Abstract
Transcription of snRNA genes by either RNA polymerase II (U1 to U5) or RNA polymerase III (U6) is dependent upon a proximal sequence element (PSE) located approximately 40 to 60 bp upstream of the transcription start site. In Drosophila melanogaster, RNA polymerase specificity is determined by as few as three nucleotide differences within the otherwise well-conserved 21-bp PSE. Previous photo-cross-linking studies revealed that the D. melanogaster PSE-binding protein, DmPBP, contains three subunits (DmPBP45, DmPBP49, and DmPBP95) that associate with the DNA to form complexes that are conformationally distinct depending upon whether the protein is bound to a U1 or a U6 PSE. We have identified and cloned the genes that code for these subunits of DmPBP by virtue of their similarity to three of the five subunits of SNAPc, the human PBP. When expressed in S2 cells, each of the three cloned gene products is incorporated into a protein complex that functionally binds to a PSE. We also find that the conformational difference referred to above is particularly pronounced for DmPBP45, herein identified as the ortholog of human SNAP43. DmPBP45 cross-linked strongly to DNA for two turns of the DNA helix downstream of the U1 PSE, but it cross-linked strongly for only a half turn of the helix downstream of a U6 PSE. These substantial differences in the cross-linking pattern are consistent with those of a model in which conformational differences in DmPBP-DNA complexes lead to selective RNA polymerase recruitment to U1 and U6 promoters.
In eukaryotes, small nuclear RNAs (snRNAs) are required for pre-mRNA splicing. Most snRNAs, such as U1, U2, U4, and U5, are synthesized by RNA polymerase II, but U6 snRNA is synthesized by RNA polymerase III (2, 3, 6, 10, 11, 18, 26). Transcription of snRNA genes by either RNA polymerase is dependent upon a proximal sequence element (PSE) located upstream of position −40 relative to the transcription start site. In the insect Drosophila melanogaster, the PSE is referred to more specifically as the PSEA to distinguish it from a second conserved element termed the PSEB (38).
Although the PSEAs of all D. melanogaster snRNA genes share extensive sequence similarity, the PSEAs of the three U6 genes present in the fly genome consistently vary at a few nucleotide positions from the PSEAs of the RNA polymerase II-transcribed snRNA genes (13). Indeed, RNA polymerase specificity can be determined by as few as three base pair differences within the otherwise well-conserved U1 and U6 PSEAs (13). As a result, the fly U1 and U6 PSEAs are not interchangeable either in vitro or in vivo (13, 22).
Nonetheless, both PSEAs are recognized by the same D. melanogaster PSE-binding protein, DmPBP (29, 33). DmPBP contains three distinct subunits (DmPBP45, DmPBP49, and DmPBP95) that can be specifically photo-cross-linked to DNA containing U1 or U6 PSEA sequences. Interestingly, the pattern of photo-cross-linking was different depending upon whether DmPBP was bound to a U1 or a U6 PSEA (33). Those results, together with the functional polymerase specificity studies, led us to propose a hypothetical model in which the U1 and U6 PSEAs act as differential allosteric effectors of the conformation of DmPBP, subsequently leading to the specific downstream recruitment of either RNA polymerase II or RNA polymerase III (5, 33).
In vertebrates, by contrast, the PSEs of U1, U2, and U6 snRNA genes are functionally interchangeable, and RNA polymerase specificity is determined by the presence of a TATA box (providing RNA polymerase III specificity) or the absence of a TATA box (resulting in RNA polymerase II specificity) (17, 21). In humans, a multiprotein complex, referred to as snRNA activator protein complex (SNAPc), PSE transcription factor (PTF), or PBP, specifically binds to the PSE (9, 24, 28, 31, 32, 36). Human SNAPc /PTF consists of five stably associated subunits: SNAP190 or PTFα (34, 36), SNAP50 or PTFβ (1, 7), SNAP43 or PTFγ (9, 37), SNAP45 or PTFδ (27, 37), and SNAP19 (8).
A search of the D. melanogaster nucleic acid database with the five human SNAP sequences revealed fly genes that code for putative proteins with regions of substantial similarity to only three of the human SNAPc subunits: SNAP43, SNAP50, and SNAP190. By expressing tagged versions of the D. melanogaster genes in S2 cells, we show that the protein products can be incorporated into functional DmPBP that binds specifically to a PSEA sequence. Moreover, experimental data allow each of the cloned gene products to be correlated unambiguously with the individual subunits previously identified by the photo-cross-linking assay. Finally, pronounced differences were observed between the cross-linking patterns of DmPBP45 to DNA downstream of U1 PSEAs and those of U6 PSEAs.
MATERIALS AND METHODS
Site-specific protein-DNA photo-cross-linking.
Thirty-six different probes, each containing a cross-linking reagent at a unique position, were used for scanning for DmPBP interactions with the DNA downstream of the U1 and U6 PSEA sequences on both the template and nontemplate strands. A cross-linker was incorporated at every second phosphate position beginning 4 bp beyond the PSEAs and extending to 22 bp downstream of the PSEAs. Probes were prepared as described by Wang and Stumph (33), based upon methods previously described by Yang and Nash (35) and Lagrange et al. (15). Briefly, DNA oligonucleotides 20 to 28 bases long were synthesized with phosphorothioate incorporated 5′ of the third nucleotide from the 5′ end. The phosphorothioate-incorporated oligonucleotides were derivatized with azidophenacyl bromide and then radiolabeled at the 5′ end by using [γ-32P]ATP and T4 polynucleotide kinase.
To make the double-stranded photo-cross-linking probes, a derivatized oligonucleotide and an unmodified upstream primer were annealed to one of four long oligonucleotides (see 81- and 82-mer sequences below) that contained either the template or the nontemplate strand sequence of the U1 or U6 PSEA. The primers were extended by using T4 DNA polymerase and the four deoxynucleoside triphosphates. The nick that remained at the 5′ end of the derivatized oligonucleotide was sealed by using T4 DNA ligase. U1 probes with cross-linking reagent in the template strand were prepared by using the following 82-mer in the reactions described above: 5′-ACGAATTCATTCTTATAATTCCCAACTGGTTTTAGCGGTACCGCCATGGAAAGGTATGGGATCCTCAATACTTCGGCATGCA-3′. To generate U1 probes with the cross-linking reagent in the nontemplate strand, the following 81-mer was used: 3′-GCTTAAGTAAGAATATTAAGGGTTGACCAAAATCGCCATGGCGGTACCTTTCCATACCCTAGGAGTTATGAAGCCGTACGA-5′. (The letters in boldface represent the nucleotides of the U1 PSEA.) To prepare cross-linking probes that contained the U6 PSEA, identical oligonucleotides were used except that they contained U6 PSEA bases at the five underlined nucleotides (see also Fig. 1A).
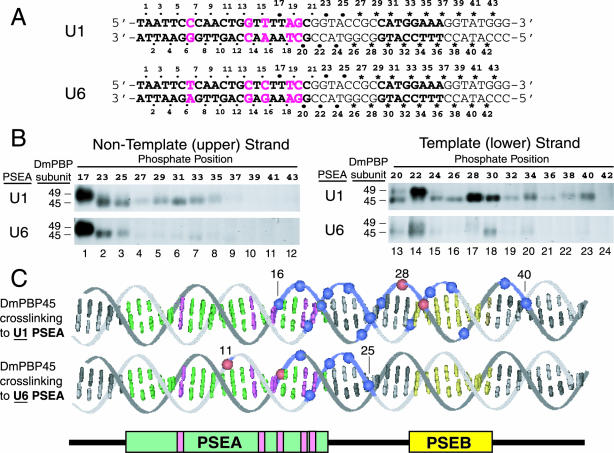
FIG. 1.
