Abstract
In human cells, telomere elongation by telomerase is repressed in cis by the telomeric protein TRF1. Tankyrase 1 binds TRF1 via its ankyrin domain and poly(ADP-ribosyl)ates it. Overexpression of tankyrase 1 in telomerase-positive cells releases TRF1 from telomeres, resulting in telomere elongation. The tankyrase 1 ankyrin domain is classified into five conserved subdomains, ARCs (ankyrin repeat clusters) I to V. Here, we investigated the biological significance of the ARCs. First, each ARC worked as an independent binding site for TRF1. Second, ARCs II to V recognized the N-terminal acidic domain of TRF1 whereas ARC I bound a discrete site between the homodimerization and the Myb-like domains of TRF1. Inactivation of TRF1 binding in the C-terminal ARC, ARC V, either by deletion or point mutation, significantly reduced the ability of tankyrase 1 to poly(ADP-ribosyl)ate TRF1, release TRF1 from telomeres, and elongate telomeres. In contrast, other ARCs, ARC II and/or IV, inactivated by point mutations still retained the biological function of tankyrase 1. On the other hand, ARC V per se was not sufficient for telomere elongation, suggesting a structural role for multiple ARCs. This work provides evidence that specific ARC-TRF1 interactions play roles in the essential catalytic function of tankyrase 1.
The ends of vertebrate chromosomes are capped by telomeres, which consist of tandem TTAGGG repeats and a complex array of binding proteins (3). In most human somatic tissues, telomeric DNA shortens after each cell cycle due to incomplete replication of the terminal sequences (the so-called end-replication problem). As a result, critically shortened telomeres act as a signal for replicative senescence (42). It is not the average-sized, but rather the shortest, telomeres that cause telomere dysfunction (19). Telomeres also protect chromosome ends from being recognized as double-strand breaks, which would elicit a checkpoint or an apoptotic response (12). Direct visualization of mammalian telomeres by electron microscopy has revealed a potentially protective structure, the t-loop, in which the single-stranded telomeric terminus folds back and invades the double-stranded region (18). Such a higher-order structure could be disrupted at critically shortened telomeres in senescent cells (23).
Telomerase is a reverse transcriptase that adds telomeric repeats to the ends of chromosomes (17, 32). It is composed of a catalytic subunit, TERT, and an RNA component, TR, that acts as a template for newly synthesized telomeric repeats (33). In mice, genetic disruption of TR, which abolishes telomerase activity, causes telomere erosion, resulting in chromosomal aberrations and functional defects in highly proliferative organs after four to six generations (4, 26). In humans, telomerase is activated in the vast majority of immortal cells, including germ lines and cancer, but not in most somatic cells (24, 41). Introduction of the TERT gene into human somatic cells extends their replicative capacity (5). On the basis of these observations, it has been thought that cancer cells bypass telomere crisis by activating telomerase.
In spite of telomerase activation, however, telomere length in most telomerase-positive cells does not increase in an uncontrolled manner but is, instead, maintained at a constant average value (10, 25). This fact indicates that a regulatory mechanism involving telomerase limits telomere elongation. In humans, this mechanism involves TRF1, a duplex telomeric repeat binding protein (8, 46). TRF1 has a C-terminal DNA binding motif that is closely related to the Myb domain of c-Myb, and it forms a homodimer via its dimerization domain (1). Homodimerization of TRF1 allows it to bind the telomeric DNA through two Myb domains located on the telomeric recognition site, 5′-YTAGGGTTR-3′ (2, 15). Overexpression of TRF1 in telomerase-positive cells results in a gradual and progressive shortening of telomeres. On the other hand, the deletion mutant TRF1(66-385), which inhibits the telomere binding of TRF1 in a dominant-negative fashion, induces telomere elongation (48). These observations suggest that TRF1 negatively regulates telomere length by inhibiting the interaction between telomerase and the telomeres.
Tankyrase 1, originally identified as a TRF1 binding protein (47), is a member of the growing family of poly(ADP-ribose) polymerases (PARPs) (43). PARPs catalyze formation of long chains of poly(ADP-ribose) onto protein acceptors using NAD+ as a substrate. The net effect of the negatively charged polymers is to drastically alter the properties of the protein acceptor. In addition to the PARP domain at the C terminus, tankyrase 1 contains a domain of 24 ankyrin (ANK) repeats, thereby making it a member of the ankyrin family of structural proteins. The 33-amino-acid ANK repeat motif mediates protein-protein interactions (38). Tankyrase 1 uses its ankyrin domain to bind TRF1 and its PARP domain to ADP-ribosylate itself and TRF1, which inhibits TRF1's ability to bind telomeric DNA in vitro (47). Consequently, overexpression of tankyrase 1 in the nucleus of telomerase-positive cells releases TRF1 from telomeres and induces their elongation, indicating that tankyrase 1 is a positive regulator of telomere length (45). As a closely related homologue (83% overall identity), tankyrase 2 also binds and poly(ADP-ribosyl)ates TRF1 (9, 22).
Members of the tankyrase binding protein family include insulin-responsive amino peptidase (IRAP), adaptor protein Grb14, a tankyrase binding protein of 182 kDa (TAB182), and a nuclear-mitotic apparatus protein (7, 29, 36, 39). However, the functional significance of the interactions is largely unknown. Remarkably, the ankyrin domains of tankyrase 1 and 2 are a common platform for interaction with those partners. Comparison of the ankyrin domains of ankyrins and tankyrases, both of which contain 24 ANK repeats, indicates no continuous homology. Instead, the ankyrin domain of tankyrases shows five discrete, highly conserved regions designated as ANK repeat clusters (ARCs) I to V (39) or ANK subdomains, which are each separated by a highly conserved LLEAAR/K motif (14, 37). Yeast two-hybrid analysis suggests that each ARC is an independent binding site for TRF1 and TAB182, with differential specificities (39). Recently, the most C-terminal ARC, ARC V (or subdomain V), has been confirmed in vitro to recognize a novel RXXPDG motif, which is shared by human TRF1 (RXXADG), TAB182, IRAP, and nuclear-mitotic apparatus protein but not Grb14 (36). However, whether other ARCs recognize such partners in vitro is still to be determined. Also, it remains unknown whether multiple ARCs are required for the biological function of tankyrase 1.
In this study, we confirmed that each ARC independently bound TRF1 in vitro and in intact cells. Inactivation of TRF1 binding at the C-terminal ARC, ARC V, either by deletion or point mutation significantly reduced the ability of tankyrase 1 to poly(ADP-ribosyl)ate TRF1, release TRF1 from telomeres, and elongate telomeres. In contrast, inactivation of other ARCs, ARC II and/or IV, by point mutations still resulted in the retention of the biological function of tankyrase 1. These results indicate that ARC V, a specific subdomain in the ankyrin domain, recruits TRF1 to the poly(ADP-ribosyl)ating reaction, which is essential for the telomere function of tankyrase 1. Meanwhile, the presence of ARC V per se was not sufficient for telomere elongation in spite of its partial ability to poly(ADP-ribosyl)ate TRF1 and release TRF1 from telomeres. These observations suggest a structural role for multiple ARCs.
