Abstract
Soggy (Sgy) and Tead2, two closely linked genes with CpG islands, were coordinately expressed in mouse preimplantation embryos and embryonic stem (ES) cells but were differentially expressed in differentiated cells. Analysis of established cell lines revealed that Sgy gene expression could be fully repressed by methylation of the Sgy promoter and that DNA methylation acted synergistically with chromatin deacetylation. Differential gene expression correlated with differential DNA methylation, resulting in sharp transitions from methylated to unmethylated DNA at the open promoter in both normal cells and tissues, as well as in established cell lines. However, neither promoter was methylated in normal cells and tissues even when its transcripts were undetectable. Moreover, the Sgy promoter remained unmethylated as Sgy expression was repressed during ES cell differentiation. Therefore, DNA methylation was not the primary determinant of Sgy/Tead2 expression. Nevertheless, Sgy expression was consistently restricted to basal levels whenever downstream regulatory sequences were methylated, suggesting that DNA methylation restricts but does not regulate differential gene expression during mouse development.
The average spacing of genes in human cells has been estimated to be 85 kb (22, 48). However, the number of closely spaced, divergently expressed genes is surprisingly high (1). In some cases, two closely spaced genes are transcribed in the same cells (coordinately expressed), whereas in other cases, only one of the two genes is transcribed in a particular cell type (differentially expressed). A common feature of these bidirectional loci is the presence of a high density of unmethylated CpG dinucleotides (CpG islands), suggesting that DNA methylation may regulate their expression during animal development. To evaluate this hypothesis, we compared expression and DNA methylation at the Soggy (Sgy)/Tead2 gene locus in mouse cells and tissues.
The Sgy/Tead2 locus provides not only an example of two closely spaced, divergently transcribed genes but a unique paradigm for differential regulation of gene expression during mammalian development. Mammals contain four highly conserved genes that were originally named transcription enhancer factor or TEF genes but which have been redesignated TEA domain or Tead genes by the mouse genome project (www.informatics.jax.org; references 14, 15, and 18 and references therein). These genes encode site-specific DNA binding proteins that, in the company of the transcriptional coactivator YAP65, can activate expression of genes in a variety of embryonic and adult cells (47). Tead2 (TEF-4), like other Tead genes, is expressed to various extents in many cells and tissues (references 17 and 18 and references therein). However, Tead2 is the only Tead gene expressed in mouse embryos during the first 7 days of development (17, 50), suggesting that it plays a unique role at the beginning of mammalian development by allowing preimplantation mouse embryos to utilize Tead-dependent promoters and enhancers (17, 24, 26). Sgy is a novel single-copy gene whose mRNA start site is located only 3.8 kb upstream of the Tead2 mRNA start site (19). The function of Sgy is unknown, but it is presumed important because of its restricted expression pattern in the adult and its partial homology to the Dickkopf gene family, potential effectors of the Wnt signaling pathway (19). Since Tead2 and Sgy are transcribed in opposite directions, their regulatory elements lie in close proximity. In fact, the same locus is found in humans on chromosome 19q13.3, except that the two mRNA start sites are separated by only 1.5 kb. Both mRNA start sites lie within CpG islands (this report). Moreover, the Sgy/Tead2 locus (chromosome 7, 23.0 cM) lies adjacent to the imprinted region of chromosome 7 corresponding to Prader-Willi/Angelmann syndromes (20), suggesting that one or both genes may be imprinted.
In the examples reported so far, Sgy and Tead2 appear to be differentially expressed; either Tead2 or Sgy is expressed in a particular cell type, but the two are never expressed together. In adult mice, Tead2 is expressed strongly in heart and lung tissues and the granulosa cells of the ovary and weakly in several other tissues. Furthermore, Tead2 and its transcriptional coactivator YAP65 are enriched in embryonic, neural, and hematopoietic stem cells (30). In contrast, Sgy appears to be expressed only in the developing spermatocytes of seminiferous tubules and in lymphocytes.
In principle, DNA methylation could govern differential gene expression at bidirectional loci by preventing expression of one gene while allowing expression of the other. Clearly, differentiated cells first appear during blastocyst formation with a separation of embryonic stem (ES) cells (inner cell mass [ICM]) from trophoblasts (cells forming the outer layer). After implantation, most CpG sequences are progressively methylated, except for those located in the promoter region of active genes (32). Thus, active promoters are frequently associated with CpG islands (44). Methylation of CpG dinucleotides is commonly correlated with loss of gene expression both in vivo and in vitro (6, 29, 40). However, hypermethylation of CpG islands in some cell lines appears to be an intrinsic property of the cell line rather than the tissue from which it originated (41). Furthermore, the absence of a change in the DNA methylation pattern of several tissue-specific genes during development either of wild-type or of DNA methyltransferase-deficient mouse embryos suggests that CpG methylation is a consequence rather than a cause of the transcription repression seen (reference 49 and references therein). In this capacity, DNA methylation may serve primarily to ensure that repressed genes remain silent. Moreover, while DNA methylation has been linked directly to X chromosome inactivation, genomic imprinting, and silencing of transposable elements, a direct role for DNA methylation in regulating gene expression during animal development has yet to be demonstrated (33, 42, 49).
We reasoned that if DNA methylation is the primary factor in determining which of two closely spaced genes is expressed in a particular cell type, there should exist a strict correlation between the methylation status of a gene's regulatory region and its expression. This correlation should exist in normal cells such as germ cells, preimplantation embryos, ES cells, splenocytes, and tissues, as well as in established cell lines. Moreover, changes in DNA methylation should accompany changes in gene expression in normal cells. We found that while DNA methylation can differentially regulate the expression of two closely linked genes such as Sgy and Tead2 in established mouse cell lines, DNA methylation is not the primary determinant of Sgy/Tead2 expression patterns during mouse development. Nevertheless, DNA methylation of downstream regulatory sequences did appear to restrict expression of the Sgy gene to basal levels in both normal cells and established cell lines.
MATERIALS AND METHODS
Cells, tissues, and nucleic acid isolation.
CCE ES cells were from StemCell Technologies. LC12, M109, CA51, and MC38 cells were from the National Cancer Institute Frederick Cancer Research and Development Center DCT Tumor Repository. All other cell lines were from the American Type Culture Collection. The lymphocytes (splenocytes) were purified from spleen tissue isolated from 10-week-old BALB/c females (21). Preimplantation embryos and oocytes were isolated from CD-1 mice (17). ES cells and embryoid bodies were cultured as previously described (37). Genomic DNA was isolated from sperm of 14-week-old CD-1 males (8) and from other cells (39), and total RNA was isolated from cells as previously described (17).
Methylation-sensitive restriction enzyme assays.
Fifteen micrograms of genomic DNA from either EL4 or TM3 cells was digested with SacI (40 to 80 U, 37°C, 16 h; Roche), precipitated with ethanol in the presence of 2.5 M ammonium acetate, resuspended in water, and combined with 100 pg of pBluescript-NE. This plasmid DNA consisted of Bluescript KS plasmid (Stratagene) modified by inserting double-stranded oligonucleotides with restriction sites for NsbI and Eco47III between the SacI and KspI sites. The appropriate buffer and enzyme were added, and the following mixture was incubated at 37°C (SmaI at 25°C) for 16 h: 5 U of Cfr10I, 12.5 U of KspI, 5 U of SmaI, 12.5 U of XhoI, 10 U of Bsh1285I, 10 U of NsbI, 2.5 U of Eco47III, and 5 U of Psp1406. The buffer recommended by the enzyme manufacturer was used. NsbI was from Fermentis, and all others were from Roche Molecular Biochemicals. DNA was then precipitated with ethanol-ammonium acetate, resuspended in Tris-EDTA (pH 8.0), fractionated by electrophoresis in 0.7% agarose (Tris-borate-EDTA buffer), transferred to Nytran N membrane (Schleicher & Schuell), and hybridized with a 32P-labeled 2.1-kb SacI/XhoI DNA fragment (see Fig. 6A) (19). The blot was stripped and reprobed with 32P-labeled plasmid DNA.
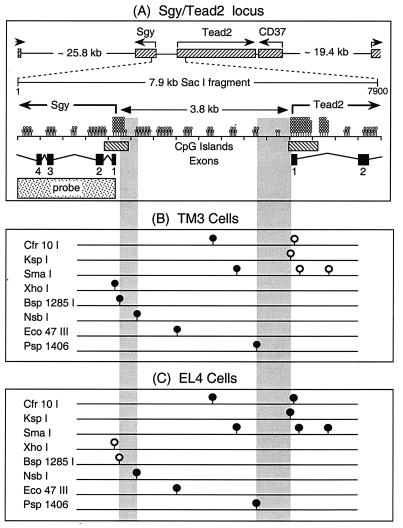
FIG. 6.