Site-specific protein-DNA photo-cross-linking of DmPBP to phosphate positions downstream of the PSEAs of D. melanogaster U1 and U6 genes. (A) Sequences of the relevant areas of the U1 and U6 photo-cross-linking probes. The sequences of the 21-bp U1 and U6 PSEAs are shown in boldface type. (Also shown in boldface type is the sequence of an 8-bp U1 PSEB, which is a second sequence fairly well conserved in D. melanogaster snRNA genes transcribed by RNA polymerase II [16, 38].) The five base pairs where the U1 and U6 PSEAs differ are indicated by magenta letters. Except for these five positions, the probes were identical in sequence to ensure that any differences in the cross-linking pattern were due solely to the five differences within the two PSEAs. The individual phosphate positions at which a cross-linker was incorporated are indicated by the dots, asterisks, and numbers above and below the sequences. The dots indicate positions for which data were reported in an earlier study (33). The asterisks indicate new positions not previously investigated; these 36 phosphate positions were individually derivatized to generate the 36 new probes used in this study. The larger dots indicate positions repeated in the present study as intensity standards and size markers to relate the new studies to the previous one. (B) Twenty-four different radiolabeled site-specific U1 probes and 24 different U6 probes were each incubated in separate reaction mixtures with DmPBP. Following UV irradiation and nuclease digestion, polypeptides that cross-linked to the DNA were detected by SDS-gel electrophoresis and autoradiography. Only the regions of the gels that correspond to the mobilities of DmPBP49 and DmPBP45 are shown. The left panels show the results of cross-linking to the nontemplate strand, and the right panels show cross-linking to the template strand. In each case, the upper panel illustrates the pattern for the U1 PSEA and the lower panel illustrates the pattern for the U6 PSEA. (C) Summary of DmPBP45 cross-linking to DNA probes that contain either the U1 PSEA or the U6 PSEA. Cross-linking data from Fig. 1B, together with previously reported data (33), are shown projected onto B-form DNA. Phosphate positions that cross-linked relatively strongly to the 45-kDa subunit of DmPBP are indicated by blue spheres, whereas the very strongest cross-linking positions are indicated by red spheres. Weak cross-links are not indicated. Base pairs that comprise the PSEA are shown in green and magenta, and those that comprise the U1 PSEB are shown in yellow. When DmPBP was bound to a U1 PSEA, phosphate positions between 16 and 40 cross-linked to DmPBP45, but when DmPBP was bound to a U6 PSEA, cross-linking to DmPBP45 was restricted to a region between phosphates 11 and 25. When the DNA is oriented as shown, the cross-links to DmPBP45 are mainly on the upper surface of the DNA helix.
Photo-cross-linking reactions and analysis were carried out as previously described (33). Briefly, derivatized DNA fragments were incubated with partially purified DmPBP, either the HA300 fraction from fruit fly embryo nuclear extracts (29) or a nickel-chelating column fraction of overexpressed His6-tagged protein from S2 cells. After 30 min at 25°C in the dark, reaction mixtures were irradiated with UV light for 5 min followed by digestions with DNase I and S1 nuclease. Samples were electrophoresed through sodium dodecyl sulfate (SDS)-polyacrylamide gels, and the cross-linked peptides were detected by autoradiography.
Identification and cloning of D. melanogaster genes that code for subunits of DmPBP.
BLAST searches of the D. melanogaster genome were carried out with the amino acid sequences of human SNAP190 (34), SNAP50 (7), SNAP45 (27), SNAP43 (9), and SNAP19 (8) as query sequences. The human SNAP190, SNAP50, and SNAP43 sequences each identified single loci in the D. melanogaster genome corresponding to FlyBase genes CG2702, CG11508, and CG9597. Full-length cDNA clones for the DmSNAP190 and DmSNAP50 genes were purchased from Research Genetics. In each case, the coding region was amplified and inserted into the plasmid pMT/V5-His-TOPO for expression in insect cells under the control of the metallothionein promoter (Invitrogen, Carlsbad, Calif.). Since no cDNA clone was available for DmSNAP43, but the gene consisted of a single exon, the coding region was amplified directly from the genomic DNA of wild-type fly embryos and cloned into pMT/V5-His-TOPO. By appropriate choice of primers, clones were prepared to express each protein by using the natural start and stop codons or to delete the stop codon so that the proteins would include the V5 epitope and His6 tags encoded by the vector sequences. Inclusion of the tags was expected to increase the molecular mass of the proteins by about 5,000 Da.
Expression of DmSNAP genes in D. melanogaster S2 cells.
The expression constructs described above were used individually or in various combinations together with pCoBLAST to cotransfect S2 cells according to conditions recommended by Invitrogen. Stably transfected cell lines were established by selection on blasticidin-containing media. Lines that expressed each of the DmSNAP genes individually (either tagged or untagged) or all three genes together in various combinations of tagged and untagged proteins (e.g., tagged DmSNAP190 and DmSNAP50 with untagged DmSNAP43, or tagged DmSNAP190 and DmSNAP43 with untagged DmSNAP50) were established. The expression of DmSNAPs was induced from the metallothionein promoter by treatment of cells for 24 h with 0.5 mM copper sulfate. Cells were then lysed, and extracts were used either for immunoblot analysis or for purification of tagged DmPBP by affinity chromatography using Invitrogen's ProBond nickel-chelating resin.
Antibodies, mobility shift assays, and immunoblots.
Anti-peptide antibodies were raised against the three peptide sequences indicated in Fig. 2. Peptides were synthesized by Sigma-Genosys or Research Genetics, conjugated to keyhole limpet hemocyanin, and used to immunize rabbits. Peptide-specific antibodies were purified by affinity chromatography on immobilized peptide columns prior to use in immunoblots. Monoclonal anti-V5 antibodies were purchased from Invitrogen.
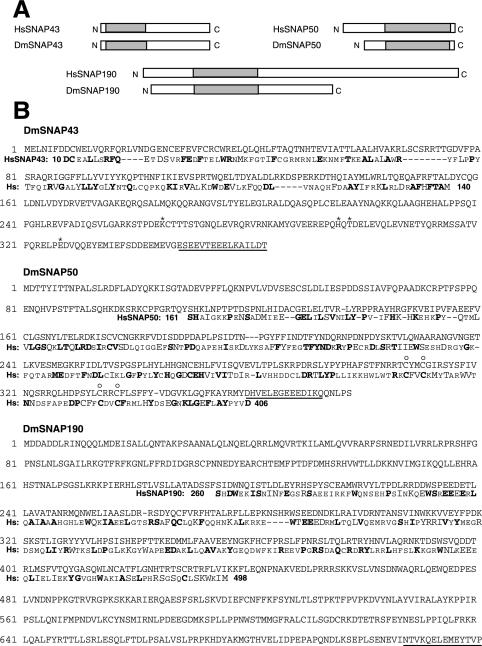
FIG.2.
