Abstract
To clarify the molecular basis of the photoperiodic induction of flowering in the short-day plant Pharbitis nil cv Violet, we examined changes in the level of mRNA in cotyledons during the flower-inductive photoperiod using the technique of differential display by the polymerase chain reaction. A transcript that accumulated during the inductive dark period was identified and a cDNA corresponding to the transcript, designated PnC401 (P. nil C401), was isolated. RNA-blot hybridization verified that levels of PnC401 mRNA fluctuated with a circadian rhythm, with maxima between 12 and 16 h after the beginning of the dark period) and minima of approximately 0. This oscillation continued even during an extended dark period but was damped under continuous light. Accumulation of PnC401 mRNA was reduced by a brief exposure to red light at the 8th h of the dark period (night-break treatment) or by exposure to far-red light at the end of the light period (end-of-day far-red treatment). These results suggest that fluctuations in levels of PnC401 mRNA are regulated by phytochrome(s) and a circadian clock and that they are associated with photoperiodic events that include induction of flowering.
The circadian rhythms identified in most eukaryotes and some prokaryotes are approximately 24-h rhythms that are governed by a circadian clock that functions autonomously (Kay and Millar, 1995). Many physiological processes in plants, such as leaf movement, stem elongation, opening of stomata, and photosynthesis, often exhibit a circadian rhythm (Thomas and Vince-Prue, 1997). In photoperiodic species of plants, flowering is regulated by the absolute duration of light and darkness during an approximately 24-h cycle (Garner and Allard, 1920; Vince-Prue, 1975), and the induction of flowering is thought to include the actions of a biological clock (Evans, 1971). Physiological studies have demonstrated the participation of the circadian clock in photoperiodic responses (Thomas and Vince-Prue, 1997). Moreover, a recent study of a mutant of Arabidopsis (elf3) revealed a close relationship between the circadian clock and photoperiodic flowering (Hicks et al., 1996).
Biochemical, molecular, and genetic studies are starting to uncover the mechanisms that underlie the induction of flowering. Several mutants of Arabidopsis with altered flowering times have been studied, and some relevant genes have been cloned and analyzed (Lee et al., 1994; Putterill et al., 1995; Peeters and Koornneef; 1996; Macknight et al., 1997). Biochemical studies of flowering in plants other than Arabidopsis have focused on the photoperiodism of flowering (Thomas and Vince-Prue, 1997). In most of the plant systems used in such studies, flowering can be induced by one photoperiodic treatment, which allows biochemical studies of the process (Bernier, 1988). Changes in biological activities, including changes in gene expression in leaves during photoperiodic treatments, have been studied in such plants (Heintzen et al., 1994a, 1994b; O'Neill et al., 1994; Ono et al., 1996; Périlleux et al., 1996). Because most plants in which photoperiodic flowering has been studied are LDPs, such as Arabidopsis, complementary studies of flowering of SDPs are particularly important at this time.
Pharbitis nil Choisy cv Violet, a SDP, is ideal for the study of the early events in the photoperiodic induction of flowering, because young, light-grown seedlings can be induced to flower quantitatively by exposure to a single dark period of 16 h (Vince-Prue and Gressel, 1985). In P. nil the phase of the rhythm that is maximally sensitive to an NB usually occurs 8 h after the end of the light period, with a second phase of maximal sensitivity occurring 24 h later (Vince-Prue and Gressel, 1985). Therefore, it has been concluded that the timing of the NB response is a manifestation of a circadian rhythm (Vince-Prue and Lumsden, 1987).
In P. nil, as in other plant systems, regulation of processes related to photoperiodically induced flowering probably occurs at a number of levels, including the level of gene expression. Results obtained with chemical inhibitors of gene expression and results of the biochemical analyses of various macromolecules suggest that changes in gene expression might participate in the generation in leaves of a state that leads to induction of flowering (Vince-Prue and Gressel, 1985; O'Neill, 1992). Actinomycin D, a potent inhibitor of RNA synthesis, completely suppresses floral induction without any visible effects on vegetative growth (Vince-Prue and Gressel, 1985). Lay-Yee et al. (1987) found an increase in the intensity of the spot of one particular protein among products of translation in vitro that had been resolved by two-dimensional PAGE. Using the technique of differential-hybridization screening of cDNA libraries, Felsheim and Das (1992), Krishna et al. (1992), Zheng et al. (1993), and O'Neill et al. (1994) generated cDNAs of cotyledonous mRNAs, the levels of which were altered by photoperiodic treatment. Steady-state levels of some of these mRNAs showed circadian oscillations, which suggested their participation in the photoperiodic induction of flowering (Zheng et al., 1993; O'Neill et al., 1994). Moreover, the level of a specific protein increased during the latter half of the flower-inductive dark period, as indicated by results of two-dimensional PAGE after labeling of polypeptides with [35S]Met in vivo (Ono et al., 1993). The level of the corresponding mRNA increased during the flower-inductive dark period and also showed circadian oscillations (Ono et al., 1996). The mRNAs mentioned above are all relatively abundant and none has been studied in further detail to prove the direct participation in the photoperiodic induction of flowering.