Methylation status of 11 CpG dinucleotides within the Sgy/Tead2 gene locus. The methylation status of a 7.9-kb SacI DNA fragment containing the Sgy and Tead2 gene start sites was characterized. (A) Schematic representation of the 23.0-cM region within chromosome 7 containing the Sgy, Tead2, and CD37 genes (GenBank accession number NW000319), with a more detailed representation of an ∼7.9-kb SacI fragment containing the Tead2/Sgy intergenic region (GenBank accession number AF274313). The Sgy gene consists of 5 exons within a 4.6-kb region, and the Tead2 gene consists of 12 exons within a 17.9-kb region (43). Indicated are the number of CpG dinucleotides (lollipops) per 0.5-kb segment (not arranged according to map position), the locations of the only two CpG islands (nucleotides 1868 to 2370 and 5941 to 6620) in the bp 1 to 7866 region, the start sites for the Sgy (nucleotide 2166) and Tead2 (nucleotide 6031) mRNAs, and the sequence used as a probe to detect specific restriction endonuclease cleavage events. (B) Methylation status of 11 CpG dinucleotides in TM3 cells, which express Tead2 but not Sgy (•, mCpG; ○, CpG). (C) Methylation status of 11 CpG dinucleotides in EL4 cells, which express Sgy but not Tead2. The transitions from unmethylated to methylated DNA (vertical shaded bars) determined from these analyses were 385 bp at the Sgy locus and 789 bp at the Tead2 locus.
RT-PCR assay.
Reverse transcription (RT)-PCR assays were carried out as follows. Five micrograms of total RNA was transcribed with Superscript II reverse transcriptase (Invitrogen) by using random primers, and then 10% of the cDNA reaction mixture (2 μl) was amplified with Platinum Taq polymerase (Invitrogen) in accordance with the manufacturer's protocol. Gene products were detected with PCR primers 1 and 2 (Tead2), primers 3 and 4 (Sgy), and primers 5 and 6 (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) (Table 1) as previously described (17). Reactions were carried out for the indicated number of cycles. One-tenth of the reaction mixture (5 μl) was then fractionated by electrophoresis in a 10% polyacrylamide gel with Tris-borate-EDTA buffer (Bio-Rad), and the DNA was visualized by staining with ethidium bromide.
TABLE 1.
PCR primers used in this study
| Primer | Sequence |
|---|---|
| 1 | CCGACATTGAGCAGAGTTTTCAGG |
| 2 | CTTCACGTCTGGAACATTCCATGG |
| 3 | ACTGAGGGTCTTGCTGCTGCT |
| 4 | GGAAGTTCCTAGGAAGGTCTC |
| 5 | CCCTTCATTGACCTCACTACATGG |
| 6 | CCTGCTTCACCACCTTCTTGATGTC |
| 7 | AAGGCCTAATCTCAGAAGC |
| 8 | CATAAACCATCACTCTTGGG |
| 9 | ACCCAGAAGACTGTGGATG |
| 10 | GATGGTATTCAAGAGAGTAGGG |
| 11 | GACTAAGAGCTGGGACAC |
| 12 | TTCTGGCCACTTGTCTTTGC |
Poly(A)+ PCR assay.
Quantitative RT-PCR assays [poly(A)+ PCR] were carried out as previously described (3, 37), with modifications. The PCR contained 5 U of Platinum Taq Polymerase (Invitrogen). One-tenth (5 μl) of the PCR product was fractionated in a 1.2% agarose gel (Tris-borate-EDTA buffer) and then transferred to a Zeta-probe GT membrane (Bio-Rad). A probe complementary to the 3′ end of Tead2 was generated by PCR amplification of full-length Tead2 cDNA (17) with primers 7 and 8 (Table 1). The Sgy probe consisted of the entire Sgy cDNA containing the 3′ untranslated region (19). GAPDH and Rex-1 probes were generated with 3 μg of total RNA from undifferentiated CCE ES cells. Reverse transcription with oligo(dT) primers was carried out as described above, and 10% of the reaction product was amplified for 25 cycles with 2.5 U of Platinum Taq polymerase (described above) and primers 9 and 10 for GAPDH or primers 11 and 12 for Rex-1 (Table 1). The amplified product (amplicon) was purified by gel electrophoresis, cloned, and sequenced. A Prime-It RmT Random Primer Labeling Kit (Stratagene) and 6,000 Ci of dCTP (Amersham) per mmol were used to label 25 ng of probe, which was then purified through a ProbeQuant G-50 Micro column (Amersham). Southern blotting-hybridization analyses were carried out as previously described (19).
Bisulfite genomic sequencing.
Bisulfite genomic sequencing was carried out as previously described (25, 35), with modifications. pGEM 3Zf(+) (100 ng; Applied Biosystems) and 20 U of SacI (Roche) were added to lysates (prepared as previously described [25]) of 80 to 100 oocytes, 75 to 375 two-cell embryos, or 20 to 200 morulae and then incubated overnight at 37°C. For all other samples, 100 μg of genomic DNA was digested with SacI in the presence of 100 ng of pGEM 3Zf(+). Bisulfite treatment and subsequent purification were carried out as already described, except that the digested genomic DNA was frozen and thawed twice (−80°C and room temperature) and then heated at 100°C for 10 min before addition of NaOH to ensure complete denaturation. One-fifth of the bisulfite-treated DNA was resuspended in 20 μl of H2O, amplified by PCR (5 min at 94°C and then 30 to 40 cycles of 30 s at 94°C, 30 s at 48°C, 1 min at 72°C, and then finally 7 min at 72°C). Except for amplicons D and F, two sets of primers were used to amplify the indicated region (Table 2). After the first round of amplification (outer primers), products were purified over a PCR purification spin column (Qiagen), and 1 to 4 μl of a 50-μl eluate was used for a second round of amplification (inner primers). PCR products were purified with a PCR purification spin column and eluted with 30 to 40 μl of H2O. The amount of DNA was estimated by fractionating a sample by agarose gel electrophoresis. A portion of the total PCR product (∼100 ng) was sequenced directly with the inner primer set and an ABI 373 or 310 sequencer.
TABLE 2.
Amplicons used for bisulfite genomic sequencing
| Amplicona | Outer primer set | Inner primer set |
|---|---|---|
| A (1415-1927) | TTCCTATTTTATATTATCATATCC | ATCTATTCAACCCAACACCTAC |
| TGATTGTGGGATTTGAAATTGT | TTTGTTATAGTTTTTTATTTATTTGT | |
| B (1873-2420) | CACATTACAAATAAATAAAAAACTA | AACAAAAACAATTTCAAATCCCA |
| AGGTATTAGGTAAAATAAATGTTTG | TATGAGTAAGGTTATTAGGTGTA | |
| C (2368-2879) | TCTACACCTAATAACCTTACTCAT | CAAACATTTATTTTACCTAATACCT |
| GATGGATGGTTGATTTTTTTGG | TAGAGAATAGTAGATAGAGAATAG | |
| D (6135-6529) | TTAATGGTTGAAGTTTTAAGGGGTT | TTAATGGTTGAAGTTTTAAGGGGTT |
| AAATCCTACTTAAAATTTATAAAT | AAATCCTACTTAAAATTTATAAAT | |
| E (5233-5471) | ATACCCTCTTCTAACCTCTA | TACATACAAACAAAACATACAA |
| TAAGTATATTGTAGTTGATTTAAG | TAAGTATATTGTAGTTGATTTAAG | |
| F (4803-5024) | CTAATTAAACCAAATACAACTC | CTAATTAAACCAAATACAACTC |
| GTATTTGGGAGGTAGAGGTA | GTATTTGGGAGGTAGAGGTA |
Nucleotide map positions are given in parentheses for each amplicon (1 = nucleotide 1 of GenBank accession no. AF274313).
Nuclease-hypersensitive site assay.
Approximately 108 cells were washed twice with ice-cold phosphate-buffered saline and then lysed in 5 ml of buffer A (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 0.2% Triton X-100, 1 mM dithiothreitol, 0.5 mM EGTA, 0.4 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml). Nuclei were recovered by sedimentation (300 × g, 5 min, 4°C), washed with buffer A without Triton X-100, and resuspended in 1 ml of a mixture containing 10 mM Tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, and 1 mM CaCl2. Aliquots (100 μl) were digested with pancreatic DNase I (0 to 2 U of RQ-RNase free DNase; Promega) for 5 min at 25°C. Digestion was stopped by addition of 20 μl of a mixture containing 10 mM Tris-HCl (pH 8.0), 60 mM EDTA, and 3% sodium dodecyl sulfate. RNase A (10 μg; Sigma) was added, and the sample was incubated for 15 min at 37°C. Proteinase K (400 μg; Roche) was added, and the sample was incubated overnight at 37°C. Genomic DNA was extracted with phenol-chloroform-isoamyl alcohol and precipitated with ethanol-ammonium acetate (39). DNA (15 μg) was digested with SacI and fractionated by electrophoresis in a 0.7% Tris-borate-EDTA agarose gel, and the Sgy/Tead2 locus was detected by Southern blotting-hybridization with the Sgy probe.