MATERIALS AND METHODS
Plasmid construction.
GST-TAB182C and GST-TRF1 fusions were constructed as described previously (39). Partial cDNA for IRAP (amino acids 2 to 109), containing a tankyrase binding motif (7), was obtained by PCR from leukocyte QuickClone cDNA (Clontech). GST fusions for IRAP and TRF1 fragments were constructed by cloning respective PCR fragments into pGEX-5X-1 (Pharmacia). LexA-ARC fusions, Myc-TRF1, and Myc-TRF1(2-210) were constructed by cloning each fragment into pCR3 (Invitrogen) and pcDNA3myc (H. Seimiya, M. Waase, and S. Smith, unpublished data). Wild-type FN-tankyrase 1 vectors were constructed by cloning a FN-tankyrase 1 fragment (45) into pcDNA3 (Invitrogen) and pLPC (40). The ARC deletion mutants were generated essentially as described previously (21), and the sequence for the coding region was confirmed by DNA sequencing. The FN-tankyrase-1/PARP-1 chimera was generated by replacing the PARP domain of tankyrase 1 (aa 1151 to 1327) with that of PARP-1 (aa 829 to 1014). ARC IImt (P438L), ARC IVmt (P753L), and ARC Vmt (P906L) were created using a QuikChange XL site-directed mutagenesis kit (Stratagene).
Preparation of GST fusion proteins.
GST fusion proteins were produced in the bacterial BL21-CodonPlus-RP strain (Stratagene) and purified with glutathione-Sepharose 4B (Pharmacia). For immunoprecipitation and the PARP assays, fusion proteins were eluted using the glutathione elution buffer containing 10 mM glutathione and 50 mM Tris-HCl (pH 8.0) and were dialyzed against phosphate-buffered saline (PBS). Protein concentrations were determined using protein assay system reagent (Bio-Rad).
In vitro binding assay.
LexA fusion, TAB182C (aa 824 to 867 and 1221 to 1729), FN-tankyrase 1, and Myc-TRF1(2-210) proteins were prepared by using a TNT T7 transcription-translation system (Promega). For GST pull-down assays, 8 μl of LexA fusion or FN-tankyrase 1 was incubated for 1 h at 4°C with GST fusion protein-bound glutathione-Sepharose (containing 1 μg of fusion protein) in TNE buffer (10 mM Tris-HCl [pH 7.8], 1% Nonidet P-40 [NP-40], 150 mM NaCl, and 1 mM EDTA). For the competitive binding assay, 10 μl of FN-tankyrase 1, 20 μl of TAB182C, and 0 to 1 μg of GST-TRF1 or GST were incubated in TNE buffer for 30 min at 4°C. The reaction mixtures were further incubated with protein G-Sepharose (Pharmacia) for 1 h at 4°C. The resulting supernatants were incubated with mouse anti-FLAG antibody (M2; Sigma) for 1 h at 4°C and incubated for an additional 1 h in the presence of protein G-Sepharose. The beads were washed with TNE buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Signals were detected by Western blot analysis (see below).
Western blot analysis.
Western blot analysis was performed (as described previously) (39) with the following primary antibodies: mouse anti-LexA (5397-1; Clontech) (20 ng/ml), rabbit anti-TAB182 (388) (0.5 μg/ml) (39), rabbit anti-GST (1:4,000) (Seimiya and Tsuruo, unpublished), rabbit anti-myc (A-14; Santa Cruz Biotechnology) (0.8 μg/ml), rabbit anti-tankyrase (H-350; Santa Cruz Biotechnology) (2 μg/ml), rabbit anti-poly(ADP-ribose) (Alexis Biochemicals) (1:10,000), rabbit anti-TRF1 (5747) (0.5 μg/ml; see below), and mouse anti-TRF2 (4A794; Imgenex) (2.5 μg/ml).
Transfection.
A standard electroporation method was used with each vector to transfect HeLa I.2.11 cells (subclones of HeLa I containing telomeres of 15 to 25 kb) (49). After incubation for 18 to 21 h at 37°C, cells were processed for further experiments.
Coimmunoprecipitation assay.
Cells were washed in ice-cold PBS and lysed in TNE buffer on ice for 30 min. After centrifugation at 12,000 × g for 10 min, the supernatant was incubated with protein G-Sepharose at 4°C overnight. The resulting supernatants were incubated with either rabbit anti-myc or normal immunoglobulin G for 1 h and further incubated for 1 h in the presence of protein G-Sepharose. The beads were washed once with PBS and four times with TNE buffer and subjected to SDS-PAGE. Western blot analysis was performed with mouse anti-LexA antibody.
Generation of affinity-purified anti-TRF1 antibody.
A Japanese white rabbit was immunized with purified GST-TRF1, following a standard protocol (Sawady Technology). The resulting immune serum, anti-GST-TRF1 5747, was collected and passed through GST-coupled Sepharose 4B. The flowthrough was further applied onto the GST-TRF1-coupled beads, and bound fractions were eluted with 0.1 M glycine (pH 2.3). After immediate neutralization with 1 M Tris (pH 9.5), the peak fractions were dialyzed using PBS.
Immunofluorescence microscopy.
Cells were fixed with 2% paraformaldehyde-PBS and permeabilized with 0.5% NP-40-PBS. Cells were blocked in PBS containing 1% bovine serum albumin and incubated with the following antibodies: mouse anti-FLAG (M2, 2 μg/ml) and affinity-purified rabbit anti-TRF1 (5747) (1 μg/ml) or rabbit anti-FLAG (Sigma) (0.45 μg/ml), mouse anti-TRF2 (4A794) (2.5 μg/ml), and mouse anti-cyclin B (Transduction Laboratories) (2.5 μg/ml). The primary antibodies were detected with fluorescein isothiocyanate- or Texas Red-conjugated donkey anti-rabbit or sheep anti-mouse immunoglobulin (Amersham Pharmacia) (1:25). DNA was stained with 0.2 μg of 4,6-diamino-2-phenylindole (DAPI)/ml. Images were acquired using an Olympus IX-71 microscope with a Photometrics Quantix camera. For some experiments, the levels of intensity of telomeric spots (TRF1 signals) in each cell were quantitated at equal exposure times below saturation; these values were normalized to the quantitated values of DAPI signals in the nuclei of respective cells. Differences in these normalized values of TRF1 signals were evaluated by an unpaired t test.
Retroviral cell lines.