Recent studies of Arabidopsis have demonstrated the importance of molecules that are present at low levels in the photoperiodic induction of flowering (Putterill et al., 1995). We also predicted the existence of rare transcripts that are related to flowering (Ono et al., 1993). Therefore, to examine this issue we decided to exploit a method for the isolation of preferentially expressed mRNAs, namely, differential display by PCR (Liang and Pardee, 1992). In this report we describe the isolation and characterization of a cDNA clone, PnC401, from P. nil. PnC401 corresponds to a single-copy gene without any similarity to known genes. Results of RNA gel-blot studies show that expression of the PnC401 gene is regulated by the circadian clock, and there is a relatively low level of the corresponding transcript. Moreover, the mode of expression of the PnC401 gene appears to reflect the photoperiodic induction of flowering.
MATERIALS AND METHODS
Plant Materials and Photoperiodic Treatments
Seeds of Pharbitis nil Choisy cv Violet (purchased from Marutane Co., Kyoto, Japan) and cv Kidachi (kindly provided by Dr. K. Yokota, Biotechnology Institute, Ibaraki, Japan) were soaked in concentrated sulfuric acid for 30 min, with occasional stirring, and then rinsed in running tap water for 1 h. Alternatively, part of the surface of each seed was slightly abraded with a file. The seeds were then soaked in a large volume of distilled water for 16 h and sown on wet vermiculite for germination. Growth conditions were set at 25 ± 1°C with illumination by continuous cool-white fluorescent light (13.6 W m−2, FLR 40H-WA lamps, Matsushita Electronics Co., Tokyo, Japan) in cultivation chambers (CU-350; Tomy Seiko Co., Tokyo, Japan). When the cotyledons had opened maximally (6 d after treatment with sulfuric acid or abrasion), the seedlings were subjected to photoperiodic treatments. Photoperiodic treatments began with a dark period, and the time elapsed was counted from the beginning of the dark period. Red light (0.6 W m−2) was obtained by passage of the above-mentioned fluorescent light through a sheet of red acrylic resin (Acrylite no. 102, Mitsubishi Rayon Co., Ltd., Tokyo, Japan) that excluded light at wavelengths below 600 nm (Saitou et al., 1992). FR light (0.8 W m−2) was obtained from a lamp (model FL20S-FR74, Toshiba Electronics Co., Tokyo, Japan) with a passage filter (Saitou et al., 1992). The cotyledons and other organs were excised, frozen in liquid nitrogen, and stored at −80°C. During the dark period tissues were harvested in complete darkness. To determine the extent of flower induction, about 10% of the plants were not harvested but were cultured under LL. After 3 weeks, the number of flower buds was counted.
Differential Display
Total RNA was isolated as described by Ozeki et al. (1990). Poly(A+) RNA was isolated by chromatography on oligo(dT)-cellulose (Pharmacia), as originally described by Aviv and Leder (1972), and was stored at −80°C for later use. Differential display by PCR was performed as described by Liang et al. (1993). Purified total RNA (0.6 μg) was reverse transcribed using one of the following primers: T12MA, T12MC, T12MG, or T12MT, where M stands for dA, dC, and dG. The solution for reverse-transcription reactions (total volume, 60 μL) contained 1 μm T12MN and 20 μm deoxyribonucleotide triphosphate, and reactions were carried out as follows. Solutions were heated at 65°C for 5 min and then at 37°C for 10 min, after which 150 units of Stratascript reverse transcriptase (Stratagene) was added. The reactions were heated at 37°C for 50 min and at 95°C for 5 min, and then the mixtures were stored at −20°C until they were used for PCR. Two microliters of the mixture after reverse transcription was used as the source of template for PCR in a reaction mixture that contained a T12MN primer in combination with an arbitrary 10-base primer (1 of 20 different primers) in the presence of 32P-labeled dCTP. The conditions for PCR were as follows: 94°C for 30 s, 40°C for 2 min, and 72°C for 30 s for 40 cycles, followed by incubation at 72°C for 5 min. Aliquots of duplicate reaction mixtures after PCR were subjected to electrophoresis on a 6% polyacrylamide gel to separate the amplified cDNAs.
