Abstract
KSHV inflammatory cytokine syndrome (KICS) is a newly described condition characterized by systemic illness as a result of systemic, lytic KSHV infection. We used Illumina sequencing to establish the DNA vironome of blood from such a patient. It identified concurrent high-level viremia of human herpesvirus (HHV) 8 and 6a. The HHV8 plasma viral load was 5,300,000 copies/ml, which is the highest reported to date; this despite <5 skin lesions and no HHV8 associated lymphoma. This is the first report of systemic HHV6a/KSHV co-infection in a patient. It is the first whole genome KSHV sequence to be determined directly from patient plasma rather than cultured or biopsied tumor material. This case supports KICS as a new clinical entity associated with KSHV.
Keywords: Kaposi sarcoma-associated herpesvirus, KSHV, HHV8, HHV6, AIDS, KICS
Introduction
Kaposi Sarcoma (KS) and the plasmablastic variant of multicentric Castleman disease (MCD) are associated with Kaposi sarcoma-associated herpesvirus (KSHV) infection and occur with increased prevalence in AIDS patients. KS remains among the leading causes of death for people living with HIV/AIDS today. It is the most frequent cancer overall in many Sub Saharan countries (e.g. Uganda) (Franceschi et al., 2008). Since skin KS lesions are relatively easy to diagnose clinically, KSHV viral load assays are seldom conducted. KSHV gene transcription in KS is variable, but predominantly restricted to the latent viral genes (Dittmer, 2003, Dittmer, 2011). Compared to diseases cause by other herpesviruses (HSV-1, CMV, EBV) systemic viremia in KS is an order of magnitude less (Lin et al., 2009). As a consequence, infectious virus has not been efficiently recovered from patients, perhaps due to comparatively low systemic viremia vis-à-vis other herpesviruses. There exists a gap in our knowledge as all known sequence information for KSHV was derived from cell line propagated or tumor derived clones, but not free virus systemically replicating in the natural host.
MCD is a hyperproliferative B cell disorder. MCD patients express high levels of viral IL6 (vIL6) and human IL6, which are thought to be responsible for the systemic illness and inflammatory symptoms (Brandt et al., 1990, Oksenhendler et al., 2000). Clinical symptoms of MCD include edema, weight loss, pleural effusion (mostly cell free, as opposed to primary effusion lymphoma), anemia, thrombocytopenia, lymphadenopathy and splenomegaly (reviewed in Polizzotto et al, 2012). An alternative clinical diagnosis is that of Systemic Inflammatory Response Syndrome (SIRS). SIRS can cause rapid death in AIDS patients. It is thought of as an extreme, uncontrolled, cytokine-mediated response to infection in CD4 deficient patients. The underlying trigger of this response is typically unknown. Thus, symptomatic treatment remains the only option.
KSHV inflammatory cytokine syndrome (KICS) has been recently been proposed as a new KSHV associated disease (Uldrick et al., 2010). The six patients originally described had no evidence of MCD despite thorough examination, and no development of MCD upon follow up. However, they presented with clinical inflammatory syndromes as seen in MCD, high human IL6 levels, and high systemic KSHV viral load. The patient described here had the highest systemic KSHV viral load on record to date, with no identifiable MCD. He fits the novel clinical description for KICS and we were able to establish the complete DNA vironome from blood.
Systemic KSHV – HHV6a co-infection was uncovered by next generation sequencing of peripheral blood mononuclear cells and plasma. We obtained a complete KSHV genome sequence from high titer, replicating virus rather than tumor associated or tissue culture propagated variants (Depledge et al. 2011, Lin et al. 2012), which in the case of KSHV tend to replicate extremely poorly and may have accumulated mutations.
Results
The patient is a 42-year old Caucasian male who was diagnosed with human immunodeficiency virus (HIV) infection in 1993 and treated with zidovudine/lamivudine and nevirapine. He discontinued medication in 2003, when his HIV-1 viral load was undetectable. He presented in December 2008 with a HIV-1 viral load of 600,000 copies/ ml and a CD4 count of 27 cells / μl, and met the clinical criteria for SIRS. The definition of SIRS is based on clinical parameters of systemic inflammation not unlike KICS, but typically does require an association with a specific infectious agent.
First, we tested for the presence of typical infectious agents. Stool for clostridium difficile and sputum for pneumocystis jirovecii were negative. Serology studies for cryptococcus, toxoplasma, mycobacteria, and syphilis as well as urine histoplasma antigen were also negative. Epstein-Barr-virus loads (EBV) and human cytomegalovirus loads (HCMV) were negligible with < 250 copies/ml and 647 copies/ml, respectively. The IL6 level was 54.1 pg / ml. In the absence of AIDS defining infections (and of CD4 T cells), we asked why would this patient show evidence of a severe, systemic inflammatory reaction?
