Abstract
The NuA4 histone acetyltransferase (HAT) multisubunit complex is responsible for acetylation of histone H4 and H2A N-terminal tails in yeast. Its catalytic component, Esa1, is essential for cell cycle progression, gene-specific regulation and has been implicated in DNA repair. Almost all NuA4 subunits have clear homologues in higher eukaryotes, suggesting that the complex is conserved throughout evolution to metazoans. We demonstrate here that NuA4 complexes are indeed present in human cells. Tip60 and its splice variant Tip60b/PLIP were purified as stable HAT complexes associated with identical polypeptides, with 11 of the 12 proteins being homologs of yeast NuA4 subunits. This indicates a highly conserved subunit composition and the identified human proteins underline the role of NuA4 in the control of mammalian cell proliferation. ING3, a member of the ING family of growth regulators, links NuA4 to p53 function which we confirmed in vivo. Proteins specific to the human NuA4 complexes include ruvB-like helicases and a bromodomain-containing subunit linked to ligand-dependent transcription activation by the thyroid hormone receptor. We also demonstrate that subunits MRG15 and DMAP1 are present in distinct protein complexes harboring histone deacetylase and SWI2-related ATPase activities, respectively. Finally, analogous to yeast, a recombinant trimeric complex formed by Tip60, EPC1, and ING3 is sufficient to reconstitute robust nucleosomal HAT activity in vitro. In conclusion, the NuA4 HAT complex is highly conserved in eukaryotes, in which it plays primary roles in transcription, cellular response to DNA damage, and cell cycle control.
Studies in recent years have identified several enzymes responsible for acetylation, methylation, ubiquitination, and phosphorylation of histones. These enzymes are directly implicated in gene regulation through modification of chromatin (58). They are often part of stable multisubunit complexes that contain protein modules thought to be important for regulation, recruitment, and other specialized functions (10). Furthermore, specific histone modifications have been shown to regulate others, both on the same N-terminal tail and in a transhistone fashion (17). These modifications can be recognized by protein domains such as bromodomains (acetylated lysine residues) and chromodomains (methylated lysine residues). Thus, posttranslational modifications of histones cannot only directly change chromatin structure but also modulate interactions of specific proteins with chromatin. Histone acetylation is the most characterized modification and is controlled by histone acetyltransferase (HAT) and histone deacetylase (HDAC) complexes, which are recruited by activators and repressors to promoter regions and regulate transcription. In Saccharomyces cerevisiae, HAT enzymes Gcn5 and Esa1 are present in two large protein complexes, SAGA and NuA4, recruited to promoter regions by transcription activators (10).
NuA4 (for nucleosome acetyltransferase of H4) is a 12-subunit HAT complex responsible for acetylation of histone H4 and H2A N-terminal tails. The catalytic subunit Esa1 is the only essential HAT in yeast and is homologous to human Tip60 (2, 12, 53). Both proteins have been linked to transcription regulation, as well as DNA double-strand break repair (6, 26; reviewed in reference 56). Other yeast subunits include Tra1, an essential ATM family cofactor implicated in recruitment by transcription activators (2, 9); Yng2, an ING1 tumor suppressor homolog required for transcription activation by p53 and DNA damage response (11, 41, 42); Arp4, an actin-related protein linked to epigenetic control of transcription (20); Eaf3, a chromodomain-containing protein related to the dosage compensation complex in Drosophila (14); and Epl1, a Drosophila E(Pc) (Enhancer of Polycomb) homolog, which is a suppressor of position-effect variegation (7, 20). NuA4 can be recruited by activators in vitro and in vivo to create a large domain of histone H4/H2A hyperacetylation and activate transcription (41, 47, 57, 59; A. Nourani, R. T. Utley, S. Allard, and J. Côté, unpublished data). At least 10 of the 12 yeast NuA4 subunits have clear mammalian homologs, suggesting the existence of a NuA4 complex in mammals (see Table 1). These homologs have been implicated in transcription activation and cell transformation by c-Myc, E2F and E1A, p53 function, coactivation of steroid receptors and NF-κB, DNA repair, apoptosis, and Alzheimer molecular onset (reviewed in references 10 and 56).
TABLE 1.
Human homologs to yeast NuA4 subunits
| Yeast NuA4 subunit | Human homolog(s) | Domain(s)a |
|---|---|---|
| Tra1 | TRRAP (transformation/transcription domain-associated protein) | PI-3 kinase/ATM |
| Eaf1 | hDomino? (p400) | SANT, HSA |
| Epl1 | Enhancer of Polycomb (EPC1, EPC-like) | EpcA |
| Eaf2 | DMAP1 (DNMT1-associated protein) | SANT |
| Arp4 | BAF53a (BRG1-associated factor) | Actin related |
| Esa1 | Tip60/Tip60b (Tat interactive protein) | Chromodomain/MYST HAT |
| Eaf3 | Mortality factor related genes (MORF4, MRG15/X) | Chromodomains |
| Act1 | Actin | |
| Yng2 | Inhibitor of growth gene family (ING1 to -5) | PHD finger |
| Eaf5 | ? | |
| Yaf9 | YEATS family (AF9, ENL, GAS41) | YEATS |
| Eaf6 | hEaf6 (FLJ11730) |
PI-3 kinase, phosphatidylinositol 3-kinase.
In the present study, we investigated the existence of NuA4 complexes in higher eukaryotes. Human Tip60 and its splice variant Tip60b/PLIP, which are both highly related to yeast Esa1 over its entire sequence, were used in affinity purification protocols from retrovirally transduced human cells. Western blot/fractionation analysis, immunoprecipitation data, and mass spectrometry analysis showed that both Tip60 variants are part of large multisubunit human NuA4 complexes with HAT activity similar to the yeast complex. We found that homologs of 11 yeast subunits are present in human NuA4 and confirmed their stable association by reciprocal tagging. In contrast, ruvB-like helicases and bromodomain-containing Brd8 are specific to the human complex even though homologous proteins are present in yeast. Chromodomain-containing MRG15 and a subset of other human NuA4 subunits, including DMAP1 (DNMT1-associated protein 1) are also present in distinct human complexes harboring different chromatin-related enzymatic activities. Furthermore, identification of ING3 as a subunit of human NuA4 supports a functional link between NuA4 and cell cycle control by tumor suppressor p53. Accordingly, we show that ING3 has strong growth inhibition activity and that Tip60 HAT activity cooperates with p53 for activation of the p21/WAF1 gene. Finally, we purified a core recombinant complex formed by Tip60, EPC1, and ING3 that is sufficient to enable strong HAT activity on nucleosomal templates. In parallel with studies in yeast, our data indicate that NuA4 is also a primary regulator of gene expression and cell cycle progression in human cells.
MATERIALS AND METHODS
Plasmids.
The Tip60 cDNA was a generous gift from G. Chinnadurai (28) and was subcloned by PCR into the KpnI and EcoRI sites of pcDNA3-FLAG. Tip60b was generated by deleting exon 5 of Tip60 by site-directed mutagenesis by using the ExSite kit (Stratagene). The Tip60 HAT-dead mutant was described previously (26). The MRG15 cDNA was amplified from a human liver cDNA bank, and the DMAP1 cDNA was amplified from a human kidney cDNA bank. Both were cloned into the BamHI and XhoI sites of pcDNA3-FLAG. The ING3 cDNA was amplified from a HeLa cDNA bank and subcloned into the BamHI site of pcDNA3-FLAG.
Retroviral vectors expressing Tip60 and Tip60b fused to an N-terminal FLAG epitope were constructed as follows. The pcDNA3-FLAG-Tip60/Tip60b constructs were digested with NcoI, blunted, and cut with XhoI and then ligated into the pRevTre vector (Clontech) digested with BamHI, blunted, and cut with SalI.
To generate the mammalian TAP-tag C-terminal fusion expression vectors, a BamHI/EcoRV fragment from pBS1479 (45) was subcloned into the BamHI/EcoRV sites of pcDNA3 to generate pcDNA3-TAP. The retroviral vector pRevTre-TAP was obtained by subcloning a BamHI/HindIII fragment from pBS1479 into the respective sites of pRevTre. The cDNAs of Tip60, MRG15, and DMAP1 fused to their N-terminal FLAG epitopes were amplified by PCR and subcloned into the BamHI site of pcDNA3-TAP and pRevTre-TAP. Details on the cloning procedure (e.g., primer sequences) are available upon request. For expression in bacteria, cDNAs were amplified from a HeLa cDNA bank and subcloned into the polycistronic vector according to a shuttle vector procedure described elsewhere (55).
Antibodies.
