Abstract
BACKGROUND
Viral respiratory infections are among the most common reasons for hospitalization of children in the United States. Our objective was to compare molecular and conventional methods in a cohort of hospitalized children with and without symptoms of respiratory viral illness (RVI).
METHODS
Retrospective cohort study of infants and toddlers hospitalized between December 2007 and March 2008 at Johns Hopkins Hospital. 569 of 641 patient visits (89%) were tested on admission. Conventional tests (immunochromatography, direct fluorescent antibody, shell vial, and tube culture) were performed on all patients and nucleic acid tests (NATs) were performed on available samples (n=306). Viruses were grouped into those routinely (Group 1) and those not routinely (Group 2) detected by conventional methods.
RESULTS
In children with RVI symptoms (N=148), NATs identified a virus in 83% specimens compared with 49% by conventional methods (p<0.001), but detected a similar percentage of specimens with Group 1 viruses (48.6% and 55.4%, p=0.13) compared with conventional tests. In children without RVI symptoms (N=158), NATs identified a virus in 41.7% specimens compared with 4.4% by conventional tests (p<0.001) and identified more Group 1 viruses (9.5% and 4.4%, p=0.03) compared with conventional tests. Group 2 viruses were identified by NATs in a similar percentage of symptomatic and asymptomatic patients (25% and 32.3%, p=0.20).
CONCLUSION
Molecular assays may have several advantages over conventional methods for detecting respiratory viruses, including improved sensitivity and rapid detection, but given the high prevalence of positive results in children without RVI symptoms, results should be interpreted cautiously.
Keywords: molecular diagnostic methods, pediatric respiratory infections, nucleic acid tests, virus isolation
INTRODUCTION
Viral respiratory infections are among the most common reasons for emergency department visits and hospitalization of children in the United States 1, 2. The economic impact of viral respiratory infections is estimated to be around $40 billion 3. Cohort studies to identify children with respiratory viral illnesses rely on testing children with common clinical symptoms, such as fever, rhinorrhea, or cough 4–6. Rapid detection of respiratory viruses can guide antiviral therapy, limit diagnostic testing which can be invasive and costly, and facilitate cohorting of patients to prevent nosocomial transmission 7–9.
Current challenges in detecting respiratory viruses include inconsistent clinical manifestations of respiratory viral illness and variable sensitivity and specificity of available diagnostic assays. Traditional methods of diagnosing respiratory viruses including direct fluorescent antibody (DFA) and enzyme immunoassay (EIA) provide results within hours, but are limited in scope, sensitivity, and positive predictive value. Viral cultures are very specific, but are time consuming and technically demanding. New molecular techniques to detect respiratory viruses are gaining popularity. Compared with traditional methods, polymerase chain reaction (PCR), or nucleic acid tests (NATs), can increase sensitivity of detecting common respiratory viruses, such as rhinovirus and enterovirus as well as identify emerging viruses, including human metapneumovirus (hMPV), coronavirus, and bocavirus. As molecular laboratory techniques become more widespread, we have the potential to detect a wide range of established and newly discovered viruses. However, interpretation of a positive result must be made cautiously in the context of a patient’s clinical illness as a positive result may signify the presence of the virus, but does not necessarily indicate that the virus is causing a patient’s clinical symptoms 10. Our objective was to compare the results of molecular techniques and conventional diagnostic methods in a cohort of hospitalized children with and without symptoms of respiratory viral illness (RVI).
MATERIALS AND METHODS
Study population and setting
Children admitted to the infant and toddler ward within 24 hours of hospitalization at The Johns Hopkins Hospital Children’s Center between December 1, 2007 and March 15, 2008 were included in the study. The infant and toddler ward cares primarily for children three years of age and younger with a wide range of underlying medical issues. The Johns Hopkins Hospital has a comprehensive 2-stage program to identify and isolate children with contagious respiratory viruses 9, 11. Beginning in 2007, all patients admitted to the infant and toddler ward had a nasopharyngeal aspirate (NPA) collected within 24 hours of admission, regardless of the presence of respiratory symptoms, as part of standard care. Complete medical records were obtained for all children from whom NPA samples were collected. We categorized patient presentations into ‘with RVI symptoms’ if they exhibited upper/lower respiratory tract symptoms consistent with a respiratory viral illness (e.g. rhinorrhea, congestion, cough, apnea, wheezing, tachypnea, hypoxia) or subtle symptoms like unexplained fever at the time of admission or within 4 days of hospitalization, and ‘without RVI symptoms’ if there were no symptoms consistent with a respiratory viral illness at the time of admission or within 4 days of hospitalization.
