiency of translation, presumably by impeding the progress of the scanning ribosome. However, under conditions in which the levels of phosphorylated eIF2α rise and levels of available ternary complex fall, these uORFs may favor the association of the transcript with active ribosomes. Examples to date of transcripts that are regulated in this manner include the transcription factors GCN4 in Saccharomyces cerevisiae and ATF4 in mammalian cells (17, 20, 31). Additionally, some mRNAs with 5′ UTRs containing internal ribosome entry site elements are also translationally up-regulated when eIF2α is phosphorylated (26).
To further clarify the role of eIF2α in apoptosis, transformation, and gene regulation, we have developed inducible and constitutive expression systems for wild-type (WT) and variant forms of eIF2α. Here we report that regulation of translation initiation through eIF2α is sufficient to inhibit viral replication in the absence of other eIF2-independent stress-responsive pathways but cannot account for the induction of apoptosis or for aspects of the broad gene regulation observed upon activation of stress-responsive kinases. Additionally, we demonstrate for the first time that a translation factor can transform human cells in collaboration with defined genetic elements hTERT and simian virus 40 large T antigen, collectively confirming the importance of translational regulation in tumorigenesis.
MATERIALS AND METHODS
Plasmid construction.
The pTREFLAG vector was derived from the pTRE vector (pUHD15-neo1) (28) by the insertion of the FLAG tag coding sequence preceded by an in-frame ATG initiation codon into the SacII/NdeI restriction sites in pTRE. pTREFLAG-eIF2αWT was derived from pTREFLAG by the insertion of the WT eIF2α sequence into the NdeI/BamHI restriction sites. pTREFLAG-eIF2αS51A and pTREFLAG-eIF2αS51D were developed from pTREFLAG-eIF2αWT by using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) and contain point mutations rendering a serine-to-alanine or serine-to-aspartic-acid change, respectively, at position 51. pFB-NeoWTeIF2α, pFB-NeoeIF2αS51D, and pFB-NeoeIF2αS51A were constructed by inserting the WT or variant eIF2α sequences into the EcoRI/BamHI restriction sites in the pFB-Neo vector (Stratagene). pFB-NeoWTRas and pFB-NeoRasV12 vectors were generated by insertion of the WT Ras or RasV12 coding sequences into the EcoRI/BamHI restriction sites in pFB-Neo. pVPack-GP and pVPack env expression vectors were purchased from Stratagene. The luciferase reporter constructs were obtained by cloning the entire 5′ leader sequence of the Fas gene into the PGL3 control vector (Promega, Madison, Wis.), in frame with the ATG start codon from the luciferase ORF.
Cell lines.
3T3 L1 cells (Clontech, Palo Alto, Calif.) stably transfected with a pTETOFF vector (pUHD15-neo1) (28) were subsequently cotransfected with the pTREFLAG-eIF2αWT, pTREFLAG-eIF2αS51A, or pTREFLAG-eIF2αS51D vector along with pTK-HYG (Clontech) by using Lipofectamine (Gibco-BRL, Grand Island, N.Y.), according to the manufacturer's protocol. Immediately upon removal of the DNA-liposome complexes, doxycycline (DOX; Sigma Chemical, St. Louis, Mo.) was added to the medium (5 μg/ml). After 48 h of recovery, cells were selected with 250 μg of G418 (Gibco-BRL)/ml and 100 μg of hygromycin (Clontech)/ml. Resistant colonies were isolated and expanded. Protein expression and inducibility of individual clones were examined by Western blotting with lysate from cells passaged for 1 week in the presence or absence of DOX. NIH 3T3 and 3T3 L1 cell lines were obtained from the American Type Culture Collection.
Immunoblot analysis.
Protein extracts from cell lines were prepared by disrupting cells in lysis buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 1 mM dithiothreitol, 2 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 100 U of aprotinin/ml, 1% NP-40) (all reagents obtained from Sigma). Supernatants were added to an equal volume of loading buffer (5% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 20% glycerol, and 150 mM Tris-HCl [pH 7.50]) and boiled for 2 min prior to being loaded on an SDS-polyacrylamide gel. After electrophoretic resolution, proteins were transferred to nitrocellulose membranes, incubated for 1 h in blocking solution (phosphate-buffered saline [PBS], 0.1% Tween 20, 10% nonfat dry milk) at room temperature, and incubated with specific monoclonal or polyclonal antibodies overnight at 4°C. Membranes were washed three times in PBS-Tween 20 and incubated with horseradish peroxidase-conjugated secondary anti-mouse or anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, West Grove, Pa.). Following a second washing, proteins were visualized by addition of chemiluminescent substrate (Pierce Chemicals, Rockford, Ill.).
FLAG M5 monoclonal antibody (MAb) and β-actin MAbs were obtained from Sigma. Antibodies to Fas, Bax, and Bcl-X and Bcl-2 were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Anti-FADD MAb was obtained from Upstate Biotechnology Inc. (Lake Placid, N.Y.). Anti-Ras MAb was purchased from Oncogene Inc. (Cambridge, Mass.).
Protein synthesis analysis.
Equal numbers of cells passaged in either the presence or the absence of DOX were split into six-well dishes and allowed to settle for 48 h. Cells were labeled with 75 μCi of [35S]methionine (Amersham Pharmacia Biotech, Piscataway, N.J.)/ml in methionine-free medium (Gibco-BRL) supplemented with 10% dialyzed fetal bovine serum (FBS; Gibco-BRL) for 30 min at 37°C. Cells were washed three times in warm PBS, detached with trypsin-EDTA, counted, and finally disrupted with lysis buffer. Lysate volumes of equivalent cell numbers were added to bovine serum albumin-methionine carrier buffer (0.002% bovine serum albumin [Pierce], 0.01% methionine [Sigma], 0.1 volume of urea sample buffer). Urea sample buffer consisted of 50% urea (Sigma), 0.2% NP-40, 0.05% Coomassie blue, and 0.5% β-mercaptoethanol. Protein bound counts were precipitated by the addition of 0.5 volume of ice-cold 50% trichloroacetic acid (TCA; Sigma) and centrifuged at 14,000 × g for 5 min at 4°C. Pellets were washed with 10% TCA at 4°C and resuspended in Soluene 350 (Packard BioScience, Boston, Mass.). Counts were analyzed by liquid scintillation.
Ternary complex immunoprecipitations.
Coimmunoprecipitations were performed by transfecting subconfluent monolayers of 293T cells (American Type Culture Collection) in six-well dishes with either a vector expressing FLAG-tagged WT eIF2α or a control plasmid expressing a nonspecific FLAG-tagged protein. Forty-eight hours following transfection, cells were lysed with lysis buffer. Whole-cell lysate was added to low-salt buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 0.2% Triton X-100, 0.1% β-mercaptoethanol) and precleared by incubation with 50 μl of protein G agarose (Invitrogen Corp., Carlsbad, Calif.) and 2.5 μl of normal rabbit immunoglobulin G (Santa Cruz Biotechnology) for 1 h at 4°C. Protein G was removed by centrifugation, and an additional 50 μl of protein G slurry was added along with 5 μl of rabbit polyclonal anti-FLAG antiserum (Sigma). Lysates were incubated for 2 h at 4°C. Protein G was pelleted by centrifugation, and the lysate was removed. Protein G pellet was washed three times with 1 ml of low-salt buffer and then heated.
RNA analysis.
Cell lines were induced in the absence of DOX for the indicated periods, and total RNA was harvested with the RNeasy minikit (Qiagen, Valencia, Calif.). RNA was incubated with an α-32P-labeled mAPO-2 or mAPO-3 probe set (Riboquant; PharMingen, San Diego, Calif.) according to the manufacturer's protocol. Following RNase treatment, protected probes were resolved using 5% polyacrylamide gels and imaged with autoradiography.
Retrovirus production.
Individual retrovirus was produced according to the manufacturer's instructions included with pVPack Moloney murine leukemia virus-based retroviral expression vectors (Stratagene). Briefly, 293T cells at 50% confluency in 10-cm-diameter dishes were transfected with 5 μg each of pVPack-GP, pVPack-Eco or pVPack-VSV-G, and pFB-Neo or pFB-Neo containing the appropriate inserted gene, with the use of Lipofectamine. Retrovirus-containing supernatants were harvested after 48 h and frozen at −80°C until needed. Target cells were subsequently infected by the addition of retrovirus-containing supernatants to the medium along with DEAE-dextran to a final concentration of 10 μg/ml. Target cells were selected 24 h following infection by the addition of 400 μg of neomycin (Gibco-BRL)/ml.
Apoptosis analysis.