The D. melanogaster genome codes for proteins with similarity to Homo sapiens SNAP43 (HsSNAP43), HsSNAP50, and HsSNAP190. (A) BLAST searches of the D. melanogaster nucleic acid database identified genes predicted to code for proteins with similarities to three of the human SNAP proteins. The rectangles indicate the relative lengths of the proteins from the N to the C termini, and the shaded areas indicate the regions of the homologous protein pairs that share greater than 26% identity and 42% similarity. (B) The amino acid sequences of the predicted DmSNAP43, DmSNAP50, and DmSNAP190 proteins are shown. Sequences from the homologous human SNAPs, in those regions where the corresponding proteins from the two species exhibit substantial similarity (shaded areas in panel A), are shown below the fly sequences. Dissimilar residues are indicated by small capital letters, chemically similar residues are indicated by large capital letters, and identical residues are indicated by boldface. The amino acid positions in the human SNAPs of the first and last residues shown are indicated by the numbers immediately before and after the human sequences (Hs). The underlined residues indicate the peptide sequences from the fly proteins that were used to generate antibodies. The sequence of the DmSNAP43 gene, cloned by us from both wild-type embryo DNA and S2 tissue culture cells, codes for a protein that differs at four amino acid positions from the sequence predicted in the D. melanogaster nucleic acid database. These four amino acids are indicated by asterisks above the DmSNAP43 sequence. The open circles above the DmSNAP50 sequence indicate four cysteines that may form a zinc finger and are conserved in homologous human, trypanosome, and roundworm proteins.
Mobility shift assays were normally carried out in 12-μl reaction volumes containing either 4 μl of DmPBP HA300 fraction from fruit fly embryos (29) or 2 to 10 μl of DmPBP purified from S2 cells by nickel-chelating chromatography. The final buffer composition was approximately 20 mM HEPES (pH 7.9), 100 mM KCl, 5 mM MgCl2, 0.01 mM ZnCl2, 2 mM EDTA, 3 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 20% (by volume) glycerol. Reaction mixtures also contained 1 μg of poly(dI-dC) · poly(dI-dC) and 1 μg of poly(dG-dC) · poly(dG-dC). The reaction mixtures were incubated at room temperature for 30 min prior to being loaded onto 5% (29:1 acrylamide-bisacrylamide) native gels, and bands were detected by autoradiography. When antibodies were added for supershifts, the antibodies were added halfway through the 30-min incubation. Competitor peptides, when included, were added at the beginning of the incubation to a final concentration of 0.1 μg/μl.
For immunoblots, 30 μg of total S2 cell protein extract was electrophoresed through SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. Untagged DmSNAP proteins were detected by using the rabbit anti-peptide antibodies described above followed by goat anti-rabbit alkaline phosphatase-conjugated secondary antibody and Western Blue-stabilized substrate (Promega) according to the manufacturer's instructions. V5-tagged proteins were detected by using anti-V5/AP antibody from Invitrogen.
RESULTS
The 45-kDa subunit of DmPBP makes extensive protein-DNA contacts downstream of the U1 PSEA (but not downstream of the U6 PSEA).
In earlier work, a high-resolution site-specific protein-DNA photo-cross-linking assay was used to map the interaction of the DmPBP subunits with the phosphate backbone of the U1 and U6 PSEAs (33). In that assay, double-stranded DNA probes that had individual phosphate positions derivatized with a photo-cross-linking agent adjacent to a 32P radiolabel were used. This method has been used by others to map the position and orientation of the general transcription factors along the DNA in the RNA polymerase II preinitiation complex (14, 15). In earlier work (33), Wang and Stumph used 50 individually labeled probes to scan through a 25-bp region of the U1 and U6 PSEAs at every second phosphate position on both the template and nontemplate strands extending to a position 4 bp beyond the 3′ end of the conserved PSEAs (Fig. 1A). It was noted that DmPBP45 and DmPBP95 cross-linked to the phosphate in the most 3′ position examined in those studies (position 25 on the nontemplate strand). Thus, despite the fact that the 21-bp PSEAs provided the sequence specificity for DNA binding, there was the possibility that one or more subunits of DmPBP might remain in close proximity to the DNA for a substantial distance downstream of the U1 or U6 PSEA.
To investigate DmPBP-DNA interactions downstream of the PSEA, we generated 36 new probes, each containing a cross-linking agent at a unique phosphate position 3′ of the U1 or U6 PSEA (Fig. 1A). In addition, a number of probes (phosphate positions 17, 23, and 25 on the nontemplate strand and positions 20, 22, and 24 on the template strand) (Fig. 1A) were used as standards of a known cross-linking pattern and intensity based upon the results of the previous study (33). Following the incubation of each probe individually with protein, reaction mixtures were irradiated with UV light to activate the cross-linking agent, subjected to extensive nuclease digestion, and run on SDS gels. Protein bands that contained a cross-linked radiolabel were detected by autoradiography.
Figure 1B shows that DmPBP49 cross-linked to these probes only at the positions previously reported: U1 phosphate positions 17 (very strongly), 20 (moderately), and 22 (very strongly) (Fig. 1B, upper panels, lanes 1, 13, and 14) and U6 phosphate positions 17 (very strongly) and 22 (weakly) (Fig. 1B, lower panels, lanes 1 and 14). DmPBP49 did not cross-link to any of the new U1 or U6 probes that scanned for interactions further in the 3′ direction (Fig. 1B, lanes 4 to 12 and 16 to 24). In other results, DmPBP95, the largest subunit, likewise did not cross-link to any positions further in the 3′ direction beyond those previously reported, except for one minor cross-link at phosphate position 27 with both the U1 and U6 probes (data not shown).
Notably, however, the DmPBP45 subunit cross-linked extensively and strongly to many of the new probes used in this study. When DmPBP was bound to a U1 PSEA, DmPBP45 cross-linked as far downstream as phosphate position 35 on the nontemplate strand and as far as position 40 on the template strand (Fig. 1B, upper panels, lanes 4 to 8 and 16 to 23). Because position 20 was the site of the strongest DmPBP45 cross-linking to a U1 PSEA observed in the previous study (33), the comparable intensities of the bands obtained with the downstream probes suggest that DmPBP45 resides in close proximity to the DNA for 20 bp beyond the end of the conserved U1 PSEA sequence. In contrast, when DmPBP was bound to a U6 PSEA, only minimal cross-linking of DmPBP45 was observed downstream of phosphate position 25 (Fig. 1B, lower panels, lanes 4 to 12 and 16 to 24). Only site 30 on the template strand of the U6 PSEA cross-linked weakly. (Figure 1B, lower right panel, is an overexposure to reveal the bands in lanes 13 and 14, which indicate weak cross-links [33].) Since all the probes contained identical DNA sequences flanking the U1 and U6 PSEAs, the ability of the DmPBP45 subunit to cross-link to the downstream DNA was determined by whether DmPBP was bound to a U1 or a U6 PSEA.