We sometimes found quantitative and qualitative differences in the patterns of bands obtained after duplicate reactions; thus, only material in bands that were differentially amplified in a consistent manner was further analyzed. The regions of the gel containing the differentially expressed cDNAs were excised from the dried gel and placed in 100 μL of distilled water. The cDNA molecules that diffused from the gel fragments were reamplified with the appropriate pair of primers. The primers used in the experiment that led to the isolation of PnC401 cDNA were T12MC (5′-TTTTTTTTTTTTMC-3′) and AP-4 (5′-GGTACTCCAC-3′). The samples after amplification were subjected to electrophoresis on a 5% polyacrylamide gel, purified, and subcloned into pBlueScript SK+ vector (Stratagene). The insert cDNAs were used as the probes for RNA gel-blot hybridization of the same samples of RNA as used for the initial reverse transcriptions.
Construction and Screening of a Library
A cDNA library was constructed from the poly(A+) RNA of SD-treated cotyledons of P. nil using a cDNA synthesis kit and a cDNA cloning kit (Amersham) in accordance with the instructions from the manufacturer. Screening of plaques and preparation of phage were also performed according to the instructions from Amersham.
Sequencing and Analysis of DNA
The nucleotide sequence of PnC401 cDNA was determined with fluorescent primers and an automated DNA sequencer (model 373A, Applied Biosystems). Nucleotide and amino acid sequences were analyzed with GENETYX-MAC software, version 8.0 (Software Kaihatsu Co., Tokyo, Japan). Databases were searched with the DDBJ BLAST system 1.4.9 (DNA Data Bank of Japan, Mishima, Shizuoka, Japan; Altschul et al., 1990) and with the ExPASy system (Molecular Biology Center at Geneva, University Hospital and University of Geneva, Switzerland).
DNA Gel-Blot Hybridization
Genomic DNA was isolated from apical buds with small leaves of P. nil as described by Rogers and Bendich (1985), and was digested with EcoRI, BamHI, and HindIII. Digested DNA was subjected to electrophoresis on an agarose gel, and bands of DNA were transferred to a nylon membrane filter (Biodyne B, Nihon Pall, Ltd., Tokyo, Japan). The DNA on the filter was allowed to hybridize with 32P-labeled PnC401 cDNA in a hybridization solution that contained 6× SSPE (1× SSPE is 0.18 m NaCl, 0.01 m sodium phosphate, and 1 mm Na2EDTA, pH 7.7), 5× Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% PVP, and 0.02% BSA), 0.5% SDS, and 150 μg mL−1 salmon- sperm DNA at 65°C for 16 h. The filter was washed twice with 2× SSPE and 0.1% SDS for 5 min at room temperature and then twice for 30 min at 65°C (low-stringency conditions). After exposure to an imaging plate for an appropriate time, the same filter was washed twice with 0.1× SSPE and 0.1% SDS for 30 min at 65°C (high-stringency conditions). For visualization of bands on the filter, we used a bioimaging analyzer with an imaging plate (BAS2000, Fuji Photo Film Co., Ltd., Tokyo, Japan).
RNA Gel-Blot Hybridization
Total RNA (20 μg) was fractionated by electrophoresis on a formaldehyde-agarose gel, and the bands of RNA were transferred to a nylon membrane filter (Biodyne B). The RNA on the filter was allowed to hybridize with 32P-labeled PnC401 cDNA in a hybridization solution that contained 50% formamide, 5× SSPE, 5× Denhardt's solution, 0.1% SDS, and 150 μg mL−1 salmon-sperm DNA at 42°C for 20 h. The filter was first washed with 2× SSC at room temperature and then with 2× SSC and 0.1% SDS at 42°C. For visualization of the bands on the filter, we again used the bioimaging analyzer. To provide an internal control, the same blot was rehybridized with the PnrRNA cDNA, which encodes the 16S rRNA of P. nil (K. Sage-Ono, unpublished data).