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) was done using the fluorescein in situ cell death detection kit (Roche, Mannheim Germany) according to the manufacturer's instructions. The annexin V-propidium iodide (PI) binding assay kit was purchased from R & D Systems (Minneapolis, Minn.). Quantitation of cell killing in response to apoptotic stimuli was done using trypan blue exclusion following 12 and 24 h of treatment. Jo-2 antibody was used from 0.1 to 1.0 μg/ml. Tumor necrosis factor (TNF) was used from 10 to 750 ng/ml. Poly(IC) was used in concentrations from 0.25 to 2 μg/ml. Soluble recombinant TRAIL was used in concentrations from 10 to 750 ng/ml.
Oncogenic collaboration.
One hundred thousand HEK cells stably transduced with large T antigen and hTERT (a gift from Robert Weinberg) (29) were transduced with retrovirus encoding WT eIF2α, eIF2α-S51A, or RasV12, according to the manufacturer's instructions. Forty-eight hours following transduction, cells were selected with 0.5 μg of puromycin (Invitrogen)/ml for 10 days. Remaining colonies were subcloned and photographed for morphology.
Anchorage-independent growth.
Cell lines were trypsinized and counted, and 500 or 5,000 cells were mixed with 1 ml of warm Dulbecco modified Eagle medium (DMEM), 10% FBS, and 0.5% low-melting-temperature agarose (LMTA). Cells were then layered onto 1 ml of DMEM-10% FBS with 0.75% LMTA that had previously been added to individual wells of a six-well plate. Finally an additional layer consisting of DMEM with 0.75% LMTA was added over the top. Cells were cultured at 37°C and 5% CO2 for approximately 3 weeks (21 days). Colony growth was scored using light microscopy.
RESULTS
Inducible expression of eIF2α variants in 3T3 L1 cells.
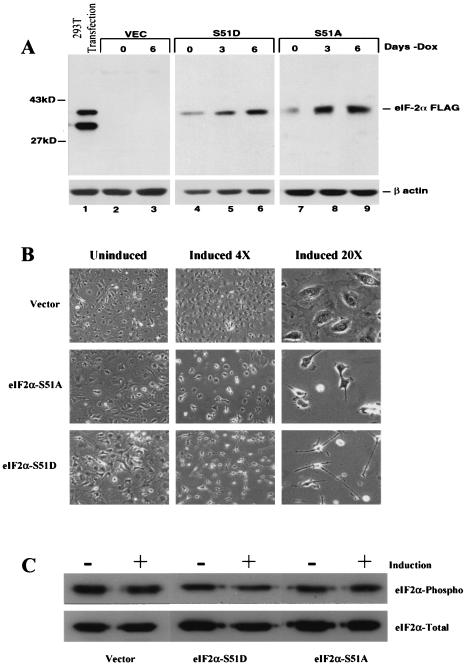
To analyze the specific effects of eIF2α phosphorylation on the cell in the absence of other eIF2-independent stress signaling cascades, we established novel 3T3 L1 cell lines that inducibly express a FLAG-tagged phosphomimetic variant of eIF2α, referred to as eIF2α-S51D, or the phosphorylation-incompetent variant eIF2α-S51A (61). Clonal 3T3 L1 cell populations were isolated which express the exogenous forms of eIF2α under the control of a DOX-repressible promoter (28). Removal of DOX from the culture medium induced a dose-dependent accumulation of both the eIF2α-S51A and the eIF2α-S51D variants that was detectable after 3 days by using a MAb specifically recognizing the FLAG epitope tag (42) (Fig. 1A). Maximal protein levels of eIF2α-S51A were observed approximately 3 days postinduction. In contrast, eIF2α-S51D accumulated more slowly, reaching apparent maximal expression after 6 days (Fig. 1A, lanes 4 to 9). These data collectively demonstrate the successful inducible expression of eIF2α variants in 3T3 L1 cells.
FIG. 1.
(A) Inducible expression of eIF2α variants in 3T3 L1 cells. Cells carrying vector alone or tetracycline-inducible plasmids expressing empty vector, FLAG eIF2α-S51D, or FLAG eIF2α-S51A were induced by the withdrawal of DOX, and whole-cell lysates were harvested at 0, 3, and 6 days postinduction. Cell extracts were analyzed for the expression of FLAG-tagged proteins by immunoblotting with a MAb directed against the FLAG epitope as described in Materials and Methods. Lane 1 is whole-cell lysate harvested from 293T cells transiently transfected for 48 h with a plasmid expressing FLAG-tagged WT eIF2α. (B) Morphological changes induced by expression of eIF2α variants. The vector-, FLAG eIF2α-S51D-, and FLAG eIF2α-S51A-expressing cell lines were induced to express the indicated variants for 6 days, and cell morphology was monitored at 0 and 6 days postinduction by phase-contrast microscopy at ×4 and ×20 magnifications. (C) Levels of total and phosphorylated eIF2α in DOX-inducible cell lines. Whole-cell lysates from 3T3 L1 DOX-inducible cells either uninduced or induced to express the indicated eIF2α variants for 6 days were harvested, electrophoresed, and probed with MAbs directed against either total eIF2α or eIF2α specifically phosphorylated on serine 51.
It was found that, following removal of DOX from the culture medium, a pronounced change in the morphology of the eIF2α-S51D- and eIF2α-S51A-expressing cell lines was evident, which began approximately 48 h postinduction (Fig. 1B). A number of clones expressing comparable levels of the two FLAG-tagged constructs were screened, and a similar morphological change was evident, indicating that the observed effect was not clone dependent (data not shown). Similar morphological changes were not observed in vector-containing cell lines (Fig. 1B), and those eIF2α-expressing clones demonstrating the most profound alteration in morphology were selected for further study. Typically cells expressing the eIF2α-S51D variant exhibited a thin and spindle-like morphology while, in contrast, cells expressing the eIF2α-S51A variant became overall smaller and developed a refractile cytoplasm (Fig. 1B) but did not form foci in culture and were not found to exhibit other hallmarks of malignancy (data not shown).
Recent reports have shown that phosphorylation of eIF2α induces a GADD34-dependent phosphatase activity that acts as a negative regulator of the stress response by specifically dephosphorylating eIF2α (55). To further characterize our inducible cell lines and more specifically to ascertain whether such a stress response was being activated by the expression of eIF2α-S51D in our cell lines, the levels of endogenous phosphorylated eIF2α were assayed using a MAb specifically recognizing phosphorylated eIF2α before, and immediately following, induction of our eIF2α variants (Fig. 1C). We were, however, unable to demonstrate any changes in the levels of the endogenous pools of phosphorylated eIF2α in response to expression of either the eIF2α-S51D or eIF2α-S51A variant.
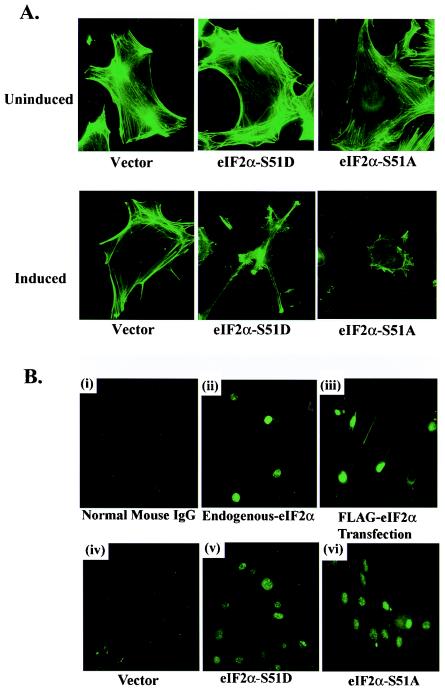
Since available data indicate that alterations in cellular morphology can be associated with changes in the organization of the underlying cytoskeleton, we therefore further characterized the altered morphology caused by the S51A and S51D variants, by examining the structure of polymerized F-actin stress fibers, before and after induction by immunoblotting and immunofluorescence (Fig. 2A). These data indicated that expression of either eIF2α-S51D or eIF2α-S51A did indeed produce changes in the organization of the polymerized actin filaments, although the levels of actin protein did not necessarily appear to fluctuate as determined by Western blotting (Fig. 1A).
FIG. 2.
(A) Polymerized F-actin filament organization in FLAG eIF2α variant-expressing cells. Vector-, FLAG S51A-, and FLAG S51D-expressing cell lines grown on glass coverslips were permeabilized and stained with fluorescein isothiocyanate-conjugated phalloidin proteins that specifically bind to polymerized F-actin filaments on day 0 or day 6 postinduction. Actin filaments were visualized by immunofluorescence microscopy. (B) Subcellular localization of endogenous and FLAG-tagged eIF2α. Staining of 3T3 L1 cells with nonspecific normal mouse immunoglobulin G was included as a negative control (i). Expression and localization of endogenous eIF2α in normal 3T3 L1 cells were determined using immunofluorescence with a MAb directed against native eIF2α (ii). Localization of FLAG-tagged eIF2α in normal 3T3 L1 cells was determined by immunofluorescence with a MAb directed against the FLAG epitope (clone M5; Sigma) following transient transfection for 48 h with a vector expressing FLAG-tagged WT eIF2α (iii). Localization of inducible FLAG-tagged eIF2α variants in empty vector-, FLAG eIF2α-S51A-, or FLAG eIF2α-S51D-expressing 3T3 L1 cell lines was determined by immunofluorescence with an antibody to the FLAG epitope following 6 days of induction in the absence of DOX (iv to vi). IgG, immunoglobulin G.