Amphotropic retroviruses were generated by transfecting pLPC/FN-tankyrase 1 plasmids into Phoenix amphotropic cells through the use of standard calcium phosphate precipitation. HTC75 cells (a clonal cell line derived from HT1080 fibrosarcoma cells) (48) were infected with the retrovirus essentially as described previously (40). Infected cells were selected with 2 μg of puromycin/ml and grown in cultures as described previously (9).
Preparation of cellular lysates.
Cells were washed with ice-cold PBS and resuspended in buffer C, consisting of 20 mM HEPES (pH 7.9), 420 mM KCl, 25% glycerol, 0.1 mM EDTA, 5 mM MgCl2, 0.2% NP-40, 1 mM dithiothreitol, and a 1:40 volume of protease inhibitor cocktails (Sigma) on ice for 30 min. After centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was collected as the whole-cell lysate. Nuclear extracts were prepared using a CelLytic NuCLEAR extraction kit (Sigma).
Southern blot analysis.
Genomic DNA was isolated as described previously (13). Telomere restriction fragments (TRF) were detected by Southern blot analysis with a 32P-labeled TTAGGG probe as previously described (11). The mean length of TRF was determined on an Atto densitoscan analyzer with scanned images of autoradiograms.
Tankyrase1 PARP assay.
The same amounts of whole-cell lysates (20 μg of protein) were subjected to Western blot analysis with anti-tankyrase antibody. The signal intensity was quantitated by densitometry (Atto densitoscan analyzer), and the relative abundance of the exogenous FN-tankyrase 1 mutant proteins was determined. In the PARP assay, lysates that contained comparable amounts of FN-tankyrase 1 mutant protein (verified by Western blot analysis as described for Fig. 8A) were incubated with anti-FLAG M2 agarose (Sigma) for 1 h at 4°C. The same amount of lysate that was used for the wild-type FN-tankyrase 1 (5 mg of protein per 10 reactions) was also used for the mock transfectant. The beads were washed with PARP reaction buffer (47) and incubated for 45 min at 25°C in the buffer containing 1.3 μM [32P]NAD+ (4 μCi) and 4 μg of GST-TRF1 per reaction. Under these conditions, the quantitative linear range of the reaction period was verified as lasting up to 1 h. Reactions were terminated by adding 20% trichloroacetic acid (TCA). Acid-insoluble proteins were collected by centrifugation, rinsed in 5% TCA, and fractionated on SDS-PAGE. ADP-ribosylated proteins were detected by autoradiography and/or phosphorimaging with a Fuji BAS imaging analyzer. Since we used an excess amount of GST-TRF1 relative to the enzyme, self-modification of FN-tankyrase 1 was negligible (data not shown). Kinetic assays were performed in the presence of NAD+ (0.1, 0.5, 1, 3, 6, and 10 mM) using identical specific radioactivity levels in all samples. Reactions were stopped with 20% TCA after 0.5, 1, 2, 3, and 4 min followed by filtration through a 96-well UniFilter-GF/C plate (Perkin Elmer). Filters were rinsed five times with 100 μl of 1% TCA, and retained radioactivity levels were determined in a TopCount liquid scintillation counter (Packard; Perkin Elmer). Under these conditions, all PARP labeling occurred within the linear range of the reaction curve for each mutant enzyme (data not shown). Michaelis-Menten constants (Km) were determined by fitting plots of ADP-ribose incorporation rate/substrate concentration values versus substrate concentration values to the Michaelis-Menten equation.
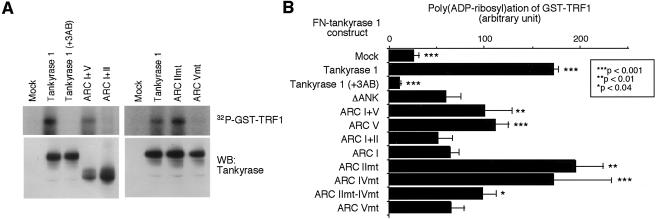
FIG. 8.
Crucial requirement of ARC V for poly(ADP-ribosyl)ation of TRF1 in vitro. (A) Poly(ADP-ribosyl)ation of TRF1 by FN-tankyrase 1 constructs in vitro. Whole-cell extracts were immunoprecipitated with anti-FLAG M2 agarose. The beads were washed and incubated with 4 μg of GST-TRF1 and 1.3 μM [32P]NAD+. +3AB, 1 mM 3-aminobenzamide (3AB) was added prior to addition of GST-TRF1. Reactions were terminated by adding TCA and fractionated by SDS-PAGE. Poly(ADP-ribosyl)ated GST-TRF1 was detected by autoradiography (upper panels). Small aliquots of the beads were subjected to Western blot analysis (WB) with anti-tankyrase antibody (lower panels). mt, mutant. (B) Quantitation of PARP activity in each construct. Radioactivity in the area corresponding to the size of GST-TRF1 was quantitated with a Fuji BAS imaging analyzer. Bars indicate the averages of values obtained in four to five independent experiments. Asterisks indicate statistically significant differences (evaluated using an unpaired t test and FN-tank-ΔANK).
RESULTS
Tankyrase 1 ARCs independently recognize their ligands.