The anti-TRRAP (α-TRRAP; T-17), α-p33ING1 (C-19), α-actin (I-19), and α-p21 (C-19) antibodies were purchased from Santa Cruz Biotechnologies. The α-Tip60 and α-Sin3A antibodies were purchased from Upstate Biotechnology. The α-DMAP1, α-p53 (Ab-6), and α-FLAG M2 antibodies were purchased from Affinity Bioreagent, Oncogene, and Sigma, respectively. These antibodies were used at a dilution of 1:500, except for α-FLAG M2 and α-p53 (1:1,000). The α-EPC1, α-MRG15, α-BAF53a, α-Tip49a(RUVBL1), α-Tip49b(RUVBL2), α-HDAC2, and α-GAS41 antibodies were generous gifts from different labs (see Acknowledgments).
Retroviral infection of cell lines and triple-affinity protein purification.
MCF7 and HeLa S3 tet-off cell lines were purchased from Clontech and cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. 293T cells were cultured in the same medium. The retroviral packaging cell line PA317 was transfected with the different pRevTre expression vectors by using the calcium phosphate method, and the viral load obtained was used to infect HeLa and MCF7 cells as described by the manufacturer (Clontech). HeLa S3 and MCF7 cells were selected for 2 weeks with 500 and 250 μg of hygromycin B/ml, respectively, and in the presence of 1 μg of doxycyclin/ml. For suspension cultures, HeLa S3 cells were cultured in Joklik modified minimum essential medium supplemented with 10% fetal calf serum.
Nuclear extracts from HeLa S3 cells transduced with an N-terminal FLAG and C-terminal TAP Tip60/DMAP1 constructs were performed as described previously (62) and adjusted to 0.1% NP-40. Approximately 100 mg of nuclear extract was precleared with 250 μl of Sepharose CL-6B (Sigma) for 45 min at 4°C, and then 500 μl of immunoglobulin G (IgG) Sepharose beads (Amersham Bioscience) was added to the extract, followed by rotation for 2 h at 4°C. The beads were then washed with 10 column volumes of IgG-Sepharose wash buffer (20 mM HEPES [pH 7.9], 10% glycerol, 300 mM KCl, 0.1% NP-40, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 2 μg of pepstatin/ml, 2 μg of leupeptin/ml, and 5 μg of aprotinin/ml) and equilibrated with TEV cleavage buffer (20 mM HEPES [pH 7.9], 10% glycerol, 150 mM KCl, 0.1% NP-40, 0.5 mM EDTA, and 1 mM DTT). Bound proteins were eluted with 500 U of TEV protease (Invitrogen) in 1.25 ml of TEV cleavage buffer for 2 h at 16°C. The beads were washed once with 750 μl of TEV cleavage buffer, and this fraction was pooled with the eluate and incubated with 25 μl of protein A-Sepharose (Amersham Bioscience) for 15 min at 4°C to eliminate IgG antibodies leaking from the IgG-Sepharose resin. The eluate was then diluted four times in calmodulin binding buffer (20 mM HEPES [pH 7.9], 10% glycerol, 150 mM KCl, 0.1% NP-40, 1 mM imidazole, 1 mM magnesium acetate, 2 mM CaCl2, and 1 mM DTT supplemented with protease inhibitors) and 7.5 μl of 1 M CaCl2 was added, along with 500 μl of calmodulin resin (Stratagene), followed by rotation for 2 h at 4°C. The beads were then washed with 10 column volumes of calmodulin binding buffer. Proteins were eluted with calmodulin elution buffer (20 mM HEPES [pH 7.9], 10% glycerol, 150 mM KCl, 0.1% NP-40, 10 mM β-mercaptoethanol, 1 mM imidazole, 1 mM magnesium acetate, and 5 mM EGTA supplemented with protease inhibitors). Five fractions of 500 μl were collected and tested for HAT activity as described previously (2). Active fractions were pooled and then incubated with 100 μl of α-FLAG M2 resin (Sigma) overnight at 4°C. The beads were washed with FLAG elution buffer (20 mM HEPES [pH 7.9], 20% glycerol, 0.1% NP-40, and 150 mM KCl supplemented with protease inhibitors) and batch eluted five times with one column volume of FLAG elution buffer containing 400 μg of 3X FLAG peptide (Sigma)/ml. Active fractions were precipitated with 9 volumes of cold acetone-triethylamine-acetic acid (90:5:5), loaded on a 10% Tris-glycine gel, and silver stained. Preparative gels for tandem mass spectrometry were stained with Sypro Ruby (Bio-Rad). Protein identification by analysis of gel slice tryptic digests on a microcapillary reversed-phase high-pressure liquid chromatography nanoelectrospray tandem mass spectrometry on a Finnigan LCQ DECA XP Plus quadrupole ion trap mass spectrometer was performed as described before (14).
Transfections and immunoprecipitations.
Nuclear extracts from MCF7 cells transduced with N-terminal FLAG Tip60 and Tip60b constructs were performed as described previously (62) and diluted to 150 mM KCl with dilution buffer (20 mM HEPES [pH 7.9], 0.1% Tween 20, 10% glycerol, 5 mM MgCl2, 1 mM DTT supplemented with protease inhibitors). The nuclear extracts (∼40 mg) were then incubated with 250 μl of α-FLAG M2 resin (Sigma) for 4 h at 4°C. The beads were then washed three times with 300 mM KCl FLAG wash buffer (20 mM HEPES [pH 7.9], 0.1% Tween 20, 10% glycerol, 5 mM MgCl2, 1 mM DTT, and protease inhibitors), followed by two washes at 150 mM KCl. Bound proteins were eluted twice with 200 μg of FLAG peptide/ml in FLAG wash buffer 150 for 1 h at 4°C. The eluate was then loaded on a Superose 6HR (Pharmacia) gel filtration column at 350 mM KCl. The chromatography was done essentially as described previously (2) except that every parameter was scaled down 10 times and data were processed on a SMART system (Pharmacia).
293T cells were transfected with 15 μg of either pcDNA3-FLAG-ING3, pcDNA3-TAP-MRG15, or the empty pcDNA3 vector by the calcium phosphate method. At 48 h posttransfection (no stably integrated cell lines could be established for these two proteins), nuclear extracts were prepared, adjusted to 150 mM NaCl, and immunoprecipitated as follows. Nuclear extracts prepared from FLAG-ING3- or FLAG-MRG15-expressing cells were precleared with protein A-Sepharose (Amersham Bioscience) for 45 min and then incubated with α-FLAG M2 resin (Sigma) overnight at 4°C. The beads were then washed three times with binding buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 0.1% NP-40, 10% glycerol, and 1 mM DTT supplemented with protease inhibitors), and bound proteins were eluted in binding buffer supplemented with 400 μg of 3X FLAG peptide (Sigma)/ml for 4 h at 4°C in batch. For the TAP purification of transiently transfected MRG15-TAP, nuclear extracts were incubated with IgG-Sepharose beads (Amersham Bioscience) overnight at 4°C. The beads were then washed three times with IPP150 buffer (45), equilibrated with TEV cleavage buffer, and proteins were eluted with TEV protease (Invitrogen). The eluate was then incubated with calmodulin resin (Stratagene) for 4 h, followed by three washes with calmodulin binding buffer. Each fraction was then tested for HAT and HDAC activity and by Western blot as described previously (2, 60).
In Fig. 3B, Nuclear extracts prepared from HeLa S3 cells stably expressing Tip60-TAP and DMAP1-TAP were bound on IgG-Sepharose beads and then eluted with TEV protease as described for MRG15-TAP. HAT assays and Western blots were performed with the TEV eluate.
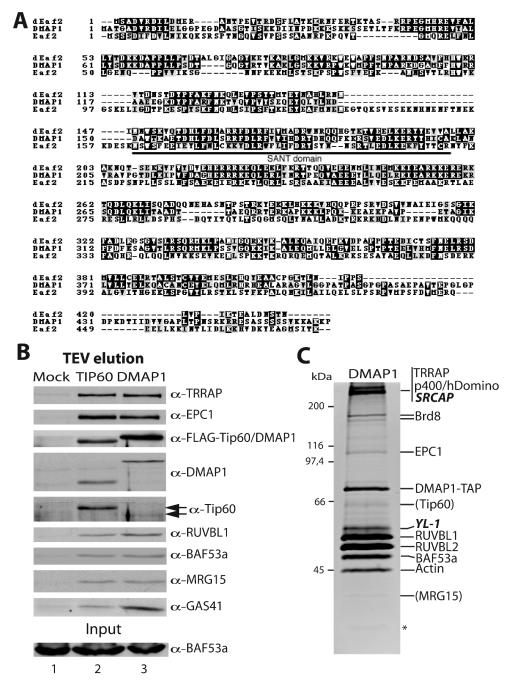
FIG. 3.