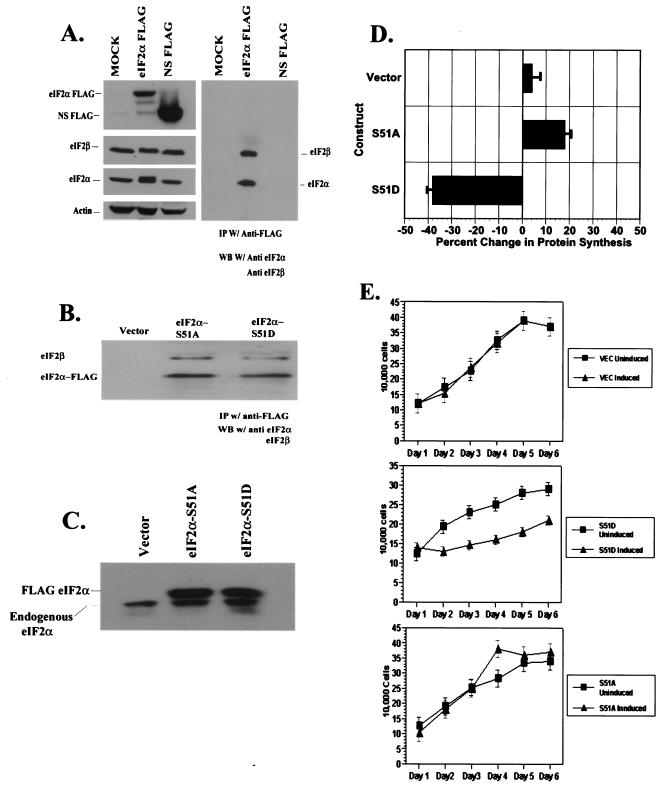
As the inducible eIF2α variants carry an 8-amino-acid N-terminal FLAG epitope tag, we next attempted to confirm that these epitope-tagged versions displayed authentic subcellular localization. The cellular localization of endogenous eIF2α was first determined using immunofluorescence staining in normal 3T3 L1 cells with monoclonal and polyclonal antibodies directed against eIF2α (Fig. 2B). Unexpectedly, endogenous eIF2α was almost exclusively localized to the nucleus (Fig. 2B, panel ii). 3T3 L1 cells transiently transfected with WT FLAG-tagged eIF2α and stained with a MAb directed against the FLAG epitope also indicated that the FLAG-tagged versions appeared exclusively nuclear (Fig. 2B, panel iii). Immunofluorescence staining of our vector-, eIF2α-S51A-, or eIF2α-S51D-expressing cell line with the anti-FLAG MAb confirmed nuclear localization of our FLAG-tagged eIF2α variants (Fig. 2B, panels iv to vi), strongly suggesting that the recombinant proteins were being authentically compartmentalized within the cell. These data are in agreement with other recent reports indicating that a large fraction of the recombinant eIF2α localizes to the nucleus (16, 41). As a control to ensure that the FLAG tag was not sterically interfering with the incorporation of eIF2α into eIF2 along with endogenous eIF2β and eIF2γ, we also transiently overexpressed either our WT FLAG eIF2α or a nonspecific FLAG-tagged protein and subsequently immunoprecipitated whole-cell lysates by using a polyclonal antiserum to the FLAG epitope. Our results showed that immunoprecipitated eIF2α specifically associated with endogenous eIF2β, indicating that FLAG-eIF2α variants are correctly combining with their natural binding partners in vivo (Fig. 3A). As additional confirmation, we then coimmunoprecipitated FLAG eIF2α and eIF2β from the inducible 3T3 L1 cell lines, establishing that recombinant eIF2α is indeed associated with at least one of the other members of the ternary complex and likely is fully functional (Fig. 3B). To ascertain the relative levels of overexpression of FLAG-tagged eIF2α compared to the endogenous protein, we separated the two forms on the basis of size by SDS-14% polyacrylamide gel electrophoresis (PAGE) and probed with a pan-eIF2α MAb (Fig. 3C). Densitometric analysis indicated that the FLAG-tagged proteins were expressed at levels two- to threefold greater than was the endogenous eIF2α. Collectively our data would indicate that the FLAG-tagged eIF2α is authentically recognized in the cell.
FIG. 3.
(A) Association of FLAG-tagged eIF2α with native ternary complexes in 293T cells. 293T cells were mock transfected or transiently transfected with plasmids encoding WT FLAG eIF2α or a nonspecific FLAG-tagged protein (NS FLAG). Forty-eight hours later, whole-cell lysates were harvested and split equally. One half was used in Western blotting to quantitate expression of transfected proteins. The blot was subsequently probed with antibodies to eIF2β and actin. The remaining lysate was used in an immunoprecipitation with a polyclonal antiserum recognizing the FLAG epitope. Immunoprecipitated proteins were resolved by SDS-PAGE and probed with antibodies recognizing eIF2α or eIF2β. (B) Association of FLAG-tagged eIF2α with native ternary complexes in inducible 3T3 L1 cells. Whole-cell lysates from empty vector-, eIF2α-S51A-, or eIF2α-S51D-expressing inducible 3T3 L1 cell lines were harvested following 6 days of induction and used in an immunoprecipitation with apolyclonal antiserum directed against the FLAG tag. (C) Quantitation of expression levels of endogenous and FLAG-tagged eIF2α in inducible 3T3 L1 cell lines. Whole-cell lysates harvested from empty vector-, eIF2α-S51A-, or eIF2α-S51D-expressing cell lines were harvested after 6 days of induction. Lysates were resolved by SDS-14% PAGE and probed with a MAb directed against eIF2α. (D) Regulation of translation rates by eIF2α variants. Triplicate samples of vector-, S51D-, or S51A-expressing inducible cell lines were passaged in the presence or absence of DOX for 6 days. Cells were transiently labeled with [35S]methionine for 15 min. Total protein from whole-cell lysates was precipitated with TCA and resuspended. Protein bound radioactive counts were measured via scintillation. Standard deviations are shown. (E) Regulation of growth rates by eIF2α variants. Equal numbers of cells from vector- and eIF2α variant-expressing cell lines were seeded in triplicate samples in 12-well dishes and passaged in the presence or absence of DOX for 6 days. Cells were trypsinized, and cell counts were determined daily.
Regulation of protein synthesis and growth rates by eIF2α variants.
To assess the capacity of our FLAG-tagged versions of the eIF2α variants to influence translation, we measured the rates of protein synthesis in each cell line in the presence of DOX (uninduced) and following 6 days of induction by using [35S]methionine labeling (Fig. 3D). As expected, vector-expressing control cells did not display significant perturbation in protein synthesis rates. However, eIF2α-S51D-expressing cells demonstrated a significant 40% decrease in synthesis rates as measured by label incorporation, while the eIF2α-S51A-expressing cell line evidenced an approximately 18% increase in protein synthesis. These data are in accord with reported values from an eIF2α-S51A knock-in mouse model that reported a 17 to 30% increase in translation rates and confirm the functional capacity of the FLAG-tagged variants (65).
As translation has been reported to influence cell growth and division, the growth characteristics of the inducible 3T3 L1 cell lines were analyzed following induction of both eIF2α variants. Cell numbers were measured in triplicate experiments for 6 days following the removal of DOX from the medium for 24 h. All cell lines remained viable and continued to divide. However, we observed a marked reduction in the growth rates of the eIF2α-S51D-expressing cells, with the reduction in cell number after 6 days approaching 35% compared to the identical cell line passaged in the presence of DOX (Fig. 3E). A slight but reproducible increase in cell number in the induced eIF2α-S51A-expressing cell lines was observed at the later time points, while no significant change was observed in the growth rates of the vector-expressing cell lines in the presence or absence of DOX (Fig. 3E). These data confirm that variant forms of eIF2α can strongly influence rates of protein synthesis and govern cell growth.
Phosphorylation of eIF2α does not induce apoptosis.