Figure 1C illustrates and summarizes the cross-linking pattern of DmPBP45 projected onto B-form DNA based upon data combined from Fig. 1B and the previous study (33). Phosphate positions that cross-linked strongly to DmPBP45 are indicated by colored spheres on the DNA backbone in Fig. 1C. Positions with weak or nondetectable cross-linking are not indicated. For both PSEAs, DmPBP45 cross-linked primarily to the upper face of the duplex when the DNA is oriented as shown in the figure. When DmPBP was bound to a U1 PSEA, DmPBP45 contacted the DNA as far 3′ as position 40. In contrast, on a U6 PSEA, no strong cross-linking was observed beyond position 25. On a U1 PSEA, the very strongest cross-links of DmPBP45 occurred downstream of the PSEA at positions 28 and 30 (upper DNA duplex), but with a U6 PSEA, the very strongest cross-links occurred at positions 11 and 16 within the PSEA (lower DNA duplex).
It is interesting that the binding of DmPBP to a U1 PSEA results in the cross-linking of DmPBP45 strongly to phosphate positions within and near to PSEB, an element that contributes to the promoter strength of the RNA polymerase II-transcribed snRNA genes of fruit flies (38). At the present time, we cannot be certain whether the cross-linking of DmPBP45 observed in the vicinity of PSEB is determined solely by the activity of the upstream U1 PSEA regardless of the downstream sequence or whether the sequence of PSEB itself may contribute to the interactions of the DNA with DmPBP45. Nonetheless, it is clear that the U6 PSEA, unlike the U1 PSEA, does not allow these close interactions to take place between DmPBP45 and the PSEB region. Thus, the interaction of DmPBP45 with the DNA is substantially different depending upon whether DmPBP is bound to a U1 or a U6 PSEA.
Identification of D. melanogaster genes that code for proteins homologous to three of the subunits of human SNAPc.
Human SNAPc consists of five distinct subunits (SNAP19, SNAP43, SNAP45, SNAP50, and SNAP190) whose genes have been cloned and characterized. We searched the D. melanogaster nucleic acid database for genes that encode proteins homologous to the human polypeptides. Highly significant matches were found for the SNAP43, SNAP50, and SNAP190 polypeptides, but no matches were found for SNAP19 and SNAP45.
Figure 2A illustrates the structural relationships of the human SNAPs and the related sequences coded in the fly genome. For convenience, we designate the D. melanogaster homologs as DmSNAP43, DmSNAP50, and DmSNAP190. Well-conserved regions of the proteins where the sequences from the two species are greater than 26% identical and 42% similar are shown in Fig. 2A. Outside of these conserved regions, there is little or no significant sequence similarity. The SNAP43 homologs are well conserved within their amino-terminal halves, whereas the carboxyl-terminal two-thirds of the SNAP50 homologs share substantial identity. The SNAP190 homologs are similar in a region that contains an unusual signature motif that consists of 4.5 Myb repeats. This region of human SNAP190 has DNA binding activity (12, 20, 23, 34).
The primary sequences of the three putative DmSNAP proteins are shown in Fig. 2B. The calculated molecular masses of the three polypeptides are 42, 43, and 84 kDa for DmSNAP43, DmSNAP50, and DmSNAP190, respectively. For purposes of comparison, Fig. 2B also shows sequences from the three homologous human proteins in those regions where they exhibit significant sequence similarity to the respective fly proteins.
The predicted DmSNAP proteins are components of native DmPBP.
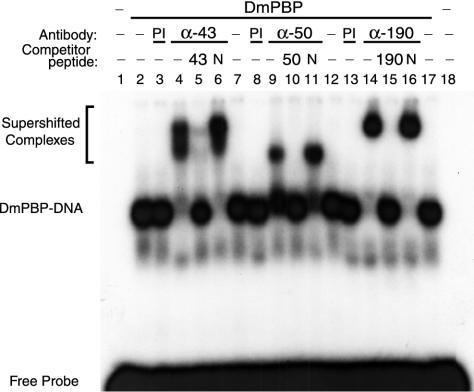
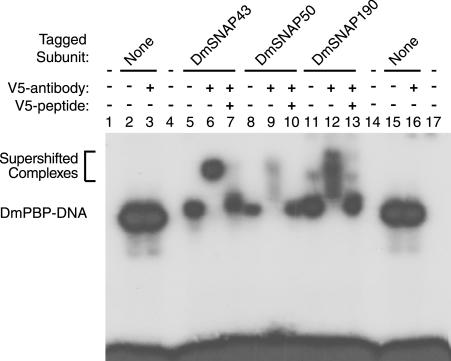
To obtain evidence that the proteins whose sequences are depicted in Fig. 2B are components of the D. melanogaster PSEA-binding protein, antibodies were raised against peptide sequences found within each of the DmSNAPs. The antibodies were then tested for their ability to supershift DmPBP-DNA complexes in a mobility shift assay. Mobility shifts were performed by using the U1 PSEA sequence and native DmPBP from fly embryos partially purified from the soluble nuclear fraction as previously described (29). Antibodies against peptides from each of the predicted DmSNAP subunits supershifted the native DmPBP-DNA complex (Fig. 3, lanes 4, 9, and 14). Supershifts were not observed when preimmune serum was used instead of the specific antibodies (Fig. 3, lanes 3, 8, and 13). Moreover, the supershifted complexes were outcompeted by excess specific peptide (Fig. 3, lanes 5, 10, and 15) but not by nonspecific peptide (lanes 6, 11, and 16). These data suggest strongly that the DmSNAP43, DmSNAP50, and DmSNAP190 genes identified in the D. melanogaster genome code for polypeptide subunits of DmPBP.
FIG. 3.
Antibodies against peptide sequences from the predicted DmSNAP proteins react with native DmPBP. Electrophoretic mobility shift assays were carried out by using DmPBP prepared from fruit fly embryos and a probe that contains U1 PSEA. Additional components were added as indicated above the individual lanes. Lanes 4 to 6, 9 to 11, and 14 to 16 contained antibodies from rabbits injected with synthetic peptides corresponding to sequences from the DmSNAP43 (α-43), DmSNAP50 (α-50), and DmSNAP190 (α-190) sequences, respectively (Fig. 2). Lanes 3, 8, and 13 contained preimmune sera (PI). Lanes 5, 10, and 15 contained excess synthetic peptide specific to the antibodies used in the same lane. Lanes 6, 11, and 16 contained nonspecific peptide (N).
Overexpression of DmSNAPs in D. melanogaster S2 cells.
We placed the DmSNAP43, DmSNAP50, and DmSNAP190 genes into insect expression vectors under the control of the inducible metallothionein promoter. Individual clones were prepared such that each protein could be expressed with or without the V5 and His6 C-terminal tags provided by the pMT/V5-His vector. These clones were cotransfected into D. melanogaster S2 cells together with a plasmid to provide blasticidin resistance. Blasticidin-resistant cell lines that could be induced to express the DmSNAP genes upon the addition of copper sulfate were selected.