This study was approved by Johns Hopkins School of Medicine Institutional Review Board with a waiver of informed consent.
Laboratory Testing
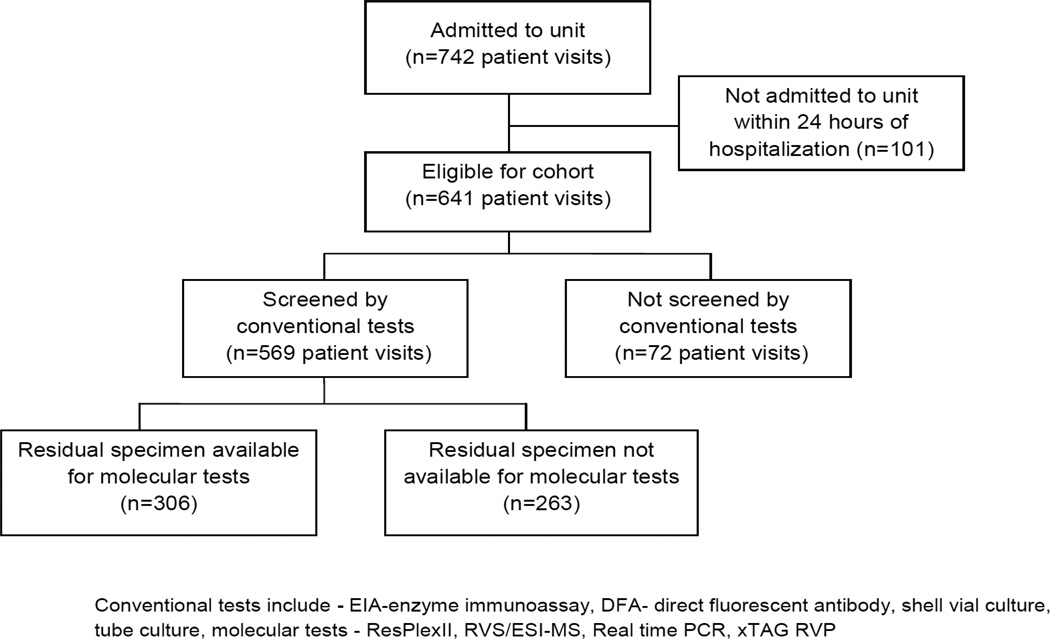
All NPA specimens were tested using conventional methods. Residual specimens stored at −80° Celsius were identified for further NATs. Figure, Supplemental Digital Content 1, http://links.lww.com/INF/B273 illustrates the number of patient admissions during which a NPA was collected and those with residual specimens available for testing. Patients with and without residual specimens were compared. Viruses were grouped into those routinely and those not routinely detected by conventional methods (Group 1 - adenovirus, influenza (Flu), respiratory syncytial virus (RSV), and parainfluenza (PIV); Group 2 - hMPV, rhinovirus, enterovirus and coronavirus).
Figure 1.
Cohort Scheme
a) Conventional diagnostic methods
NPAs were initially tested using immunochromatography to identify RSV, Flu A, Flu B as previously described 11. Negative specimens were further tested by DFA staining, shell vial culture and tube culture to identify RSV, Flu A, Flu B, PIV 1, 2, or 3, adenovirus and hMPV (See Fig., Supplemental Digital Content 1, http://links.lww.com/INF/B273 for details).
b) Molecular diagnostic methods
Available specimens were tested by NATs as described below. If the results obtained from two NATs were discordant, then the specimen was tested by a third NAT with the exception of rhinoviruses and enteroviruses as discussed below. A confirmed molecular result required virus detection by two NATs.