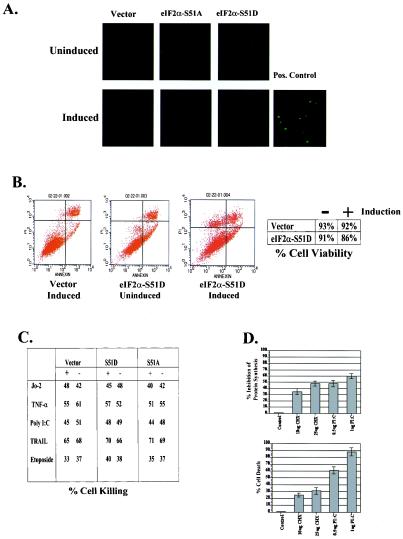
Previous investigations into eIF2α have shown that transient transfection of Cos cells with the eIF2α-S51D variant is capable of inducing apoptosis (68). In contrast, the eIF2α-S51A variant has been shown to be protective against apoptosis in some systems, implying a role for this translation factor in the regulation of cell death (27). To assess the capacity of eIF2α to regulate apoptosis in the absence of eIF2α-independent stress-responsive pathways, we assayed for the ability of eIF2α-S51D expression alone to induce apoptosis in our 3T3 L1 cells. However, TUNEL of induced and uninduced vector-, eIF2α-S51A-, and eIF2α-S51D-expressing cell lines did not demonstrate any significant induction of apoptosis attributable to overexpression of the eIF2α-S51D construct (Fig. 4A). Agonistic anti-Fas antibody was used as a positive control for the induction of apoptosis (Fig. 4A). As the TUNEL assay lacks sensitivity against cells that are in later stages of apoptosis and have detached from the culture dish, we subsequently assayed for cell killing by utilizing annexin V binding and PI staining. After induction for 4 days, the tissue culture medium was changed and the cells were induced for an additional 3 days. At this point cells from the tissue culture supernatant and those remaining attached were stained and analyzed by flow cytometry. Using this methodology, we observed only a slight decrease in viability in the induced eIF2α-S51D-expressing cells (Fig. 4B). Curiously, these dead cells accumulated as a PI singly staining population, indicating that they were in the very late stages of apoptosis or had undergone necrosis. However, eIF2α-S51D-dependent cell death represented only a 5% decrease in viability compared to uninduced cells, implying that phosphorylation of eIF2α does not potently induce spontaneous apoptosis (Fig. 4B).
FIG. 4.
(A) Expression of eIF2α-S51D does not induce apoptosis in 3T3 L1 cells. Equal numbers of cells from vector-, S51A-, and S51D-expressing inducible cell lines were seeded onto glass coverslips in 12-well dishes and passaged in the presence or absence of DOX for 6 days. Cells were TUNEL stained and imaged with fluorescence microscopy to detect fragmented nuclear DNA. One microgram of agonistic anti-Fas (Jo-2) antibody per milliliter was added to vector cells for 24 h as a positive control. (B) Annexin V-PI staining of 3T3 L1 cell lines. Vector- and eIF2α-S51D-expressing cell lines were left uninduced or induced for 6 days. All cells remaining attached to the substrate and those that had detached into the medium were harvested and pooled. Cells were subsequently stained with annexin V-PI and analyzed by flow cytometry to detect apoptotic death. (C) eIF2α variant expression does not sensitize cells to apoptotic death. Duplicate samples of vector-, S51A-, or S51D-expressing cell lines in the induced or uninduced states were treated with the indicated inducers of apoptosis for 24 h. Cells were subsequently harvested, and cell viability was determined by trypan blue exclusion. (D) General inhibition of protein synthesis does not initiate apoptosis. Normal 3T3 L1 cells were treated with the indicated concentrations of CHX or transfected with poly(IC). Six hours later protein synthesis rates were determined by metabolic labeling with [35S]methionine for 15 min. Protein bound counts were precipitated with TCA, resuspended, and quantitated by scintillation. Following 24 h of treatment cells were harvested, and cell viability was determined by trypan blue exclusion.
The lack of obvious apoptosis induced by the eIF2α-S51D variant led us to examine whether elevated levels of phosphorylated eIF2α might potentiate or protect eukaryotic cells from death induced by common proapoptotic stimuli. To investigate this possibility, we subjected our cell lines to treatment with a variety of prodeath agents including agonistic anti-mouse Fas antibody (Jo-2), murine TNF alpha (TNF-α), poly(I · C), soluble recombinant TRAIL, and the DNA-damaging drug etoposide. However, we did not observe a differential killing in any of our cell lines with or without induction in response to any of the agents that were used (Fig. 4C). Multiple concentrations of each agent yielded similar results (Fig. 4C and data not shown). The unexpected absence of cell death caused by expression of eIF2α-S51D led us to ask whether inhibition of translation in itself is sufficient to induce apoptosis or whether additional signaling events are required. To test this hypothesis, we treated 3T3 L1 cells with the general protein synthesis inhibitor cycloheximide (CHX) and with poly(IC), a potent activator of the eIF2α kinase PKR, in doses sufficient to reduce protein synthesis to rates roughly 50% of physiologic levels. Following 24 h of each treatment, we assayed for cell viability. Although CHX and poly(IC) reduced protein synthesis to an equivalent degree, the dsRNA-treated cells demonstrated a markedly decreased cell viability compared to CHX-treated populations (Fig. 4D). This result argues that, at least in the 3T3 L1 background, apoptosis is not initiated by a reduction in protein synthesis rates per se.
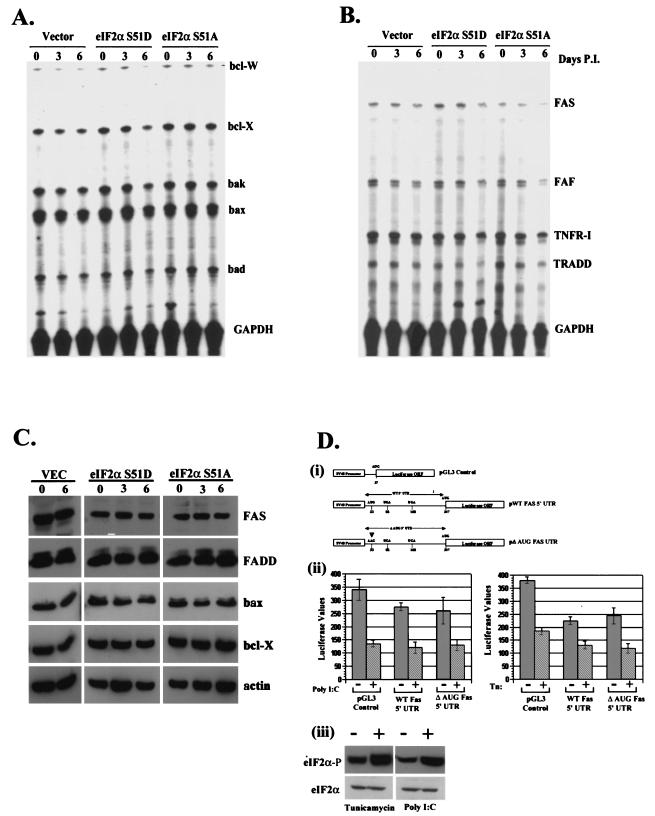
Previous work in our laboratory has demonstrated pronounced regulation of key apoptotic genes in 3T3 L1 cells, such as Fas, in response to overexpression of WT or mutant PKR (5). To assess whether PKR-dependent regulation of apoptotic proteins is a result of differential regulation of translation initiation, we examined the RNA and protein levels of these same genes in our inducible cell system including Fas, Bax, and Bcl-2. However, no difference in gene expression at the transcriptional or translational level was observed for the genes assayed (Fig. 5A to C). The Fas death receptor in particular has been identified previously as a dsRNA-responsive gene and has been shown to be regulated by PKR activity (5, 22). Although we did not observe an overt change in Fas expression levels in our inducible cell system, we confirmed this by obtaining the 5′ UTR of the Fas gene, which contains a uORF, and fused it to the ORF of firefly luciferase. We then assayed for possible subtle contributions of the UTR to translational regulation which may be dependent upon levels of phosphorylated eIF2α (17). In addition, a second construct was generated in which the putative start codon in the 5′ UTR was mutated to eliminate the uORF (Fig. 5D, panel i). These constructs, along with a control vector, were stably transfected into 3T3 L1 cells, and the pooled populations were treated with poly(IC) and the glycosylation inhibitor tunicamycin. Both of these agents have been previously shown to be powerful stimulators of eIF2α phosphorylation (5, 31). Phosphorylation of eIF2α was confirmed by Western blotting with whole-cell lysates harvested from 3T3 L1 cells probed with antibodies directed against phosphorylated eIF2α (Fig. 5D, panel iii). However, we did not observe any preferential translation of the constructs which contained the intact uORF under conditions in which eIF2α was phosphorylated (Fig. 5D, panel ii). These observations indicate that the PKR-induced translational regulation of Fas previously reported likely functions through pathways other than through the phosphorylation of eIF2α.