RESULTS
Identification of cDNAs of Transcripts, the Levels of which Increased during the Inductive Dark Period
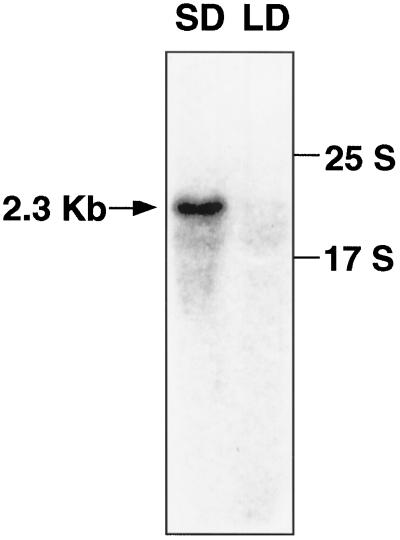
To gain some insight into the molecular basis of the photoperiodic induction of flowering in P. nil, we attempted to isolate cDNAs of mRNAs, the transcription of which was induced during an inductive dark photoperiod by differential display with PCR (Liang and Pardee, 1992). We compared cotyledonous mRNAs at 12 and 16 h under SD conditions and at 0 h under LD conditions (LL), as measured from the beginning of photoperiodic treatment. Initially, we observed a significant number of relevant bands of different cDNAs (more than 300, about 3% of all bands detected). Among them, 32 reproducible bands of cDNA fragments that exhibited the appropriate differences were excised, reamplified by PCR, and used as the probes for RNA gel-blot hybridization analysis in a comparison of the relative levels of transcript in SD- and LD-treated cotyledons. Most cDNA fragments (22 bands) gave no signals on RNA gel-blot hybridization, and nine cDNA fragments did not show the expected results. The remaining cDNA, designated PnC401, which hybridized to a transcript of about 2.3 kb, preferentially accumulated in SD-treated cotyledons (Fig. 1). To obtain a full-length cDNA clone, the cDNA fragment was used as a probe for screening of a cDNA library of SD-treated cotyledons. Because only two PnC401 cDNAs were obtained in a screening of 2 × 105 plaques, PnC401 mRNA appeared to be a rare transcript, even in SD-treated cotyledons.
Figure 1.
Preferential accumulation of PnC401 mRNA in dark-treated cotyledons. Seedlings of cv Violet were grown under LL for 6 d after germination of seeds and then transferred to SD conditions (16 h of dark; flower-inductive conditions) or LD conditions (LL; noninductive conditions). Cotyledons were harvested at the 12th and 16th h of each photoperiodic treatment. Total RNA (20 μg per lane) was fractionated by gel electrophoresis and allowed to hybridize to a 3′ fragment of PnC401 cDNA.
Sequence Analysis of PnC401 cDNA
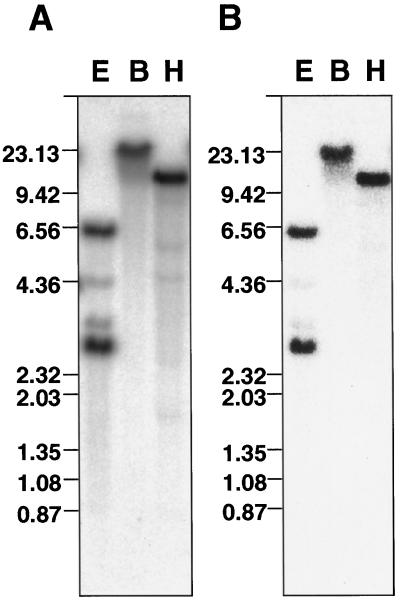
We determined the nucleotide sequence of PnC401 cDNA and the deduced amino acid sequence. The cDNA consisted of 2363 bp and contained a 173-bp untranslated leader sequence followed by a 1995-bp open reading frame that encoded a putative polypeptide of 665 amino acids with a molecular mass of 74 kD and a predicted pI of 9.0. The nucleotide sequence and the deduced amino acid sequence were used in a search of databases by the DDBJ BLAST system 1.4.9 (Altschul et al., 1990). The results revealed only negligible similarity to some previously characterized genes and proteins. However, the search of databases did reveal the strong similarity of PnC401 to just one Arabidopsis expressed sequence tag clone. The middle region of the partial sequence (224 bp) of the Arabidopsis clone (241B6T7; Newman et al., 1994) was about 65% identical to PnC401 at both the nucleotide and amino acid sequence levels and showed about 90% similarity in amino acid sequence. We isolated and sequenced the corresponding gene from Arabidopsis and named it AtC401 (data not shown). Southern-blot analysis of genomic DNA from P. nil indicated that PnC401 corresponded to a single-copy gene (Fig. 2). Similar results were obtained for AtC401 in Arabidopsis (data not shown). A search of protein databases using the ExPASy system showed that the deduced PnC401 protein included no known protein motifs and that it is possible that this protein is located in the cytoplasm.