ResPlexII assay
The ResPlexII panel v2.0 (Qiagen, Germantown, MD) provides differential detection of 18 viral gene targets using QIAplex and xMAP technologies. The viral targets detected were RSV A and B, Flu A and B, PIV 1–4, hMPV, coxsackieviruses/echoviruses, rhinovirus, adenovirus B and E, coronaviruses NL63, HKU1, 229E, and OC 43, and bocavirus. ResPlexII panel amplification and detection were performed as per manufacturer’s instructions. The software does not define a cut-off for positivity. Several studies have used a variety of cutoff values for analysis. In our study, ResPlexII results were initially analyzed using four cutoff values- three times background, ≥50, ≥100, or ≥150 MFI values. The three times background cut-off was selected because it demonstrated excellent overall agreement with the other methods and it allowed inter-run data normalization by accounting for run-to-run variability in background fluorescence. For the purposes of this study, Flu A and B were not differentiated, and coronaviruses were combined due to small numbers. Also, RSV A and B were not differentiated and bocavirus was not included in the analysis. Bocaviruses can be detected with ResPlexII and detection reagents were initially available in the RVS/MS panel but were dropped from the product during the course of this study. Due to the change in the RVS/MS product and to the unresolved diagnostic utility of bocavirus testing, we elected not to perform an analysis of bocavirus detection by ResPlexII.
Respiratory Virus Surveillance kit with electrospray ionization mass spectrometry (RVS/MS)
RVS/MS (Ibis Biosciences, Inc., a part of Abbott Molecular, Des Plaines, IL) detects RSV, Flu A and B, PIV 1–4, hMPV, adenoviruses A-F, coronaviruses NL63, HKU1, 229E and OC43. Amplification products were analyzed using the T5000 universal biosensor. Mass measurements of the complimentary single-strands determine the nucleotide compositions which are then compared with a known database to establish virus identification.
c) Additional molecular diagnostic methods
Two individual real-time reverse transcriptase PCR (RT-PCR) assays for enteroviruses 12 and rhinoviruses 13 were used as comparators for ResPlexII because these viruses were not detected by RVS/MS. Discordant results for enteroviruses and rhinoviruses were not performed using a third NAT. Discordant results between ResPlex II and RVS/MS for adenovirus and hMPV were adjudicated using real-time PCR. xTAG Respiratory Viral Panel (RVP) (Luminex, Austin Texas) was used to adjudicate discrepancies between ResPlexII and RVS/MS for RSV, 229E, PIV4, and HKU1.
Statistical Analysis
Data were maintained in Microsoft Access (Microsoft, 2007, Bellevue, WA) and were analyzed using Stata, version 11.0 (Stata Corp., College Station, TX). Demographic and clinical variables were described as frequencies and percents or medians and interquartile ranges (IQR). We used the two sample test of proportions for comparing characteristics in patients with and without residual specimens as well as comparing positive results by molecular and conventional tests. Two-tailed p-values of <0.05 were considered statistically significant.
RESULTS
In our cohort, 569 of 641 patient visits (89%) underwent NPA testing on admission. Of all children tested, 311 (55%) patients were admitted for or with RVI symptoms. Conventional testing methods detected a respiratory virus in 29% of children, including 27% of those with symptoms of RVI and 2% of those without symptoms of RVI. Of those tested by conventional methods, 306 specimens were available for molecular testing. We compared those with and without a residual specimen and found that the groups were similar with regard to demographic characteristics, but patients with residual samples were more likely to have had a virus detected by conventional tests compared with those without a residual sample (29 vs. 12%, p<0.001, Table 1). Groups with and without residual specimens were equally likely to have RVI symptoms (56.3% vs. 53.2%, p=0.47). Of the 306 residual specimens tested, 62% were positive by NATs, while only 26% were positive by conventional tests (p<0.001) (Table 2).
Table 1.
Characteristics of patients with and without residual specimens available for molecular testing
| Variables | Patient visits without a Residual Specimen (n=263) |
Patient visits with a Residual Specimen (n=306) |
All Patient visits (n=569) |
|---|---|---|---|
| Age in years (Median, [IQR]1) | 1 (0–2) | 1 (0–2) | 1 (0–2) |
| Male (n %) | 150 (57) | 182 (59.4) | 332 (58.3) |
| Race, n (%) | |||
| Caucasian | 102 (41.6) | 148 (45.4) | 250 (43.9) |
| African American | 107 (43.7) | 143 (43.9) | 250 (43.9) |
| Other | 36 (14.7) | 35 (10.6) | 71 (12.2) |
| Length of Stay in days (Median, IQR) | 2 (1–4) | 2 (1–4) | 2 (1–4) |
| Virus2, n (%) | |||
| Adenovirus | 5 (2.0) | 5 (1.5) | 10 (1.8) |
| Influenza A | 9 (3.7) | 16 (4.9) | 25 (4.4) |
| Influenza B | 1 (0.4) | 1 (0.3) | 2 (<0.01) |
| Parainfluenza 1 | 0 | 1 (0.3) | 1 (<0.01) |
| Parainfluenza 2 | 1 (0.4) | 0 | 1 (<0.01) |
| RSV | 13 (5.3) | 64 (19.6) | 77 (13.5) |
| hMPV | 0 | 7 (2.1) | 7 (1.0) |
| None | 216 (88.2) | 232 (71.2) | 448 (78.7) |
| SYMPTOMS, n (%) | |||
| Symptoms of RVI illness | 148 (56.3) | 163 (53.2) | 311 (54.67) |
IQR- Interquartile Range
Reported viruses were detected by conventional methods
Table 2.