Excisional axillary lymph node biopsy showed effacement of the expected architecture, with general lymphocyte depletion, increased numbers of mature-appearing interfollicular plasma cells, and disrupted lymphoid follicles typical of HIV lymphadenitis (histologic pattern B). There were no aggregates of plasmablastic mononuclear cells. Moreover, reactive germinal centers and those showing hyaline vascular Castleman features were not identified. Kappa and lambda in situ hybridization demonstrated overall polytypic staining of the plasma cells, and no monoclonal B cell population was identified by thorough flow cytometric analysis. Thus, lymphoma and multicentric Castleman disease (MCD) were excluded. KSHV latency associated nuclear antigen (LANA) expression was largely confined to the residual follicles, in contrast to the pattern observed in MCD (Figure 1, panel A and B) (Chadburn et al. 2008, Schmidt et al. 2008). Lambda light chain staining showed overlap with LANA localization, consistent with recent reports that suggest preferential KSHV infection of lambda-expressing B cells (Hassman et al, 2011). No overlap of EBV EBER and KSHV LANA expression was observed.
Figure 1. Patient was positive for Kaposi Sarcoma, as confirmed by autopsy, immunohistochemistry, and PCR.
(A) LANA (red) and hematoxilin (blue) stain of a lymph node biopsy at 100x, (B) and at 400x. (C–D, arrows) KS lesions in lung at autopsy, (E) Gel of PCR products confirming KSHV positivity. Isolated PBMC DNA was amplified with primers located within the KSHV genes: (1) LANA, (2) vFLIP, (3) K14, (4) vGPCR, (5) Kaposin, (6) Orf69 and GAPDH (“+”) as a reference gene. No primer controls for each sample are indicated by “.” For comparison, KSHV positive PEL DNA was used (“pos”), and a non-template control (NTC) was included as a negative control (“neg”). Molecular weight markers are shown at the beginning and end.
The patient ultimately died of disease-related complications. Autopsy confirmed KS of the oral mucosa, skin, small bowel, and lung (Figure 1, panel C, D). However, KS could not explain the massive hepatosplenomegaly, severe anemia and thrombocytopenia thought to be secondary to sequestration.
The presence of KSHV in plasma was initially detected by DNA PCR using primers directed against multiple regions of the viral genome, specifically the open reading frames LANA, vFLIP, vGPCR, Kaposin and orf69 (Figure 1, panel E). We chose multiple genes to increase the specificity of our assay. These primers were previously validated on clinical and experimental samples (Dittmer, D.P., 2011, 2003). As positive control for DNA isolation from clinical specimens we used gapdh primers. The positive control was total DNA from BCBL-1 cells and the negative control sample was water. All primers amplified the predicted size fragment from the patient sample and positive control, but not the negative control (the very faint bands in lane 1 and 3 represent primer dimers, which were smaller than the predicted size). No band was observed in the ‘no primer control’ lane (Figure 1, panel E, lane “.”). As this assay is a conventional endpoint PCR assay, it is not quantitative. The brightness of the individual bands is dependent on their size as well as accumulation. The three longest PCR products correspond to KSHV genes LANA, vGPCR and gapdh (Figure 1, panel E, lanes 1, 4, “+”). The four very short PCR products yielded shorter and thus less bright products (Figure 1, panel E, lanes 2, 3, 5, 6). This demonstrates that KSHV was present in PBMC of this patient.
To detect free virus, we determined the plasma viral load of KSHV. The patient had a plasma KSHV viral load of 5,300,000 copies/ml (Viracor-IBT Laboratories, MO). This represents the highest KSHV viral load on record (Table 1). By comparison KS patients (Polstra et al., 2003, Sayer et al., 2011) typically have a mean KSHV viral load of 1000 copies / ml (range 151 – 26,915), PEL patients (Simonelli et al., 2009) of 10,284 copies / ml (range 2,558 – 36,300) and MCD (Simonelli et al., 2009) of 4,400 copies / ml (range 600 – 1,678,000). KS malignancies are AIDS defining, but rarely associated with high-level virus replication. Average KSHV plasma viral loads are 1–2 log10 units lower than EBV or HCMV viral loads even in symptomatic patients (Laney et al., 2007, Whitby et al., 1995). Hence, for this exceptional case we considered that the patient succumbed to acute KSHV viremia.