Figure 2.
DNA gel-blot analysis of the PnC401 gene. Genomic DNA was prepared from cv Violet, digested with EcoRI (E), BamHI (B), and HindIII (H), and subjected to DNA gel-blot hybridization. Full-length PnC401 cDNA was used as the probe. A, Low-stringency conditions. B, High-stringency conditions. Numbers at left indicate mobilities of markers with lengths in kilobases.
Circadian Oscillations of Levels of PnC401 mRNA
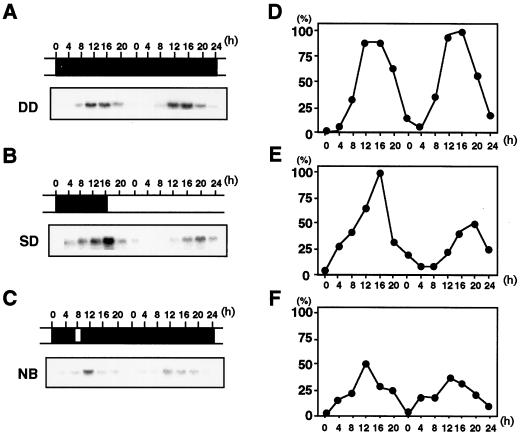
We performed northern-blot analysis of mRNAs during various photoperiodic treatments for 2 d (Fig. 3). Seedlings were grown for 6 d under LL and then transferred to specific photoperiodic conditions (DD, SD, and NB). PnC401 mRNA was not detected under LL or before the dark treatment. When seedlings were transferred to DD and SD conditions, the level of PnC401 mRNA increased and then decreased during the 24-h dark period, reaching a maximum at the 12th to 16th h of darkness (Fig. 3, A and B). NB treatment, a 10-min exposure to red light at the 8th h of dark treatment, reduced the extent of accumulation of mRNA (Fig. 3C). Under DD conditions, the level of PnC401 mRNA exhibited circadian oscillations, reaching a maximum at the 12th and 16th h of darkness (Fig. 3A). However, constant light after a 16-h inductive dark period under SD conditions reduced the size of the second peak, with a lag phase of approximately 4 h (Fig. 3B).
Figure 3.
Effects of various photoperiodic treatments on the level of PnC401 mRNA in cotyledons. Seedlings of cv Violet were grown under LL for 6 d and then subjected to various photoperiodic treatments: A, DD; B, SD (16 h of darkness); or C, NB (10-min exposure to light at the 8th h of darkness). The cotyledons of 15 plants were harvested every 4 h during photoperiodic treatments and used for extraction of RNA. Total RNA (20 μg per lane) was fractionated by gel electrophoresis and allowed to hybridize to full-length PnC401 cDNA. Uniformity of the loading and transfer of the RNA was confirmed by reprobing the blots with a cDNA fragment for 16S rRNA of P. nil. Relative levels of PnC401 mRNA were calculated from the intensity of each radioactive band, as determined with an imaging plate: D for A, E for B, and F for C.
Effects of Irradiation with Red and FR Light
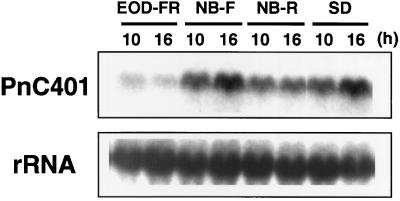
The effects of phytochromes on time keeping have been studied extensively in P. nil, in which brief exposure to red light at the 8th h after the beginning of the dark period inhibits flowering (NB), and brief exposure to FR light at the beginning of darkness (EOD) inhibits flowering (Vince-Prue and Gressel, 1985; Bernier, 1988). We examined changes in the steady-state level of PnC401 mRNA after such light treatments (Fig. 4). Irradiation with FR light at EOD strongly inhibited the accumulation of PnC401 mRNA. With NB irradiation with red light, the level of accumulation of PnC401 mRNA was reduced. By contrast, NB with FR light did not affect the accumulation of PnC401 mRNA. These results were closely correlated with the physiological data. In other words, the extent of inhibition of the accumulation of the mRNA reflected the extent of floral inhibition (data not shown).
Figure 4.