RESULTS
Coordinate expression of Sgy and Tead2 in totipotent cells of preimplantation embryos.
To determine whether Sgy and Tead2 are expressed differentially or coordinately at the beginning of mouse development, the number of copies of Tead2 mRNA was determined with an RT-PCR assay based on uniform amplification of the 3′-terminal region of all poly(A)+ mRNAs (31). The amplified products were then quantified by hybridization with 32P-labeled DNA probes that were specific for either Tead2 or Sgy mRNA. The data were expressed first as counts per minute bound per ovum or embryo (Fig. 1A and D) and then as the number of mRNA copies per ovum or embryo (Fig. 1B and E), calculated as previously described (31).
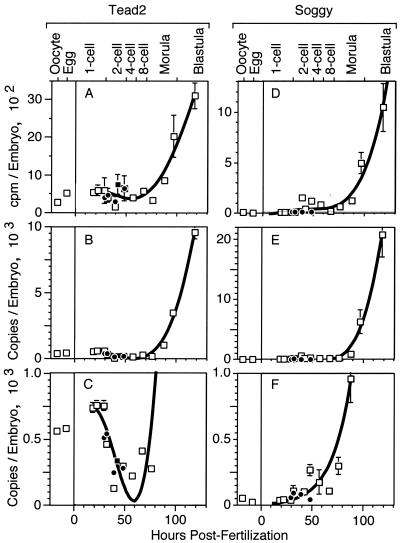
FIG. 1.
Tead2 and Sgy expression in mouse oocytes and preimplantation embryos. One-cell embryos were isolated from pregnant females and cultured in vitro to allow development up to the blastocyst stage (□). Some one-cell embryos were cultured in the presence of α-amanitin to prevent transcription (•). Some two-cell embryos were isolated from pregnant females (▪). RT-PCR was used to amplify the entire population of poly(A)+ mRNA from mouse ova and embryos under conditions that preserve the relative abundance of each mRNA in the cDNA population (31). Three to eight samples were used per stage. Data were fitted to a fourth-order polynomial with the standard error of the mean indicated. A 32P-labeled probe specific for Tead2 (A) or Sgy (D) was hybridized with this cDNA population, and the number of counts per minute per ovum or embryo was recorded. The data in panels A and D were used to calculate the number of copies of mTead2 (B) or Sgy (E) mRNA as previously described (31). The scale used in panels B and E was expanded to facilitate comparison of the early stages in development (C and F). The data for Tead2 were reproduced from reference 17; in the process, an arithmetical error discovered in the Tead2 copy number was corrected.
The results revealed that only Tead2 was expressed in oocytes but that Tead2 and Sgy were expressed concurrently and to similar levels during zygotic gene expression in preimplantation mouse embryos. Sgy poly(A)+ mRNA was first detected in two-cell and four-cell embryos (Fig. 1F), and it then increased to about 20,000 copies per blastocyst (Fig. 1E). Tead2 poly(A)+ mRNA was present in oocytes and unfertilized eggs at about 500 copies per cell (Fig. 1C; standard RT-PCR data in reference 17). Following fertilization, the level of Tead2 mRNA decreased until the late two- or four-cell stage and then increased rapidly (Fig. 1C). Since Tead2 mRNA levels in one- and two-cell embryos were insensitive to α-amanitin (a specific inhibitor of RNA polymerase II), most of this mRNA was inherited from the egg. Tead2 and Sgy mRNAs accumulated dramatically from the eight-cell stage to the blastocyst stage, consistent with their transcription from zygotic genes. In blastocysts, the level of both mRNAs was about 10,000 to 20,000 copies per embryo (Fig. 1B), about 1.5 to 3% of the level of β-actin mRNA (31). Thus, the level of Tead2 in blastocysts was 20-fold greater than in oocytes or 50-fold greater than in two- and four-cell embryos. Comparisons of preimplantation embryos with oocytes (Fig. 1) and ICMs with blastocysts (Table 3) suggested that while the two genes are expressed at similar levels in blastocysts, Tead2 is expressed preferentially in the ICM while Sgy is expressed preferentially in the trophectoderm. This suggested that Sgy and Tead2 became differentially expressed as totipotent embryonic cells differentiated into specific cell types.
TABLE 3.
Differential expression of Tead2 and Sgy in blastocystsa
| Gene | Blastocyst | ICM | Trophectoderm | Trophectoderm/ ICM ratio |
|---|---|---|---|---|
| Tead2 | 321 ± 36 | 187 ± 50 | 134 | 0.7 |
| Sgy (expt 1) | 213 ± 27 | 34 ± 5 | 179 | 5.3 |
| Sgy (expt 2) | 1,868 ± 442 | 218 ± 29 | 1,650 | 7.6 |
Assays were done as described in the legend to Fig. 1, and mean counts per minute ± the standard error of the mean were obtained from six blastocysts and six ICMs. Trophectoderm = blastocyst − ICM.
To test this hypothesis, the levels of Tead2 and Sgy poly(A)+ mRNA were measured in mouse ES cells before and after they were induced to differentiate into embryoid bodies by removal of leukemia inhibition factor and transfer to bacterial petri dishes. ES cells are derived from the ICMs in blastocysts, and they are capable of developing into a complete embryo when transplanted back into the blastocoel cavity. Embryoid bodies contain a variety of specific cell types that appear during myogenesis, angiogenesis, hematopoiesis, neurogenesis, and cardiogenesis (36, 37).
Sgy and Tead2 were expressed coordinately in ES cells, but Tead2 was expressed to a greater extent than Sgy (Fig. 2, zero time point), suggesting that differential expression of these two genes begins when the two distinct cell lineages of the blastocyst, trophectoderm and ICM, are produced (Table 3). In addition, within 1 to 2 days after induction of ES cell differentiation, Sgy expression was selectively repressed, and within 5 days, Tead2 expression was selectively stimulated (Fig. 2). These changes were accompanied by repression of Rex-1 expression (Fig. 2) and by morphological transformation of ES cells into embryoid bodies (data not shown), changes that have been reported previously (37, 38). The ubiquitous GAPDH gene was expressed continuously at high levels during this period of time, as previously reported (27), and was therefore used as a standard reference throughout subsequent studies.
FIG. 2.
Sgy and Tead2 expression in mouse ES cells and embryoid bodies. ES cells were cultured for the indicated number of days in the absence of leukemia inhibitory factor (LIF) after being transferred to petri dishes in order to induce cell differentiation. Quantitative poly(A)+ PCR assays for the indicated mRNA were carried out by repeatedly stripping and reprobing the same blot. Both short and long exposures of the same blots are provided to facilitate comparisons.
In a previous study, we did not detect coordinate expression of Sgy and Tead2 in any of the cells or tissues examined, including pluripotent embryonic carcinoma F9 cells (19). Since this study used Northern blotting-hybridization analysis (Northern analysis) with appropriate gene-specific 32P-labeled DNA probes to detect Sgy and Tead2, a direct comparison of ES cells and some of the cells and tissues from this previous study was carried out by Northern analysis (Fig. 3A). The results confirmed those previously reported and extended them to show that Northern analysis also did not detect the presence of Sgy mRNA in ES cells. Apparently, the threshold of the Sgy gene probe was insufficient to detect low levels of Sgy RNA.
FIG. 3.

Sgy and Tead2 expression in mouse cells and tissues. (A) Total RNA (20 μg) was analyzed by Northern blotting-hybridization analysis (17). (B) Total RNA was isolated from the indicated cell or tissue (39) and assayed for Sgy, Tead2, or GAPDH mRNA by RT-PCR. Identity was based both on sequence specificity of primers and on amplicon size. Water was used for a mock RT-PCR. Numbers of PCR cycles are indicated on the right.
The results described above revealed that Sgy and Tead2 are expressed concurrently at the beginning of mouse development and only begin to be differential expressed when cell differentiation begins during embryonic development.
Differential expression of Sgy and Tead2 in differentiated cells.
PCR-based assays [RT-PCR and poly(A)+ PCR] revealed three levels of gene expression in mouse cells and tissues (summarized in Table 4): off, basal level, and on. Cells in which RNA was not detected by PCR were considered not to express the gene. For example, Sgy was not expressed in either oocytes (Fig. 1) or TM3 cells (Fig. 3B), and Tead2 was not expressed in either MPC-11 or EL4 cells (Fig. 3B). Cells in which RNA could only be detected by RT followed by >25 cycles of PCR, and in which expression was not detected by Northern analysis, were considered to express the gene at basal levels. For example, Sgy was expressed at basal levels in MPC-11, F9, and ES cells (Fig. 3). Cells in which RNA could be detected by both PCR and Northern analyses were considered to fully express the gene. For example, Sgy was fully expressed in EL4 cells and testis tissue (Fig. 3).
TABLE 4.