Patient visits with a positive result by conventional or molecular methods
| Patient visits with RVI Symptoms (n=148) |
Patient visits without RVI symptoms (n=158) |
All patient visits (n=306) |
|
|---|---|---|---|
| Conventional Testing1 | 72 (48.6%) | 7 (4.4%) | 79 (25.8%) |
| Group 1 viruses | 72 (48.6%)3 | 7 (4.4%)4 | 79 (25.8%) |
| <48hrs (EIA, DFA, shell vial culture) | 64 (43.2%) | 5 (3.2 %) | 71 (23.2%) |
| >48hrs (tube culture) | 8 (5.4%) | 2 (1.2%) | 10 (3.7%) |
| Group 2 viruses only | - | - | - |
| Molecular NATs2 | 123 (83.1%) | 66 (41.7%) | 189 (61.7%) |
| Group 1 viruses | 85 (57.4%)3 | 15 (9.5%)4 | 100 (32.7%) |
| Group 2 viruses only | 38 (25%)5 | 51 (32.3%)5 | 89 (29.1%) |
RVI – respiratory viral illness; EIA - enzyme immunoassay; DFA - direct fluorescent antibody
EIA, DFA, shell vial culture, tube culture
ResPlexII, RVS/MS, Real-time PCR, xTAG RVP
Test of significance for detection of Group1 viruses comparing conventional vs. NATs in patients with RVI symptoms, p= 0.13
Test of significance for detection of Group1 viruses comparing conventional vs. molecular methods in patients without RVI symptoms, p= 0.03
Test of significance for detection of Group 2 viruses by molecular methods comparing patients with and without RVI symptoms, p= 0.20
Comparison of Conventional and Molecular Testing in Patients with RVI Symptoms
In children with RVI symptoms (N=148), NATs identified a virus in 83% compared to 49% by conventional methods (p<0.001, Table 2). This increased detection was largely due to the 25% of patients identified with a Group 2 virus that were not detected by conventional methods. Conventional and molecular assays detected a similar percent of patients with Group 1 viruses (48.6% v 57.4%, p=0.13). However, when comparing positive results from NATs and conventional tests that provide results within 48 hours (EIA, DFA, shell vial culture), then NATs increased virus detection (57.4% v 43.2%, p=0.01).
Comparison of Conventional and Molecular Testing in Patients without RVI Symptoms
In children without RVI symptoms (N=158), NATs identified a virus in 41.7% compared to 4.4% by conventional methods (p<0.001). Similar to patients with RVI symptoms, this increased detection was largely due to the 32% of patients identified with a Group 2 virus only. NATs also identified more Group 1 viruses compared to conventional testing in these patients (9.5% v 4.4% p=0.03). The most prevalent Group 2 virus in asymptomatic patients was rhinovirus/ enterovirus which was detected in 34 specimens by ResPlexII. Group 2 viruses were identified by NATs in a similar percentage of symptomatic and asymptomatic patients (25% and 32.3%, p=0.20).
Comparison of NATs in Patients with and without RVI Symptoms
In children with RVI symptoms (Table 3), neither ResPlexII nor RVS/MS detected a significantly higher percentage of Group 1 viruses than conventional tests (p=0.73 and p=0.56 respectively). Most of the additional positive results from RVS/MS were from detection of adenovirus, however only 6 of the 9 specimens were confirmed for adenovirus by 2 NATs. Similarly, the additional positive results by ResPlexII were mostly from detection of hMPV, but in this case 11 of the 12 samples positive for hMPV by ResPlexII were confirmed by two molecular NATs.
Table 3.