Table 1.
Comparison of viral loads for KSHV-associated diseased, as compared to this case.
| Disease | Min (copies/mL) | Max (copies/mL) | Mean (copies/mL) |
|---|---|---|---|
| KSa | 151 | 26,915 | 1,000 |
| PELb | 2,558 | 36,300 | 10,284 |
| MCDb | 600 | 1,678,000 | 4,400 |
| Patient | - | - | 5,300,000 |
KS, Kaposi sarcoma, ( Polstra et al., 2003)
PEL, primary effusion lymphoma, MCD, multicentric Castleman disease, reference (Simonelli et al., 2009)
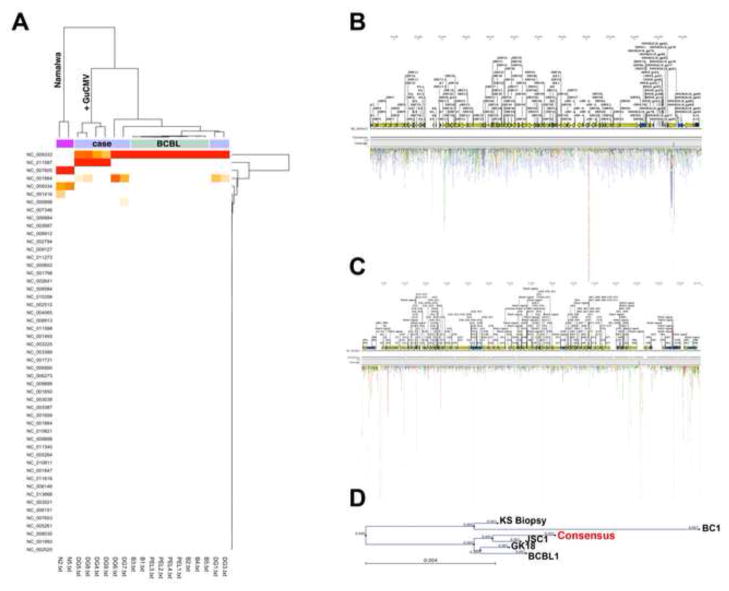
We turned to Illumina-based sequencing to determine the complete vironome and complete genome sequence of this KSHV isolate. Total DNA was isolated from white blood cells. No attempts were made to enrich for viral sequences. Illumina™ sequencing was performed by sonication of DNA, primer ligation and tethered amplification, followed by DNA synthesis to yield >35 base reads. To determine the vironome, sequencing reads were aligned to the reference genomes of all known viruses. Combining 5 individual sequencing runs we obtained a total of 145,179,479 reads of 36 bp (paired), 50bp (single) and 75 bp (paired and single) from samples of this patient. A discovery score (D score) based on the total number of aligned reads and coverage for each viral genome was computed and subjected to hierarchical clustering. Sequencing reads from samples of known herpesvirus genome status (two copy EBV - integrated cell line Namalwa, PEL cell line BCBL-1) were included to calibrate the virus discovery algorithm. In four of the sequencing runs for this patient, purified Guinea Pig CMV DNA was spiked in as an internal control, and was recovered in sequence alignment for these samples (Genbank ID: AB592928).
The clustering results are displayed as heatmap and dendrograms (Figure 2, panel A). Positivity is displayed as by color (white, orange, red), with deeper red color indicating a higher D score. D score is a weighted combination of coverage, genome-size and number of hits. For the 24 human chromosomes we calculated at 95%CI of −0.03 to 0.07 log10 copies/ cell, i.e. two copies per cell. Our initial screen identified sequences for 648 genbank entries, which were classified as either fungal, viral, protozoal or bacterial. However, most were present at ≤ −2 log10 genome copies/ cell, i.e. fewer than 1 copy per cell. Several bacterial pathogens, including toxoplasma, were tested for, but could not be confirmed by standard tests. Mitochondrial genomes were present at 1.52 log10 copies/ cell, and KSHV was present at 0.23 log10 copies/ cell, i.e. at > 1 copy per cell.
Figure 2. Nextgen sequencing analysis reveals KSHV and HHV6a coinfection.
(A) Heatmap and dendrogram of unsupervised single linkage clustering using a Manhattan distance matrix. Reads from individual Illumina lanes were aligned to 3608 whole viral genomes in genbank. For each alignment we computed a score based on the number of hits, coverage and length of the target sequence.