SANT domain-containing protein DMAP1 is a stable subunit of the human NuA4 complex. (A) Amino acid sequence alignment between yeast NuA4 subunit Eaf2, human DMAP1, and a Drosophila homologous protein (dEaf2, accession number AAF57436). The conserved SANT domain region is indicated. (B) DMAP1 associates with the same set of proteins as Tip60. Nuclear extracts from FLAG-Tip60-TAP, FLAG-DMAP1-TAP, or mock-transduced HeLa S3 cells were partially purified over IgG-Sepharose beads, and the TEV eluate was analyzed by Western blotting as in Fig. 1B. (C) Triple affinity purification of DMAP1 identifies distinct non-HAT complex(es). The DMAP1-containing complexes were purified as in Fig. 2 and analyzed on a gel by quantitative Sypro Ruby staining. Proteins identified by Western analysis or tandem mass spectrometry are labeled on the right. New proteins not identified in the Tip60 purification are in italics. The positions of MRG15 and Tip60 based on Western analysis of the previous fraction are given in parentheses. The asterisk indicates a nonspecific band. A significant amount of GAS41 protein was also detected but was run out of the gel presented here (see Western signal in panel B). The numbers of specific peptide sequences obtained by tandem mass spectrometry analysis performed as described for Fig. 2 are as follows: 6 for TRRAP, 8 for p400/hDomino, 7 for SRCAP (2,971 aa), 4 for Brd8, 13 for DMAP1, 4 for YL-1 (364 aa), 10 for RUVBL1, 14 for RUVBL2, 5 for BAF53a, and 5 for actin.
Colony formation assay.
NIH 3T3 cells were transfected with 15 μg of pcDNA3-FLAG vector alone or carrying ING3 cDNA by the calcium phosphate method. At 48 h posttransfection, the cells were diluted serially into Dulbecco modified Eagle medium supplemented with 10% calf serum plus 800 μg of G418/ml. After 2 weeks of selection, resistant colonies were fixed, stained with Giemsa, and counted. This experiment was repeated three times, with the same results. Analysis of ING3 expression by Western blot was performed on nuclear extracts prepared from an aliquot of the initially transfected cells, as well as from a pool of G418-resistant cells.
RT-PCR.
To determine endogenous levels of p21/WAF1, MDM2, and GADD45 mRNAs, 293T cells were transfected with different combinations of cytomegalovirus (CMV)-driven p53 expression vector (1 μg) and Tip60 constructs (pcDNA3) (19 μg) by the calcium phosphate method. At 24 h posttransfection, cells were irradiated with 15 Gy using a 60Co source and lysed 5 h later, and total RNA was extracted by using Trizol reagent (Invitrogen). After reverse transcription (RT), PCR amplification of the target genes was done with the specific sets of primers (sequence available on request). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and p53 mRNAs were used as internal controls. MDM2, p53, and GAPDH were amplified for 21 cycles; p21/WAF1 and GADD45 were amplified for 23 cycles.
Recombinant Piccolo NuA4.
For analysis of the minimal domain requirement for nucleosomal HAT activity, full-length Tip60, Tip60b, ING3, ING3(1-300), and EPC1(1-400) proteins were expressed in Escherichia coli by using modified T7-based expression vectors. These polycistronic expression vectors allow coexpression of multiple proteins in bacteria and purification of stable complexes (55). Proteins and complexes were partially purified over Talon cobalt affinity resin via engineered hexahistidine tags at the C terminus of Tip60(b). HAT assays were performed by using long oligonucleosomes prepared from HeLa cells and normalized for the amount of Tip60/Tip60b. To confirm stable association, the core (Piccolo) hNuA4 HAT complex was purified from E. coli extracts by using a polycistronic expression vector that coexpresses EPC1(1-400) with a C-terminal hexahistidine tag, ING3(1-300) with a C-terminal Strep-II tag and untagged full-length Tip60b. Soluble extracts prepared from cells induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) were purified successively by Talon cobalt metal affinity chromatography (Clontech) and Strep-Tactin affinity chromatography (IBA GmbH).
RESULTS
Human Tip60 and its splice variant Tip60b/PLIP are homologous to yeast Esa1 over the entire length of the protein.
In search of what could be functionally equivalent to yeast NuA4 in human cells we used the yeast catalytic HAT subunit in sequence homology search. The closest Esa1 homolog is Tip60, a human protein with similar HAT activity preference for histone H4 N termini as a recombinant protein (63). Tip60 has been functionally linked to activation of a number of genes by transcription activators, namely, steroid nuclear receptors and NF-κB, and to the cellular response to DNA damage and apoptosis (reviewed in references 10 and 56). Tip60 has been purified as a multisubunit complex harboring HAT activity toward histone H4 and H2A in chromatin (26). The gene coding for Tip60 also produces a splicing variant, Tip60b or PLIP, which has been linked to cPLA2 signaling (51). Both proteins are homologous to Esa1 over the entire length of the yeast protein, spanning over the MYST and chromo domains (Fig. 1A). Interestingly, the 52 amino acids encoded by exon 5 and missing in Tip60b are also missing in the yeast Esa1 protein. This sequence is often responsible for the detection of Tip60 in numerous two-hybrid screens with heterologous proteins (56). In an effort to study the relationship between Tip60(b) and yeast Esa1, we produced human cell lines transduced by retrovirus encoding tagged-version of Tip60 and Tip60b under the control of a Tet-regulated CMV promoter. This approach allowed us to obtain cells producing near physiological levels of both tagged proteins, decreasing the risk of natural protein complex disruption by overproduction of exogenous proteins.
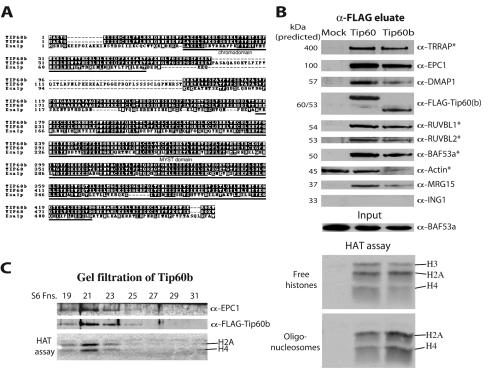
FIG. 1.
Tip60 and Tip60b are associated with homologs of yeast NuA4 complex subunits. (A) Amino acid sequence alignment between the yeast Esa1, human Tip60, and Tip60b/PLIP histone acetyltransferases. The chromodomain and MYST HAT family homology regions are indicated. (B) Nuclear extracts from mock, FLAG-Tip60, and FLAG-Tip60b-transduced MCF7 cells were immunoprecipitated with α-FLAG resin, eluted with FLAG peptide, and analyzed by Western blotting with antibodies against protein homologous to yeast NuA4 subunits. Native FLAG eluates were also tested in HAT assays on free histones and chromatin substrates. Specific association of actin with Tip60(b) cannot be concluded from this experiment since it is also detected in the control lane. Proteins previously shown to be associated with Tip60 are marked with an asterisk (26). (C) Tip60b is part of a large HAT protein complex. The Tip60b FLAG eluate from panel B was purified over a calibrated Superose 6HR gel filtration column, and fractions were analyzed by Western blotting and HAT assay. Tip60b, EPC1, and H4/H2A HAT activity coelute as a single high-molecular-mass complex of ∼1.8 MDa. Molecular mass standards eluted as follows: void volume, fraction 19; 670 kDa, fraction 28; 158 kDa, fraction 32; 44 kDa, fraction 35; and 17 kDa, fraction 37.
Tip60 and Tip60b immunoprecipitate from nuclear extracts as large nucleosomal HAT complexes containing several homologs of yeast NuA4 subunits.