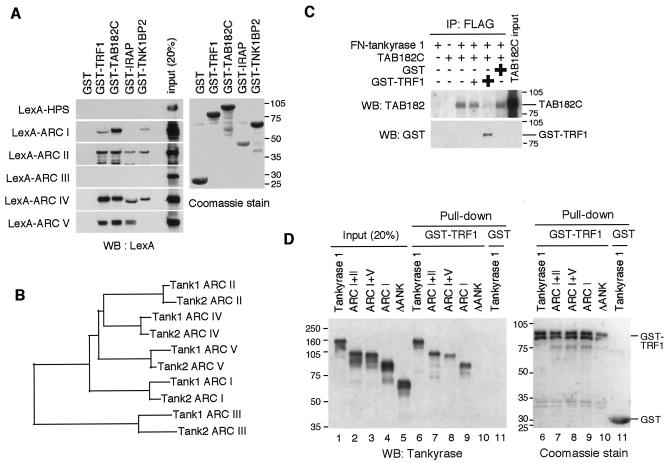
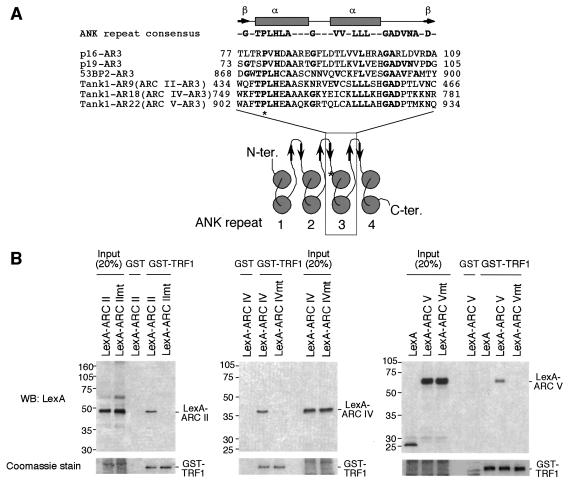
Our previous results from a yeast two-hybrid analysis suggest that isolated ARCs in tankyrase 1 can bind TRF1 and TAB182 (39). To determine whether each ARC actually binds its ligands in vitro, we performed a pull-down assay of LexA-ARC fusion proteins (Fig. 1A) with a series of glutathione S-transferase (GST)-fused tankyrase binding proteins. As shown in Fig. 2A, LexA-ARC I was pulled down by GST-TRF1, GST-TAB182C, and GST-TNK1BP2 (identified as a positive clone through two-hybrid screen for tankyrase 1 binding proteins; H. Seimiya and S. Smith, unpublished data). On the other hand, the presence of another tankyrase binding protein, GST-IRAP (7), or GST alone did not result in the pulling down of LexA-ARC I. None of the GST fusions pulled down the control, LexA-HPS. Meanwhile, ARC II and ARC IV recognized all the fusion proteins but not GST. Consistent with a previous report (36), ARC V bound to GST-TRF1, GST-TAB182C, and GST-IRAP, each of which contained a tankyrase binding RXXPDG motif. On the other hand, ARC V did not recognize GST-TNK1BP2, which did not have the motif. These results indicate that ARCs work as independent binding sites for their ligands in vitro. Unexpectedly, ARC III did not work as a binding site in vitro (not, at least, for any ligands described here). The primary amino acid sequence of each ARC may reflect its binding specificity. Thus, phylogenetic analysis of ARCs in tankyrase 1 and 2 revealed that ARCs II and IV were most closely related whereas ARCs I and V were relatively unique. Compared with the other four ARCs, ARC III was less conserved (Fig. 2B).
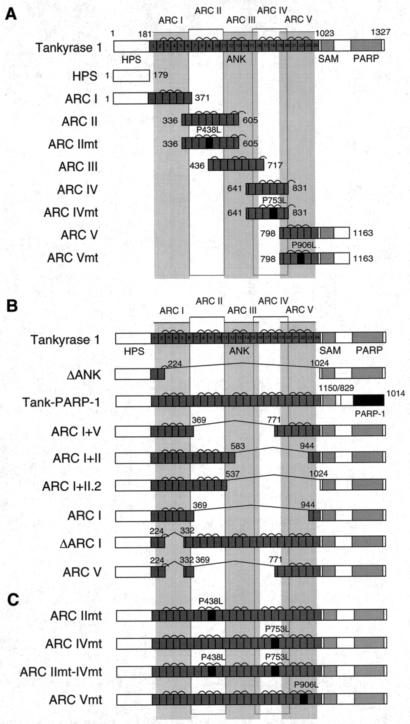
FIG. 1.
Schematic view of tankyrase 1 constructs used in the experiments. (A) LexA-ARC constructs used as described for Fig. 2, 3, and 5. (B and C) FN-tankyrase 1 constructs used as described for Fig. 2D, 4, and 6 to 8. These constructs contain a FLAG epitope tag and an NLS at the N terminus (data not shown). The numbers indicate the positions of amino acid residues. HPS, region containing homopolymeric runs of His, Pro, and Ser; ANK, ankyrin domain; SAM, multimerization domain homologous to the sterile alpha motif; PARP, PARP catalytic domain; ARC, ANK repeat cluster. Bridges above two adjacent ANK repeats indicate the presence of a conserved histidine, contributing to interrepeat stabilization.
FIG. 2.
Tankyrase 1 ARCs work as independent ligand binding sites. (A) In vitro pull-down assay of LexA-ARCs with GST-tankyrase 1 binding proteins. The indicated GST fusions (bound to beads) were incubated with in vitro-translated LexA-ARCs. The bead-bound LexA fusions were detected by Western blot (WB) analysis (left panel). GST fusions were visualized by Coomassie blue staining (right panel). Molecular mass markers (in kilodaltons) are indicated at the right. (B) Phylogenetic tree of ARCs in tankyrase 1 and 2. Amino acid sequences for ARCs were aligned with ClustalW software (http://www.ddbj.nig.ac.jp/E-mail/clustalw-e.html). The tree was created using DendroMaker software (http://www.cib.nig.ac.jp/dda/timanish/dendromaker/home.html). (C) Interaction of tankyrase 1 with TAB182 is competitively inhibited by TRF1. In vitro-translated FN-tankyrase 1 and TAB182C were incubated with increasing amounts of GST-TRF1 (0, 0.1, or 1 μg) or GST (5 μg). FN-tankyrase 1 was immunoprecipitated (IP) with anti-FLAG antibody, and bead-bound TAB182C was detected by Western blot analysis. The filter was reprobed with anti-GST antibody. (D) The presence of a single functional ARC is sufficient for the interaction of tankyrase 1 with TRF1. Deletion mutants were prepared by in vitro translation and subjected to a pull-down assay with GST-TRF1. The bead-bound proteins were detected by Western blot analysis (left panel). The results of Coomassie staining of GST-TRF1 are also shown (right panel). Molecular mass markers (in kilodaltons) are indicated at the left of each panel.
Given that ARC V recognizes a similar RXXPDG motif in TRF1 and TAB182 and that the same set of ARCs bound TRF1 and TAB182, it is likely that binding sites for TRF1 and TAB182 overlap in each ARC. Consistent with this idea was the finding that interaction of TAB182C with FN-tankyrase 1 (full-length tankyrase 1 tagged with a FLAG epitope and a nuclear localization signal (NLS) at the N terminus for further transfection experiments; see below) was competitively blocked by GST-TRF1 (Fig. 2C).
TRF1 has discrete binding sites for multiple ARCs.