(B) Distribution of reads that aligned to the KSHV (NC_003409) genome. Open reading frames are depicted as arrows.
(C) Distribution of reads that aligned to the HHV6a (NC_001664) genome.
(D) Phylogenetic tree demonstrating alignment of generated consensus sequence for KSHV isolated from patient PBMC, in comparison to previously reported KSHV sequences. BCBL1 sequence was determined from BCBL1 DNA inserted into BAC36 and cloned. GK18 sequence is from cosmid-cloned DNA originating from a classic KS patient. JSC1 sequence is derived from cellular DNA cloned into BAC219. KS biopsy sequence is derived from DNA from KS biopsies, partially digested with Sau3A, cloned into lambda phage, and sequenced. BC1 DNA was sequenced from the PEL-derived cell line, and was cloned into lambda phage and cosmids and sequenced. Our patient sequence is indicated as Consensus.
In total, 22,455 reads aligned to KSHV, resulting in a 9.23 fold median genome coverage (Figure 2, panel B). The presence of HHV6a (but not HHV6b or HHV7) was also revealed upon sequence analysis. This was surprising, as infection with HHV6a was never suspected or tested for. Upon retesting of patient plasma we determined the HHV6 viral load to be 25, 471 copies/mL. We have investigated the presence of HHV6 (a or b) in 17 additional HIV patients. 5 of these patients had KS, while the remaining 12 were KS negative, but positive for another type of malignancy. None of the patients were positive for HHV6a, and 1 patient was positive for HHV6b, with approximately 1000cp/mL. This patient was KS negative, and was positive for Melanoma, a non-AIDS defining malignancy. This is consistent with the prevalence in the general population, where HHV6b is much more common than HHV6a, which is rarely found in patients (Boutolleau et al., 2006).
A total of 23,300 reads mapped to HHV6a, yielding 7.65-fold median coverage (Figure 2, panel C). Since coverage is a function of copy number and genome size this demonstrates for the first time HHV8 and HHV6a co-replication in a patient. Both KSHV and HHV6a DNA sequences were found in roughly equal proportion in cell free plasma, where the KSHV plasma viral load was > 106. This would argue that the HHV6a signal, too, stemmed from replication virus rather than being horizontally transmitted.
We established uniformity of genome coverage as a second criterion for virus identification. All reads mapped randomly across the entire length of the HHV8 and HHV6a genomes (Figure 2, panel B, C). Reads that aligned to more than one position, i.e. to repeat region, were randomly placed at only a single position. In contrast to HHV6a, the few reads that mapped to HHV6b genome, all clustered within the terminal repeat region (data not shown), indicating insufficient coverage to demonstrate biologically relevant infection.
We derived the whole genome sequence for this KSHV strain by reference-guided assembly (Genbank ID JQ619843). The KSHV consensus sequence was compared to the KSHV reference sequence (Genbank ID: NC_009333). We did not identify novel DNA insertions or orfs. We identified small InDels in specific genes (the polymorphic regions of K1 and K15) and in repeat regions, which is a result of the short read length of the Illlumina platform, but not deletions of entire coding regions or gross aberrations. Thus no one gene could be linked to the highly replicative phenotype. These are annotated in the sequence submission.
To demonstrate relatedness of this primary KSHV isolate to tumor cell line derived viral sequences, we used ClustalW (Felsenstein, 1989) using bootstrap analysis with Neighbor-Joining. The pairwise distances between all the aligned whole genome sequences were computed using a maximum likelihood model, the number of replications for bootstrap analysis was 1000. The result is displayed using PHYLIP (R Development Core Team, 2012) and demonstrated high sequence conservation expected of double-stranded DNA viruses (Figure 2D). This is the sequence of a primary, highly replicative KSHV isolate.
It is difficult to derive genome-wide conclusions from a comparison of just six genomes. We did note extraordinary conservation in primary nucleotide and amino acid sequences among all isolates. For the unique regions of the genome, approximately one nucleotide change was present per kb and less than one amino acid per coding regions, as expected for DNA viruses. The most divergent sequence was the original BC-1, which was obtained by Saenger sequencing; and which may not have had the extensive genome coverage afforded by NextGen based sequencing. The Sau3AKS sequence has deletions in K15 compared to all other sequences, which may be the result of phage library prep and propagation in E.coli. We observed variations and gaps in the repeat regions (LIR1, DR1-6, and TR). These could be the result of misalignment, as short read – based protocols are known to generate incomplete alignments in repeat regions, or they could be the result of genome evolution. For instance, a 36 bp gap in K1 was conserved in this sequence, GK18, JSC1 and Sau3A; a 9bp deletion in DR3 was conserved in JSC1, BCBL1 and Sau3A. We did not have enough coverage for a de-novo assembly to unambiguously identify small DIPs or SNPs for this particular isolate.