We prepared nuclear extracts from MCF7 transduced cell lines and performed immunoprecipitations with FLAG antibodies, followed by peptide elution in native conditions. Western analysis confirmed the previously reported association of Tip60 with TRRAP, an ATM-related cofactor required for c-Myc and E2F transcription/cell transformation potential, BAF53a, an actin related protein also required for cell transformation by c-Myc and RUVBL1/2, two helicases highly related to Holliday junction movement responsible helicases in bacteria, also required for c-Myc function (26, 33, 44, 61) (Fig. 1B). This experiment also identified three new proteins interacting with Tip60 in vivo that are also homologs of yeast NuA4 subunits (Table 1). EPC1 is homologous to yeast Epl1 and has been linked to transcription regulation and to the oncogenic function of the RET finger protein (52). MRG15 contains two chromodomains, is homologous to yeast Eaf3, and has been implicated in c-Myb gene expression and cellular senescence when mutated (5, 14, 31). DMAP1 contains a SANT domain, is homologous to the essential yeast protein Eaf2 (Esa1-associated factor 2) and has been found associated with DNA methyltransferase-1 and replication foci in vivo (48; A. Auger, D. Cronier, L. Galarneau, A. Nourani, R. Utley, and J. Côté, unpublished data). HAT assays on free histones and oligonucleosomes with the eluted fractions showed specificity very similar to what is obtained with yeast NuA4, i.e., H3, H2A, and H4 on free histones but only H2A and H4 on chromatin (Fig. 1B). Gel filtration analysis of these fractions also showed that Tip60(b)-associated proteins coelute with the HAT activity as a single large protein complex (>1 MDa; Fig. 1C). Importantly, the results obtained with Tip60b are indistinguishable from those of Tip60 (Fig. 1B to C; data not shown). This suggests that both Tip60 isoforms are part of very similar if not identical HAT complexes.
Triple-affinity purification of Tip60 identifies multiple stably associated proteins homologous to yeast NuA4 subunits.
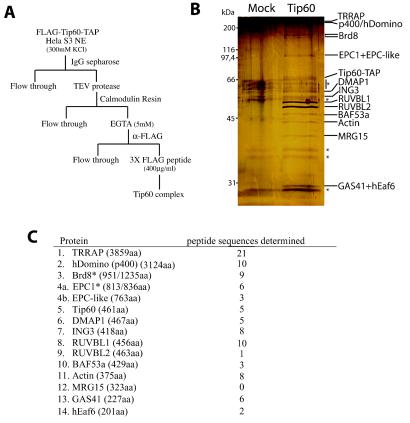
To fully characterize the Tip60-containing HAT complex, we produced transduced HeLa cell line expressing a triple-tagged version of Tip60. The protein harbors a FLAG epitope at its N terminus and a tandem affinity purification (TAP [45]) tag at its C terminus. The TAP tag allows two-step affinity purification by binding to IgG-Sepharose, TEV protease native elution, binding to calmodulin resin, followed by EGTA elution. Finally, the EGTA eluate is bound to anti-FLAG resin followed by peptide elution (Fig. 2A). This protocol allowed us to obtain near homogenous material that could be analyzed by silver staining and tandem mass spectrometry (Fig. 2B). The amount of purified material obtained was relatively small presumably because the Tip60 protein is highly regulated through MDM2-dependent targeting to the proteasome (29). Tip60-associated proteins identified by Western analysis and/or mass spectrometry are labeled on the right of the gel in Fig. 2B and listed in Fig. 2C (an example of Western analysis is shown in Fig. 3B, lane 2). These data demonstrate that all previously reported and new proteins identified in Fig. 1 were confirmed as stable Tip60-associated molecules. A splice variant, as well as a paralog of EPC1, were found, suggesting that this key subunit could create distinct functional complexes, e.g., in different cell types or during development (Fig. 2C). New proteins identified by mass spectrometry include GAS41, a protein related to AF9/ENL leukemogenic factors, amplified in early glioma, and essential for cell viability (16, 65) (Fig. 2C). GAS41 is homologous over the entire length of the protein to yeast Yaf9, a stable subunit of the NuA4 complex in yeast (Table 1) (30; H. Zhang, D. O. Richardson, R. T. Utley, Y. Doyon, J. Côté, and B. R. Cairns, unpublished data). ING3 is a member of the inhibitor of growth (ING) family of growth regulators which encompass the tumor suppressor ING1 and are linked to p53 function in transcription and apoptosis (15). ING3 itself is a candidate tumor suppressor that modulates p53-mediated transcription and apoptosis (23, 38). ING3 is homologous to yeast NuA4 subunit Yng2, one of the three ING proteins in yeast, which is required for p53-dependent transcription in yeast through specific recruitment of NuA4 activity (41, 42; see below). Finally, an uncharacterized 201-amino-acid protein (FLJ11730) was identified in the purified complex which shows clear homology to the yeast NuA4 subunit Eaf6 (Esa1-associated factor-6; N. Lacoste, S. Allard, and J. Côté, unpublished data). Since nine of the proteins that copurified with Tip60 are homologous to subunits of the yeast NuA4 HAT complex, this strongly suggests that the complex we have purified is the human counterpart of the yeast complex.
FIG. 2.
Purification of the Tip60-associated proteins identifies the human NuA4 HAT complex. (A) Scheme of the protocol for triple-affinity (using TAP [protein A/calmodulin binding peptide] and FLAG tags) purification of the Tip60-containing complex. (B) Silver-stained gel of mock-treated and triple-affinity-purified material. The Tip60-containing complex was purified from N-terminal FLAG- and C-terminal TAP-tagged Tip60-transduced HeLa cells and then analyzed by silver staining. An extract from nontransduced cells was used as control. Specific bands not present in the control and identified by mass spectrometry and/or Western analysis are labeled on the right. Nonspecific bands are labeled with asterisks. (C) Tip60-associated proteins were identified by tandem mass spectrometry. Tryptic digestion of gel slices from a sample as in panel B were analyzed by tandem mass spectrometry. The proteins identified are listed with the number of different peptide sequences detected for each of them (their lengths in amino acids are indicated in parentheses). Each protein is homologous to a yeast NuA4 subunit, with the exception of Brd8 and RUVBL1/2. MRG15, did not produce peptide hits but was identified by Western analysis. Proteins with asterisks indicate the detection of protein products from splice variants. One mass spectrometry hit was specific for the shorter isoforms of both Brd8 (also known as p120) and EPC1 (also known as EPC2). p400 is named hDomino because of the close homology with mouse and Drosophila Domino proteins (43). Identified human protein FLJ11730 is named hEaf6 because of the homology with the yeast NuA4 subunit Eaf6. The accession number for the EPC1 paralog named EPC-like is NP_056445.
ruvB-like helicases and a bromodomain-containing protein are specific to the human complex but have evolutionary conserved links with yeast NuA4.
The identification of ruvB-like helicases in the initial purification report on Tip60 laid doubts on the functional equivalence to Esa1 as the yeast NuA4 complex does not contain such subunits (26). The subsequent identification of another associated factor, SWI2/SNF2-like ATPase-containing p400/hDomino, added to these doubts since yeast NuA4 also does not contain this protein domain (19; Allard et al., unpublished). Here we confirmed the presence of both ruvB-like helicases and p400/hDomino in the human complex (Fig. 2). As previously reported, RUVBL1/2 are overstoichiometric subunits compared to Tip60 presumably because of their hexameric ring structure within the complex (26). p400/hDomino has been functionally and physically associated to cellular transformation by oncoprotein E1A and transcription regulation (19). In Drosophila the domino gene has been characterized as an enhancer of polycomb mutations and a weak suppressor of position-effect variegation (49). Upon closer analysis of p400/hDomino amino acid sequence we realized that, aside from the central SWI2-related domain (amino acid [aa] 1051 to 1625), three additional domains are present which are also found in the yeast NuA4 subunit Eaf1 (Esa1-associated factor 1, also named Vid21 in databases). These include the HSA (helicase/SANT-associated [13], aa 764 to 835), SANT (1) (aa 2328 to 2395), and C-terminal Q-rich domains and are similarly arranged (N to C termini) in Eaf1. A highly charged region just downstream of the HSA domain is also found in both Eaf1 and p400/hDomino (data not shown; Auger et al., unpublished). These homology regions suggest that p400/hDomino could be the human functional homolog of yeast Eaf1 in NuA4, bringing the number of conserved subunits to 11 of 12. Strikingly, the SWI2-related domain specific to the human protein is found to be responsible for association of the ruvB-like helicases (19). Thus, the absence of such domain in yeast NuA4 also explains the lack of ruvB-like helicases in the yeast complex. Interestingly, we recently characterized a yeast protein complex sharing 4 subunits with NuA4 and containing Swr1, a SWI2-like ATPase related to the one present in p400/hDomino, and Rvb1/2, two ruvB-like helicases (Auger et al., unpublished).
The presence of Brd8, a bromodomain-containing factor, is also specific to the human NuA4 complex. This gene produces at least two splicing isoforms, the longer (1,235 aa) harboring two bromodomains (aa 705 to 813 and aa 1101 to 1209) and the shorter (951 aa) containing only one. These two variants are most likely present in hNuA4 since the Brd8 mass spectrometry hits correspond to a protein doublet ∼150 kDa and include a peptide specific to the shorter form (data not shown). The shorter isoform, also named p120, was shown to interact with the thyroid and 9-cis-retinoic acid receptors (TR and RXR) and coactivate both in a ligand-dependent fashion (34, 35). Interestingly, the larger double-bromodomain isoform has regions homologous to the double bromodomain-containing protein Bdf1 in yeast. Bdf1 has been shown to preferentially bind hyperacetylated histone H4 through its bromodomains, and its deletion is lethal when combined with a mutant allele of ESA1 (32). The presence of Brd8 in human NuA4 complexes could reflect the Bdf1-Esa1 functional interaction in yeast.