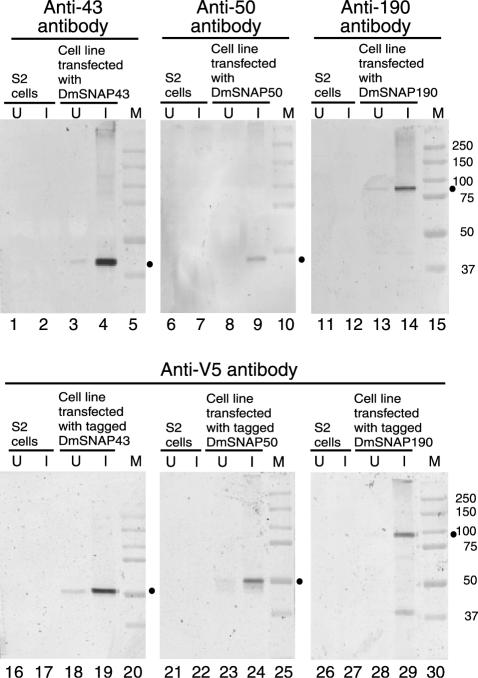
The upper panels of Fig. 4 show immunoblots in which the untagged versions of the proteins were detected with antibodies specific for each protein. In normal S2 cells, the expression levels of the endogenous proteins were too low to be detectable (lanes 1, 2, 6, 7, 11, and 12). However, each of the stably transfected cell lines expressed an increased level of a protein of an appropriate molecular weight following induction with copper sulfate (compare lanes 3 and 4, 8 and 9, and 13 and 14). (The anti-DmSNAP50 antibodies were of lower quality than the anti-DmSNAP43 and anti-DmSNAP190 antibodies, resulting in a weaker signal in the immunoblots of Fig. 4 and a weaker supershift in the mobility shift assays of Fig. 3.)
FIG. 4.
Inducible overexpression of DmSNAP proteins in S2 cells. We generated S2 cell lines that were stably transfected with one of the DmSNAP genes under the control of the metallothionein promoter. Lysates from control S2 cells, or from stably transfected S2 cells, either without (lanes U) or with (lanes I) copper induction were analyzed by immunoblotting. Untagged DmSNAPs were detected by using specific antipeptide antibodies (upper panels) (Anti-43, Anti-50, and Anti-190 refer to antibodies to DmSNAP43, DmSNAP50, and DmSNAP190, respectively), whereas epitope-tagged versions of the proteins were analyzed by using anti-V5 antibodies (lower panels). Dots indicate the positions of bands that correspond to the gene products analyzed in each individual panel. Lanes M, molecular size markers.
The lower panels of Fig. 4 show similar immunoblots but with protein extracts from cell lines expressing V5/His6 epitope-tagged versions of the proteins. The addition of the tags is expected to increase the molecular mass of each protein by about 5,000 Da. In each case, anti-V5 antibody detected copper-induced bands that had appropriately reduced mobilities relative to those of the respective untagged proteins (compare the mobilities relative to the markers in the upper and lower panels of Fig. 4).
Tagged DmSNAPs are incorporated into functional DmPBP.
Stably transfected cell lines that overexpressed all three of the DmSNAPs within the same cells were next prepared. In each case, one of the DmSNAPs was expressed as the V5/His6-tagged version of the polypeptide, but the remaining two DmSNAPs remained untagged. However, all DmSNAPs were under the control of the metallothionein promoter. Following induction, His6-tagged proteins and protein complexes were subjected to nickel-chelating chromatography. The partially purified protein fractions were then used in mobility shift assays with a DNA fragment containing a U1 PSEA (Fig. 5).
FIG. 5.
Epitope-tagged DmSNAP proteins can be incorporated into DmPBP that functionally binds to DNA. Electrophoretic mobility shift assays were performed with DmPBP obtained from three different stably transfected S2 cell lines that each overexpressed all three DmSNAP subunits, but as indicated above the appropriate lanes (5 to 7, 8 to 10, and 11 to 13), only one of the subunits in each cell line possessed a V5 tag. Lanes 2, 3, 15, and 16 contained untagged DmPBP from fruit fly embryos. Anti-V5 antibody and V5 competitor peptide were added to reaction mixtures as indicated above the lanes.
The nickel affinity-purified fraction from each of the cell lines, whether expressing tagged DmSNAP43, tagged DmSNAP50, or tagged DmSNAP190, formed protein-DNA complexes that were of similar mobilities (Fig. 5, lanes 5, 8, and 11). Each tagged complex had a mobility just slightly less than that of the complex formed with natural DmPBP isolated from embryos (Fig. 5, lanes 2 and 15), probably due to the extra amino acids at the C termini of the tagged subunits. An addition of anti-V5 antibody to the reaction mixtures had no effect on the mobilities of complexes that contained untagged DmPBP from embryos (Fig. 5, lanes 3 and 16). On the other hand, the anti-V5 antibodies supershifted the complexes that formed with S2 cell DmPBP that contained individually tagged subunits (Fig. 5, lanes 6, 9, and 12). Because the DmPBP-PSEA complexes were either quantitatively supershifted (Fig. 5, lanes 6 and 12) or sometimes partially disrupted (lane 9) by the anti-V5 antibodies, it can be inferred that nearly all of the DmPBP present in the fractions isolated by nickel chromatography contain a tagged subunit. These data indicate, first, that the DmSNAP43, DmSNAP50, and DmSNAP190 genes each code for an integral subunit of DmPBP, and second, that the V5/His6-tagged DmSNAP proteins can be incorporated into DmPBP capable of binding to DNA that contains PSEA.
Correlating the DmSNAP gene products with the three proteins previously identified in the photo-cross-linking assay.
Data presented above indicate that we have identified and expressed fruit fly genes that code for three distinct polypeptide components of DmPBP. Moreover, the molecular masses of the proteins encoded by the cloned genes are in reasonable agreement with those of the three polypeptides detected in the photo-cross-linking assay. Those results by themselves, however, could not prove a one-to-one correspondence. We therefore further examined the relationship between the cloned genes and the proteins detected by photo-cross-linking.
The DmSNAP43 and DmSNAP50 genes code for proteins of very similar predicted sizes (42 and 43 kDa, respectively). Thus, it was not known whether the DmSNAP43 and DmSNAP50 genes code for DmPBP45 and DmPBP49, respectively, or vice versa. It was even a possibility that one or both of the genes potentially could encode additional components of DmPBP not detected by the photo-cross-linking assay. On the other hand, it seemed very likely that the DmSNAP190 gene, which codes for a protein with a calculated molecular mass of 84 kDa, was the gene for DmPBP95. DmPBP95 cross-links over the entire length of the PSEA, and the DmSNAP190 gene product was expected to cross-link to DNA because it contains 4.5 Myb repeats, similar to the region that constitutes the DNA-binding domain of human SNAP190 (12, 20, 23, 34). Moreover, human SNAP190/PTFα, like DmPBP95, has been experimentally cross-linked to DNA (36).
To delineate the one-to-one relationships of the cloned D. melanogaster genes with the three polypeptides previously detected in the photo-cross-linking assay, we carried out photo-cross-linking experiments with DmPBP that contained tagged subunits. Because the extra amino acids at the C terminus of the tagged proteins increase their molecular mass by about 5,000 Da, tagged proteins exhibit slower mobilities on SDS gels than those of the corresponding untagged proteins (Fig. 4). We took advantage of this property to identify the gene products that cross-link to a particular phosphate position.