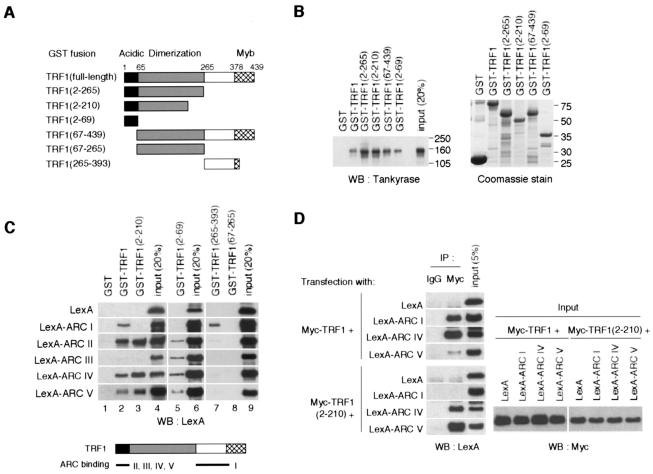
The existence of multiple ARCs as TRF1 binding sites suggests that each ARC recognizes a distinct site in TRF1. To examine this possibility, we first performed an in vitro binding assay of FN-tankyrase 1 with a series of GST-TRF1 fusions (Fig. 3A). In consistency with the results described in a previous report (47), the N-terminal acidic domain of TRF1 (amino acids [aa] 2 to 69) was able to pull down FN-tankyrase 1 (Fig. 3B). Strikingly, a TRF1 fragment that lacked the acidic domain (aa 67 to 439) still bound FN-tankyrase 1, suggesting another binding site for tankyrase 1. Thus, we performed a further binding assay of each ARC (Fig. 1A) with GST-TRF1 fusions. As shown in Fig. 3C, GST-TRF1(2-69) (lane 5) recognized ARCs II, III, IV, and V but not I (again, ARC III did not bind the full-length or aa 2 to 210 of TRF1) (lanes 2 and 3). Meanwhile, GST-TRF1(265-393) recognized ARC I but not ARCs II to V (lane 7). None of the ARCs were recognized by GST-TRF1(67-265) (lane 8). Confirming the in vitro data, transiently expressed ARCs I, IV, and V worked as independent binding sites for Myc-TRF1 in intact cells (Fig. 3D). We could not detect LexA-ARCs II and III in the lysates, probably due to unexpected lability of the proteins when expressed in the cells (data not shown). Again, ARCs IV and V recognized Myc-TRF1(2-210) whereas ARC I did not. Together, these results indicate that ARCs II, III, IV, and V bind the N-terminal acidic domain whereas ARC I binds a domain between the homodimerization and the Myb-like domains of TRF1 (Fig. 3C, bottom panel).
FIG. 3.
Tankyrase 1 ARCs recognize discrete domains of TRF1. (A) GST-TRF1 constructs used in the experiments. The numbers indicate the positions of amino acid residues. (B) Pull-down assays were performed with in vitro-translated FN-tankyrase 1 and various GST-TRF1 fusions. Bead-bound FN-tankyrase 1 was detected by Western blot (WB) analysis (left panel). GST fusions were visualized by Coomassie blue staining (right panel). Molecular mass markers (in kilodaltons) are indicated to the right of each panel. (C) Specificity of ARCs for TRF1 binding. Pull-down assays were performed with in vitro-translated LexA-ARCs and recombinant GST-TRF1 fusions. (Top panel) The bead-bound LexA-fusions were detected as described for Fig. 2A. (Bottom panel) Deduced ARC binding sites in TRF1. (D) ARCs bind TRF1 in intact cells. LexA-ARCs and Myc-TRF1 or Myc-TRF1(2-210) were transiently expressed in HeLa I.2.11 cells, and the lysates were immunoprecipitated (IP) as indicated. The bead-bound LexA fusions were detected by Western blot analysis.
ARC V is important for the TRF1-releasing activity of tankyrase 1.
Tankyrase 1 releases TRF1 from telomere DNA in a PARP activity-dependent manner (9, 27, 45). Here we refer to this effect as the “TRF1-releasing activity” of tankyrase 1. We examined whether multiple ARCs were required for the TRF1-releasing activity and which ARC, if any, was the most important. Figure 1B shows FN-tankyrase 1 deletion mutants in which specific ARCs were truncated by the deletion of actual exons in frame (except for ARC I+II.2) that were deduced from the cDNA and the genomic sequences. Essentially, all the proteins with one or more ARCs bound TRF1 (representative data are shown in Fig. 2D, lanes 6 to 9); however, FN-tank-ΔANK could not bind TRF1 (left panel, lane 10). Consistent with these observations, we found that all the proteins except FN-tank-ΔANK bound GST-TAB182C (data not shown). These results demonstrate that the presence of a single ARC is sufficient for interaction of tankyrase 1 with TRF1 or TAB182.
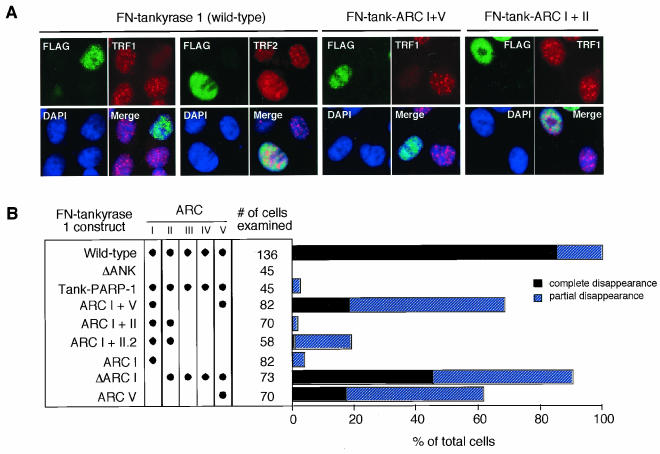
Having confirmed TRF1 binding, we assessed the TRF1-releasing activity of the mutants. Since most of the overexpressed tankyrase 1 is accumulated in the cytoplasm (44) and tankyrases may have an innate role for some cytoplasmic events (7, 22, 29, 36, 39), we introduced an NLS at the N terminus of tankyrase 1 (45). This modification allowed us to see the effect of the presence of exogenous tankyrase 1 (FN-tankyrase 1) exclusively in the nucleus. In consistency with previous reports (9, 45), we saw that overexpression of wild-type FN-tankyrase 1 in HeLa I.2.11 cells resulted in the disappearance of the telomere-like dots of TRF1 but not TRF2, which does not bind tankyrase 1 (47) (Fig. 4). In contrast, neither the FN-tank-ΔANK nor the tank-PARP-1 chimera, in which the tankyrase 1 PARP domain was replaced with that of PARP-1 (Fig. 1B), exhibited any activity. These results indicate that both TRF1 binding and an isozyme-specific PARP activity are required for TRF1-releasing activity. Among other mutants, none exhibited activity comparable with that of the wild type in spite of their abilities to bind TRF1. This could be due mainly to the fact that they were deletion mutants and not intact proteins. In fact, FN-tank-ARC I+II or I was almost completely inactivated. Nevertheless, several mutants that had intact ARC V (FN-tank-ARC I+V, V, and ΔARC I) maintained partial activity (17 to 45% of transduced cells showed the complete disappearance of the TRF1 dots). In similarity to the results seen with wild-type FN-tankyrase 1, all the mutants showed diffuse staining patterns in the nucleus. There were no substantial differences in the expression levels between the wild type and the mutants or noticeable degradation products in the cellular lysates (data not shown). These observations suggest a major requirement of ARC V for the TRF1-releasing activity of tankyrase 1. To explore the possibility that the distance of ARC V from the SAM-PARP domain is important for the TRF1-releasing activity, we also examined FN-tank-ARC I+II.2, in which the distance of ARC II from the SAM-PARP domain was designed to be similar to that of ARC V in the wild-type tankyrase 1 (Fig. 1B). Although it exhibited a slightly higher activity than original FN-tank-ARC I+II, the TRF1 signal was not affected at all in the majority of the transfected cells (Fig. 4B). Thus, ARC V did not appear to be replaceable with respect to other ARCs. Given that ARC V showed a relatively unique amino acid sequence and specificity for ligand binding (Fig. 2A and B), it seems that compared with those of ARC II or ARC IV, the sequence of ARC V is important and the distance of ARC from the SAM-PARP domain per se is not sufficient for tankyrase 1 activity.