Effects of exposure to red and FR light on the level of PnC401 mRNA in cotyledons. Seedlings were grown under LL for 6 d and then exposed to 16 h of darkness with or without various kinds of light exposure, as follows: EOD-FR, a 10-min exposure to FR light just before darkness; NB-F, a 5-min exposure to FR light at the 8th h of darkness; NB-R, a 5-min exposure to red light at the 8th h of darkness (NB); and SD, 16th h of darkness. Total RNA (20 μg) was isolated from cotyledons at the 10th and 16th h of each treatment and analyzed. Uniformity of the loading and transfer of the RNA was confirmed by reprobing the blots with the cDNA fragment for 16S rRNA of P. nil.
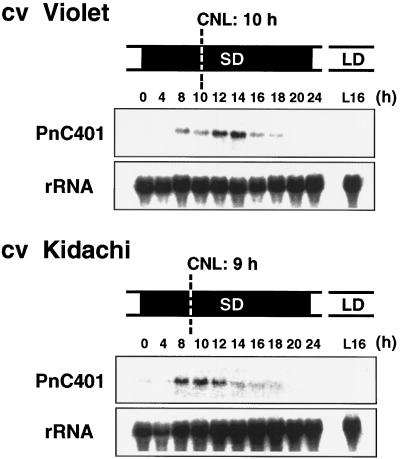
Effects of a Varietal Difference between Cultivars in CNL on the Timing of the Peak Level of PnC401 mRNA
CNL for flowering in P. nil differs among cultivars. To confirm that the peak level of PnC401 mRNA at 12 and 16 h was related to CNL, we examined another cultivar of P. nil. CNL of P. nil cv Kidachi is 8 to 9 h and is about 1 h shorter than that of cv Violet (Imamura, 1967). As shown in Figure 5, the level of PnC401 mRNA in the cotyledons of cv Kidachi began to increase at 8 h of SD treatment and reached a maximum at 12 and 14 h of SD treatment, 2 h earlier than the levels in the cv Violet.
Figure 5.
Effects of a varietal difference in CNL on the level of PnC401 mRNA. CNL of cv Violet is about 10 h and that of cv Kidachi is about 9 h at 25°C. The cotyledons of 15 plants of each cultivar were harvested at indicated times during inductive darkness (SD) or LL (LD). Total RNA (20 μg per lane) was fractionated by gel electrophoresis and allowed to hybridize to full-length PnC401 cDNA. Uniformity of the loading and transfer of the RNA was confirmed by reprobing the blots with cDNA for 16S rRNA of P. nil.
Organ-Specific Accumulation of PnC401 mRNA
We examined the organ-specific accumulation of PnC401 mRNA by RNA-blot hybridization (Fig. 6). PnC401 mRNA was detected mainly in “induced” cotyledons and leaves that had flower-inducing ability. By contrast, other organs, namely roots, stems, and petioles, contained lower amounts of PnC401 mRNA.
Figure 6.
Levels of PnC401 mRNA in different organs. Total RNA was isolated from various organs of seedlings of cv Violet. Lanes: R, root; S, stem; P, petiole; C, cotyledon; and L, first leaf. Organs were harvested at the peak time (at the 16th h of dark treatment) in the circadian oscillation of levels of the transcript in cotyledons. Total RNA (20 μg per lane) was fractionated by gel electrophoresis and allowed to hybridize to full-length PnC401 cDNA.
DISCUSSION
Using differential display by PCR, we isolated a cDNA clone designated PnC401, which corresponded to a single-copy gene in the SDP cv Violet. A search of databases revealed that the predicted protein encoded by PnC401 is a novel protein with negligible homology to previously characterized proteins. Although we have no direct evidence related to the function of PnC401, northern analysis of PnC401 mRNA yielded some clues. The steady-state levels of PnC401 mRNA reflected the state of the plant with respect to photoperiodic flowering. PnC401 mRNA accumulated preferentially in cotyledons and leaves, which are the organs responsible for the photoperiodic induction of flowering in P. nil. The level of PnC401 mRNA increased transiently during flower-inductive darkness and showed circadian oscillations when the dark period was extended. A varietal difference in CNL, which determines the minimal length of the dark period for photoperiodic induction of flowering, influenced the timing of the peak level of PnC401 mRNA. Moreover, interruption by red light at the 8th h of flower-inductive darkness (NB) and exposure to FR light at the end of the light period (EOD) reduced or inhibited the accumulation of PnC401 mRNA. These results are in harmony with physiological data related to the photoperiodic induction of flowering in P. nil (Imamura, 1967; Vince-Prue and Gressel, 1985), and suggest the possible participation of the PnC401 gene in photoperiodic events that include floral induction.