Cell-specific expression of the mouse Sgy and Tead2 genesa
| Gene expression and mouse cells | Classification | Soggy | Tead2 | Data |
|---|---|---|---|---|
| Sgy-specific expression | ||||
| Developing spermatocytes | Spermatocytes | + | − | Reference 19 |
| Round spermatids | Spermatocytes | + | − | cDNA library |
| Splenocytes | B lymphocytes, T lymphocytes | + | − | Fig. 4 |
| EL4 | T lymphocytes | + | − | Fig. 3, 4, 1111 |
| YAC-1 | MuLVb lymphoma | + | − | Fig. 4 |
| P815 | Mastoma | + | − | Fig. 4 |
| A20 | B lymphoma | + | − | Fig. 4 |
| MPC-11 | B lymphoma | ± | − | Fig. 3, 11 |
| CH1 | B lymphoma | ± | − | Fig. 4 |
| Tead2-specific expression | ||||
| TM3 | Leydig cells | − | + | Fig. 3, 44, 11 |
| TM4 | Sertoli cells | − | + | Fig. 4 |
| NIH 3T3 | Embryo fibroblasts | − | + | Fig. 4 |
| LL/2 | Lung carcinoma | − | + | Fig. 4 |
| LLC | Lung carcinoma | − | + | Fig. 4 |
| CA51 | Colon carcinoma | − | + | Fig. 4 |
| MC38 | Colon carcinoma | − | + | Fig. 4 |
| B16 | Melanoma | − | + | Fig. 4 |
| C127I | Mammary epithelial cells | − | + | Fig. 4 |
| Oocytes | Germ cells | − | + | Fig. 1 |
| Eggs | Germ cells | − | + | Fig. 1 |
| Embryoid bodies | Multiple cell types | − | + | Fig. 2 |
| Sgy and Tead2 expression | ||||
| Two-cell embryos to morulae | Totipotent blastomeres | + | + | Fig. 1 |
| Blastulae | ICM, trophoblasts | + | + | Fig. 1, Table 3 |
| Trophectoderm | Trophoblasts | + | + | cDNA libraries |
| F9 | Embryonic carcinoma | ± | + | Fig. 3 |
| ES | Embryonic stem cells | ± | + | Fig. 2, 3 |
Gene-specific RNA was assayed by RT-PCR, by the poly(A)+ PCR assay, by in situ hybridization (19), or by analysis of cDNA libraries (National Center for Biotechnology Information Unigene). Most of the tissues analyzed expressed various levels of Tead2, while testis tissue was the only tissue that strongly expressed Sgy (19; data not shown).
MuLV, murine leukemia virus.
The results from PCR-based assays confirmed that either Sgy or Tead2 was expressed in differentiated cells and tissues but not both (Fig. 3 and 4; Table 4). Tead2 was expressed to various extents in a variety of tissues (15, 17, 54, 55), most notably in lung and heart tissues, in cells derived from these tissues (Table 4), in oocytes (Fig. 1), and in embryoid bodies (Fig. 2). In contrast, Sgy was expressed only in lymphocytes isolated from the spleen (splenocytes), in a variety of lymphocytic cell lines such as the T-lymphocyte cell line EL4, and in testis tissue, where its expression was localized to developing spermatocytes (Fig. 3 and 4) (19). Tead2 was not detected in any cell of hematopoietic origin even when RT-PCR was carried out for 35 or more cycles (Fig. 5; data not shown). Moreover, the absence of a functional Tead2 protein was confirmed in EL4 cells by the fact that they did not support Tead-dependent transcription unless both Tead2 and its transcriptional coactivator YAP65 were provided by transfection with appropriate expression vectors (47). The trace amount of Tead2 RNA detected in testis tissue by RT-PCR presumably originated from Leydig and Sertoli cells, as represented by TM3 and TM4 cells, respectively (Fig. 3 and 4).
FIG. 4.

Sgy and Tead2 expression in mouse cells and tissues before and after treatment with 5AC. Splenocytes (Sc) are a lymphocyte population isolated from spleen tissue. Where indicated, cells were cultured in 1 μM 5AC for 48 h before RNA isolation. Total RNA was analyzed for Tead2, Sgy, and GAPDH expression by RT-PCR.
FIG. 5.

Effects of 5AC and TSA on Sgy expression in mouse TM3 (A) and MPC-11 (B) cells. Where indicated, cells were cultured in 1 μM 5AC for 48 h, in 1 μM TSA (Wako) for 24 h, or in 1 μM 5AC for 24 h and then in 1 μM 5AC and TSA for 24 h. Total RNA was isolated and used to determine gene expression levels by RT-PCR (A and B) or poly(A)+ PCR (C) assays.
Repression of Sgy gene expression by DNA methylation.
Sequence analysis (44) of a 10-kb fragment containing the Sgy/Tead2 locus (GenBank accession number AF274313) revealed only two CpG islands (observed CpG/expected CpG ratio = 0.7), one at the Sgy mRNA start site and one at the Tead2 mRNA start site (Fig. 6; see also Fig. 10 and 11). Therefore, one mechanism by which cell differentiation could cause differential gene expression is by methylating either the Sgy or the Tead2 promoter region. To test this hypothesis, established cell lines were cultured in 5′-aza-deoxycytosine (5AC) to inhibit DNA methyltransferase and thereby generate unmethylated DNA. 5AC did not reduce expression of either Sgy or Tead2 in cells that normally expressed that gene (Fig. 4 and 5). However, 5AC did induce Sgy expression in several cell lines that did not normally express the Sgy gene (CA51, MC38, C127I, and CH1). This suggested that these cells were permissive for Sgy expression but that the Sgy gene was repressed by DNA methylation. Furthermore, 5AC did not induce Sgy expression in all cells (e.g., lung LL/2 and LLC cells), and it did not induce Tead2 expression in any of the lymphocytic cells. This suggested either that some aspect of chromatin structure may repress the genes in these cell types or that these cells lack one or more of the transcription factors required for expression of these genes.
FIG. 10.
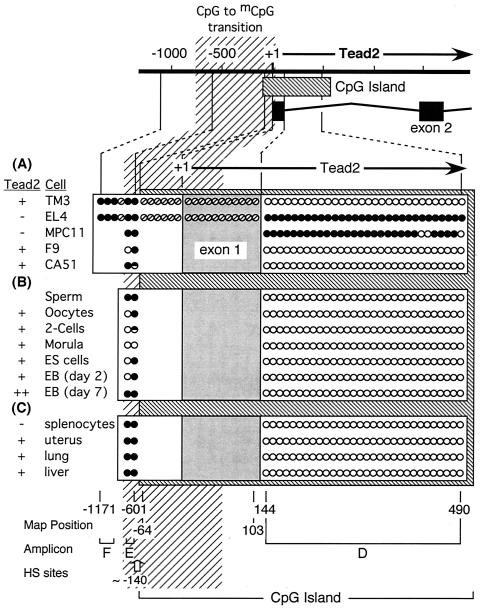
Methylation status of 52 CpG dinucleotides at the Sgy gene locus in mouse cells and tissues. Bisulfite genomic sequencing analysis was applied to three DNA fragment amplicons (A, B, and C [Table 2]) that encompass nucleotides +699 to −655 in the indicated cells and tissues (open circles = CpG; filled circles = mCpG; half-filled circles = mixed population). The CpG-to-mCpG transition mapped in Fig. 9 is indicated. (A) Four established cell lines. (B) CA51 cells before (CA51) and after (CA51*) treatment with 5AC as described in the legend to Fig. 4. (C) Germ cells, preimplantation embryos, ES cells, and ES cells undergoing differentiation (embryoid bodies [EB] at days 2 and 7). (D) Splenocytes and tissues. Nucleotide positions of the first and last CpG in each box, the number of base pairs encompassed by each box, and the positions of DNase I-hypersensitive (HS) sites (Fig. 8) are indicated.
FIG. 11.
Methylation status of 36 CpG dinucleotides at the Tead2 gene locus in mouse cells and tissues. Bisulfite genomic sequencing analysis was applied to the Tead2 locus. The status of some CpGs (divided circles) was not determined, because primers were not found that would amplify bisulfite-treated DNA in this region. The methylation status of CpG's from −601 to −1171 (amplicon F [Table 2]) was determined for TM3 and EL4 cells. The shaded vertical bar indicates the CpG-to-mCpG transition defined in Fig. 6. (A) Five established cell lines. (B) Same as Fig. 10C. (C) Same as Fig. 10D.
To determine whether or not induction of Sgy gene expression was accompanied by demethylation of the Sgy gene locus, the DNA methylation status of CA51 cells was analyzed by bisulfite genomic sequencing (described below for Fig. 9). The Sgy gene locus in CA51 cells was fully methylated, but the Sgy gene promoter and exon 1 and intron 1 were partially demethylated in 5AC-treated CA51 cells (see Fig. 10B), suggesting that DNA methylation of the Sgy promoter region repressed Sgy gene transcription in these cells.