Rather than directly testing for each potential pathogen sequentially, NextGen sequencing technology allowed us to detect a broad range of viruses, bacteria, fungi, and parasites, resulting in the detection of additional factors that may be playing a role in the disease - in this case, HHV6a.
Discussion
This report highlights NextGen sequencing technology on patient material to identify infectious agents in a case of fatal inflammation reaction of unknown origin. Rather than testing for each potential pathogen individually, sequencing of total DNA in plasma and PBMC allowed us to screen for a broad range of viruses, as well as bacteria, fungi, and parasites (data not shown), thus resulting in the detection of additional factors that may be playing a role in patient illness- in this case, HHV6a viremia. This novel application of Illumina sequencing to assist in patient diagnosis proved to be a useful tool in determining the cause of illness and death in this patient.
This represents the first documentation of systemic KSHV/HHV6 co-replication in a patient. Primary HHV6 infection is mostly subclinical and HHV6 has near 100% sero prevalence in the US. HHV6a is primarily associated with exanthema subitum (Roseola). It has been detected in end-stage AIDS patients, but these instances are extremely rare. HHV6a DNA has been detected in a single archival case of a PEL biopsy before (Asou et al., 2000) but not as free, replicating virus. HHV6a is also found in forms of reactive lymphoid hyperplasia, such as Crohn’s disease, histiocytic necrotizing lymphadenitis or Kikuchi-Fujimoto disease in high prevalence areas, although present studies are insufficient to support a conclusive association of HHV6a with these or other forms of rare reactive lymphoid hyperplasia (Bouvard et al., 2009). HHV6a can cause KSHV reactivation from PEL cell lines in culture, as can HCMV (Lu et al., 2005, Vieira et al., 2001).
Whether, in turn, KSHV can stimulate HHV6a replication and/or its pathogenesis is presently unknown. Both viruses encode as well as induce cytokines and chemokines, including human IL6 and vIL6, which can induce inflammatory symptoms. We surmise that systemic replication of both viruses, rather than KSHV-associated neoplasia, led to the clinical symptoms. The clinical description of SIRS as a cytokine storm is consistent with that of KICS, which was recently proposed as a new KSHV associated clinical entity and which is defined by high cytokine production, particularly of IL-6 and the presence of KSHV, but a negative diagnosis for MCD. We propose that co-infection may have added to the intensity of KICS. Further studies are necessary to assess whether HHV6a co infection is present in other KICS patients, or if it is necessary for the development of KICS.
Conclusions
We have described a fatal case of systemic KSHV and HHV6a co-viremia in an AIDS KICS patient. Using Next generation sequencing, we were able to generate the first KSHV sequence directly from patient sample, without implementing cloning techniques. This novel sequence has been deposited into Genbank.
Materials and Methods
DNA extraction
Genomic DNA was isolated from white blood cells or plasma using Wizard SV™ kit (Promega Inc.). All PCR products were of the expected size by gel electrophoresis. No amplification was observed without template. Detailed methods were published previously (Chugh et al., 2010, Dittmer, 2011). The primer sequences were: (LANA78 5′ - GGAAGAGCCCATAATCTTGC, 5′ - GCCTCATACGAACTCGAGGT (204 bp); KS10076-1 (vFLIP) 5′ - ATGCCCTAATGTCAATGCTT, 5′ - GCGATAGTGTTGGGAGTGTG (90 bp); K14F 5′ - TGGTGGGCCTATTTGGGATA, 5′ - GATGCACCGCCCTGCTT (60 bp); orf74F-spl (vGPCR) 5′ - TGGCCCAAACGGAGGATCCTAG, 5′ -GATGATGAATCCTGGAATGAAACT (232 bp); kaposin 5′ - CAACAGATAATGGGCCTTGT, 5′ - AGGTGCAAGCAGACATAATTG; ORF69 5′ - TGCAGTGCAGGTACACACCA, 5′ - GCATCTCGTCGGTGCAGTCT (61 bp); hu gapdhF 5′ - GAAGGTGAAGGTCGGAGTC, 5′ - GAAATCCCATCACCATCTTC (225 bp). Amplicon sizes are based on alignment to HHV8 reference genome NC_009333 and NM_002046 for gapdh.