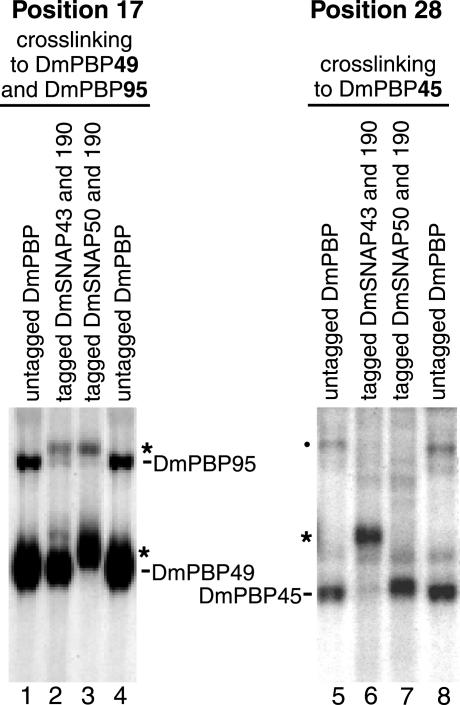
We prepared S2 cell lines that stably coexpressed V5/His6-tagged versions of DmSNAP43 and DmSNAP190 together with untagged DmSNAP50 or, alternatively, coexpressed tagged DmSNAP50 and tagged DmSNAP190 together with untagged DmSNAP43. Extracts were prepared from the two cell lines, and the tagged complexes were isolated by nickel affinity chromatography. These tagged DmSNAP complexes were then used for photo-cross-linking to U1 probes that contained the cross-linking agent at either phosphate position 17 of the upper strand or at phosphate position 28 of the lower strand. Position 17 was selected because previous data (33) indicated that position 17 cross-links strongly to DmPBP95 and very strongly to DmPBP49, but it does not cross-link to DmPBP45. In contrast, phosphate position 28 of the U1 probe cross-links exclusively and specifically to DmPBP45 (Fig. 1).
The left panel of Fig. 6 shows results obtained with the DNA probe that contained a cross-linker at position 17 of the nontemplate strand. When the reactions were carried out with untagged DmPBP (Fig. 6, lanes 1 and 4), two bands that represent cross-linking to the DmPBP95 and DmPBP49 subunits were observed as expected. Lane 2 of Fig. 6 shows results obtained when we used protein isolated from S2 cells expressing tagged DmSNAP43 and tagged DmSNAP190 but untagged DmSNAP50. In this case, the DmPBP95 band shifted to a higher position, suggesting that the DmSNAP190 gene indeed codes for the polypeptide termed DmPBP95. On the other hand, the mobility of the DmPBP49 band was unchanged, strongly suggesting that the DmSNAP43 gene does not code for DmPBP49. The results shown in Fig. 6, lane 3, support these conclusions and provide evidence that DmPBP49 is encoded by the DmSNAP50 gene. When the protein complex contained tagged DmSNAP50 and tagged DmSNAP190 but untagged DmSNAP43, the DmPBP49 band (as well as the DmPBP95 band) demonstrated a reduced mobility (Fig. 6, lane 3). This is the result expected if the DmSNAP50 gene codes for DmPBP49 and the DmSNAP190 gene codes for DmPBP95.
FIG. 6.
Correlation of the DmSNAP gene products with the DmPBP subunits detected by photo-cross-linking. Photo-cross-linking was carried out with probes derivatized with a cross-linker at phosphate position 17 (probe reacts with DmPBP49 and DmPBP95 [lanes 1 to 4]) or at position 28 (probe reacts with DmPBP45 [lanes 5 to 8]). Protein components in the reaction mixtures were as follows: untagged DmPBP from embryos (lanes 1, 4, 5, and 8); protein from stably transfected S2 cell lines expressing V5/His6-tagged DmSNAP43, tagged DmSNAP190, and untagged DmSNAP50 (lanes 2 and 6); or protein from stably transfected S2 cell lines expressing V5/His6-tagged DmSNAP50, tagged DmSNAP190, and untagged DmSNAP43 (lanes 3 and 7). The asterisks indicate the positions of cross-linking products with reduced mobility due to the C-terminal polypeptide extension that contains the V5 and His6 epitopes. The dot indicates a nonspecific band observed only in the embryo protein preparation that is not competed by excess unlabeled probe (data not shown).
The identities of the DmSNAP43 and DmSNAP50 gene products were confirmed by the complementary experiment results shown in the right panel of Fig. 6. In this case, the U1 DNA probe contained a cross-linker at position 28 of the template strand, a position that cross-links exclusively to the DmPBP45 subunit (Fig. 1B). When the DmSNAP43 gene carried the tag, the band corresponding to DmPBP45 ran with a slower mobility (Fig. 6, lane 6). However, when DmSNAP50 was tagged, the migration of the band was unchanged (Fig. 6, lane 7). Taken together, the experimental results shown in Fig. 6 provide convincing evidence that (i) the DmSNAP43 gene codes for DmPBP45, (ii) the DmSNAP50 gene codes for DmPBP49, and (iii) the DmSNAP190 gene codes for DmPBP95.
DISCUSSION
The three subunits of DmPBP identified by photo-cross-linking are orthologous to three subunits of human SNAPc.
DmPBP contains three subunits (DmPBP45, DmPBP49, and DmPBP95) that can be photo-cross-linked to DNA (33). A comparison of the D. melanogaster genome with the sequences of the five known human SNAPc subunits revealed significant matches to SNAP43, SNAP50, and SNAP190. Antibodies produced against polypeptides predicted by the sequences of the fly genes specifically supershifted native DmPBP-PSEA complexes. Furthermore, the expression of the fly genes in S2 cells indicated that tagged versions of the predicted DmSNAPs could be incorporated into functional DmPBP capable of binding to DNA. By using DmPBP that contained different tagged subunits, it was possible to correlate the gene products with the proteins previously identified by photo-cross-linking. Results of these experiments allow us to conclude that DmPBP45 and human SNAP43, DmPBP49 and human SNAP50, and DmPBP95 and human SNAP190 are orthologous pairs of proteins.
Phylogenetic and structural features of the PBP subunits.
A unique structural motif in both human and fly SNAP190/DmPBP95 comprises 4.5 Myb repeats (34). In truncated SNAP190, this region is capable of binding to DNA, although with a low level of specificity (12, 20, 34). Outside of this region, there is no obvious similarity between the human and fly proteins, and the overall length of the fly protein is only about half that of its human ortholog. Interestingly, the D. melanogaster U1 and U6 PSEA sequences used in our studies each contain a consensus Myb recognition element at positions 9 to 13 (sequence AACNG) (25, 30). This sequence occurs within a region of the U1 and U6 PSEAs that cross-links to DmPBP95 (33).
DmPBP49 and human SNAP50 are similar to each other, primarily in their C-terminal regions. This part of both proteins contains a putative zinc finger that may be involved in protein-DNA or protein-protein interactions. Experimentally, SNAP50 has been cross-linked to a mammalian PSEA and is therefore presumed to reside in close proximity to the DNA (7). DmPBP49 cross-links to a U1 PSEA primarily between positions 13 and 22 but to a U6 PSEA between positions 12 and 17. Thus, the position of the putative zinc finger, if involved in DNA binding, may shift depending upon which D. melanogaster DNA sequence DmPBP is bound to. Recently, a protein orthologous to SNAP50 was also identified as a subunit of PBP-1, a three-subunit protein that binds to a regulatory sequence 60 to 70 bp upstream of trypanosome spliced leader RNA genes (4). A putative Caenorhabditis elegans protein also exhibits substantial similarity to the human, fly, and trypanosome proteins (1). Thus, the SNAP50 orthologs are representatives of a very ancient protein apparently involved in the transcription of snRNA and snRNA-like genes in widely divergent eukaryotes. Less phylogenetic information is available about SNAP43 and its orthologs. The most highly conserved region of the human and fly SNAP43 proteins is toward their amino termini (Fig. 2). In human SNAP43, this region is involved in interacting with human SNAP50 (12, 19). Thus, the most conserved domains of these two proteins may be involved in interacting with each other.