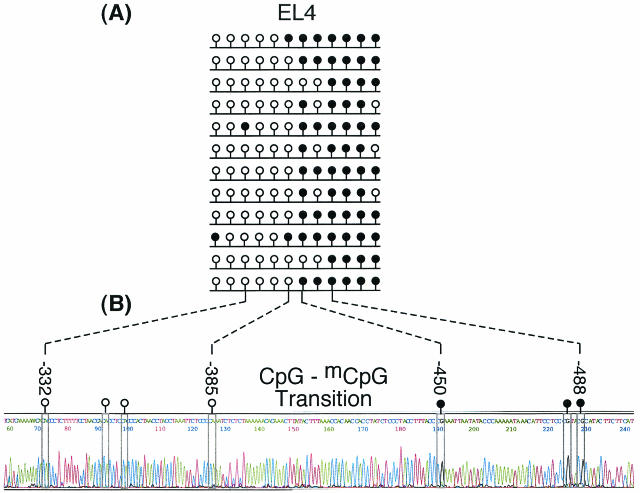
FIG. 9.
Transition between unmethylated and methylated DNA upstream of the Sgy gene mRNA start site. Bisulfite genomic sequencing analysis was applied to a single 449-bp DNA fragment from position −679 to position −230 in EL4 cells. (A) Twelve random clones were isolated from a PCR amplicon and sequenced. Their methylation status is shown. (B) About 10% of the total PCR amplicon was sequenced directly to obtain the methylation status of the entire population. Seven of the 12 CpGs in this sequence are shown as an example. Nucleotides appear as color-coded peaks in the electropherogram. CpG dinucleotides are enclosed by rectangles, and their methylation status is indicated by an open (CpG) or closed (mCpG) lollipop.
DNA methylation and chromatin deacetylation act synergistically to repress Sgy expression.
Previous studies have suggested that some silent genes can be reactivated to basal-level expression by treating cells with 5AC to induce demethylation of regulatory sequences but that further reactivation requires a combination of 5AC and trichostatin A (TSA), an inhibitor of histone deacetylase that induces hyperacetylation of core histones (2). These results support the hypothesis that mCpG dinucleotides recruit histone deacetylase, which changes chromatin structure to a transcriptionally repressive state (16, 28). In our studies, some cells, such as TM3 cells, did not express Sgy and contained a fully methylated Sgy promoter region (described below). Other cells, such as MPC-11 cells, expressed Sgy at a basal level and contained an unmethylated promoter (described below) that exhibited DNase I-hypersensitive sites characteristic of active promoters (see Fig. 8). To determine whether or not these levels of gene activity were dictated solely by the extent of DNA methylation, TM3 and MPC-11 cells were treated either with 5AC or with a combination of 5AC and TSA.
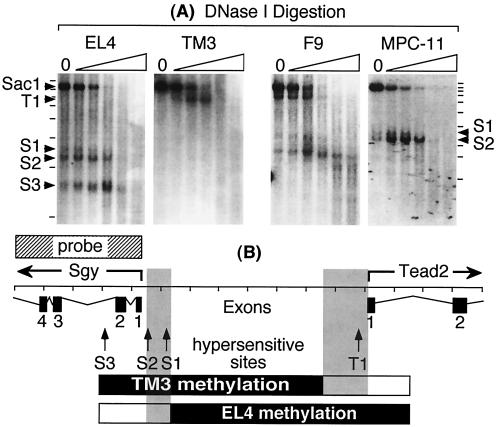
FIG. 8.
Detection of DNase I-hypersensitive sites. (A) Nuclei were isolated from EL4, MPC11, F9, or TM3 cells and digested with increasing amounts of DNase I. No DNase I was added to lane 0. Genomic DNA was purified, digested with SacI, fractionated by gel electrophoresis, and visualized with a 32P-labeled DNA probe (Fig. 6A) by blotting-hybridization. The positions of an ∼7.8-kb SacI fragment and fragments generated because of the presence of hypersensitive sites are indicated by arrows. DNase I-hypersensitive sites (S1, S2, and S3) in the Sgy gene were located at approximately map positions −430, −120, and +615, respectively. Hypersensitive site T1 in the Tead2 gene was located at approximately position −140. (B) Map positions of the Sgy and Tead2 mRNA start sites, DNase I-hypersensitive sites, and methylated regions in TM3 and EL4 cells (see Fig. 6A).
The results revealed that Sgy expression in TM3 cells could be increased to basal levels by 5AC alone (Fig. 5A and C) and that this stimulation was accompanied by partial demethylation of the Sgy promoter (data not shown). TSA also activated Sgy expression to basal levels in TM3 cells, but TSA and 5AC together stimulated expression more than either treatment alone (Fig. 5A and C). However, in MPC-11 cells that already expressed Sgy at basal levels, 5AC alone had no effect whereas TSA alone stimulated Sgy expression (Fig. 5B and C). These data suggest that DNA methylation and histone deacetylation act synergistically to repress Sgy gene expression in mammalian cells.
Differential methylation of Sgy and Tead2 genes in established cell lines.
To determine whether or not differential gene expression is accompanied by differential DNA methylation, the methylation status of the Sgy/Tead2 locus was determined by measuring its sensitivity to methylation sensitive restriction endonucleases (34). Eleven specific sites within a 7.9-kb SacI DNA fragment that encompassed the Sgy/Tead2 locus (Fig. 6A) were examined in TM3 and EL4 cells. TM3 cells expressed Tead2 but not Sgy, whereas EL4 cells expressed Sgy but not Tead2. Genomic DNA was mixed with an unmethylated plasmid DNA control and then digested with the indicated endonuclease. The DNA products were fractionated by gel electrophoresis, attached to a membrane, and then hybridized either with an Sgy-specific 32P-labeled DNA probe (Fig. 6A) or with a plasmid-specific probe. In each sample, the unmethylated plasmid DNA was cleaved completely by the indicated enzyme (Fig. 7B), confirming that digestion was complete. Therefore, in those samples where the cellular DNA was digested completely (Fig. 7A), the indicated restriction site was not methylated (Cfr10I, KspI, and SmaI in TM3 cells [Fig. 6B]; XhoI and Bsh1285I in EL4 cells [Fig. 6C]). In those samples where genomic DNA was either not digested or partially digested (Psp1406 in TM3 cells), the indicated site was either completely or partially methylated, respectively. Partial methylation meant that only a fraction of the genomes (i.e., cells) in the population was methylated at this site.
FIG. 7.
Digestion of genomic DNA with methylation-sensitive restriction endonucleases (RE). (A) DNA from either TM3 or EL4 cells was digested with SacI and then with the indicated methylation-sensitive restriction endonuclease (see Fig. 6B and C). DNA digestion products were fractionated by gel electrophoresis and visualized with a 32P-labeled DNA probe (Fig. 6A) by blotting-hybridization. (B) The extent of DNA cleavage in each genomic DNA sample was monitored by cleavage of an unmethylated plasmid DNA added as an internal standard. Arrows indicate the positions of undigested SacI DNA fragments. The size(s) of the expected DNA product(s) from each digestion is indicated at the bottom of each lane, while boldface values indicate the sizes of the DNA fragments observed. The endonucleases used were Cfr10I (C), KspI (K), SmaI (S), XhoI (X), Bsh1285I (B), NsbI (N), Eco47III (E), and Psp1406 (P).
The results revealed a striking inverse correlation between DNA methylation and gene expression: the gene that was not expressed was methylated in each cell line, whereas the gene that was expressed was unmethylated in each cell line (Fig. 6B and C). Thus, DNA methylation accompanied the selective inactivation of either the Tead2 or the Sgy gene. Furthermore, the transitions between methylated and unmethylated DNA (Fig. 6, vertical shaded bars) indicated the presence of sharply defined boundaries in which all of the CpGs on one side were methylated while all of the CpGs on the other side were not (diagrammed in Fig. 8B).
DNase I-hypersensitive sites were associated only with the active gene.
Regulatory sequences for transcriptionally active genes commonly contain nuclease-hypersensitive sites that result from the presence of site-specific DNA binding proteins (9). To determine whether or not such sites exist in the Sgy/Tead2 locus, nuclei were isolated and digested with increasing concentrations of pancreatic DNase I. The results (Fig. 8A) revealed three hypersensitive sites (S1, S2, and S3) at the Sgy gene locus in cells that expressed Sgy at high levels (e.g., EL4) and two hypersensitive sites (S1 and S2) in cells that expressed Sgy at basal levels (e.g., F9 and MPC-11 cells). Conversely, cells that did not express Sgy did not exhibit DNase I-hypersensitive sites in the Sgy gene region (e.g., TM3 cells). Similarly, cells that expressed Tead2 (TM3 and F9 cells) contained at least one DNase I-hypersensitive site (T1) just upstream of the Tead2 mRNA start site, whereas cells that did not express Tead2 (EL4 and MPC-11 cells) did not exhibit any hypersensitive sites in the Tead2 gene region. These data are consistent with the presence of site-specific DNA binding proteins in the promoters of active genes but not in the promoters of silent genes. Moreover, the S3 site in EL4 cells in the Sgy locus suggests the presence of a regulator element downstream of the Sgy mRNA start that is required for full Sgy expression.