FIG. 4.
Tankyrase 1 deletion mutants retaining ARC V still show TRF1-releasing activity. (A) Effects of transient overexpression of FN-tankyrase 1 constructs on telomeric localization of TRF1 in HeLa I.2.11 cells. Cells were transfected with vectors as indicated. FN-tankyrase 1 constructs, TRF1, and TRF2 were detected by indirect immunofluorescence staining with anti-FLAG M2 (green), anti-TRF1 5747 (red), and anti-TRF2 4A794 (red) antibodies, respectively. DAPI staining of DNA is shown in blue. (B) Quantitation of the TRF1-releasing activity. Cells displaying overexpression of the FN-tankyrase 1 constructs were classified into three categories (depending on the appearance of TRF1 dots): complete disappearance, partial disappearance (i.e., observed but obviously decayed compared with those in surrounding nontransduced cells), and no effect (i.e., intensity comparable with that of mock- and nontransduced cells). Closed circles indicate the existence of intact ARCs.
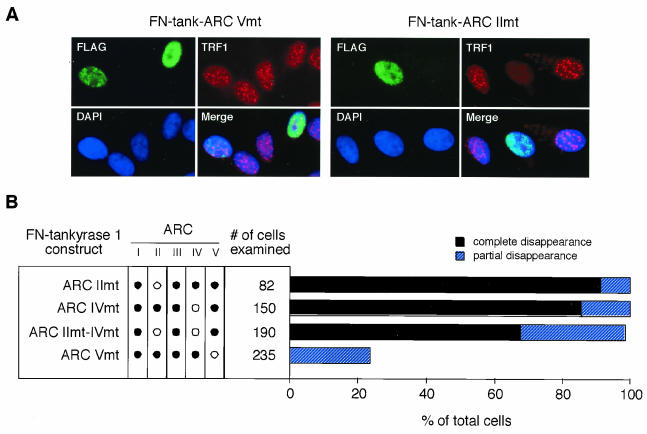
Next, we tried to inactivate the TRF1 binding activity of ARC V. Typically, an individual ANK repeat has an L-shaped structure consisting of a β hairpin followed by two α-helices that pack in an antiparallel fashion (Fig. 5A) (38). We introduced a point mutation in the third ANK repeat of each ARC, which resulted in the alteration of the fifth proline into leucine. This proline is important in maintaining the ANK repeat structure, and its replacement by leucine abolishes the function despite the near-native level of the secondary structure (28, 50). A relevant mutation has been described for the ankyrin domain of p16INK4a, a cyclin D-dependent kinase inhibitor, as a somatic mutation derived from familial melanoma (20). The resulting mutant p16INK4a is incapable of binding its partner, cyclin-dependent kinase 4 (35). As expected, LexA-ARC II, IV, or V mutants harboring the Pro→Leu substitution (Fig. 1A) did not bind TRF1 in vitro (Fig. 5B). Thus, we introduced the same point mutations into the full-length FN-tankyrase 1 and examined the TRF1-releasing activity of the resulting constructs, FN-tank-ARC IImt, IVmt, and Vmt (Fig. 1C). As shown in Fig. 6, both FN-tank-ARC IImt and IVmt had activities comparable with those of the wild type. Furthermore, the double mutant, FN-tank-ARC IImt-IVmt, still retained activity although the ratio of “complete disappearance” was slightly reduced. On the other hand, FN-tank-ARC Vmt showed significant reduction in the level of the activity. We found no substantial differences in the expression levels between the constructs or noticeable degradation products in the cellular lysates (data not shown). Taken together, these observations indicate an important role for ARC V in the TRF1-releasing activity of tankyrase 1.
FIG. 5.
ARC point mutations that disrupt the TRF1 binding. (A) (Top panel) Secondary structure of ANK repeat. Arrows and rectangles indicate the approximate positions of the β-strands and α-helices, respectively. (Middle panel) Alignment of the third ANK repeat (AR) in several ANK family proteins and tankyrase 1 ARCs II, IV, and V. Conserved residues are in boldface type. (Bottom panel) Topological diagram of the secondary-structure elements of typical ARC. Circles indicate the α-helices with helix axes perpendicular to the plane of the figure. The asterisk indicates the conserved proline, which was replaced with leucine in this study. (B) Replacement of the conserved proline with leucine abolished the TRF1 binding of ARCs. In vitro-translated LexA-ARC mutants (mt) were subjected to a pull-down assay with GST-TRF1 or GST (as described for Fig. 2A). The bead-bound LexA fusions were detected by Western blot analysis (WB) (top panels). The results of Coomassie blue staining of GST-TRF1 are also shown (bottom panels).
FIG. 6.
Inactivation of ARC V abolishes the TRF1-releasing activity of tankyrase 1. (A) Effects of transient overexpression of the point mutant FN-tankyrase 1 on telomeric localization of TRF1 in HeLa I.2.11 cells. Cells were transfected with vectors as indicated. FN-tankyrase 1 constructs (green), TRF1 (red), and DNA (blue) were detected as described for Fig. 4A. (B) Quantitation of the TRF1-releasing activity. Cells displaying overexpression of the FN-tankyrase 1 constructs were classified into three categories as described for Fig. 4B. Closed and open circles indicate wild-type and Pro→Leu mutant (mt) ARCs, respectively.
ARC V is required but not sufficient for telomere elongation by tankyrase 1.