FIG. 5.
(A) Phosphorylation of eIF2α does not regulate the transcription of key apoptotic genes. Total RNA was harvested from vector-, S51A-, and S51D-expressing cell lines after 0, 3, or 6 days of induction and quantified by an RNase protection assay with radiolabeled probes derived from the indicated genes. (B) Total RNA was harvested from cells as described for panel A and quantitated by an RNase protection assay with radiolabeled probes derived from the indicated cytoplasmic apoptotic signaling genes. (C) eIF2α phosphorylation does not affect the level ofapoptotic proteins. Total-cell lysates harvested from vector-, S51A-, or S51D-expressing inducible cell lines on 0, 3, or 6 days postinduction were resolved by SDS-PAGE and probed with antibodies to the indicated proteins. (D) Translational regulation of Fas requires elements in addition to the 5′ UTR. Pooled populations of normal 3T3 L1 cells stably expressing luciferase, luciferase fused in frame with the WT Fas 5′ UTR, or luciferase fused to a mutant Fas 5′ UTR (i) were transfected with either 10 μg of poly(IC) or 10 μg of tunicamycin/ml to induce phosphorylation of eIF2α. Relative luciferase counts were determined after 3 h in the presence or absence of treatment (ii). Additionally, whole-cell lysates derived from normal 3T3 L1 cells or cells treated with tunicamycin or poly(IC) were resolved by SDS-PAGE and probed with antibodies recognizing total or phosphorylated eIF2α (iii). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; P.I., postinduction; SV40, simian virus 40.
Inhibition of viral replication by eIF2α.
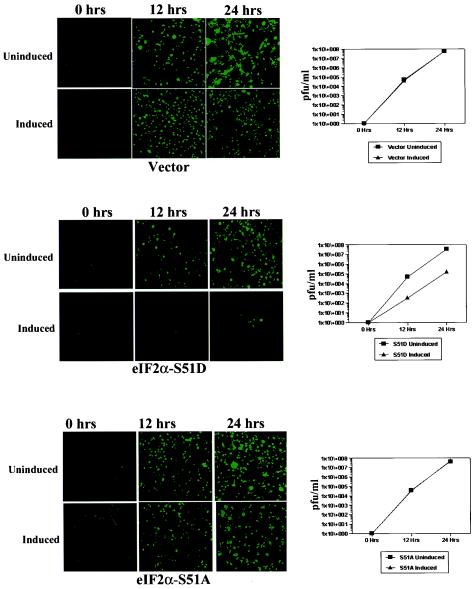
Earlier experiments performed in our laboratory have shown the necessity for functional PKR in the innate immune response to vesicular stomatitis virus (VSV) (6). However, the downstream substrates of PKR that potentiate the observed antiviral immunity in mammalian cells have never been definitively shown, and while eIF2α remains the most widely studied substrate for this kinase, other proteins have been shown to be in vitro and in vivo substrates for PKR (45, 64). To rigorously demonstrate the sufficiency of phosphorylated eIF2α in the inhibition of viral replication in mammalian tissue, we have utilized a version of the VSV that has been engineered to express the green fluorescent protein (VSV-GFP) (24). Uninduced and 6-day-induced 3T3 L1 cell lines were infected with VSV-GFP at a multiplicity of infection (MOI) of 0.1 and were monitored during the course of viral replication over the following 24 h. By observing GFP fluorescence, we were able to examine the relative rates of viral protein synthesis. These experiments indicated that eIF2α-S51D-expressing cells showed a profoundly diminished production of viral proteins over the 24-h time course (Fig. 6). Similar results were observed at a higher MOI (data not shown). In contrast, neither the vector-expressing control cells nor the eIF2α-S51A-expressing cells showed a DOX-dependent alteration of viral replication. To definitively assay viral yield and to control for possible GFP-specific effects of eIF2α-S51D expression, we determined VSV titers at the 12- and 24-h time points after infection. These analyses indicated that expression of the eIF2α-S51D variant resulted in a dramatic 100-fold reduction in VSV-GFP titer at all times measured. In agreement with our GFP fluorescence observations, no reduction of viral titer was observed in the vector- or eIF2α-S51A-expressing cells (Fig. 6). The failure of the S51A construct to enhance viral replication may be due to the fact that the S51A is not a true dominant-negative mutation. While eIF2α-S51A is phosphorylation incompetent, eIF2α bearing the S51A substitution nonetheless requires eIF2B-mediated GDP-GTP exchange in order to function in protein synthesis. In the context of a viral infection of cells expressing the eIF2α-S51A mutation, activated PKR may still phosphorylate enough of the endogenous eIF2α to sequester eIF2B and inhibit translation of viral transcripts.
FIG. 6.
Phosphorylation of eIF2α is sufficient to mediate antiviral immunity. Triplicate samples of uninduced or induced vector-, S51A-, or S51D-expressing cell lines were infected with VSV-GFP at an MOI of 0.1. Viral GFP intensity was measured at 0-, 12-, and 24-h time points by fluorescence microscopy. Additionally, cell culture supernatants were harvested at 0, 12, and 24 h, and viral replication was determined by plaque titration with BHK cells.
eIF2α as a regulator of malignant transformation.
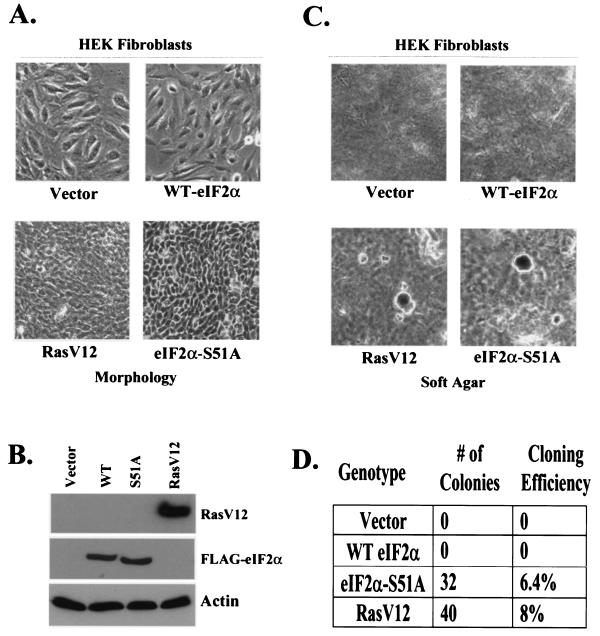
A number of proteins such as eIF4G and eIF4F and eEF1 have been shown to be capable of influencing the transformation of mammalian cells (1, 47, 49). Overexpression of the eIF2α-S51A variant in particular has been shown to transform NIH 3T3 cells (23). However, although we observed distinct morphological changes in our 3T3 L1 cells upon induction of the eIF2α-S51A variant in all clones tested, we did not observe evidence of cellular transformation. To complement these studies, we subsequently retrovirally transduced the eIF2α-S51A into normal 3T3 L1 cells to produce cell lines constitutively expressing this variant. Following this approach, we were able to obtain a large number of colonies after selection with neomycin. Most colonies expressing recombinant eIF2α-S51A exhibited a reduced cell size, but none became transformed, fully consistent with results from the inducible cell system (Fig. 7A). As 3T3 L1 cells retain expression of the INK4 tumor suppressor locus, unlike NIH 3T3 cells, this product may be functioning to inhibit cellular transformation (66). Thus, the eIF2α-S51A variant was transduced by retrovirus into the NIH 3T3 background in an attempt to recapitulate previously reported phenotypes (23). Expression of the transduced constructs in NIH 3T3 and 3T3 L1 cell lines was confirmed by Western blotting (Fig. 7B). Interestingly, colonies transfected with eIF2α-S51A were readily obtained after 7 days of selection in neomycin that displayed all of the well-known hallmarks of transformation. Such NIH 3T3 cells expressing S51A had a marked reduction in cell size and a dramatic reduction in doubling time (Fig. 7C and data not shown). In addition, these cell lines exhibited reduced adherence to tissue culture dishes and were able to sustain anchorage-independent growth (Fig. 7C). Thus, 3T3 L1 cells do not undergo cellular transformation in response to overexpression of eIF2α-S51A, while NIH 3T3 cells are rapidly transformed.
FIG. 7.
(A) eIF2α-S51A can transform NIH 3T3 but not 3T3 L1 cell lines. Normal 3T3 L1 cells were transduced with retrovirus carrying the indicated genes. Stable colonies were selected following selection in G418. Morphology was assessed by phase-contrast microscopy. To measure transformation, two colonies per construct were selected and plated in 0.5% soft agar for 21 days. (B) Stable expression of eIF2α variants in 3T3 L1 cell lines was determined by Western blotting with a MAb directed against the FLAG tag. (C) NIH 3T3 cells were retrovirally transduced and assayed for transformation as described for panel A. (D) Stable expression of RasV12 or indicated eIF2α variants was confirmed by Western blotting with appropriate MAbs. Cloning efficiency for each cell line was determined following growth in soft agar for 21 days, by dividing the colony number by the total number of cells plated.