SANT-domain protein DMAP1 links NuA4 to DNA replication and is also present in distinct protein complexes with SWI2-related ATPase activities.
To confirm the stable association of the identified proteins within a human NuA4 complex, we produced transduced cell lines expressing a TAP-tagged version of DMAP1 (Fig. 3). DMAP1 is homologous to yeast Eaf2 (also named God1 in databases) over the entire length of the protein (Fig. 3A) and provides a connection between the NuA4 HAT and the process of DNA replication. DMAP1 was shown to interact with DNA methyltransferase 1 (DNMT1) and associates with DNA replication foci in vivo in a DNMT1-dependent manner (48). When nuclear extract from TAP-tagged DMAP1-expressing cells was fractionated over IgG-Sepharose and analyzed by Western blots, the same set of copurifying polypeptides was identified as for Tip60 (Fig. 3B). The fraction was also tested for HAT activity and demonstrated specificity identical to the Tip60 fraction (data not shown). On the other hand, the Western signal for Tip60 in the TAP-DMAP1 fraction is very weak, suggesting a heterogeneous population of DMAP1 complexes (Fig. 3B, compare lanes 2 and 3). This idea was confirmed upon triple affinity purification of DMAP1-associated proteins, followed by tandem mass spectrometry analysis. Figure 3C shows a protein gel of the purified material stained with quantitative Sypro Ruby red dye. While the pattern of bands appeared similar to the purified Tip60-TAP complex, the relative amounts varied greatly. Though DMAP1-TAP purification yielded significantly more material, much lower specific activity was observed in the HAT assay (data not shown). Furthermore, Tip60 and MRG15 proteins are underrepresented such that they were only detectable in previous fractions. When the purified material was fractionated by gel filtration as in Fig. 1C a larger elution profile of EPC1 was observed, whereas DMAP1 signal peaked at slightly smaller size, arguing for more than one protein complex (data not shown). This was confirmed by the identification of two new proteins that were not found in the Tip60-TAP purified material. SRCAP (Snf2-related CBP activator protein) is another large protein (2,971 aa) and contains a SWI2-related ATPase domain very close to the one present in p400. However, SRCAP does not contain HSA and SANT domains. SRCAP has been found previously to bind and cooperate with the CREB-binding protein (CBP) to activate transcription (27, 36). YL-1 is a nuclear protein that is able to act as a specific cell transformation suppressor when overexpressed (25). Altogether these data indicate that DMAP1 is present in the NuA4 complex and at least one other distinct non-HAT complex with SWI2-related ATPase activity. The distinct complex(es) contain(s) other subunits shared with NuA4, including TRRAP, p400/hDomino, Brd8, RUVBL1/2, BAF53a, actin, and GAS41. This is in agreement with published work on p400/hDomino that identified a TRRAP/p400 complex lacking Tip60 and HAT activity (19). The function of these distinct complexes containing SWI2-related ATPase activities remains to be investigated.
Chromodomain protein MRG15 implicates NuA4 in cell proliferation control and is also present in an HDAC complex.
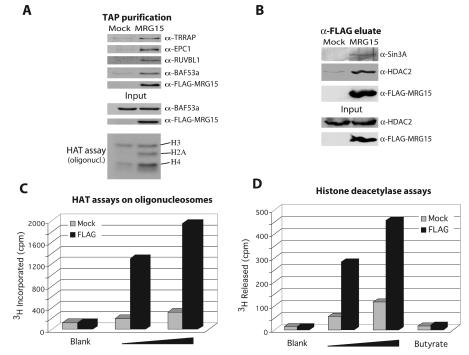
MRG15 is highly related to yeast Eaf3 and again links the NuA4 complex to the control of cell proliferation (14, 31). Its N-terminal chromodomain is truncated in a cell senescence-inducing mutant (MORF4; see reference 5). Fractionation of MRG15-TAP extract over IgG-Sepharose and calmodulin resins also confirms the stable association of MRG15 within the human NuA4 complex since the same set of proteins and HAT activity were observed (Fig. 4A and C). On the other hand, the activity was quite low which could be explained by the fact that MRG15 yeast homologs (Eaf3 and Schizosaccharomyces pombe Alp13) have also been found as part of Sin3/HDAC complexes (22, 39; Auger et al., unpublished). MRG15 itself has been found to interact with mSin3A and implicated in transcription repression (64). We confirmed that human MRG15 is also a subunit of a Sin3/HDAC complex by Western blot, immunoprecipitation, and HDAC assays (Fig. 4B and D). Immunoprecipitation of MRG15 from nuclear extract brings down Sin3A and HDAC2 (Fig. 4B), and the pelleted material contains significant amount of sodium butyrate-sensitive HDAC activity (Fig. 4D). These results, along with the literature, indicate that MRG15 is a stable subunit of both HAT and HDAC complexes, suggesting a common specific role in regulating interaction with chromatin, most likely through its chromodomains.
FIG. 4.
The chromodomain-containing MRG15 protein is present in both hNuA4 HAT and Sin3A/HDAC complexes. (A) MRG15 copurifies with specific subunits of the hNuA4 complex and nucleosomal H4/H2A HAT activity. Nuclear extracts from 293T cells transiently transfected with MRG15-TAP or mock transfected with pcDNA3-TAP were sequentially purified over IgG-Sepharose and calmodulin beads, analyzed by Western blotting, and nucleosomal HAT assay. Some nonspecific HAT activity was detected in the mock purification of this experiment, presumably due to high-copy transfected plasmid, but specific H4/H2A HAT activity is clearly seen in the tagged MRG15 fraction. (B) MRG15 coimmunoprecipitates with a Sin3A/HDAC complex. Nuclear extracts from 293T cells transiently transfected with FLAG-MRG15 or mock transfected with pcDNA3-FLAG were immunoprecipitated with α-FLAG resin, eluted with FLAG peptides, and analyzed by Western blot. (C) Increasing amounts of the native FLAG eluates from the experiment in panel B (1 and 2 μl) were tested for HAT activity with oligonucleosomes as the substrate. (D) The same eluates were also tested for HDAC activity by using in vivo-labeled purified free core histones. The specificity of the detected activity was verified by incubation with the HDAC inhibitor sodium butyrate (25 mM).
ING3 confirms NuA4 function in p53-dependent transcription and cell cycle control.
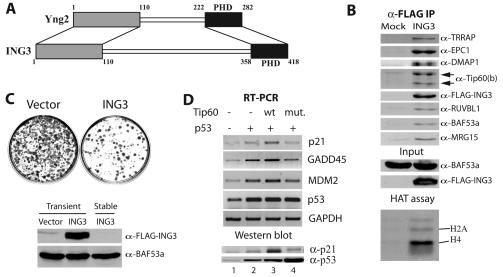
During our preliminary sequence analysis of potential human homologs of yeast Yng2, we did not suspect ING3 as the human functional counterpart. This was due to the fact that ING3 is larger than the other ING family members and contains a central serine-rich region with no obvious homology. Upon reexamination, the first and last 100 aa of ING3 are in fact the best human match for the equivalent regions in Yng2 (Fig. 5A). The C-terminal region contains the PHD finger domain, while the N-terminal region is responsible for anchoring Yng2 in the yeast NuA4 complex, an association required for Esa1-dependent acetylation of chromatin in vitro and in vivo (7, 41). Again, fractionation of cells expressing an epitope-tagged version of ING3 identified the same set of copurifying polypeptides and HAT activity compared to tagged-Tip60 (Fig. 5B). Importantly, Tip60b was also detected, further confirming that both Tip60 isoforms are part of similar if not identical complexes.
FIG. 5.