Site-specific transition from unmethylated to methylated DNA.
To define accurately the transitions from unmethylated to methylated DNA, the methylation status of each cytosine in the transition loci was determined by bisulfite genomic sequencing (34). Bisulfite-induced deamination converts C to U in single-stranded DNA. Subsequent amplification by PCR translates each uracil into thymidine. Thus, CpG dinucleotides are converted into TpG dinucleotides on one strand and CpA dinucleotides on the complementary strand. Cytosines are not converted by bisulfite if they are either methylated or reside in double-stranded DNA (34). In the work described here, the possibility that unconverted cytosines resulted from regions of undenatured DNA was eliminated in two ways. First, only cytosines within CpG dinucleotides were resistant to bisulfite; all of the cytosines in CpC, CpA, and CpT dinucleotides were converted to U. Second, PCR primers were designed to select against any unconverted DNA that may have been present as a result of incomplete denaturation (34).
Bisulfite genomic sequencing can be analyzed in two ways: either individual DNA molecules from the PCR amplification product are cloned and sequenced (Fig. 9A), or the entire PCR amplification product is sequenced (Fig. 9B). The first method reveals the methylation status of individual genomes, whereas the second method determines the average methylation status of a cell population. In addition, it avoids pitfalls inherent in the cloning and sequencing of individual genomes (5, 34). Therefore, since the results of the two approaches were comparable, the second method was used routinely in order to directly observe the average methylation status of thousands of individual genomes.
With a single DNA fragment, a sharp transition between unmethylated and methylated DNA was detected 646 bp upstream of the Sgy transcription start site in EL4 cells (Fig. 9). Here, the last CpG and the first mCpG were separated by only 64 bp. Upstream of this transition site, all of the CpGs were methylated (Fig. 9, 10A, and 11A). Downstream of this transition site, all of the CpGs were unmethylated, at least to +691 (Fig. 9, and 10A). These data were consistent with those gathered at methylation-sensitive restriction endonuclease sites (Fig. 6C).
The CpG-to-mCpG transition in the Tead2 locus was not mapped with the same accuracy, because primers were not found that would amplify bisulfite-treated DNA in the transition site. Nevertheless, the available bisulfite data (Fig. 11A, amplicons D and E), together with restriction endonuclease data (Fig. 6B), indicate that a sharp transition also exists somewhere within a 167-bp region that encompasses the Tead2 mRNA start site (Fig. 11). These transitions presumably mark the upstream boundary of the active promoter.
In normal cells, gene activity was restricted, but not determined, by DNA methylation.
To determine whether or not the three levels of Sgy gene transcription described above (off, basal, and on) are related to DNA methylation, the methylation status of all 52 CpG dinucleotides within a 1,337-bp region encompassing the Sgy gene regulatory region was determined by bisulfite genomic sequencing of DNA from a variety of cells and tissues, and the data were related to Sgy gene expression (Fig. 10). The same analysis was also carried out for 36 of the 53 CpG dinucleotides within a 1,726-bp region encompassing the Tead2 locus (Fig. 11). Unfortunately, despite numerous attempts, we were not able to amplify the bisulfite-treated DNA product from the 167-bp segment containing exon 1 and 17 CpGs. Fortunately, we were able to analyze the 30 CpGs in the remaining 387-bp portion of the Tead2 CpG island, and this served to clearly distinguish methylated from unmethylated promoter regions. These results revealed that DNA methylation may restrict Sgy gene expression by limiting it to basal levels but that DNA methylation could not be the primary mechanism for preventing either Sgy or Tead2 expression during animal development, because the promoter regions in normal cells and tissues were unmethylated, even when the gene was silent.
The Sgy and Tead2 promoter regions were unmethylated in all cells that expressed the gene, consistent with the hypothesis that DNA methylation would repress gene expression. This was true for normal cells, as well as for established cell lines. For example, in both splenocytes (a mixture of T and B lymphocytes isolated from spleen tissue; Fig. 10D) and EL4 cells (a cell line derived from a T-cell lymphoma; Fig. 10A), the Sgy gene locus was unmethylated and the Sgy gene was expressed to similar levels (Fig. 4). In fact, all cells that expressed Sgy, at either basal or fully activated levels, contained an Sgy promoter region that was unmethylated. Several of these genomes also exhibited a sharp transition between unmethylated and methylated DNA at positions −385 to −450, similar to EL4 cells (Fig. 10; data not shown). Similar, but less complete, data were obtained for the Tead2 promoter region (Fig. 6 and 11).
In surprising contrast, neither the Sgy nor the Tead2 promoter region was methylated in any cells that did not express the gene, revealing that DNA methylation is not the primary determinant in silencing these genes. While established cell lines that did not express Sgy contained fully methylated Sgy gene regions (TM3 and CA51 cells, Fig. 10A and B), normal cells that did not express Sgy did not contain a methylated Sgy promoter (oocytes, Fig. 10C). Similarly, established cell lines that did not express Tead2 contained fully methylated Tead2 gene regions (EL4 and MPC-11 cells, Fig. 11A). However, splenocytes also did not express Tead2, even though their Tead2 promoter was unmethylated (Fig. 11C). Therefore, while all methylated promoters were silenced, DNA methylation was not required to silence either gene. This conclusion was confirmed by differentiation of ES cells into embryoid bodies. Sgy expression was completely repressed by day 2 and then again expressed by day 7 (Fig. 2), but the Sgy promoter remained unmethylated during the entire period (Fig. 10C).
DNA methylation did appear to restrict Sgy expression in both normal cells (ES cells and uterus, lung, and liver tissues [Fig. 10C and D]) and established cell lines (MPC11, F9, and CA51* cells [Fig. 10A and B]) because Sgy expression was inversely related to the extent of DNA methylation downstream of the promoter region. Only a basal level of Sgy expression was detected when sequences downstream of the promoter were methylated. These sequences included about half of the CpG island encompassing the Sgy mRNA start site. The DNase I-hypersensitive site at +615 (S3) was absent from MPC-11 and F9 cells, although the sites at −120 (S1) and −430 (S2) were present (Fig. 8). These data suggest that the site near +615 marks the location of an Sgy gene regulatory element that is sensitive to DNA methylation. Thus, cells containing low levels of Sgy transcripts always contained an unmethylated Sgy gene promoter, whereas in cells that contained high levels of Sgy transcripts, the unmethylated region was extended downstream of exon 2 and exhibited an additional DNase I-hypersensitive site.
DISCUSSION
DNA methylation is clearly required for normal embryonic development. In the absence of DNA methyltransferase 1, ES cells proliferate normally but die upon differentiation. Consequently, embryos lacking this enzyme are delayed in development and do not survive past mid-gestation (23). Similarly, mice lacking MBD-3, a protein that binds specifically to mCpG dinucleotides, also die during early embryogenesis (10). The precise reasons for these effects, however, and the identity of the factors that control the pattern of DNA methylation during gametogenesis and early development are largely unknown.
Moreover, a direct role for DNA methylation in regulating gene expression during animal development has yet to be demonstrated (2, 33, 42). On the one hand, methylation of promoter sequences can repress gene expression by interfering with binding of proteins required for transcription through recruitment of histone deacetylase and other transcription repressors (16, 28). On the other hand, the promoters of several tissue-specific genes are not methylated in some tissues in which they are inactive, and they remain inactive under conditions in which global demethylation causes up regulation of imprinted loci (49). These results suggest that while DNA methylation can repress gene expression, DNA methylation is not the primary mechanism that regulates gene expression during animal development.
The studies described here addressed this conundrum by determining whether or not there is a strict correlation between the methylation status of a gene's regulatory region and its expression in both normal mouse cells and established cell lines. The Sgy/Tead2 locus was chosen for this study, because these two closely linked genes each contain a CpG island and became differentially expressed concurrent with the onset of cell differentiation and DNA methylation. The results described here, however, reveal that while DNA methylation can repress Sgy expression in established cell lines and restrict its expression to basal levels during mouse development, DNA methylation per se is not the mechanism primarily responsible for repressing either Sgy or Tead2 expression during development.
Differential gene expression is developmentally acquired.