Respiratory viruses detected by conventional and molecular methods in patients with and without symptoms of respiratory viral illness (RVI)
| Viruses in patients with RVI symptoms (n=148) | ||||
|---|---|---|---|---|
| Virus | Conventional No. (confirmed1) |
ResPlex II No. (confirmed1) |
RVS/ MS No. (confirmed1) |
Confirmed by 2 molecular assays2 |
| Adenovirus | 2 (2) | 3 (3) | 8 (6) | 6 |
| Influenza | 13 (11) | 14 (13) | 13 (13) | 13 |
| PIV | 1 (1) | 3 (1) | 1 (1) | 1 |
| RSV | 56 (53) | 55 (53) | 53 (51) | 54 |
| Group 1 total | 72 (67) | 75 (70) | 77(71) | 74 |
| hMPV | 7 (6) | 12 (11) | 5 (5) | 10 |
| Coronavirus | NT | 7 (6) | 7 (6) | 6 |
| Rhinovirus Enterovirus |
NT | 41 (41) | NT | 41 |
| Group 2 total | 7 (6) | 60 (59) | 11 (11) | 58 |
| Viruses in patients without RVI symptoms (n=158) | ||||
| Adenovirus | 3 (2) | 3 (3) | 9 (9) | 9 |
| Influenza | 3 (1) | 3 (3) | 3 (3) | 3 |
| PIV | 0 | 1 (0) | 0 | 0 |
| RSV | 1 (1) | 2 (2) | 3 (3) | 3 |
| Group 1 total | 7 (4) | 9 (8) | 15 (13) | 15 |
| hMPV | 0 | 1 (1) | 0 | 1 |
| Coronavirus | NT | 6 (5) | 7 (5) | 5 |
| Rhinovirus Enterovirus |
NT | 34 (32) | NT | 32 |
| Group 2 total | 0 (0) | 41 (38) | 6(5) | 38 |
RVI – respiratory viral illness; n- number ; NT- Not tested
result confirmed by an alternate NAT
Confirmed by any 2 NATs- Either concordant by 2 multiplex assay, or discordant result confirmed by alternate NAT (two positive tests could include ResPlexII and RVS/MS or ResPlexII and RT-PCR or RVS/MS and RT-PCR or xTAG RVP and RVS/MS or xTAG RVP and ResPlexII)
In children without RVI symptoms (Table 3), RVS/MS detected a higher percentage of Group 1 viruses compared to conventional tests. Most of the additional positive results from RVS/MS, were from detection of adenovirus, but in this case, all 9 samples positive for adenovirus by RVS/MS were confirmed by a second molecular assay.
Detection of Viral Co-Infections
Of the 306 children with available specimens, molecular tests detected a single virus in 144 (47.1%) specimens, while 56 (18.3%) specimens revealed coinfections with more than one virus. No viruses were detected in the remaining 106 (34.6%) specimens. Conventional tests did not detect any co-infections in our cohort. Of these 56 coinfections, Rhinovirus was identified in 41 specimens. A greater proportion of those with viral coinfections presented with RVI symptoms (41 vs. 15, p<0.001).
DISCUSSION
Our results demonstrate that NATs increased rapid detection of respiratory viruses in symptomatic patients, but also uncovered a high prevalence of positive test results for respiratory viruses in children without RVI symptoms. Molecular assays have several advantages over conventional methods for detecting respiratory viruses, including improved sensitivity, relatively rapid results, and automated processing, but their results should be interpreted cautiously, particularly when clinical symptoms are not suggestive of an RVI 14–17. Case-control studies in the past have detected respiratory viruses in asymptomatic children18, 19, but to our knowledge, this is the first cohort study comparing results from multiple molecular assays and conventional tests in infants and toddlers with and without RVI symptoms.
In our cohort, molecular tests identified a respiratory virus in 62% of patients. Our inclusion of children who presented without symptoms of RVI may explain our lower detection rates than previously reported for respiratory viruses using molecular assays 4, 20. In children with RVI symptoms, NATs identified a respiratory virus in 83% of NPA specimens, as compared with 49% by conventional tests. In previous studies in children with acute respiratory viral illnesses, 93–95% of nasal aspirates were found positive by molecular NATs, as compared with 20–35% by conventional methods 4, 20. In our cohort, molecular tests did not increase the overall detection of Group 1 viruses compared with conventional tests in children with RVI symptoms. This increased sensitivity of conventional tests may be attributed to the use of tube culture for virus detection in our laboratory. However, when compared with the results from conventional testing available within 48 hours, there was an increase in detection of Group 1 viruses by NATs. These findings have confirmed previous reports that molecular tests increase rapid identification of conventional viruses like RSV, influenza, PIVs and adenoviruses in patients with respiratory illness 4, 21–23.