Illumina Sequencing
Illumina™ sequencing was performed by sonication of DNA, primer ligation and tethered amplification, followed by DNA synthesis to yield 35 base reads. The high viral load allowed us to use total DNA without virus-specific enrichment (e.g. Kwok et al. 2012). To prepare the library, DNA was sonicated, then ends were repaired and adenylated. Paired end adapters were ligated to DNA fragments and purified via gel electrophoresis excision and QIAGEN Gel Extraction Kit (QIAGEN, part #28704). Ligated products were amplified using PCR prior to sequencing run. Sequencing was performed using a variety of conditions: Paired end 2×76 cycles. Additional runs include 50bp single reads, 36 bp paired reads, and 76bp single reads. The reads were aligned to human, viral (Genbank ID: NC_003409 for KSHV, NC_001664 for HHV6a) and microbial genomes using CLCbio software (CLCbio Inc.) and EMBOSS (Rice et al., 2000).
By computing the number of sequence reads that aligned to a given pathogen genome against the genome size, we estimated the relative frequency of the pathogen. For human DNA, we obtained on average 0.005 reads/ bp; more for mitochondrial genomes, since there are multiple mitochondria per cell genome. For KSHV and for HHV6a we obtained almost equal coverage with 0.003 reads/ bp. In contrast, HHV6b coverage was 10 fold lower (5×10−4 reads/bp), as was EBV (3×10−5 read/ bp) and (HCMV 8×10−6 read/ bp) coverage. Our discovery score D takes is a linear combination of the number of hits, the target genome and coverage. It was calculated for each genbank entry and each sample and subjected to unsupervised clustering using R v2.8.0 (Maindonald and Braun, 2007).
Phylogenetic comparison
To compare our KSHV isolate to existing whole genome data, we used clustalW and the following sequences: BCBL1 (Genbank ID: HQ404500), JSC1 (Genbank ID: GQ994935), KS biopsy (Genbank ID: U93872), BC1 (Genbank ID: KSU75698), and GK18 (Genbank ID: NC_009333). Our patient sequence is indicated as Consensus and submitted as Genbank ID: JQ619843.
Immunohistochemistry
A formalin fixed, paraffin-embedded lymph node was sectioned and dehydrated through a series of ethanol washes. Sections were then stained using a rat monoclonal anti-LANA antibody (Advanced Biotechnologies Inc.) diluted at 1:1000, and Vectastain™ (Vectorlabs Inc.) kit, counterstained with hematoxylin, and rehydrated through a series of xylene washes before being fixed with a xylene-based mounting solution. Images were collected on a LEICA DM LS microscope with DFC480 camera.
Acknowledgments
This work was supported by NIH grants DE018304, CA109232, NCI Supplemental funding to the Center for AIDS research (CfAR, AI50410), and the University Cancer Research Fund (UCRF). K.T. is supported by T32 GM07092-34 and by a grant to the University of North Carolina at Chapel Hill from Howard Hughes Medical Institute (HHMI) through the Med into Grad Initiative. We thank Seth Matheson for lung images, Yuri Fedoriw for additional pathology and Melissa Miller for HHV6 assay.
List of Abbreviations
- KSHV
Kaposi Sarcoma associated Herpesvirus
- KICS
KSHV inflammatory cytokine syndrome
- MCD
Multicentric Castleman Diseasel
- HHV
human herpesvirus
- HIV
human immunodeficiency virus
- SIRS
systemic inflammatory response syndrome
- EBV
Epstein barr virus
- HCMV
human cytomegalovirus
- LANA
Latency associated nuclear antigen
- KS
Kaposi Sarcoma
- HSV-1
herpes simplex virus 1
- vIL-6
viral interleukin 6
Footnotes
The authors declare that they have no competing interests.
Authors’ contributions:
KT-drafted manuscript and conducted IHC; DY conducted sequence analysis DR- conducted KSHV PCR; JP, AL, NM, VM, SM, CvH – identified the case, evaluated clinical information and provided pathology information. DPD conceived the project, conducted data analysis and co-wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kristen M. Tamburro, Email: tamburro@med.unc.edu.
Dongmei Yang, Email: dy0009@unc.edu.
Jessica Poisson, Email: JPoisson@unch.unc.edu.
Yuri Fedoriw, Email: GFedoriw@unch.unc.edu.
Debasmita Roy, Email: debasmita_roy@med.unc.edu.
Amy Lucas, Email: ASLucas@unch.unc.edu.
Sang-Hoon Sin, Email: hoon_sin@med.unc.edu.