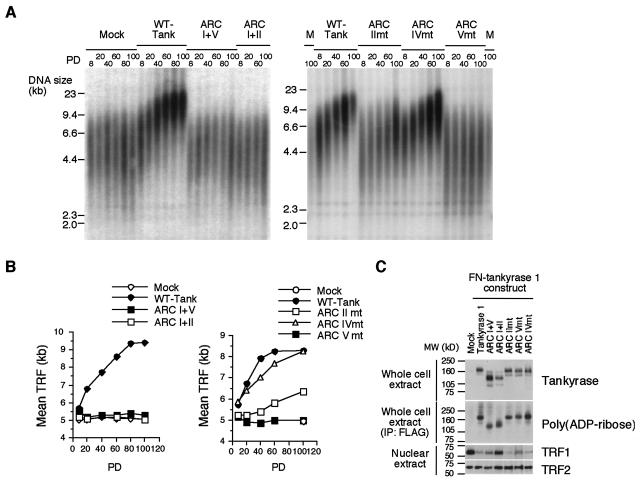
We next examined whether the mutant tankyrase 1 could mimic telomere elongation induced by overexpression of the wild-type tankyrase 1. Stable HTC75 fibrosarcoma cell lines expressing the FN-wild type or mutant tankyrase 1 were cultivated for up to 100 population doublings (PD). As shown in Fig. 7A, overexpression of the wild-type FN-tankyrase 1 induced telomere elongation. Telomeres showed progressive elongation at rates of 51 ± 10 bp per PD (Fig. 7B), in similarity to previous results (9, 45). Eventually, the telomeres were stabilized, indicating that a feedback mechanism was operating. Overexpression of FN-tank-ARC IImt, ARC IVmt, or ARC IImt-IVmt also induced telomere elongation (Fig. 7A, right panel, 7B, right panel, and data not shown). Compared with the wild-type FN-tankyrase 1, FN-tank-ARC IVmt elongated telomeres at relatively similar rates (32 ± 3 bp/PD) whereas FN-tank-ARC IImt and ARC IImt-IVmt exhibited reduced rates (17 ± 2 bp/PD and 20 ± 2 bp/PD, respectively). We repeated the same experiments two to three times for each point mutant and obtained similar results (data not shown). In contrast, overexpression of FN-tank-ARC Vmt had no effect on telomere length during the course of cultivation. Together, these results indicate that ARC V is required for telomere elongation by FN-tankyrase 1. Meanwhile, we found that the presence of ARC V itself was not sufficient for telomere elongation (Fig. 7A, left panel, and 7B, left panel). Thus, overexpression of FN-tank-ARC I+V did not elongate telomeres in spite of its partial TRF1-releasing activity. The mutant proteins were overexpressed to an extent similar to that seen with the wild type (Fig. 7C). Percentages of overexpressed cells among populations between the cell lines were similar (80 to 95%) (data not shown).
FIG. 7.
Differential requirements of multiple ARCs for telomere elongation by tankyrase 1. (A) Effects of overexpression of FN-tankyrase 1 constructs on telomere length in HTC75 cells. TRF at the indicated PD were detected by Southern blot analysis. The results of two representative experiments are shown. M, mock; WT-Tank, wild-type FN-tankyrase 1; mt, mutant. (B) Graphic representations of telomere length change. The plots represent the mean TRF values derived the results presented in panel A. (C) Western blot analysis of whole-cell or nuclear extracts (20 μg of proteins per lane). Blots were probed with antibodies as indicated. For detection of self-poly(ADP-ribosyl)ation, lysates were immunoprecipitated (IP) with anti-FLAG antibody and the pellets were subjected to Western blot analysis with anti-poly(ADP-ribose). MW, molecular mass.
In consistency with the TRF1-releasing activity and telomere elongation results, wild-type FN-tankyrase 1, FN-tank-ARC IImt and IVmt, and ARC IImt-IVmt induced loss of TRF1 (6, 9, 45) (Fig. 7C and data not shown). FN-tank-ARC I+V, but not I+II, partially reduced the TRF1 level. Of note, FN-tank-ARC Vmt also diminished the TRF1 level, which could be attributed to a residual level of TRF1-releasing activity in HeLa I.2.11 cells (Fig. 6). Further, we performed an indirect immunofluorescence stain analysis of TRF1 in the stable FN-tank-ARC Vmt-expressing HTC75 cells. Semiquantitative analysis of the signal intensity revealed that the cells exhibited a slightly weak TRF1 signal (75% of mock cells; P < 0.05), but the signal was much stronger than that seen with the wild-type FN-tankyrase 1-expressing cells (27% of mock cells [P < 0.0001 for FN-tankyrase 1-expressing cells and mock, and P < 0.0001 for FN-tankyrase 1- and FN-tank-ARC Vmt-expressing cells]; data not shown). Together, these observations indicate that FN-tank-ARC Vmt retains (to a small extent) the ability to release TRF1 from telomeres. As expected, TRF2 was not affected by overexpression of any constructs described herein. Similar levels of self-poly(ADP-ribosyl)ation (45, 47) of each mutant were observed but did not correlate with telomere elongation. Together, these results support an important role for ARC V in the telomere function of tankyrase 1.
ARC V is essential for efficient poly(ADP-ribosyl)ation of TRF1 in vitro.
Telomere function of tankyrase 1 depends on its PARP activity (9, 27, 45). Thus, we next examined this activity of each mutant with TRF1 in vitro. The wild-type or mutant FN-tankyrase 1 in HTC75 cells was affinity purified and incubated with GST-TRF1 and [32P]NAD+ as a substrate. As shown in Fig. 8, the wild-type FN-tankyrase 1 poly(ADP-ribosyl)ated GST-TRF1. This activity was inhibited by 3-aminobenzamide, a PARP inhibitor, in a dose-dependent manner (up to 1 mM) (Fig. 8 and data not shown) and was not detected on GST alone (reference 39 and data not shown). An immunocomplex from mock cells did not show the activity. FN-tank-ΔANK showed a residual activity, but we assumed it was within a background level. Since each FN-tankyrase 1 mutant protein retained the sterile alpha motif (SAM) domain (Fig. 1B and C), it is possible that a portion of endogenous tankyrase 1 formed complexes with the mutant proteins via SAM-SAM homophilic interaction (14) and was therefore coimmunoprecipitated with anti-FLAG antibody. Although such endogenous tankyrase 1 was not detected, at least by Western blot analysis (Fig. 8A), this situation might give some background signal in subsequent PARP assays. Alternatively, TRF1 may interact weakly with tankyrase 1 via another domain (e.g., the HPS [a region containing homopolymeric runs of His, Pro, and Ser] or SAM domains) although such interaction was not detected in the binding assay (Fig. 2A and D). Among the deletion mutants, FN-tank-ARC I+V and V exhibited partial activity but others did not. Among the point mutants, FN-tank-ARC IImt and IVmt retained activity comparable with that of the wild type whereas FN-tank-ARC IImt-IVmt exhibited partial activity. An important note is that FN-tank-ARC Vmt was significantly inactivated. We performed further kinetic assays for representative constructs, including wild-type FN-tankyrase 1, FN-tank-ARC I+V, FN-tank-ARC IImt, and FN-tank-ARC Vmt, with GST-TRF1 and NAD+ as the substrate. Km values of 0.9 mM for wild-type FN-tankyrase 1, 7.7 mM for FN-tank-ARC I+V, and 1.3 mM for FN-tank-ARC IImt were determined. As for FN-tank ARC-Vmt, we could not determine a reliable Km value since its enzymatic activity level was very low (data not shown). These results are consistent with those seen with the TRF1-releasing activity and demonstrated that ARC V was essential for efficient poly(ADP-ribosyl)ation of TRF1.