As there was a discrepancy in our observations concerning cellular transformation induced by eIF2α-S51A, we performed oncogenic collaboration experiments in a more defined, human genetic background with previously described HEK cells that have been stably transfected with the large T antigen and hTERT (29). Such HEK cells are immortal and require a single additional genetic element to become fully transformed. Retroviral transduction of WT eIF2α into these cells produced no obvious morphological changes (Fig. 8A). While transduction of eIF2α-S51D produced some colonies following selection, we were not able to expand these into cell lines presumably due to aberrancies in protein synthesis affecting cell growth (data not shown). In contrast, transduction of eIF2α-S51A or RasV12 produced colonies with a reduced cell size and a refractile cytoplasm (Fig. 8A). Expression of RasV12, a mutation that has been previously shown to transform human or mouse tissue, and the FLAG-eIF2α constructs was confirmed by Western blotting (Fig. 8B). Transformation of these cells was confirmed by their ability to sustain anchorage-independent growth, while WT eIF2α-expressing cells were unable to grow in soft agar (Fig. 8C). Thus, dysregulation of eIF2α activity is able to collaborate in the transformation of primary human cells. The results of the colony-forming assay are quantitated in Fig. 8D.
FIG. 8.
(A) eIF2α-S51A can collaborate in the transformation of primary human tissue. HEK cells stably transduced with simian virus 40 large T antigen and hTERT (29) were subsequently transduced with either RasV12 or eIF2α-S51A. Following selection in puromycin, two colonies per construct were examined by phase-contrast microscopy to determine morphology. (B) Expression of the indicated constructs in individual HEK clones was confirmed by Western blotting. (C) eIF2α-S51A and RasV12 confer anchorage-independent growth on HEK cells. Transformation of HEK clones expressing RasV12 or eIF2α-S51A was confirmed by plating in 0.5% soft agar for 21 days. (D) Soft agar cloning efficiency for each HEK cell line was determined by dividing the number of colonies observed by the total number of cells plated.
DISCUSSION
Considerable evidence exists to indicate that in mammalian cells hormonal, mitogenic, and stress response signaling cascades can regulate the process of protein synthesis through posttranslational modification of eukaryotic translation initiation factors (eIFs) (34, 39). The eIF2 complex, perhaps the most comprehensively studied of the translation initiation factors, is a substrate for a number of highly specific stress-responsive kinases such as PERK, PKR, heme-regulated inhibitor kinase, and GCN2 (63). Nutrient deprivation, inadequate reticular protein processing, arsenical exposure, oxidative stress, and dsRNA all lead to activation of one or more of these kinases and to the subsequent inhibition of translation via phosphorylation of eIF2α on serine 51 (57). In addition to limiting protein synthesis, signaling cascades triggered in response to stress often result in complex cellular phenotypes that may include cell cycle arrest, regulation of gene expression at the transcriptional and translational levels, and/or induction of apoptosis (10, 13, 32). The precise role, if any, that translational control through eIF2α plays in these responses is unclear. Phosphorylation of eIF2α by stress-responsive kinases such as GCN2 has, however, been shown in yeast to be sufficient to account for the pronounced inhibition of translation that accompanies amino acid deprivation (17, 21, 38). Additionally, eIF2α has also been demonstrated to be a transcript-specific regulator of protein expression for the transcription factors GCN4 in yeast and ATF4 in mammals (19, 31).
To examine those processes that are subject to eIF2α regulation and to further explore the relationship between eIF2α regulation and the stress-responsive kinases, we developed clonal, inducible, 3T3 L1 cell lines expressing FLAG-tagged versions of the phosphomimetic eIF2α variant (eIF2α-S51D) and the phosphorylation-insensitive eIF2α-S51A variant. Each of these variants was successfully overexpressed two- to threefold over endogenous levels of native eIF2α and was incorporated into eIF2; though it remains formally possible that the FLAG tag is affecting eIF2α activity in subtle ways, we have seen no evidence for this. Through this approach, we observed a reproducible and distinct change in the cellular morphology of both the eIF2α-S51D- and the eIF2α-S51A-expressing cell lines. The pathways regulated downstream of eIF2α that are responsible for this cytoskeletal reorganization remain to be elucidated. Nevertheless, the distinct morphologies produced by the S51D and the S51A variants suggest that they may be delivering qualitatively different signals. For example, regulation of cellular morphology by components of the translation initiation machinery has been reported previously (15). In particular, antisense RNA against the mRNA cap binding protein eIF4E in HeLa cells has been shown to result in altered cell shape and growth patterns (15). Additionally, it has been demonstrated that eIF4E is capable of modulating signaling by the ras oncogene, which is itself a potent regulator of the activity of the Rho family of small GTPases (58). Signaling through such GTPases is an extensively characterized mechanism for regulation of morphology in response to extracellular signals such as platelet-derived growth factor and insulin (58). Whether eIF2α also specifically influences these GTP-dependent pathways regulating morphology remains to be determined.
The eIF2α-S51D-expressing cells also demonstrated a marked reduction in growth rate, suggesting a connection between cell cycle regulation and translation initiation. Previous studies examining the activation of the ER-resident eIF2α kinase PERK have revealed that ER stress induces a transient G1 cell cycle arrest with kinetics similar to those of eIF2α phosphorylation (9, 10). The kinase activity of PERK was shown to be essential for this arrest, strongly implying that eIF2α phosphorylation plays a role in this process. Elevated levels of phosphorylated eIF2α were subsequently shown to inhibit translation of cyclin D1 (36, 60, 66). Inducible expression of the eIF2α-S51D variant in our system may be exerting a similar effect, albeit at low levels since to date we have been unable to demonstrate a significant perturbation of the cell cycle profile in exponentially growing populations of the inducible eIF2α-S51D-expressing cell lines (data not shown). In contrast, expression of the eIF2α-S51A variant in our 3T3 L1 system did result in a very mild increase in growth rates equivalent to approximately 15%. This is in agreement with data derived from constitutive overexpression of S51A in the immortalized murine fibroblast cell line NIH 3T3, where a 20% increase in growth rates and an approximately threefold increase in translation rates were observed (23). Similarly, fibroblasts isolated from a knock-in mouse model carrying two copies of the S51A allele exhibited translation rates which were elevated a modest 18 to 35% (65). Such knock-in mice die within hours after birth due to hypoglycemia caused by deterioration of pancreatic beta islet cells. Thus, translation regulation by eIF2α may directly or indirectly govern the abundance of a selected set of proteins involved in cell growth and division.
The failure of the eIF2α-S51A mutant to cause transformation in our inducible 3T3 L1 system is unlikely to be the result of insufficient expression levels given that we obtained significant overexpression of our mutants in our inducible cell lines. Additionally, retroviral transduction of eIF2α-S51A into 3T3 L1 cells also failed to produce transformed colonies. In contrast, NIH 3T3 cells were readily transformed by eIF2α-S51A. Presumably, 3T3 L1 cells are more refractory to transformation than the NIH 3T3 cells are, given that the 3T3 L1 cells retain expression of the p16 tumor suppressor and exhibit a longer doubling time (9). The capacity of the eIF2α-S51A variant to collaborate in transformation with large T antigen and hTERT in HEK cells suggests that stimulation of translation initiation may be delivering mitogenic signals similar to those transmitted by activated Ras (46). To our knowledge, this is the first time that a translation initiation factor has been shown in a defined genetic system to collaborate in the transformation of human tissue.
Stress which produces activation of eIF2α kinases, such as liposome-mediated transfection of synthetic dsRNA [poly(IC)], is known to initiate an apoptotic cascade (5, 30, 32). Transient expression of eIF2α-S51A has been reported to partially ameliorate dsRNA- and TNF-α-induced apoptosis, while, in contrast, expression of the eIF2α-S51D variant has been reported to exert apoptotic activity (27, 68). We were, however, unable to demonstrate a marked induction of apoptosis in response to DOX-dependent induction of eIF2α-S51D in our 3T3 L1 system (Fig. 3). We did observe the transition of approximately 5% of the S51D-expressing population to a late-apoptotic-necrotic population. While this is a small fraction of the total cell number, it does represent an approximately 50-fold increase over the same population in the uninduced cells. Thus, it appears that in our inducible system the eIF2α-S51D mutant causes a modest elevation in background cell death but does not induce global cell death. Surprisingly, apoptosis in response to well-characterized stimuli was unaltered by expression of either of the eIF2α variants. This is in apparent contradiction to an earlier report in which eIF2α-S51A was shown to partially inhibit TNF-α- and PKR-mediated toxicity in NIH 3T3 cells (27, 68). This discrepancy may be due to the fact that the NIH 3T3 cells are malignantly transformed by eIF2α-S51A and, as a result, may have sustained additional genetic lesions that inactivate components of the apoptotic signaling cascades.