Nadia Malouf, Email: malouf@med.unc.edu.
Vincent Moylan, Email: vmoylan@unch.unc.edu.
Blossom Damania, Email: blossom_damania@med.unc.edu.
Stephan Moll, Email: stephan_moll@med.unc.edu.
Charles van der Horst, Email: cvdh@med.unc.edu.
Dirk P. Dittmer, Email: dirk_dittmer@med.unc.edu.
References
- Asou H, Tasaka T, Said JW, Daibata M, Kamada N, Koeffler HP. Co-infection of HHV-6 and HHV-8 is rare in primary effusion lymphoma. Leuk Res. 2000;24:59–61. doi: 10.1016/s0145-2126(99)00144-7. [DOI] [PubMed] [Google Scholar]
- Boutolleau D, Duros C, Bonnafous P, Caiola D, Karras A, Castro ND, Ouachee M, Narcy P, Gueudin M, Agut H, Gautheret-Dejean A. Identification of human herpesvirus 6 variants A and B by primer-specific real-time PCR may help to revisit their respective role in pathology. J Clin Virol. 2006;35:257–263. doi: 10.1016/j.jcv.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V WHO International Agency for Research on Cancer Monograph Working Group. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009;10:321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- Brandt SJ, Bodine DM, Dunbar CE, Nienhuis AW. Dysregulated interleukin 6 expression produces a syndrome resembling Castleman’s disease in mice. J Clin Invest. 1990;86:592–599. doi: 10.1172/JCI114749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadburn A, Hyjek EM, Tam W, Liu Y, Rengifo T, Cesarman E, Knowles DM. Immunophenotypic analysis of Kaposi sarcoma herpesvirus (KSHV; HHV-8)-infected B cells in HIV+ multicentric Castleman diseases (MCD) Histopathology. 2008;53:513–24. doi: 10.1111/j.1365-2559.2008.03144.x. [DOI] [PubMed] [Google Scholar]
- Chugh P, Tamburro K, Dittmer DP. Profiling of pre micro RNAs and microRNAs using quantitative real-time PCR (qPCR) arrays. J Vis Exp. 2010;(46):2210. doi: 10.3791/2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge DP, Palser AL, Watson SJ, Lai IY, Gray ER, Grant P, Kanda RK, Leproust E, Kellam P, Breuer J. Specific capture and whole-genome sequencing of viruses from clinical samples. PLoS One. 2011;6:e27805. doi: 10.1371/journal.pone.0027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer DP. Restricted Kaposi’s sarcoma (KS) herpesvirus transcription in KS lesions from patients on successful antiretroviral therapy. MBio. 2011;2:e00138–11. doi: 10.1128/mBio.00138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer DP. Transcription profile of Kaposi’s sarcoma-associated herpesvirus in primary Kaposi’s sarcoma lesions as determined by real-time PCR arrays. Cancer Res. 2003;63:2010–2015. [PubMed] [Google Scholar]
- Felsenstein J. Mathematics vs. Evolution: Mathematical Evolutionary Theory Science. 1989;246:941–942. doi: 10.1126/science.246.4932.941. [DOI] [PubMed] [Google Scholar]
- Franceschi S, Maso LD, Rickenbach M, Polesel J, Hirschel B, Cavassini M, Bordoni A, Elzi L, Ess S, Jundt G, Mueller N, Clifford GM. Kaposi sarcoma incidence in the Swiss HIV Cohort Study before and after highly active antiretroviral therapy. Br J Cancer. 2008;99:800–804. doi: 10.1038/sj.bjc.6604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassman LM, Ellison TJ, Kedes DH. KSHV infects a subset of human tonsillar B cells, driving proliferation and plasmablast differentiation. J Clin Invest. 2011;121:752–68. doi: 10.1172/JCI44185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok H, Tong AH, Lin CH, Lok S, Farrell PJ, Kwong DL, Chiang AK. Genomic sequencing and comparative analysis of epstein-barr virus genome isolated from primary nasopharyngeal carcinoma biopsy. PLoS One. 2012;7:e36939. doi: 10.1371/journal.pone.0036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney AS, Cannon MJ, Jaffe HW, Offermann MK, Ou CY, Radford KW, Patel MM, Spira TJ, Gunthel CJ, Pellett PE, Dollard SC. Human herpesvirus 8 presence and viral load are associated with the progression of AIDS-associated Kaposi’s sarcoma. AIDS. 2007;21:1541–1545. doi: 10.1097/QAD.0b013e3282202b7d. [DOI] [PubMed] [Google Scholar]