In children without RVI symptoms, NATs identified a respiratory virus in 42% of NPA specimens, as compared with 4% by conventional testing. Although NATs increased the detection of Group 1 viruses compared with conventional tests in these children, the majority of viruses identified in children without RVI symptoms were Group 2 viruses, including rhinovirus, enterovirus, coronavirus and hMPV. Interestingly, molecular assays detected an almost equal percentage of Group 2 viruses in both symptomatic and asymptomatic children, suggesting that these viruses may commonly be present in the respiratory tracts of young children. This has been shown previously with rhinovirus 18, 24–26. In our cohort, almost half of the NPA specimens (45.3%) that were positive for rhinovirus/enterovirus were obtained from children without RVI symptoms. Rhinovirus has been detected in 32.3% of asymptomatic children and has been shown to persist in 50% of cases for up to 2 weeks beyond an acute infection.
Our findings bring to light some important strengths and challenges of NATs. We have confirmed prior reports that molecular tests increase sensitivity and rapidity of detection when compared with traditional methods 4, 14–17, 27. However, increased sensitivity may not always improve clinical outcomes and influence clinician behavior. A few randomized trials have studied the impact of viral testing on patient management 28–30. Bonner et al showed no differences in management for children with respiratory viral illness, except for influenza. Another randomized trial of influenza rapid testing showed that the use of a rapid test did not influence clinician behavior 31. Molecular testing can increase detection of Group 2 viruses like rhinovirus and enterovirus, but in the absence of available therapeutics, unless a positive test limits antibiotic usage or further diagnostic testing, the testing results may not change the course of treatment or improve patient outcomes. Recent research suggests that there may be an association between rhinovirus induced wheezing and childhood asthma 32–34, but the role and availability of antiviral therapy in these children remains in question. Rapid detection of respiratory viruses is used to isolate contagious patients to prevent transmission of viruses between hospitalized patients. Increased detection of PCR positive but culture negative patients may have negative consequences, such as unnecessary isolation of hospitalized patients. Further studies are needed to assess the impact of these diagnostic tests on patient outcomes as more clinical labs begin to use molecular tests to detect respiratory viruses.
Several points are worth noting with regard to our data. First, an analysis of patients with and without residual specimen demonstrated that those with residual specimen were more likely to have a positive conventional test result. However, the groups with and without available specimens were otherwise similar and had equal proportions of children with RVI symptoms, so our study population and findings should reflect the entire the cohort. Second, there is no standard method for detection of respiratory viruses. Because NATs can have false positives, we compared results from 2 large multiplex NATs, and found overall good concordance. When these NATs were discordant, we used a third molecular assay such as singleplex RT-PCR or x-TAG RVP to confirm the result. Also, ResPlexII demonstrated some cross reactivity in detecting rhinovirus and enterovirus and hence their positive results are reported in combination as rhinovirus/enterovirus. Finally, this is a single institution study of hospitalized patients and our findings may not be generalizable to other institutions and settings, but the general advantages and disadvantages of molecular testing still likely remain.
Results from our study confirm previous reports that molecular tests increase detection of respiratory viruses in children with respiratory illness, but they also detect viruses in many children without RVI symptoms. Detection of respiratory viruses in asymptomatic children is common and poses a challenge in diagnostic interpretation of molecular NATs18, 19. Further studies should focus on defining cut-off values that aid with interpreting positive results with more accuracy and reliability35. Clinicians should be encouraged to interpret results based on variations in patient populations and characteristics, clinical presentation, seasonal occurrence, and local virus epidemiology. Careful interpretation of testing results is needed to make complicated decisions that will guide individual patient’s treatment and management.
Acknowledgments
The authors thank Dr. Charlotte Gaydos, Christina Newman, and members of The Johns Hopkins Hospital Department of Hospital Epidemiology and Infection Control and The Johns Hopkins Hospital Clinical Virology Laboratory for their support and expertise, and Dr. Pranita Tamma for review of the manuscript. Reagent support was kindly provided by Qiagen and Ibis Biosciences/Abbott Molecular.