Previous work in our laboratory using 3T3 L1 cell lines inducibly overexpressing human PKR has also shown that activation of overexpressed PKR by dsRNA transfection results in an apparent 10-fold translational induction in the levels of the Fas death receptor protein (5). It is, therefore, plausible that translational regulation of key apoptotic genes may occur in response to dsRNA. Such PKR-dependent induction might function through a mechanism similar to ATF4 regulation via untranslated ORFs in the 5′ UTR of the ATF4 transcript. However, we were unable to explain the potent regulation of Fas in response to activation of PKR by dsRNA through phosphorylation of eIF2α. Additionally, we could not demonstrate a role for the 5′ uORF found in the human Fas transcript. Finally, no experimental evidence was found to support the translational up-regulation of any of the other key proapoptotic genes examined. While it is possible that our system may not accurately reflect the levels of phosphorylated eIF2α that occur under normal conditions of transient physiologic stress, our present data indicate a greater likelihood that the dsRNA-dependent regulation of Fas and apoptosis occurs independently of eIF2α through other dsRNA signaling pathways.
Finally, while popular dogma concerning the role of PKR in innate immunity suggests that it functions primarily by inhibiting translation through the phosphorylation of eIF2α in the presence of replicating virus, this has not been shown definitively. In this report, we find that, while expression of the phosphomimetic eIF2α-S51D is singularly sufficient to suppress viral replication, phosphorylated eIF2α does not play a significant role in inducing programmed cell death. Our data collectively indicate that regulation of translation through eIF2α is an important component of innate immunity to viral infection, the dysregulation of which can contribute to tumorigenesis in human cells.
Acknowledgments
We gratefully acknowledge Robert Weinberg for the contribution of the immortalized HEK cell lines. We also thank Yoshi Nakanishi for the Fas 5′ UTR constructs and Scott Kimball for antibodies to eIF2α and eIF2β.
REFERENCES
- 1.Anand, N., S. Murthy, G. Amann, M. Wernick, L. A. Porter, I. H. Cukier, H. C. Collins, J. W. Gray, J. Diebold, D. J. Demetrick, and J. M. Lee. 2002. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat. Genet. 31:301-305. [DOI] [PubMed] [Google Scholar]
- 2.Anthony, T. G., J. R. Fabian, S. R. Kimball, and L. S. Jefferson. 2000. Identification of domains within the epsilon-subunit of the translation initiation factor eIF2B that are necessary for guanine nucleotide exchange activity and eIF2B holoprotein formation. Biochim. Biophys. Acta 1492:56-62. [DOI] [PubMed] [Google Scholar]
- 3.Asano, K., and A. G. Hinnebusch. 2001. Protein interactions important in eukaryotic translation initiation. Methods Mol. Biol. 177:179-198. [DOI] [PubMed] [Google Scholar]
- 4.Asano, K., A. Shalev, L. Phan, K. Nielsen, J. Clayton, L. Valasek, T. F. Donahue, and A. G. Hinnebusch. 2001. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 20:2326-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balachandran, S., C. N. Kim, W. C. Yeh, T. W. Mak, K. Bhalla, and G. N. Barber. 1998. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 17:6888-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balachandran, S., P. C. Roberts, L. E. Brown, H. Truong, A. K. Pattnaik, D. R. Archer, and G. N. Barber. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129-141. [DOI] [PubMed] [Google Scholar]
- 7.Barber, G. N., J. Tomita, A. G. Hovanessian, E. Meurs, and M. G. Katze. 1991. Functional expression and characterization of the interferon-induced double-stranded RNA activated P68 protein kinase from Escherichia coli. Biochemistry 30:10356-10361. [DOI] [PubMed] [Google Scholar]
- 8.Boileau, G., P. Butler, J. W. Hershey, and R. R. Traut. 1983. Direct cross-links between initiation factors 1, 2, and 3 and ribosomal proteins promoted by 2-iminothiolane. Biochemistry 22:3162-3170. [DOI] [PubMed] [Google Scholar]
- 9.Brewer, J. W., and J. A. Diehl. 2000. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl. Acad. Sci. USA 97:12625-12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brewer, J. W., L. M. Hendershot, C. J. Sherr, and J. A. Diehl. 1999. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. USA 96:8505-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J. J., J. S. Crosby, and I. M. London. 1994. Regulation of heme-regulated eIF2 alpha kinase and its expression in erythroid cells. Biochimie 76:761-769. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J. J., M. S. Throop, L. Gehrke, I. Kuo, J. K. Pal, M. Brodsky, and I. M. London. 1991. Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2 alpha (eIF2α) kinase of rabbit reticulocytes: homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF2α kinase. Proc. Natl. Acad. Sci. USA 88:7729-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemens, M. J. 2001. Initiation factor eIF2α phosphorylation in stress responses and apoptosis. Prog. Mol. Subcell. Biol. 27:57-89. [DOI] [PubMed] [Google Scholar]
- 14.Clemens, M. J., B. Safer, W. C. Merrick, W. F. Anderson, and I. M. London. 1975. Inhibition of protein synthesis in rabbit reticulocyte lysates by double-stranded RNA and oxidized glutathione: indirect mode of action on polypeptide chain initiation. Proc. Natl. Acad. Sci. USA 72:1286-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Benedetti, A., S. Joshi-Barve, C. Rinker-Schaeffer, and R. E. Rhoads. 1991. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol. Cell. Biol. 11:5435-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeGracia, D. J., J. M. Sullivan, R. W. Neumar, S. S. Alousi, K. R. Hikade, J. E. Pittman, B. C. White, J. A. Rafols, and G. S. Krause. 1997. Effect of brain ischemia and reperfusion on the localization of phosphorylated eukaryotic initiation factor 2 alpha. J. Cereb. Blood Flow Metab. 17:1291-1302. [DOI] [PubMed] [Google Scholar]
- 17.Dever, T. E. 1997. Using GCN4 as a reporter of eIF2 alpha phosphorylation and translational regulation in yeast. Methods (Duluth) 11:403-417. [DOI] [PubMed] [Google Scholar]
- 18.Dever, T. E. 1999. Translation initiation: adept at adapting. Trends Biochem. Sci. 24:398-403. [DOI] [PubMed] [Google Scholar]
- 19.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545-556. [DOI] [PubMed] [Google Scholar]
- 20.Dever, T. E., J. J. Chen, G. N. Barber, A. M. Cigan, L. Feng, T. F. Donahue, I. M. London, M. G. Katze, and A. G. Hinnebusch. 1993. Mammalian eukaryotic initiation factor 2 alpha kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc. Natl. Acad. Sci. USA 90:4616-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585-596. [DOI] [PubMed] [Google Scholar]
- 22.Donze, O., J. Dostie, and N. Sonenberg. 1999. Regulatable expression of the interferon-induced double-stranded RNA dependent protein kinase PKR induces apoptosis and fas receptor expression. Virology 256:322-329. [DOI] [PubMed] [Google Scholar]
- 23.Donze, O., R. Jagus, A. E. Koromilas, J. W. Hershey, and N. Sonenberg. 1995. Abrogation of translation initiation factor eIF2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 14:3828-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez, M., M. Porosnicu, D. Markovic, and G. N. Barber. 2002. Genetically engineered vesicular stomatitis virus in gene therapy: application for treatment of malignant disease. J. Virol. 76:895-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filipowicz, W., Y. Furuichi, J. M. Sierra, S. Muthukrishnan, A. J. Shatkin, and S. Ochoa. 1976. A protein binding the methylated 5′-terminal sequence, m7GpppN, of eukaryotic messenger RNA. Proc. Natl. Acad. Sci. USA 73:1559-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerlitz, G., R. Jagus, and O. Elroy-Stein. 2002. Phosphorylation of initiation factor-2 alpha is required for activation of internal translation initiation during cell differentiation. Eur. J. Biochem. 269:2810-2819. [DOI] [PubMed] [Google Scholar]