Both Sgy and Tead2 were expressed coordinately and in equivalent amounts during the activation of zygotic genes from the 2-cell stage to the morula stage (compacted 8- to 32-cell embryos) in preimplantation mouse embryos (Fig. 1), consistent with the lack of DNA methylation in their promoter regions (Fig. 10 and 11). Differential expression appeared only with the onset of cell differentiation and DNA methylation. Sgy was overexpressed in the trophoblasts but underexpressed in the ICM (Table 3), in totipotent ES cells (Fig. 2) derived from the ICM, and in pluripotent F9 cells (Fig. 3) derived from an embryonic carcinoma. Moreover, ES cells induced to undergo differentiation into embryoid bodies rapidly repressed Sgy expression and then stimulated Tead2 expression (Fig. 2). Only one of the two genes was expressed in 10 different tissues (testis, ovary, uterus, kidney, muscle, liver, lung, spleen, brain, and heart [15, 17, 19, 54, 55]), in 4 normal cell types (spermatocytes, splenocytes, oocytes, and embryoid bodies), as well as in 15 different established cell lines (summarized in Table 4). Moreover, the gene chosen for expression was independent of cell immortalization or cell transformation. For example, normal lymphocytes (splenocytes), immortalized lymphocytes (EL4 cells), and transformed lymphocytes (YAC-1, A20, MPC-11, and CH1) all expressed the Sgy gene exclusively. Thus, Sgy and Tead2 are differentially expressed in most mammalian cells, but differential expression of these two genes is developmentally acquired when cell differentiation begins.
DNA methylation can repress gene expression at the Sgy/Tead locus in established cell lines.
One mechanism that could account for the developmental acquisition of differential gene expression (i.e., repressing one of two closely linked but divergently transcribed genes) during cell differentiation is DNA methylation. Only two CpG islands exist within a 10-kb region encompassing the Sgy/Tead2 locus, one at the Sgy mRNA start site and one at the Tead2 mRNA start site (Fig. 6A). Analyses of these two regions and the intervening sequences by methylation-sensitive restriction endonucleases (Fig. 6 and 7) and by bisulfite genomic sequencing (Fig. 9 to 11) in established cell lines TM3 and EL4 revealed DNA methylation patterns consistent with the hypothesis that DNA methylation can differentially repress expression of Sgy and Tead2 during mouse development.
In fact, none of the cells in which either the Sgy or the Tead2 promoter region was methylated expressed that gene. In these cases, gene-specific RNA was not detected either by Northern analysis or by RT-PCR. In those cases, such as CA51 cells, in which Sgy expression could be restored by treatment with 5AC alone (Fig. 4), recovery of Sgy expression was accompanied by demethylation of its promoter region (Fig. 10B). In those cases, such as TM3 cells, in which recovery of Sgy expression required treatment with both 5AC and TSA (Fig. 5), Sgy expression was accompanied by only partial demethylation of its promoter region (data not shown), as previously reported for other genes that could be reactivated by the same regimen (4). Taken together, these results revealed an inverse correlation between the extent of DNA methylation in the promoter region and the extent of gene expression, consistent with a role for DNA methylation in repressing gene activity during development.
DNA methylation restricts Sgy expression to basal levels during mouse development.
Methylation of downstream sequences appeared to restrict Sgy expression to basal levels during mouse development. Full Sgy expression was observed only when both the promoter region and the downstream hypersensitive site (S3 in Fig. 8) were unmethylated in either normal cells such as splenocytes, two-cell embryos, and morulae or in established cell lines such as EL4 cells (Fig. 10). However, expression was restricted to basal levels when the promoter region was unmethylated but the S3 site was methylated in either normal cells such as ES cells, embryoid bodies, and uterus, lung, and liver cells or in established cell lines such as MPC-11, F9, and CA51 cells following treatment with 5AC (Fig. 10).
A similar result occurs in the skeletal alpha-actin gene promoter, where a subset of CpG dinucleotides are preferentially methylated in nonexpressing tissues (51). These data are consistent with previous studies showing that the extent of transcriptional suppression by DNA methylation in a plasmid-encoded reporter gene depends on the density of mCpG dinucleotides (12) and on the location of the methylated region (13). Maximum repression occurs when both the promoter and downstream regions of the gene are methylated.
The sequences downstream of the Sgy mRNA start site that contain hypersensitive site S3 appear to contain an enhancer or other regulatory element. Sequences from this region stimulated expression of a plasmid-encoded reporter gene driven by a viral promoter and bound a protein(s) present in EL4 cells but not in MPC-11 cells (data not shown). Interestingly, S3 corresponds to a repetitive SINE/B4 element located within intron 2. Such repetitive Alu elements are associated with insulator activity (53), and insulator activity can be regulated by DNA methylation (11). Therefore, sequences within intron 2 and their methylation status may determine whether Sgy is expressed at basal levels or fully activated. An analogous situation may exist with the Tead2 gene, where an enhancer has been identified within intron 1 of the Tead2 gene (45; data not shown).
DNA methylation is not the primary determinant of Sgy/Tead2 expression during mouse development.
If DNA methylation is the primary mechanismdetermining differential expression at the Sgy/Tead2 locus, then the gene that is not expressed should always contain a methylated promoter region. Surprisingly, the promoter regions of both genes were unmethylated in all of the primary cells and tissues examined. These included sperm cells, oocytes, two-cell embryos, morulae, ES cells, primary embryo fibroblasts, embryoid bodies, and splenocytes and uterus, lung, and liver tissues. Furthermore, in three of these examples, one of the two genes was silent despite the fact that its promoter region was unmethylated. (i) Oocytes expressed Tead2 but not Sgy, although the Sgy promoter region was unmethylated. Sgy expression began with zygotic gene expression following fertilization. (ii) The mixture of T and B lymphocytes isolated from spleen tissue did not express Tead2, despite the fact that the Tead2 promoter region was unmethylated. (iii) ES cells expressed Sgy at basal levels, but when they were induced to differentiate into embryoid bodies, Sgy gene expression was rapidly repressed by day 2 without increasing the extent of DNA methylation in the Sgy promoter region. Therefore, DNA methylation is not the primary determinant of Sgy/Tead2 expression during mouse development. The absence of DNA methylation at promoter regions is required but not sufficient for gene activity. The only silent Sgy or Tead2 genes that were also methylated were found in established cell lines, consistent with previous observations that hypermethylation of CpG islands is an intrinsic property of cultured cell lines rather than a general mechanism for regulating gene activity during animal development (41).
Regulation of zygotic gene activation.
The results described here suggest that the regulatory regions of genes such as Sgy and Tead2 that are destined to be transcribed during zygotic gene activation are not methylated in either sperm cells or oocytes. In oocytes, both the Sgy and Tead2 promoter regions were unmethylated. In sperm cells, the entire Sgy gene CpG island and sequences upstream to the Tead2 gene were largely unmethylated. This pattern was also found in two-cell embryos and in morulae, consistent with previous reports that, with the exception of imprinted genes (reference 52 and references therein), a global demethylation of the mouse genome occurs upon fertilization (32). Sequences downstream of exon 2 remained methylated, revealing that sequences in this region are immune to the global demethylation that occurs during preimplantation development (33). DNA methylation downstream of the Sgy mRNA start site increased as two-cell embryos underwent development to the ES cell stage and further increased as ES cells differentiated into embryoid bodies.
What is the purpose of these demethylation-remethylation events in preimplantation embryos? If the primary function of DNA methylation is to suppress expression of parasitic repeat sequences (49), then it is curious that oocytes and cleavage stage embryos contain an abundance of mRNA transcripts for B1 and B2 repeat elements (Alu repeats in humans) (46), consistent with the notion that global demethylation would result in an increase in the transcription of repeated sequences. Thus, oocytes and early embryos can tolerate expression of these potentially harmful sequences during the first 3 to 4 days of development. One general consequence of global demethylation is to remove one of the major obstacles to binding of proteins to DNA. These proteins may include repressor proteins whose binding is eliminated by methylation (7). With the exception of the five proteins known to bind specifically to mCpG dinucleotides, the affinity for DNA of all other DNA binding proteins appears to be reduced by methylation of their DNA binding sites. Thus, demethylation may facilitate remodeling of sperm chromatin into somatic cell chromatin in one-cell embryos, and it may allow more subtle remodeling to take place in both paternal and maternal genomes during the preimplantation period.
Acknowledgments
We thank the members of the Laboratory of Molecular Growth and Regulation, NICHD/NIH, for helpful discussions and comments.
This work was supported in part by a grant from NIH/NCRR (RR15253) awarded to K.L.