Funding: This study was funded primarily by a grant from the R Baby Foundation. Additional funding came from The Johns Hopkins Clinical Research Career Development Grant 1KL2RR025006-01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research, and its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures: A.M. receives grant support from Sage Products and BioMerieux. The authors have no conflicts of interest.
REFERENCES
- 1.Weber MW, Mulholland EK, Greenwood BM. Respiratory syncytial virus infection in tropical and developing countries. Trop Med Int Health. 1998;3(4):268–280. doi: 10.1046/j.1365-3156.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 2.Shay DK, Holman RC, Newman RD, Liu LL, Stout JW, Anderson LJ. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA. 1999;282(15):1440–1446. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- 3.Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch Intern Med. 2003;163(4):487–494. doi: 10.1001/archinte.163.4.487. [DOI] [PubMed] [Google Scholar]
- 4.Freymuth F, Vabret A, Cuvillon-Nimal D, et al. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol. 2006;78(11):1498–1504. doi: 10.1002/jmv.20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jartti T, Lehtinen P, Vuorinen T, et al. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis. 2004;10(6):1095–1101. doi: 10.3201/eid1006.030629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aberle JH, Aberle SW, Pracher E, Hutter HP, Kundi M, Popow-Kraupp T. Single versus dual respiratory virus infections in hospitalized infants: impact on clinical course of disease and interferon-gamma response. Pediatr Infect Dis J. 2005;24(7):605–610. doi: 10.1097/01.inf.0000168741.59747.2d. [DOI] [PubMed] [Google Scholar]
- 7.Barenfanger J, Drake C, Leon N, Mueller T, Troutt T. Clinical and financial benefits of rapid detection of respiratory viruses: an outcomes study. J Clin Microbiol. 2000;38(8):2824–2828. doi: 10.1128/jcm.38.8.2824-2828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dundas NE, Ziadie MS, Revell PA, et al. A lean laboratory: operational simplicity and cost effectiveness of the Luminex xTAG respiratory viral panel. J Mol Diagn. 2011;13(2):175–179. doi: 10.1016/j.jmoldx.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karanfil LV, Conlon M, Lykens K, et al. Reducing the rate of nosocomially transmitted respiratory syncytial virus. Am J Infect Control. 1999;27(2):91–96. doi: 10.1016/s0196-6553(99)70087-8. [DOI] [PubMed] [Google Scholar]
- 10.Pavia AT. Viral infections of the lower respiratory tract: old viruses, new viruses, and the role of diagnosis. Clin Infect Dis. 2011;52(Suppl 4):S284–S289. doi: 10.1093/cid/cir043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milstone AM, Perl TM, Valsamakis A. Epidemiology of respiratory viruses in children admitted to an infant/toddler unit. Am J Infect Control. 2011 doi: 10.1016/j.ajic.2011.05.024. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Lai KK, Cook L, Wendt S, Corey L, Jerome KR. Evaluation of real-time PCR versus PCR with liquid-phase hybridization for detection of enterovirus RNA in cerebrospinal fluid. J Clin Microbiol. 2003;41(7):3133–3141. doi: 10.1128/JCM.41.7.3133-3141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Holloway B, Dare RK, et al. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46(2):533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freymuth F, Vabret A, Galateau-Salle F, et al. Detection of respiratory syncytial virus, parainfluenzavirus 3, adenovirus and rhinovirus sequences in respiratory tract of infants by polymerase chain reaction and hybridization. Clin Diagn Virol. 1997;8(1):31–40. doi: 10.1016/s0928-0197(97)00060-3. [DOI] [PubMed] [Google Scholar]
- 15.Eugene-Ruellan G, Freymuth F, Bahloul C, Badrane H, Vabret A, Tordo N. Detection of respiratory syncytial virus A and B and parainfluenzavirus 3 sequences in respiratory tracts of infants by a single PCR with primers targeted to the L-polymerase gene and differential hybridization. J Clin Microbiol. 1998;36(3):796–801. doi: 10.1128/jcm.36.3.796-801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinberg GA, Erdman DD, Edwards KM, et al. Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J Infect Dis. 2004;189(4):706–710. doi: 10.1086/381456. [DOI] [PubMed] [Google Scholar]