Absence of SNAP19 and SNAP45 orthologs from DmPBP.
Several lines of evidence lead us to believe that polypeptides orthologous to human SNAP19 and SNAP45 are not present in DmPBP. First, searches with the human SNAP19 and SNAP45 sequences did not reveal any significant matches in the Drosophila database. Second, the three subunits identified in the photo-cross-linking assay have each been accounted for as orthologs of SNAP43, SNAP50, and SNAP190. Third, SNAP45 is incorporated into SNAPc by interaction with a short region near the C terminus of SNAP190 (19, 23), a region that is absent from DmPBP95 (Fig. 2A). Fourth, SNAP19, while contributing to the stability of the interaction between SNAP43 and SNAP190 (8), is not essential (20, 23). Finally, the DNA-binding and basal transcription activities of SNAPc can be reconstituted from the SNAP43, SNAP50, and SNAP190 subunits alone in the absence of SNAP19 and SNAP45 (12, 19, 20, 23). Thus, the core PSE-binding activity that is sufficient for basal transcription in both vertebrates and insects appears to reside in only three subunits (i.e., SNAP43, SNAP50, and SNAP190 and the orthologous proteins in flies). It is possible that SNAP19 and SNAP45 may play regulatory roles (23) unique to mammalian or vertebrate systems.
Architectural arrangement of PBP subunits on U1 and U6 snRNA gene promoters.
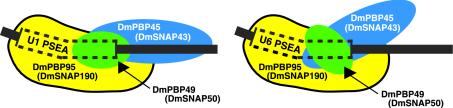
We have used a powerful, high-resolution, site-specific protein-DNA photo-cross-linking assay to identify the surfaces of the DNA where each of the three cloned subunits of DmPBP interacts with the phosphate backbone both within and downstream of the U1 and U6 PSEAs. A schematic model incorporating the results from the present and previous studies (5, 33) is shown in Fig. 7. DmPBP95 (DmSNAP190) resides in close proximity to the DNA over the entire length of the PSEA and about 5 nucleotides downstream. DmPBP49 (DmSNAP50) cross-links strongly to phosphates between positions 12 and 22 of the U1 PSEA, but comparably strong cross-linking is seen only between positions 12 and 17 of the U6 PSEA (33). The third subunit, DmPBP45 (DmSNAP43) displays profoundly different cross-linking patterns when DmPBP is bound to the U1 and U6 PSEAs (Fig. 1). On a U1 PSEA, DmPBP45 cross-links to phosphates between positions 16 and 40, but on a U6 PSEA, it cross-links to phosphates between positions 11 and 25. Since the DNA is bent similarly in DmPBP complexes with the U1 and U6 PSEAs (toward the face of the DNA that interacts with DmPBP45) (5), the data suggest that DmPBP assumes a different conformation depending upon whether it is bound to a U1 or a U6 PSEA sequence. We postulate that in the insect D. melanogaster, this difference in conformation provides at least one essential step in the process that ultimately leads to the recruitment of different general transcription factors and RNA polymerases to U1 and U6 promoters.
FIG. 7.
Schematic model of the subunits of DmPBP (DmSNAPc) bound in alternative conformations to D. melanogaster U1 and U6 PSEAs. The model is based upon past (5, 33) and present data. The three subunits of the PSEA-binding protein are shown in the approximate relative positions where they interact with the phosphate backbone of the DNA. In each case, the DNA is bent similarly toward the face of the DNA that interacts with DmPBP45 (DmSNAP43) (5). If this is considered to be the upper face of the PSEA, DmPBP49 (DmSNAP50) interacts primarily with the front face of the 3′ half of the PSEA, and DmPBP95 (DmSNAP190) interacts primarily with the front face of the 5′ half of the PSEA but with the bottom face of the 3′ half of the PSEA.
In mammals and other vertebrates, the U1 and U6 PSEAs are interchangeable without prejudice with respect to RNA polymerase specificity. Although different mechanisms are apparently involved in determining the RNA polymerase specificity of snRNA genes in vertebrates and insects, the three subunits of DmPBP (now referred to interchangeably as DmSNAPc) are orthologous to the three subunits of human SNAPc that are essential for basal activity. It therefore seems reasonable to expect that the translational and rotational arrangement of the orthologous human SNAP subunits on DNA will generally be similar to the pattern exhibited by the fly subunits. Our photo-cross-linking data indicate that the subunits that occupy the most 3′ positions along the DNA are DmPBP45 and DmPBP95 (Fig. 7). This positions them appropriately to interact with a TATA-binding protein (TBP) or a TBP-related factor bound to the DNA approximately 25 to 30 bp upstream of the transcription start site. Interestingly, the orthologous human subunits, SNAP43 and SNAP190, can interact with TBP in vitro (12, 20, 37). Additional experiments will be required in both systems to validate the comparisons.
Acknowledgments
We are very grateful to Yan Wang for helpful hints and suggestions for preparing and using the photo-cross-linking probes. We thank Adrian Elcock of the San Diego Super Computer Center for assistance with molecular graphics.
This work was supported by National Science Foundation grant MCB-0131151 and in part by the California Metabolic Research Foundation. C.L. is a recipient of the Arne N. Wick Predoctoral Research Fellowship from the California Metabolic Research Foundation.
REFERENCES
- 1.Bai, L., Z. Wang, J.-B. Yoon, and R. G. Roeder. 1996. Cloning and characterization of the β subunit of human proximal sequence element-binding transcription factor and its involvement in transcription of small nuclear RNA genes by RNA polymerases II and III. Mol. Cell. Biol. 16:5419-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahlberg, J. E., and E. Lund. 1988. The genes and transcription of the major small nuclear RNAs, p. 38-70. In M. L. Birnstiel (ed.), Structure and function of major and minor small nuclear ribonucleoprotein particles. Springer Verlag KG, Heidelberg, Federal Republic of Germany.