Some genes and cDNAs have been isolated from P. nil in studies of the molecular mechanisms of the photoperiodic induction of flowering (Zheng et al., 1993; O'Neill et al., 1994; Ono et al., 1996). However, in the present study we did not detect any cDNAs related to these reported genes and cDNAs. Because all of the cDNAs reported previously corresponded to mRNAs that were expressed to some extent under noninductive conditions, namely under LL, the method that we used for isolation of cDNAs might have excluded these cDNA clones. Our results appear to support the superiority of the isolation method that we used compared with other methods, as was also concluded by other researchers (Wan et al., 1996).
The level of PnC401 mRNA showed a transient increase during flower-inductive darkness at the 12th and 16th h of darkness, and successive peaks were observed at intervals of approximately 24 h when the dark period was extended (Fig. 3A). Therefore, the PnC401 gene seems to be one of the CCGs. Recently, several CCGs were discovered in plants and analyzed (for reviews, see McClung and Kay, 1994; Beator and Kloppstech, 1996). Most of them appear to be light regulated and their circadian rhythmicity continues during an extended light period rather than in extended darkness. However, the CCGs cloned from P. nil (HMG1, PN1, PN9, and PnGLP) were found to be dark regulated; their circadian rhythmicity continued in extended darkness rather than in an extended light period (Zheng et al., 1993; O'Neill et al., 1994; Ono et al., 1996). We do not know the reason for these differences between CCGs of P. nil and other plants. The differences between SDPs and LDPs, as well as the specific characteristics of the genus Pharbitis, might be involved. To explain the differences in the regulation of CCGs, studies on the same CCGs in different plant systems will be needed.
The finding of a homolog of PnC401 in Arabidopsis might be important for future elucidation of the function of PnC401, and for attempts to explain the differences in the regulation of CCGs in P. nil and other plants. AtC401 cDNA was very similar to PnC401 cDNA and represented a single-copy gene. Studies in Arabidopsis should allow us to obtain mutants of the AtC401 gene and to clarify its relationship to other genes that are related to photoperiodicity and flowering. We have already mapped the AtC401 gene on a chromosome of Arabidopsis and are currently searching for relevant mutants. We are analyzing changes in the level of AtC401 mRNA under different physiological conditions using wild-type plants and some photoperiodic mutants. Moreover, we are currently transforming Arabidopsis and P. nil with PnC401 cDNA for overexpression and antisense repression studies. Together, these studies should provide more information about the function and participation of the PnC401 gene in photoperiodism and in the induction of flowering.
ACKNOWLEDGMENTS
The authors are grateful to Dr. K. Yokota (Plant Biotechnology Institute, Ibaraki Agricultural Center, Ibaraki, Japan) for kindly providing seeds of P. nil cv Kidachi and to Mr. M. Kawakami for his assistance with harvesting cotyledons.
Abbreviations:
- CCG
circadian clock-controlled gene
- CNL
critical night length
- DD
continuous darkness
- EOD
end of day
- FR
far-red
- LD
long day
- LDP
long-day plant
- LL
continuous light
- NB
night breakSD, short day
- SDP
short-day plant
Footnotes
This work was supported in part by a Grant-in-Aid for Special Research Areas (grant no. 07281103; Genetic Dissection of Sexual Differentiation and Pollination Processes in Higher Plants) from the Ministry of Education, Science, Culture, and Sports, Japan, and by a Grant-in-Aid for the “Research for the Future” Program (grant no. JSPS-RFTF97L00601) from the Japan Society for the Promotion of Science.