The PHD finger protein ING3 implicates human NuA4 in p53-dependent transcription activation and growth control. (A) Diagram of homology regions between yeast Yng2 and human ING3 (>30% identity, 48% similarity). (B) ING3 coimmunoprecipitates with human NuA4 subunits and histone H4/H2A HAT activity. Nuclear extracts from 293T cells transiently transfected with FLAG-ING3 or mock transfected with pcDNA3-FLAG were immunoprecipitated with α-FLAG resin, eluted with FLAG peptides, and analyzed by Western blot and HAT assay. Note that both Tip60 HAT isoforms are detected. (C) Chronic overexpression of ING3 suppresses cell growth. NIH 3T3 cells were transfected with FLAG-ING3 or pcDNA3-FLAG, diluted serially, and selected for 2 weeks with G418. Resistant colonies were stained with Giemsa and analyzed for ING3 expression by Western blot on protein extracts. G418-resistant clones have lost ING3 expression (BAF53a signal is used as internal control). (D) Tip60 stimulates p53-dependent transcription after gamma irradiation. RT-PCR analysis of the endogenous p21/WAF1, MDM2, and GADD45 mRNAs in 293T cells transfected with different combinations of CMV-driven p53, wild-type Tip60 (wt) and Tip60 HAT-dead mutant (mut.) expression plasmids. All cells were gamma irradiated with 15 Gy. p53 and GAPDH mRNA signals are used as internal controls. Protein extracts were made in the same conditions and analyzed by Western blotting with α-p21 and α-p53 antibodies to show the significant increase of p21/WAF1 protein in the presence of wild-type Tip60 but not with the mutant, even though p53 was highly expressed (lower panel).
Since ING3 is part of a family of growth regulator proteins, we tested this activity by colony formation assay after transfection and selection for 2 weeks. Figure 5C shows representative Giemsa staining of petri dishes which clearly demonstrates that ING3 has strong growth suppressor activity (<20% of colony formation compared to empty vector). Western analysis of the limited viable clones obtained suggested that they lost expression of the ING3 transgene (Fig. 5C, lower panel). Tumor suppressor ING1 has been suggested to interact with p53 in vivo and regulate its transcription potential (21). ING2 was also shown to affect p53-dependent transcription by modulation of its acetylation level (37). In a recent independent study, overexpression of ING3 was shown to inhibit growth in a p53-dependent fashion and stimulate p53-dependent transcription (38). We investigated if ING3's role in p53 function is through its association with Tip60 in human NuA4. It is important to note that Tip60 is unable to directly acetylate p53 in vitro (data not shown). We transfected cells with p53 in the absence or presence of Tip60 or Tip60 HAT-dead mutant (26), irradiated them with 15 Gy, and analyzed by RT-PCR the transcription of the endogenous p53-regulated p21/WAF1, GADD45, and MDM2 genes (Fig. 5D). In these conditions p53 transactivates the p21/WAF1 gene as part of the process of DNA repair checkpoint. Transfection of p53 alone was found to stimulate p21/WAF1, GADD45, and MDM2 gene transcription by 1.4-, 2.7-, and 3.7-fold, respectively (lane 2). In the presence of exogenous Tip60, p21/WAF1, GADD45, and MDM2 transcription was further stimulated by 1.9-, 1.6-, and 1.5-fold, respectively, arguing that Tip60 works as a coactivator of p53-driven transcription (lane 3). When the HAT mutant Tip60 protein was expressed, this stimulation was lost, and the level of p21/WAF1, GADD45, and MDM2 transcription dropped to levels just slightly lower than with p53 alone, suggesting a small dominant-negative effect (98, 79, and 88% of p53 alone signal, respectively; Fig. 5D, lane 4). These variations are relatively small but have significant effects since Western analysis of the p21 protein in the same conditions shows a clear increase of protein level in the presence of Tip60 but not with the mutant form (Fig. 5D, lower panel). Importantly, the Tip60 protein itself, like p53, is highly and directly regulated by the MDM2 ubiquitin ligase (29), which could explain in part the relatively small effects detected by RT-PCR. These data indicate that the human NuA4 complex plays a role in p53-dependent transcription in vivo, in agreement with the presence of the ING3 subunit and previous reports implicating TRRAP-containing HAT complexes (3, 4). This is also consistent with our study in the yeast system where we showed direct interaction between NuA4 and p53 activation domain that allows recruitment to promoter regions, localized histone H4 hyperacetylation, and transcription activation (42).
A recombinant trimeric complex formed by Tip60, EPC1, and ING3 is sufficient to enable strong HAT activity toward chromatin substrates.
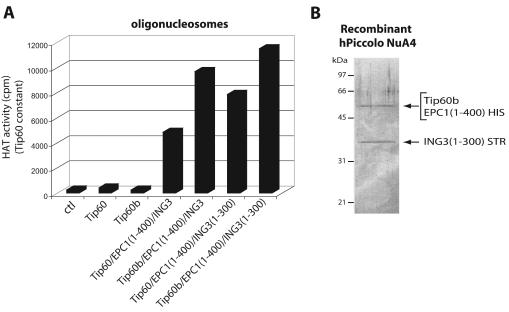
During our studies on the yeast NuA4 complex, we found that nucleosomal HAT activity depends on a core complex of Esa1, Epl1, and Yng2 named Piccolo NuA4 (picNuA4). Native picNuA4 complex was also found in cell extracts and could be responsible for global nontargeted acetylation of chromatin in vivo (7). The first 380 aa of Epl1 physically bridge together Esa1 and Yng2, where Yng2 is required for robust acetylation of chromatin substrates (Esa1 alone can only acetylate free histones) (7; W. Selleck, J. Côté, and S. Tan, unpublished data). To investigate the structural and enzymatic relationship between yeast and human NuA4 complexes, we used a polycistronic bacterial expression vector (55) to coproduce recombinant forms of 6XHIS-tagged Tip60 or Tip60b, nontagged EPC1(1-400), and ING3. After single-step affinity purification, we used these recombinant proteins in HAT assays with oligonucleosomes (Fig. 6A). As previously shown (26), Tip60 by itself is unable to acetylate chromatin substrates in vitro, and the same can now be said for Tip60b. On the other hand, when these are coproduced with EPC1(1-400) and ING3, potent nucleosomal acetyltransferase activities are detected (Fig. 6A). Analogous to what has been found with the yeast complex, the ING3 PHD finger region is not required for nucleosome acetyltransferase activity [ING3(1-300) construct]. To confirm that a stable trimeric complex was formed, we coexpressed in bacteria His6-tagged EPC1(1-400), STR-tagged ING3(1-300), and untagged Tip60b in bacteria. After successive affinity purification with both HIS and STR tags, purified material was analyzed on gel (Fig. 6B). The presence of all three proteins was confirmed by Coomassie blue staining, Western blot, and HAT assay (data not shown). These data demonstrate that Piccolo NuA4 structure and activity are also conserved from yeast to humans. Thus, the nucleosomal HAT activity associated with human Tip60 proteins is based on the same structural and enzymatic features as the yeast Esa1 protein, with a primary role for EPC and ING subunits in the NuA4 complexes.
FIG. 6.
Conservation of the core nucleosomal acetyltransferase trimeric complex, Piccolo NuA4, from yeast to humans. (A) Coexpression of EPC1 and ING3 with His-tagged Tip60 and Tip60b in bacteria enable their HAT activity on chromatin substrates. Tip60(b) were affinity purified from bacteria expressing each protein alone or in conjunction with EPC1 conserved N-terminal domain [EPC1(1-400)] and ING3 with or without its C-terminal PHD finger [ING3(1-300)]. Relative nucleosomal HAT activity of affinity-purified HIS-tagged Tip60 and Tip60b were measured by liquid scintillation counting of HAT assays on oligonucleosomes while the amount of Tip60(b) was kept constant (evaluated by Western blot). Note that Tip60 and Tip60b are not able to acetylate chromatin by themselves and that ING3 PHD finger is not required for nucleosomal HAT activity. Average counts per minute (cpm) from four different HAT assays are shown (ctl, 276 ± 74; Tip60, 425 ± 162; Tip60b, 273 ± 25; Tip60/EPC1(1-400)/ING3, 4,872 ± 271; Tip60b/EPC1(1-400)/ING3, 9,714 ± 531; Tip60/EPC1(1-400)/ING3(1-300), 7,876 ± 873; Tip60b/EPC1(1-400)/ING3(1-300), 11,522 ± 1,417). (B) Tip60, EPC1 and ING3 form a stable trimeric complex when coexpressed in bacteria. Protein extract from bacteria coexpressing EPC1(1-400) with a C-terminal His6 tag, ING3 (1-300) with a C-terminal Strep-II tag and untagged full-length Tip60b was successively affinity purified on Talon cobalt and Strep-Tactin resins and analyzed on gel after Coomassie blue staining. The presence of all three proteins was confirmed by Western blot and HAT activity (not shown).