- Lin L, Lee JY, Kaplan LD, Dezube BJ, Noy A, Krown SE, Levine AM, Yu Y, Hayward GS, Ambinder RF. Effects of chemotherapy in AIDS-associated non-Hodgkin’s lymphoma on Kaposi’s sarcoma herpesvirus DNA in blood. J Clin Oncol. 2009;27:2496–2502. doi: 10.1200/JCO.2008.20.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Puetter A, Coco J, Xu G, Strong MJ, Wang X, Fewell C, Baddoo M, Taylor C, Flemington EK. Detection of murine leukemia virus in the Epstein-Barr virus-positive human B-cell line JY, using a computational RNA-Seq-based exogenous agent detection pipeline, PARSES. J Virol. 2012;86:2970–2977. doi: 10.1128/JVI.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Zeng Y, Huang Z, Huang L, Qian C, Tang G, Qin D. Human herpesvirus 6 activates lytic cycle replication of Kaposi’s sarcoma-associated herpesvirus. Am J Pathol. 2005;166:173–183. doi: 10.1016/S0002-9440(10)62242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maindonald JH, Braun J. Volume 10 of Cambridge Series on Statistical and Probabilistic Mathematics. Cambridge University Press; 2007. Data Analysis And Graphics Using R: An Example-based Approach. [Google Scholar]
- Oksenhendler E, Carcelain G, Aoki Y, Boulanger E, Maillard A, Clauvel JP, Agbalika F. High levels of human herpesvirus 8 viral load, human interleukin-6, interleukin-10, and C reactive protein correlate with exacerbation of multicentric castleman disease in HIV-infected patients. Blood. 2000;96:2069–2073. [PubMed] [Google Scholar]
- Polizzotto MN, Uldrick TS, Hua D, Yarchoan R. Clinical Manifestations of Kaposi sarcoma herpesvirus lytic activation: multicentric Castleman disease (KSHV-MCD) and the KSHV inflammatory syndrome. Frontiers in Microbiology. 2012;3:73. doi: 10.3389/fmicb.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstra AM, van den Burg R, Goudsmit J, Cornelissen M. Human Herpesvirus 8 Load in Matched Serum and Plasma Samples of Patients with AIDS-Associated Kaposi’s Sarcoma. J Clin Microbiol. 2003;41:5488–5491. doi: 10.1128/JCM.41.12.5488-5491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing 2012 [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Sayer R, Paul J, Tuke PW, Hargreaves S, Noursadeghi M, Tedder RS, Grant P, Edwards SG, Miller RF. Can plasma HHV8 viral load be used to differentiate multicentric Castleman disease from Kaposi sarcoma? Int J STD AIDS. 2011;22:585–589. doi: 10.1258/ijsa.2011.010464. [DOI] [PubMed] [Google Scholar]
- Schmidt SM, Raible A, Kortüm F, Mayer F, Riessen R, Adam P, Gregor M, Bissinger AL. Successful treatment of multicentric Castleman’s disease with combined immunochemotherapy in an AIDS patient with multiorgan failure. Leukemia. 2008 Sep;22(9):1782–5. doi: 10.1038/leu.2008.54. [DOI] [PubMed] [Google Scholar]
- Simonelli C, Tedeschi R, Gloghini A, Talamini R, Bortolin MT, Berretta M, Spina M, Morassut S, Vaccher E, De Paoli P, Carbone A, Tirelli U. Plasma HHV-8 viral load in HHV-8-related lymphoproliferative disorders associated with HIV infection. J Med Virol. 2009;81:888–896. doi: 10.1002/jmv.21349. [DOI] [PubMed] [Google Scholar]
- Uldrick TS, Wang V, O’Mahony D, Aleman K, Wyvill KM, Marshall V, Steinberg SM, Pittaluga S, Maric I, Whitby D, Tosato G, Little RF, Yarchoan R. An interleukin-6-related systemic inflammatory syndrome in patients co-infected with Kaposi sarcoma associated herpesvirus and HIV but without Multicentric Castleman disease. Clin Infect Dis. 2010;51:350–358. doi: 10.1086/654798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, O’Hearn P, Kimball L, Chandran B, Corey L. Activation of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) lytic replication by human cytomegalovirus. J Virol. 2001;75:1378–1386. doi: 10.1128/JVI.75.3.1378-1386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby D, Howard MR, Tenant Flowers M, Brink NS, Copas A, Boshoff C, Hatzioannou T, Suggett FE, Aldam DM, Denton AS. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi’s sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]