The nucleotide sequence reported in this paper can be found in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. D85101.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aviv H, Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci USA. 1972;69:1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beator J, Kloppstech K. Significance of circadian gene expression in higher plants. Chronobiol Int. 1996;13:319–339. doi: 10.3109/07420529609012657. [DOI] [PubMed] [Google Scholar]
- Bernier G. The control of floral evocation and morphogenesis. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:175–219. [Google Scholar]
- Evans LT. Flower induction and the florigen concept. Annu Rev Plant Physiol. 1971;22:365–394. [Google Scholar]
- Felsheim RF, Das A. Structure and expression of a heat-shock protein 83 gene of Pharbitis nil. Plant Physiol. 1992;100:1764–1771. doi: 10.1104/pp.100.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res. 1920;18:553–606. [Google Scholar]
- Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. A light-and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. Plant J. 1994a;5:799–813. doi: 10.1046/j.1365-313x.1994.5060799.x. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. Circadian oscillation of a transcript encoding a germin-like protein that is associated with cell walls in young leaves of the long-day plant Sinapis alba L. Plant Physiol. 1994b;106:905–915. doi: 10.1104/pp.106.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- Imamura S (1967) Photoperiodic induction and the floral stimulus. In S Imamura, ed, Physiology of Flowering in Pharbitis nil. Japanese Society of Plant Physiologists, Tokyo, pp 15–28
- Kay SA, Millar AJ. New models in vogue for circadian clocks. Cell. 1995;83:361–364. doi: 10.1016/0092-8674(95)90113-2. [DOI] [PubMed] [Google Scholar]
- Krishna P, Felsheim RF, Larkin JC, Das A. Structure and light-induced expression of a small heat-shock protein gene of Pharbitis nil. Plant Physiol. 1992;100:1772–1779. doi: 10.1104/pp.100.4.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay-Yee M, Sachs RM, Reid MS. Changes in cotyledon mRNA during floral induction of Pharbitis nil cv. Violet. Planta. 1987;171:104–109. doi: 10.1007/BF00395073. [DOI] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM. Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell. 1994;6:75–83. doi: 10.1105/tpc.6.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Averboukh L, Pardee AB. Distribution and cloning of eukaryotic mRNAs by means of differential display refinements and optimization. Nucleic Acids Res. 1993;21:3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C and others. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- McClung CR, Kay SA (1994) Circadian rhythms in Arabidopsis thaliana, In EM Meyerowitz, CR Somerville, eds, Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 615–637
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M and others. Genes galore. A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill SD. The photoperiodic control of flowering: progress toward understanding the mechanism of induction. Photochem Photobiol. 1992;56:789–801. [Google Scholar]
- O'Neill SD, Zhang XS, Zheng CC. Plant Physiol. 1994;104:569–580. doi: 10.1104/pp.104.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Sage-Ono K, Yasui M, Okazaki M, Harada H. Changes in polypeptides in Pharbitis cotyledons during the first flower-inductive photoperiod. Plant Sci. 1993;89:135–145. [Google Scholar]
- Ono M, Sage-Ono K, Inoue M, Kamada H, Harada H. Transient increase in the level of mRNA for a germin-like protein in leaves of the short-day plant Pharbitis nil during the photoperiodic induction of flowering. Plant Cell Physiol. 1996;37:855–861. doi: 10.1093/oxfordjournals.pcp.a029022. [DOI] [PubMed] [Google Scholar]
- Ozeki Y, Matui K, Sakuta M, Matsuoka M, Ohashi Y, Kano-Murakami Y, Yamamoto N, Tanaka Y. Differential regulation of phenylalanine ammonia-lyase genes during anthocyanin synthesis and by transfer effect in carrot cell suspension cultures. Physiol Plant. 1990;80:379–387. [Google Scholar]
- Peeters AJM, Koornneef M. Semin Cell Dev Biol. 1996;7:381–389. [Google Scholar]
- Périlleux C, Ongena P, Bernier G. Changes in gene expression in the leaf of Lolium temulentum L. Ceres during the photoperiodic induction of flowering. Planta. 1996;200:32–40. [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol. 1985;5:69–76. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- Saitou T, Kamada H, Harada H. Light requirement for shoot regeneration in horseradish hairy roots. Plant Physiol. 1992;99:1336–1341. doi: 10.1104/pp.99.4.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in Plants, Ed 2. San Diego, CA: Academic Press; 1997. [Google Scholar]
- Vince-Prue D. Photoperiodism in Plants. London: McGraw-Hill; 1975. [Google Scholar]
- Vince-Prue D, Gressel J (1985) Pharbitis nil. In AH Halevy, ed, Handbook of Flowering, Vol IV. CRC Press, Boca Raton, FL, pp 47–81
- Vince-Prue D, Lumsden PJ (1987) Inductive events in the leaves: time measurement and photoperception in the short-day plant Pharbitis nil. In JG Atherton, ed, Manipulation of Flowering. Butterworths, London, pp 255–269
- Wan JS, Sharp SJ, Poirier GMC, Wagaman PC, Chambers J, Pyati J, Hom YL, Galindo JE, Huvar A, Peterson PA and others. Cloning differentially expressed mRNAs. Nature Biotechnol. 1996;14:1685–1691. doi: 10.1038/nbt1296-1685. [DOI] [PubMed] [Google Scholar]
- Zheng CC, Bui AQ, O'Neill SD. Abundance of an mRNA encoding a high mobility group DNA-binding protein is regulated by light and an endogenous rhythm. Plant Mol Biol. 1993;23:813–823. doi: 10.1007/BF00021536. [DOI] [PubMed] [Google Scholar]