- 3.Dahlberg, J. E., and E. Lund. 1991. How does III × II make U6? Science 254:1462-1463. [DOI] [PubMed] [Google Scholar]
- 4.Das, A., and V. Bellofatto. 2003. RNA polymerase II-dependent transcription in trypanosomes is associated with a SNAP complex-like transcription factor. Proc. Natl. Acad. Sci. USA 100:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardin, S. B., C. J. Ortler, K. J. McNamara-Schroeder, and W. E. Stumph. 2000. Similarities and differences in the conformation of protein-DNA complexes at the U1 and U6 snRNA gene promoters. Nucleic Acids Res. 28:2771-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry, R. W., E. Ford, R. Mital, V. Mittal, and N. Hernandez. 1998. Crossing the line between RNA polymerases: transcription of human snRNA genes by RNA polymerases II and III. Cold Spring Harbor Symp. Quant. Biol. 63:111-120. [DOI] [PubMed] [Google Scholar]
- 7.Henry, R. W., B. C. Ma, C. L. Sadowski, R. Kobayashi, and N. Hernandez. 1996. Cloning and characterization of SNAP50, a subunit of the snRNA-activating protein complex SNAPc. EMBO J. 15:7129-7136. [PMC free article] [PubMed] [Google Scholar]
- 8.Henry, R. W., V. Mittal, B. C. Ma, R. Kobayashi, and N. Hernandez. 1998. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 12:2664-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry, R. W., C. L. Sadowski, R. Kobayashi, and N. Hernandez. 1995. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerases II and III. Nature 374:653-656. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291-1308. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez, N. 2001. snRNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276:26733-26736. [DOI] [PubMed] [Google Scholar]
- 12.Hinkley, C. S., H. A. Hirsch, L. P. Gu, B. LaMere, and R. W. Henry. 2003. The small nuclear RNA-activating protein 190 Myb DNA binding domain stimulates TATA box-binding protein-TATA box recognition. J. Biol. Chem. 278:18649-18657. [DOI] [PubMed] [Google Scholar]
- 13.Jensen, R. C., Y. Wang, S. B. Hardin, and W. E. Stumph. 1998. The proximal sequence element (PSE) plays a major role in establishing the RNA polymerase specificity of Drosophila U-snRNA genes. Nucleic Acids Res. 26:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, T. K., T. Lagrange, Y. H. Wang, J. D. Griffith, D. Reinberg, and R. H. Ebright. 1997. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc. Natl. Acad. Sci. USA 94:12268-12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagrange, T., T.-K. Kim, G. Orphanides, Y. W. Ebright, R. H. Ebright, and D. Reinberg. 1996. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photo-cross-linking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc. Natl. Acad. Sci. USA 93:10620-10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo, P. C. H., and S. M. Mount. 1990. Drosophila melanogaster genes for U1 snRNA variants and their expression during development. Nucleic Acids Res. 18:6971-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobo, S. M., and N. Hernandez. 1989. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell 58:55-67. [DOI] [PubMed] [Google Scholar]
- 18.Lobo, S. M., and N. T. Hernandez. 1994. Transcription of snRNA genes by RNA polymerases II and III, p. 127-159. In R. C. Conaway and J. W. Conaway (ed.), Transcription: mechanisms and regulation. Raven Press, New York, N.Y.
- 19.Ma, B., and N. Hernandez. 2001. A map of protein-protein contacts within the small nuclear RNA-activating protein complex SNAPc. J. Biol. Chem. 276:5027-5035. [DOI] [PubMed] [Google Scholar]
- 20.Ma, B., and N. Hernandez. 2002. Redundant cooperative interactions for assembly of a human U6 transcription initiation complex. Mol. Cell. Biol. 22:8067-8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattaj, I. W., N. A. Dathan, H. D. Parry, P. Carbon, and A. Krol. 1988. Changing the RNA polymerase specificity of U snRNA gene promoters. Cell 55:435-442. [DOI] [PubMed] [Google Scholar]
- 22.McNamara-Schroeder, K. J., R. F. Hennessey, G. A. Harding, R. C. Jensen, and W. E. Stumph. 2001. The Drosophila U1 and U6 gene proximal sequence elements act as important determinants of the RNA polymerase specificity of snRNA gene promoters in vitro and in vivo. J. Biol. Chem. 276:31786-31792. [DOI] [PubMed] [Google Scholar]
- 23.Mittal, V., B. C. Ma, and N. Hernandez. 1999. SNAPc: a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev. 13:1807-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy, S., J.-B. Yoon, T. Gerster, and R. G. Roeder. 1992. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol. Cell. Biol. 12:3247-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogata, K., S. Morikawa, H. Nakamura, A. Sekikawa, T. Inoue, H. Kanai, A. Sarai, S. Ishii, and Y. Nishimura. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639-648. [DOI] [PubMed] [Google Scholar]
- 26.Parry, H. D., D. Scherly, and I. W. Mattaj. 1989. Snurpogenesis: the transcription and assembly of U snRNP components. Trends Biochem. Sci. 14:15-19. [Google Scholar]
- 27.Sadowski, C. L., R. W. Henry, R. Kobayashi, and N. Hernandez. 1996. The SNAP45 subunit of the small nuclear RNA (snRNA) activating protein complex is required for RNA polymerase II and III snRNA gene transcription and interacts with the TATA box binding protein. Proc. Natl. Acad. Sci. USA 93:4289-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadowski, C. L., R. W. Henry, S. M. Lobo, and N. Hernandez. 1993. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 7:1535-1548. [DOI] [PubMed] [Google Scholar]
- 29.Su, Y., Y. Song, Y. Wang, L. Jessop, L. C. Zhan, and W. E. Stumph. 1997. Characterization of a Drosophila proximal-sequence-element-binding protein involved in transcription of small nuclear RNA genes. Eur. J. Biochem. 248:231-237. [DOI] [PubMed] [Google Scholar]
- 30.Tanikawa, J., T. Yasukawa, M. Enari, K. Ogata, Y. Nishimura, S. Ishii, and A. Sarai. 1993. Recognition of specific DNA sequences by the c-myb protooncogene product: role of three repeat units in the DNA-binding domain. Proc. Natl. Acad. Sci. USA 90:9320-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waldschmidt, R., I. Wanandi, and K. H. Seifart. 1991. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 10:2595-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wanandi, I., R. Waldschmidt, and K. H. Seifart. 1993. Mammalian transcription factor PBP. Characterization of its binding properties to the proximal sequence element of U6 genes. J. Biol. Chem. 268:6629-6640. [PubMed] [Google Scholar]
- 33.Wang, Y., and W. E. Stumph. 1998. Identification and topological arrangement of Drosophila proximal sequence element (PSE)-binding protein subunits that contact the PSEs of U1 and U6 small nuclear RNA genes. Mol. Cell. Biol. 18:1570-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong, M. W., R. W. Henry, B. Ma, R. Kobayashi, N. Klages, P. Matthias, M. Strubin, and N. Hernandez. 1998. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol. Cell. Biol. 18:368-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, S.-W., and H. A. Nash. 1994. Specific photo-cross-linking of DNA-protein complexes: identification of contacts between integration host factor and its target DNA. Proc. Natl. Acad. Sci. USA 91:12183-12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon, J.-B., S. Murphy, L. Bai, Z. Wang, and R. G. Roeder. 1995. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol. Cell. Biol. 15:2019-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon, J.-B., and R. G. Roeder. 1996. Cloning of two proximal sequence element-binding transcription factor subunits (γ and δ) that are required for transcription of small nuclear RNA genes by RNA polymerases II and III and interact with the TATA-binding protein. Mol. Cell. Biol. 16:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamrod, Z., C. M. Tyree, Y. Song, and W. E. Stumph. 1993. In vitro transcription of a Drosophila U1 small nuclear RNA gene requires TATA box-binding protein and two proximal cis-acting elements with stringent spacing requirements. Mol. Cell. Biol. 13:5918-5927. [DOI] [PMC free article] [PubMed] [Google Scholar]