- 27.Gil, J., J. Alcami, and M. Esteban. 1999. Induction of apoptosis by double-stranded-RNA-dependent protein kinase (PKR) involves the α subunit of eukaryotic translation initiation factor 2 and NF-κB. Mol. Cell. Biol. 19:4653-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gossen, M., and H. Bujard. 1995. Efficacy of tetracycline-controlled gene expression is influenced by cell type. BioTechniques 19:213-216. [PubMed] [Google Scholar]
- 29.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 30.Han, A. P., C. Yu, L. Lu, Y. Fujiwara, C. Browne, G. Chin, M. Fleming, P. Leboulch, S. H. Orkin, and J. J. Chen. 2001. Heme-regulated eIF2α kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20:6909-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 32.Harding, H. P., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5:897-904. [DOI] [PubMed] [Google Scholar]
- 33.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. (Erratum, 398:90.) [DOI] [PubMed] [Google Scholar]
- 34.Hershey, J. W. 1989. Protein phosphorylation controls translation rates. J. Biol. Chem. 264:20823-20826. [PubMed] [Google Scholar]
- 35.Hinnebusch, A. G. 1994. The eIF2 alpha kinases: regulators of protein synthesis in starvation and stress. Semin. Cell Biol. 5:417-426. [DOI] [PubMed] [Google Scholar]
- 36.Imoto, M., Y. Doki, W. Jiang, E. K. Han, and I. B. Weinstein. 1997. Effects of cyclin D1 overexpression on G1 progression-related events. Exp. Cell Res. 236:173-180. [DOI] [PubMed] [Google Scholar]
- 37.Kimball, S. R. 1999. Eukaryotic initiation factor eIF2. Int. J. Biochem. Cell Biol. 31:25-29. [DOI] [PubMed] [Google Scholar]
- 38.Kimball, S. R. 2001. Regulation of translation initiation by amino acids in eukaryotic cells. Prog. Mol. Subcell. Biol. 26:155-184. [DOI] [PubMed] [Google Scholar]
- 39.Kimball, S. R., J. R. Fabian, G. D. Pavitt, A. G. Hinnebusch, and L. S. Jefferson. 1998. Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2α. Role of the alpha- and delta-subunits of eiF2b. J. Biol. Chem. 273:12841-12845. [DOI] [PubMed] [Google Scholar]
- 40.Kimball, S. R., N. K. Heinzinger, R. L. Horetsky, and L. S. Jefferson. 1998b. Identification of interprotein interactions between the subunits of eukaryotic initiation factors eIF2 and eIF2B. J. Biol. Chem. 273:3039-3044. [DOI] [PubMed] [Google Scholar]
- 41.Kimball, S. R., R. L. Horetsky, D. Ron, L. S. Jefferson, and H. P. Harding. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284:C273-C284. [DOI] [PubMed] [Google Scholar]
- 42.Knappik, A., and A. Pluckthun. 1994. An improved affinity tag based on the FLAG peptide for the detection and purification of recombinant antibody fragments. BioTechniques 17:754-761. [PubMed] [Google Scholar]
- 43.Kostura, M., and M. B. Mathews. 1989. Purification and activation of the double-stranded RNA-dependent eIF2 kinase DAI. Mol. Cell. Biol. 9:1576-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozak, M. 1987. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 196:947-950. [DOI] [PubMed] [Google Scholar]
- 45.Kumar, A., J. Haque, J. Lacoste, J. Hiscott, and B. R. Williams. 1994. Double-stranded RNA-dependent protein kinase activates transcription factor NF-κB by phosphorylating IκB. Proc. Natl. Acad. Sci. USA 91:6288-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lazaris-Karatzas, A., M. R. Smith, R. M. Frederickson, M. L. Jaramillo, Y. L. Liu, H. F. Kung, and N. Sonenberg. 1992. Ras mediates translation initiation factor 4E-induced malignant transformation. Genes Dev. 6:1631-1642. [DOI] [PubMed] [Google Scholar]
- 47.Lazaris-Karatzas, A., and N. Sonenberg. 1992. The mRNA 5′ cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol. Cell. Biol. 12:1234-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manchester, K. L. 1987. eIF2B and the exchange of guanine nucleotides bound to eIF2. Int. J. Biochem. 19:245-251. [DOI] [PubMed] [Google Scholar]
- 49.Mayeur, G. L., and J. W. Hershey. 2002. Malignant transformation by the eukaryotic translation initiation factor 3 subunit p48 (eIF3e). FEBS Lett. 514:49-54. [DOI] [PubMed] [Google Scholar]
- 50.Merrick, W. C. 1990. Overview: mechanism of translation initiation in eukaryotes. Enzyme 44:7-16. [DOI] [PubMed] [Google Scholar]
- 51.Merrick, W. C. 1992. Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 56:291-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meurs, E. F., Y. Watanabe, S. Kadereit, G. N. Barber, M. G. Katze, K. Chong, B. R. Williams, and A. G. Hovanessian. 1992. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J. Virol. 66:5804-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muthukrishnan, S., G. W. Both, Y. Furuichi, and A. J. Shatkin. 1975. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature 255:33-37. [DOI] [PubMed] [Google Scholar]
- 54.Nika, J., S. Rippel, and E. M. Hannig. 2001. Biochemical analysis of the eIF2β gamma complex reveals a structural function for eIF2α in catalyzed nucleotide exchange. J. Biol. Chem. 276:1051-1056. [DOI] [PubMed] [Google Scholar]
- 55.Novoa, I., H. Zeng, H. P. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153:1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pestova, T. V., V. G. Kolupaeva, I. B. Lomakin, E. V. Pilipenko, I. N. Shatsky, V. I. Agol, and C. U. Hellen. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98:7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petryshyn, R., F. Rosa, R. Fagard, D. Levin, and I. M. London. 1984. Control of protein synthesis in human reticulocytes by heme-regulated and double-stranded RNA dependent eIF2 alpha kinases. Biochem. Biophys. Res. Commun. 119:891-899. [DOI] [PubMed] [Google Scholar]
- 58.Polunovsky, V. A., A. C. Gingras, N. Sonenberg, M. Peterson, A. Tan, J. B. Rubins, J. C. Manivel, and P. B. Bitterman. 2000. Translational control of the antiapoptotic function of Ras. J. Biol. Chem. 275:24776-24780. [DOI] [PubMed] [Google Scholar]
- 59.Proud, C. G. 2001. Regulation of eukaryotic initiation factor eIF2B. Prog. Mol. Subcell. Biol. 26:95-114. [DOI] [PubMed] [Google Scholar]
- 60.Quelle, D. E., R. A. Ashmun, S. A. Shurtleff, J. Y. Kato, D. Bar-Sagi, M. F. Roussel, and C. J. Sherr. 1993. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 7:1559-1571. [DOI] [PubMed] [Google Scholar]
- 61.Ramaiah, K. V. A., M. V. Davies, J.-J. Chen, and R. J. Kaufman. 1994. Expression of mutant eukaryotic initiation factor 2 α subunit (eIF2α) reduces inhibition of guanine nucleotide exchange activity of eIF2B mediated by eIF2α phosphorylation. Mol. Cell. Biol. 14:4546-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roy, A. L., D. Chakrabarti, and N. K. Gupta. 1987. Protein synthesis in rabbit reticulocytes: Mg2+-inhibition of ternary complex (Met-tRNA(f) · eIF2 · GTP) formation by reticulocyte eIF2. Biochem. Biophys. Res. Commun. 146:114-120. [DOI] [PubMed] [Google Scholar]
- 63.Samuel, C. E. 1993. The eIF2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268:7603-7606. [PubMed] [Google Scholar]
- 64.Saunders, L. R., D. J. Perkins, S. Balachandran, R. Michaels, R. Ford, A. Mayeda, and G. N. Barber. 2001. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 276:32300-32312. [DOI] [PubMed] [Google Scholar]
- 65.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 66.Sherr, C. J. 2001. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2:731-737. [DOI] [PubMed] [Google Scholar]
- 67.Shi, Y., K. M. Vattem, R. Sood, J. An, J. Liang, L. Stramm, and R. C. Wek. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18:7499-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Srivastava, S. P., K. U. Kumar, and R. J. Kaufman. 1998. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273:2416-2423. [DOI] [PubMed] [Google Scholar]
- 69.Tolan, D. R., J. W. Hershey, and R. T. Traut. 1983. Crosslinking of eukaryotic initiation factor eIF3 to the 40S ribosomal subunit from rabbit reticulocytes. Biochimie 65:427-436. [DOI] [PubMed] [Google Scholar]
- 70.Wek, S. A., S. Zhu, and R. C. Wek. 1995. The histidyl-tRNA synthetase-related sequence in the eIF2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15:4497-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams, B. R. 1999. PKR; a sentinel kinase for cellular stress. Oncogene 18:6112-6120. [DOI] [PubMed] [Google Scholar]
- 72.Wong, C., M. Luedi, J. W. Hershey, and O. G. Issinger. 1976. Phosphorylation in vitro of eukaryotic initiation factors IF-E2 and IF-E3 by protein kinases. J. Biol. Chem. 251:7675-7681. [PubMed] [Google Scholar]