REFERENCES
- 1.Adachi, N., and M. R. Lieber. 2002. Bidirectional gene organization: a common architectural feature of the human genome. Cell 109:807-809. [DOI] [PubMed] [Google Scholar]
- 2.Baylin, S. B., M. Esteller, M. R. Rountree, K. E. Bachman, K. Schuebel, and J. G. Herman. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10:687-692. [DOI] [PubMed] [Google Scholar]
- 3.Brady, G., and N. N. Iscove. 1993. Construction of cDNA libraries from single cells. Methods Enzymol. 225:611-623. [DOI] [PubMed] [Google Scholar]
- 4.Cameron, E. E., K. E. Bachman, S. Myohanen, J. G. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103-107. [DOI] [PubMed] [Google Scholar]
- 5.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eden, S., and H. Cedar. 1994. Role of DNA methylation in the regulation of transcription. Curr. Opin. Genet. Dev. 4:255-259. [DOI] [PubMed] [Google Scholar]
- 7.Eden, S., M. Constancia, T. Hashimshony, W. Dean, B. Goldstein, A. C. Johnson, I. Keshet, W. Reik, and H. Cedar. 2001. An upstream repressor element plays a role in Igf2 imprinting. EMBO J. 20:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Maarri, O., K. Buiting, et al. 2001. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat. Genet. 27:341-344. [DOI] [PubMed] [Google Scholar]
- 9.Felsenfeld, G., J. Boyes, J. Chung, D. Clark, and V. Studitsky. 1996. Chromatin structure and gene expression. Proc. Natl. Acad. Sci. USA 93:9384-9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrich, B., J. Guy, B. Ramsahoye, V. A. Wilson, and A. Bird. 2001. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 15:710-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmgren, C., C. Kanduri, G. Dell, A. Ward, R. Mukhopadhya, M. Kanduri, V. Lobanenkov, and R. Ohlsson. 2001. CpG methylation regulates the Igf2/H19 insulator. Curr. Biol. 11:1128-1130. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh, C. L. 1994. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 14:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irvine, R. A., I. G. Lin, and C. L. Hsieh. 2002. DNA methylation has a local effect on transcription and histone acetylation. Mol. Cell. Biol. 22:6689-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacquemin, P., D. Depetris, M. G. Mattei, J. A. Martial, and I. Davidson. 1999. Localization of human transcription factor TEF-4 and TEF-5 (TEAD2, TEAD3) genes to chromosomes 19q13.3 and 6p21.2 using fluorescence in situ hybridization and radiation hybrid analysis. Genomics 55:127-129. [DOI] [PubMed] [Google Scholar]
- 15.Jacquemin, P., J. J. Hwang, J. A. Martial, P. Dolle, and I. Davidson. 1996. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J. Biol. Chem. 271:21775-21785. [DOI] [PubMed] [Google Scholar]
- 16.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko, K. J., E. B. Cullinan, K. E. Latham, and M. L. DePamphilis. 1997. Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development 124:1963-1973. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko, K. J., and M. L. DePamphilis. 1998. Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Dev. Genet. 22:43-55. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko, K. J., and M. L. DePamphilis. 2000. Soggy, a spermatocyte-specific gene, lies 3.8 kb upstream of and antipodal to TEAD-2, a transcription factor expressed at the beginning of mouse development. Nucleic Acids Res. 28:3982-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelsey, G., and W. Reik. 1998. Analysis and identification of imprinted genes. Methods 14:211-234. [DOI] [PubMed] [Google Scholar]
- 21.Kruisbeek, A. M. 2002. Isolation and fractionation of mononuclear cell populations, p. 3.1.1-3.1.5. In A. M. K. J. E. Coligan, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, Inc., New York, N.Y.
- 22.Lander, E. S., L. M. Linton, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 23.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Salas, E., E. Linney, J. Hassell, and M. L. DePamphilis. 1989. The need for enhancers in gene expression first appears during mouse development with formation of a zygotic nucleus. Genes Dev. 3:1493-1506. [DOI] [PubMed] [Google Scholar]
- 25.McDonald, L. E., and G. F. Kay. 1997. Methylation analysis using bisulfite genomic sequencing: application to small numbers of intact cells. BioTechniques 22:272-274. [DOI] [PubMed] [Google Scholar]
- 26.Melin, F., M. Miranda, N. Montreau, M. L. DePamphilis, and D. Blangy. 1993. Transcription enhancer factor-1 (TEF-1) DNA binding sites can specifically enhance gene expression at the beginning of mouse development. EMBO J. 12:4657-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, C. L., and J. M. Polak. 2002. Differentiating embryonic stem cells: GAPDH, but neither HPRT nor beta-tubulin is suitable as an internal standard for measuring RNA levels. Tissue Eng. 8:551-559. [DOI] [PubMed] [Google Scholar]
- 28.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 29.Pikaart, M. J., F. Recillas-Targa, and G. Felsenfeld. 1998. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 12:2852-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramalho-Santos, M., S. Yoon, Y. Matsuzaki, R. C. Mulligan, and D. A. Melton. 2002. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298:597-600. [DOI] [PubMed] [Google Scholar]
- 31.Rambhatla, L., B. Patel, N. Dhanasekaran, and K. E. Latham. 1995. Analysis of G protein alpha subunit mRNA abundance in preimplantation mouse embryos using a rapid, quantitative RT-PCR approach. Mol. Reprod. Dev. 41:314-324. [DOI] [PubMed] [Google Scholar]
- 32.Razin, A., and R. Shemer. 1995. DNA methylation in early development. Hum. Mol. Genet. 4:1751-1755. [DOI] [PubMed] [Google Scholar]
- 33.Reik, W., W. Dean, and J. Walter. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089-1093. [DOI] [PubMed] [Google Scholar]
- 34.Rein, T., M. L. DePamphilis, and H. Zorbas. 1998. Identifying 5-methylcytosine and related modifications in DNA genomes. Nucleic Acids Res. 26:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rein, T., H. Zorbas, and M. L. DePamphilis. 1997. Active mammalian replication origins are associated with a high-density cluster of mCpG dinucleotides. Mol. Cell. Biol. 17:416-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rizzino, A. 2002. Embryonic stem cells provide a powerful and versatile model system. Vitam. Horm. 64:1-42. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, S. M., M. Kennedy, J. M. Shannon, and G. Keller. 2000. A transitional stage in the commitment of mesoderm to hematopoiesis requiring the transcription factor SCL/tal-1. Development 127:2447-2459. [DOI] [PubMed] [Google Scholar]
- 38.Rogers, M. B., B. A. Hosler, and L. J. Gudas. 1991. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development 113:815-824. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Siegfried, Z., S. Eden, M. Mendelsohn, X. Feng, B. Z. Tsuberi, and H. Cedar. 1999. DNA methylation represses transcription in vivo. Nat. Genet. 22:203-206. [DOI] [PubMed] [Google Scholar]
- 41.Smiraglia, D. J., L. J. Rush, et al. 2001. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum. Mol. Genet. 10:1413-1419. [DOI] [PubMed] [Google Scholar]
- 42.Smith, S. S. 2000. Gilbert's conjecture: the search for DNA (cytosine-5) demethylases and the emergence of new functions for eukaryotic DNA (cytosine-5) methyltransferases. J. Mol. Biol. 302:1-7. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, K., M. Yasunami, Y. Matsuda, T. Maeda, H. Kobayashi, H. Terasaki, and H. Ohkubo. 1996. Structural organization and chromosomal assignment of the mouse embryonic TEA domain-containing factor (ETF) gene. Genomics 36:263-270. [DOI] [PubMed] [Google Scholar]
- 44.Takai, D., and P. A. Jones. 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA 99:3740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanoue, Y., M. Yasunami, K. Suzuki, and H. Ohkubo. 2001. Identification and characterization of cell-specific enhancer elements for the mouse ETF/Tead2 gene. Biochem. Biophys. Res. Commun. 289:1010-1018. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, K. D., and L. Piko. 1987. Patterns of mRNA prevalence and expression of B1 and B2 transcripts in early mouse embryos. Development 101:877-892. [DOI] [PubMed] [Google Scholar]
- 47.Vassilev, A., K. J. Kaneko, H. Shu, Y. Zhao, and M. L. DePamphilis. 2001. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15:1229-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venter, J. C., M. D. Adams, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 49.Walsh, C. P., and T. H. Bestor. 1999. Cytosine methylation and mammalian development. Genes Dev. 13:26-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Q., and K. E. Latham. 2000. Translation of maternal messenger ribonucleic acids encoding transcription factors during genome activation in early mouse embryos. Biol. Reprod. 62:969-978. [DOI] [PubMed] [Google Scholar]
- 51.Warnecke, P. M., and S. J. Clark. 1999. DNA methylation profile of the mouse skeletal alpha-actin promoter during development and differentiation. Mol. Cell. Biol. 19:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warnecke, P. M., J. R. Mann, M. Frommer, and S. J. Clark. 1998. Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics 51:182-190. [DOI] [PubMed] [Google Scholar]
- 53.Willoughby, D. A., A. Vilalta, and R. G. Oshima. 2000. An Alu element from the K18 gene confers position-independent expression in transgenic mice. J. Biol. Chem. 275:759-768. [DOI] [PubMed] [Google Scholar]
- 54.Yasunami, M., K. Suzuki, T. Houtani, T. Sugimoto, and H. Ohkubo. 1995. Molecular characterization of cDNA encoding a novel protein related to transcriptional enhancer factor-1 from neural precursor cells. J. Biol. Chem. 270:18649-18654. [DOI] [PubMed] [Google Scholar]
- 55.Yockey, C. E., and N. Shimizu. 1998. cDNA cloning and characterization of mouse DTEF-1 and ETF, members of the TEA/ATTS family of transcription factors DNA. Cell Biol. 17:187-196. [DOI] [PubMed] [Google Scholar]