- 17.Rovida F, Percivalle E, Zavattoni M, et al. Monoclonal antibodies versus reverse transcription-PCR for detection of respiratory viruses in a patient population with respiratory tract infections admitted to hospital. J Med Virol. 2005;75(2):336–347. doi: 10.1002/jmv.20276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singleton RJ, Bulkow LR, Miernyk K, et al. Viral respiratory infections in hospitalized and community control children in Alaska. J Med Virol. 2010;82(7):1282–1290. doi: 10.1002/jmv.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prill MM, Iwane MK, Edwards KM, et al. Human coronavirus in young children hospitalized for acute respiratory illness and asymptomatic controls. Pediatr Infect Dis J. 2012;31(3):235–240. doi: 10.1097/INF.0b013e31823e07fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stempel HE, Martin ET, Kuypers J, Englund JA, Zerr DM. Multiple viral respiratory pathogens in children with bronchiolitis. Acta Paediatr. 2009;98(1):123–126. doi: 10.1111/j.1651-2227.2008.01023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coiras MT, Perez-Brena P, Garcia ML, Casas I. Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested-PCR assay. J Med Virol. 2003;69(1):132–144. doi: 10.1002/jmv.10255. [DOI] [PubMed] [Google Scholar]
- 22.Syrmis MW, Whiley DM, Thomas M, et al. A sensitive, specific, and cost-effective multiplex reverse transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J Mol Diagn. 2004;6(2):125–131. doi: 10.1016/S1525-1578(10)60500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puppe W, Weigl JA, Aron G, et al. Evaluation of a multiplex reverse transcriptase PCR ELISA for the detection of nine respiratory tract pathogens. J Clin Virol. 2004;30(2):165–174. doi: 10.1016/j.jcv.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Iwane MK, Prill MM, Lu X, et al. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J Infect Dis. 2011;204(11):1702–1710. doi: 10.1093/infdis/jir634. [DOI] [PubMed] [Google Scholar]
- 25.Nokso-Koivisto J, Pitkaranta A, Blomqvist S, et al. Viral etiology of frequently recurring respiratory tract infections in children. Clin Infect Dis. 2002;35(5):540–546. doi: 10.1086/341773. [DOI] [PubMed] [Google Scholar]
- 26.Jartti T, Lehtinen P, Vuorinen T, Koskenvuo M, Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72(4):695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- 27.Jartti T, Jartti L, Peltola V, Waris M, Ruuskanen O. Identification of respiratory viruses in asymptomatic subjects: asymptomatic respiratory viral infections. Pediatr Infect Dis J. 2008;27(12):1103–1107. doi: 10.1097/INF.0b013e31817e695d. [DOI] [PubMed] [Google Scholar]
- 28.Bonner AB, Monroe KW, Talley LI, Klasner AE, Kimberlin DW. Impact of the rapid diagnosis of influenza on physician decision-making and patient management in the pediatric emergency department: results of a randomized, prospective, controlled trial. Pediatrics. 2003;112(2):363–367. doi: 10.1542/peds.112.2.363. [DOI] [PubMed] [Google Scholar]
- 29.Byington CL, Castillo H, Gerber K, et al. The effect of rapid respiratory viral diagnostic testing on antibiotic use in a children's hospital. Arch Pediatr Adolesc Med. 2002;156(12):1230–1234. doi: 10.1001/archpedi.156.12.1230. [DOI] [PubMed] [Google Scholar]
- 30.Abanses JC, Dowd MD, Simon SD, Sharma V. Impact of rapid influenza testing at triage on management of febrile infants and young children. Pediatr Emerg Care. 2006;22(3):145–149. doi: 10.1097/01.pec.0000202454.19237.b0. [DOI] [PubMed] [Google Scholar]
- 31.Iyer SB, Gerber MA, Pomerantz WJ, Mortensen JE, Ruddy RM. Effect of point-of-care influenza testing on management of febrile children. Acad Emerg Med. 2006;13(12):1259–1268. doi: 10.1197/j.aem.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 32.Kotaniemi-Syrjanen A, Vainionpaa R, Reijonen TM, Waris M, Korhonen K, Korppi M. Rhinovirus-induced wheezing in infancy--the first sign of childhood asthma? J Allergy Clin Immunol. 2003;111(1):66–71. doi: 10.1067/mai.2003.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gern JE. Rhinovirus and the initiation of asthma. Curr Opin Allergy Clin Immunol. 2009;9(1):73–78. doi: 10.1097/ACI.0b013e32831f8f1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson DJ, Gangnon RE, Evans MD, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178(7):667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansen RR, Wieringa J, Koekkoek SM, et al. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011;49(7):2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]