DISCUSSION
Collectively, the data presented here using three different human cell types clearly demonstrate the presence of NuA4 HAT complexes in higher eukaryotes. The subunit identifications indicate a strikingly high level of structural conservation, i.e., 11 of 12 subunits conserved from yeast to humans. We also showed that chromatin modification by the complex functions through a highly conserved enzymatic core complex, i.e., Piccolo NuA4. Finally, we presented evidence supporting a role for the human NuA4 complex in the control of cell proliferation in part by cooperation with p53 in transcription regulation of cell cycle control genes. Interestingly, the previously proposed role of Tip60 in gamma irradiation induced DNA damage response is also conserved since Esa1 is required for DNA double-strand break repair in yeast (6, 26).
Human NuA4 subunit identifications highlighted multiple protein domains that have been linked to chromatin function, all encompassed in the same multiprotein complex. Besides the MYST HAT domain of Tip60, hNuA4 contains chromodomains that have been proposed to be methylated-histone or RNA-binding modules, bromodomains that can be acetylated-histone binding modules (reviewed in reference 17), SANT domains that have been proposed to bind histone tails (8, 54), an ATPase domain of the SWI2 family of chromatin remodelers (58), an actin-related globular domain that may have histone chaperone activity (20, 50), a PHD finger domain which is commonly found in chromatin modifying complexes and a phosphatidylinositol 3-kinase domain related to ATM/ATR, which phosphorylate histone H2A.X in response to DNA damage (46). The presence of the essential GAS41 protein, a member of the AF9/ENL-related (YEATS) family, also confirmed the exclusive association of these proteins to transcription/chromatin-modifying complexes, including yeast NuA4, NuA3, Sas2, SWI/SNF, TFIID/mediator/TFIIF, and human SWI/SNF complexes (40, 56). We showed that the Enhancer of Polycomb homology domain of human EPC1, like Epl1 in yeast (7), is a conserved functional key for histone acetylation since it bridges the MYST HAT with the ING protein to enable potent nucleosome histone acetyltransferase activity (Fig. 6). Interestingly, a splice variant and a distinct protein highly related to EPC1 were also found in the human NuA4 complexes, suggesting the possibility of functionally different complexes within the same cell or specific to cell types, development stages.
Identification of a bromodomain-containing subunit in human NuA4 distinguished it from its yeast counterpart. Brd8 links NuA4 to ligand-dependent transcription regulation by the thyroid hormone receptor (35) and could be implicated in local retention on chromatin after initial recruitment of NuA4 (24). Interestingly, Brd8 is related to yeast Bdf1, which has been shown to functionally interact with Esa1 and preferentially bind acetylated histone H4 (32). Genetic interactions have also been detected between yeast NuA4 and other bromodomain-containing proteins (N. Bouchard and J. Côté, unpublished). Another protein that initially appeared specific to the human complex is p400/hDomino. We propose that in fact yeast NuA4 subunit Eaf1 is the functional homolog of p400/hDomino since they share four regions of homology, including SANT and HSA domains (A. Auger, D. Cronier, L. Galarneau, A. Nourani, R. T. Utley, and J. Côté, unpublished data). However, Eaf1 lacks the SWI2-related ATPase domain of p400/hDomino. Interestingly, the remaining proteins specific to the human NuA4 complex are the ruvB-like helicases (RUVBL1/2), which are likely implicated in the DNA repair function of the complex. We speculate that their absence in the yeast complex is explained by the lack of the SWI2 domain in Eaf1 since they were shown to depend on this domain for interaction with p400/hDomino (19). In fact, we recently characterized a separate non-HAT yeast complex that shares four subunits with NuA4 and contains a SWI2-related protein, Swr1, and ruvB-like helicases, Rvb1/2 (A. Auger, D. Cronier, L. Galarneau, A. Nourani, R. T. Utley, and J. Côté, unpublished data). This raises the possibility that the human NuA4 complex is functionally equivalent to a fusion of two distinct complexes in yeast, one harboring HAT activity and the other involved in ATP-dependent chromatin remodeling.
Our finding of DMAP1 in human NuA4 implicates the complex in DNA replication since this protein has been found associated to the major DNA methyltransferase at replication foci (48). On the other hand, we did not detect DNMT1 signals in our NuA4 purification (data not shown). Thus, the interaction could be transient or DMAP1 may have other roles outside of NuA4. Indeed, we found that DMAP1 is also present in one or more distinct protein complexes (Fig. 3C). The complex or complexes lack HAT activity but contain ruvB-like helicases and SWI2-related ATPase subunits. p400/hDomino was previously shown to be part of a non-HAT complex containing other proteins that are also found with Tip60 (19). We now show that one of these proteins is DMAP1. Another SWI2-related ATPase was also identified in DMAP1 purification, SRCAP. This large protein is related to p400/hDomino over the SWI2 domain but lacks the SANT and HSA domains. It was shown to be involved in transcription regulation by CBP (27, 36).
The presence of MRG15 in human NuA4 confirms its importance in the control of cell proliferation. Mortality factor-4 is a truncated version of MRG15 lacking the N-terminal chromodomain that induces senescence in a number of cell lines (5). We show that, like homologs in lower eukaryotes (14, 39), MRG15 is present in both NuA4 HAT and Sin3/HDAC complexes. This certainly reflects its key role in the interaction of these complexes with chromatin, most likely through the chromodomain. Implication of human NuA4 in the control of cell division was recently highlighted by a study showing recruitment of Tip60 and p400/hDomino to promoter regions by Myc in vivo (18). Our identification of candidate tumor suppressor ING3 further supports an important role for NuA4 in cell cycle control. Furthermore, ING3 also links hNuA4 to p53 function in transcription and apoptosis (15, 38). Accordingly, we show that Tip60 can affect p53-dependent transcription in vivo, a result reminiscent of our study with the yeast complex (41, 42). It will be interesting to continue the structural and functional characterization of human NuA4 complexes since they are poised to play essential roles in such diverse nuclear functions as gene regulation, DNA repair, and cell cycle control.
Acknowledgments
We are grateful to G. Chinnadurai for Tip60 cDNA, B. Séraphin for the TAP system plasmids, Y. Makino for anti-Tip49a/b(RUVBL1/2), Y. Matsuoka for anti-MRG15, A. Munnia and E. Meese for anti-GAS41, M. Takahashi for anti-EPC1, W. Wang for anti-BAF53a and anti-HDAC2, and A. Anderson for the p53 expression vector. We thank Marie Martineau for help in cloning, Marc Bergeron for the human kidney cDNA bank, Frédéric Beaulieu for gamma irradiation of cells, Vincent Roy and Manuel Caruso for help with retrovirus, Josée Lavoie's lab for help and advice in cell culture and transfections, Rhea Utley for critical reading of the manuscript, and members of our lab for fruitful discussions.
This study was supported by grants from the Cancer Research Society, Inc., GénomeCanada/GénomeQuébec, and the Canadian Institutes of Health Research (CIHR) to J.C. and from the National Institutes of Health to S.T. Y.D. was a Natural Sciences and Engineering Research Council graduate student and currently holds a CIHR/Canada Graduate scholarship. S.T. is a Pew Scholar in the Biomedical Sciences. J.C. is a CIHR Investigator.
REFERENCES
- 1.Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. [PubMed] [Google Scholar]
- 2.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Côté. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ard, P. G., C. Chatterjee, S. Kunjibettu, L. R. Adside, L. E. Gralinski, and S. B. McMahon. 2002. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol. Cell. Biol. 22:5650-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 5.Bertram, M. J., N. G. Berube, X. Hang-Swanson, Q. Ran, J. K. Leung, S. Bryce, K. Spurgers, R. J. Bick, A. Baldini, Y. Ning, L. J. Clark, E. K. Parkinson, J. C. Barrett, J. R. Smith, and O. M. Pereira-Smith. 1999. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 19:1479-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 7.Boudreault, A. A., D. Cronier, W. Selleck, N. Lacoste, R. T. Utley, S. Allard, J. Savard, W. S. Lane, S. Tan, and J. Côté. 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, L. A., M. R. Langer, K. A. Crowley, S. Tan, J. M. Denu, and C. L. Peterson. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10:935-942. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 10.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Côté. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 11.Choy, J. S., and S. J. Kron. 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 22:8215-8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19:2515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doerks, T., R. R. Copley, J. Schultz, C. P. Ponting, and P. Bork. 2002. Syst. identification of novel protein domain families associated with nuclear functions. Genome Res. 12:47-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, A., R. T. Utley, A. Nourani, S. Allard, P. Schmidt, W. S. Lane, J. C. Lucchesi, and J. Côté. 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 276:3484-3491. [DOI] [PubMed] [Google Scholar]
- 15.Feng, X., Y. Hara, and K. Riabowol. 2002. Different HATS of the ING1 gene family. Trends Cell Biol. 12:532-538. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, U., D. Heckel, A. Michel, M. Janka, T. Hulsebos, and E. Meese. 1997. Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I. Hum. Mol. Genet. 6:1817-1822. [DOI] [PubMed] [Google Scholar]
- 17.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 18.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 20.Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Héliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Côté. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5:927-937. [DOI] [PubMed] [Google Scholar]
- 21.Garkavtsev, I., I. A. Grigorian, V. S. Ossovskaya, M. V. Chernov, P. M. Chumakov, and A. V. Gudkov. 1998. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 391:295-298. [DOI] [PubMed] [Google Scholar]
- 22.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 23.Gunduz, M., M. Ouchida, K. Fukushima, S. Ito, Y. Jitsumori, T. Nakashima, N. Nagai, K. Nishizaki, and K. Shimizu. 2002. Allelic loss and reduced expression of the ING3, a candidate tumor suppressor gene at 7q31, in human head and neck cancers. Oncogene 21:4462-4470. [DOI] [PubMed] [Google Scholar]
- 24.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 25.Horikawa, I., H. Tanaka, Y. Yuasa, M. Suzuki, M. Shimizu, and M. Oshimura. 1995. Forced expression of YL-1 protein suppresses the anchorage-independent growth of Kirsten sarcoma virus-transformed NIH 3T3 cells. Exp. Cell Res. 220:11-17. [DOI] [PubMed] [Google Scholar]
- 26.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 27.Johnston, H., J. Kneer, I. Chackalaparampil, P. Yaciuk, and J. Chrivia. 1999. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J. Biol. Chem. 274:16370-16376. [DOI] [PubMed] [Google Scholar]
- 28.Kamine, J., B. Elangovan, T. Subramanian, D. Coleman, and G. Chinnadurai. 1996. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology 216:357-366. [DOI] [PubMed] [Google Scholar]
- 29.Legube, G., L. K. Linares, C. Lemercier, M. Scheffner, S. Khochbin, and D. Trouche. 2002. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 21:1704-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Masson, I., D. Y. Yu, K. Jensen, A. Chevalier, R. Courbeyrette, Y. Boulard, M. M. Smith, and C. Mann. 2003. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell. Biol. 23:6086-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung, J. K., N. Berube, S. Venable, S. Ahmed, N. Timchenko, and O. M. Pereira-Smith. 2001. MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J. Biol. Chem. 276:39171-39178. [DOI] [PubMed] [Google Scholar]
- 32.Matangkasombut, O., and S. Buratowski. 2003. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 11:353-363. [DOI] [PubMed] [Google Scholar]
- 33.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 34.Monden, T., M. Kishi, T. Hosoya, T. Satoh, F. E. Wondisford, A. N. Hollenberg, M. Yamada, and M. Mori. 1999. p120 acts as a specific coactivator for 9-cis-retinoic acid receptor (RXR) on peroxisome proliferator-activated receptor-gamma/RXR heterodimers. Mol. Endocrinol. 13:1695-1703. [DOI] [PubMed] [Google Scholar]
- 35.Monden, T., F. E. Wondisford, and A. N. Hollenberg. 1997. Isolation and characterization of a novel ligand-dependent thyroid hormone receptor-coactivating protein. J. Biol. Chem. 272:29834-29841. [DOI] [PubMed] [Google Scholar]
- 36.Monroy, M. A., D. D. Ruhl, X. Xu, D. K. Granner, P. Yaciuk, and J. C. Chrivia. 2001. Regulation of cAMP-responsive element-binding protein-mediated transcription by the SNF2/SWI-related protein, SRCAP. J. Biol. Chem. 276:40721-40726. [DOI] [PubMed] [Google Scholar]
- 37.Nagashima, M., M. Shiseki, K. Miura, K. Hagiwara, S. P. Linke, R. Pedeux, X. W. Wang, J. Yokota, K. Riabowol, and C. C. Harris. 2001. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc. Natl. Acad. Sci. USA 98:9671-9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagashima, M., M. Shiseki, R. M. Pedeux, S. Okamura, M. Kitahama-Shiseki, K. Miura, J. Yokota, and C. C. Harris. 2003. A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene 22:343-350. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama, J., G. Xiao, K. Noma, A. Malikzay, P. Bjerling, K. Ekwall, R. Kobayashi, and S. I. Grewal. 2003. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 22:2776-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nie, Z., Z. Yan, E. H. Chen, S. Sechi, C. Ling, S. Zhou, Y. Xue, D. Yang, D. Murray, E. Kanakubo, M. L. Cleary, and W. Wang. 2003. Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner. Mol. Cell. Biol. 23:2942-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nourani, A., Y. Doyon, R. T. Utley, S. Allard, W. S. Lane, and J. Côté. 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21:7629-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nourani, A., L. Howe, M. G. Pray-Grant, J. L. Workman, P. A. Grant, and J. Côté. 2003. Opposite role of yeast ING family members in p53-dependent transcriptional activation. J. Biol. Chem. 278:19171-19175. [DOI] [PubMed] [Google Scholar]
- 43.Ogawa, H., T. Ueda, T. Aoyama, A. Aronheim, S. Nagata, and R. Fukunaga. 2003. A SWI2/SNF2-type ATPase/helicase protein, mDomino, interacts with myeloid zinc finger protein 2A (MZF-2A) to regulate its transcriptional activity. Genes Cells 8:325-339. [DOI] [PubMed] [Google Scholar]
- 44.Park, J., M. A. Wood, and M. D. Cole. 2002. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol. Cell. Biol. 22:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 46.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 47.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 48.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 49.Ruhf, M. L., A. Braun, O. Papoulas, J. W. Tamkun, N. Randsholt, and M. Meister. 2001. The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development 128:1429-1441. [DOI] [PubMed] [Google Scholar]
- 50.Shen, X., R. Ranallo, E. Choi, and C. Wu. 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12:147-155. [DOI] [PubMed] [Google Scholar]
- 51.Sheridan, A. M., T. Force, H. J. Yoon, E. O'Leary, G. Choukroun, M. R. Taheri, and J. V. Bonventre. 2001. PLIP, a novel splice variant of Tip60, interacts with group IV cytosolic phospholipase A2, induces apoptosis, and potentiates prostaglandin production. Mol. Cell. Biol. 21:4470-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimono, Y., H. Murakami, Y. Hasegawa, and M. Takahashi. 2000. RET finger protein is a transcriptional repressor and interacts with enhancer of polycomb that has dual transcriptional functions. J. Biol. Chem. 275:39411-39419. [DOI] [PubMed] [Google Scholar]
- 53.Smith, E. R., A. Eisen, W. Gu, M. Sattah, A. Pannuti, J. Zhou, R. G. Cook, J. C. Lucchesi, and C. D. Allis. 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 55.Tan, S. 2001. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21:224-234. [DOI] [PubMed] [Google Scholar]
- 56.Utley, R. T., and J. Côté. 2003. The MYST family of histone acetyltransferases. Curr. Top. Microbiol. Immunol. 274:203-236. [DOI] [PubMed] [Google Scholar]
- 57.Utley, R. T., K. Ikeda, P. A. Grant, J. Côté, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 58.Vaquero, A., A. Loyola, and D. Reinberg. 2003. The constantly changing face of chromatin. Sci. Aging Knowl. Environ. 2003:RE4. [Online.] [DOI] [PubMed] [Google Scholar]
- 59.Vignali, M., D. J. Steger, K. E. Neely, and J. L. Workman. 2000. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 19:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wade, P. A., P. L. Jones, D. Vermaak, and A. P. Wolffe. 1999. Purification of a histone deacetylase complex from Xenopus laevis: preparation of substrates and assay procedures. Methods Enzymol. 304:715-725. [DOI] [PubMed] [Google Scholar]
- 61.Wood, M. A., S. B. McMahon, and M. D. Cole. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5:321-330. [DOI] [PubMed] [Google Scholar]
- 62.Workman, J. L., I. C. A. Taylor, R. E. Kingston, and R. G. Roeder. 1991. Control of class II gene transcription during in vitro nucleosome assembly. Methods Cell. Biol. 35:419-447. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto, T., and M. Horikoshi. 1997. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J. Biol. Chem. 272:30595-30598. [DOI] [PubMed] [Google Scholar]
- 64.Yochum, G. S., and D. E. Ayer. 2002. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and transducin-like Enhancer of Split. Mol. Cell. Biol. 22:7868-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimmermann, K., K. Ahrens, S. Matthes, J. M. Buerstedde, W. H. Stratling, and L. Phi-van. 2002. Targeted disruption of the GAS41 gene encoding a putative transcription factor indicates that GAS41 is essential for cell viability. J. Biol. Chem. 277:18626-18631. [DOI] [PubMed] [Google Scholar]