DISCUSSION
In this study, we have demonstrated that multiple ARCs in tankyrase 1 work as independent binding sites for TRF1 in vitro and in intact cells (Fig. 2 and 3). The independence of each ARC is consistent with the fact that many ANK family proteins have only a single ARC, comprising 4 to 7 ANK repeats, as a ligand binding site (38). Ankyrins, as well as tankyrases, have 24 ANK repeats; ankyrin binding with its ligand (an anion exchanger) seems to be distinct from that of tankyrase 1 with TRF1, however, since ankyrin requires 12 repeats for ligand binding (30). This contrast stems from the presence of the mutually distinct structures of the ankyrin domains in tankyrases and ankyrins (39). Thus, ankyrins have almost perfect 33-amino-acid repeats that form a contiguous spiral stack and provide an extensive surface area for protein interactions (31). In contrast, the ANK repeats of tankyrases do not seem to form such three-dimensional structures since they bear a number of insertions and deletions between ARCs (39).
Among the five ARCs, ARC V displayed an essential role for (i) poly(ADP-ribosyl)ation of TRF in vitro, (ii) release of TRF1 from telomeres, and (iii) progressive elongation of telomeres (Fig. 4 and 6 to 8). Of interest, ARC V seems to prefer the N-terminal fragment of TRF1 rather than the full-length protein, which can form a homodimer although this preference is not exclusive (Fig. 3C and D). Whether this feature of ARC V is functionally associated with the biological activity of tankyrase 1 remains to be elucidated. Since ARC V is positioned at the C terminus of the ankyrin domain, it may have a greater opportunity for presenting TRF1 to the neighboring PARP domain. In this case, poly(ADP-ribosyl)ation of TRF1 may be mediated by a “catalytic center” consisting of ARC V and a PARP domain, both of which are hinged by a SAM domain. According to this model, FN-tank-ARC Vmt has a dysfunctional catalytic center, making it completely inactive. Meanwhile, FN-tank-ARC IImt, IVmt, and IImt-IVmt retain the catalytic centers, making them capable of poly(ADP-ribosyl)ating TRF1.
Nevertheless, the presence of the deletion mutant, FN-tank-ARC I+V, which retained the catalytic center, did not elongate telomeres at all (Fig. 7). This observation suggests an essential function of the other ARCs that may be distinct from TRF1 binding. Division of the tankyrase ankyrin domain into five ARCs is also seen in other species from insects to vertebrates (14). Interestingly, two adjacent ARCs are linked by a characteristic LLEAAR/K motif (14), which is well conserved and often observed in various metabolic enzymes that bind NAD+ and ATP. Since the FN-tank-ARC I+V lacks two of the four LLEAAR/K motifs, it is possible that this mutant protein cannot recruit NAD+ efficiently to the poly(ADP-ribosyl)ation of TRF1. Actually, the Km value of FN-tank-ARC I+V for NAD+ was approximately ninefold higher than that of the wild-type tankyrase 1 (data not shown). Meanwhile, tankyrases can multimerize through their SAM domains (references 14 and 37; H. Seimiya and T. Tsuruo, unpublished observation), suggesting that tankyrases act as scaffolds that regulate assembly of larger protein complexes. It is possible that the tankyrase ankyrin domain, consisting of multiple ARCs, plays an essential role as a structural frame of the scaffold. Alternatively, the existence of multiple ARCs may enhance access of tankyrase 1 to the specific area on the telomere where TRF1 release is crucial for telomerase-dependent synthesis of the telomeric repeats. In fact, although the findings with respect to release of TRF1 from telomeres (Fig. 6), loss of TRF1 protein (Fig. 7C), and poly(ADP-ribosyl)ation of TRF1 (Fig. 8) were almost identical for wild-type FN-tankyrase 1- and FN-tank-ARC IImt-transduced cells, the mutant displayed reduced rates of telomere elongation (Fig. 7A and B). According to this scenario, it seems possible that FN-tank-ARC IImt-overexpressed cells still had residual levels of TRF1 at crucial loci in the telomeres but with a signal too weak to be detected by indirect immunofluorescence stain analysis and indistinguishable by Western blot analysis.
Tankyrase 1 resides with TRF1 on telomeres (9, 44, 47), indicating that there are conditions in vivo under which tankyrase 1 forms a complex with TRF1 but does not poly(ADP-ribosyl)ate it. In such a telomere complex, ARC V might be free from TRF1, putting tankyrase 1 in its resting state. Otherwise, interaction of ARC V with TRF1 may lead to ADP-ribosylation of the latter, resulting in dissociation of the complex from telomeres. In that sense, it is possible that interaction of ARC V with TRF1 is regulated in the cell cycle or in other context-dependent manners. Another tankyrase binding protein, TAB182, interacted with ARC V in vitro (Fig. 2) but not in yeast (39), suggesting a context-dependent interaction of ARC V with ligands (this might be also the case with ARC I, since it binds TAB182 in vitro but not in yeast). Interestingly, transient overexpression of FN-tank-ARC V (or I+V) in HeLa I.2.11 cells (Fig. 4) resulted in the display of discrepant effects on the telomeric TRF1 in each single cell. The telomeric TRF1 dots completely disappeared in a fraction of the overexpressed cells (17%), whereas in others (39%) they were not affected at all. This observation supports the hypothesis of a context dependence of the TRF1-releasing activity. Thus, we stained the FN-tank-ARC V-transfected cells with anti-cyclin B. Cyclin B is not present in the G1 phase, accumulates in the cytoplasm during S and G2 phases, and translocates to the nucleus before nuclear envelope breakdown during prophase (34). Among the cells observed (n = 499, including nontransfected fractions), 83.0% were cyclin B negative (G1 to early S phases) whereas 13.8 and 3.2% expressed cyclin B in the cytoplasm and in the nucleus, respectively. When the cells were costained with anti-FLAG antibody, 81.8% of FLAG-positive cells (n = 303) were cyclin B negative whereas 17.5 and 0.7% expressed cyclin B in the cytoplasm and in the nucleus, respectively. These two results reveal quite similar distribution characteristics and verify that transient expression of FN-tank-ARC V was not restricted to specific stages of the cell cycle. Next, the cells were stained with anti-cyclin B and anti-TRF1 antibodies. Of 53 cells that lost the telomeric dots of TRF1, none expressed cyclin B either in the cytoplasm or in the nucleus (Seimiya and Tsuruo, unpublished). These results suggest that the release of TRF1 from telomeres in FN-tank-ARC V-expressing cells occurred during G1 to mid-S phases but not at late S to G2/M phases. So far, we cannot conclude from the physiological contexts whether TRF1 dissociates from telomeres during such periods. Unlike PARP-1, tankyrase 1 is not activated by damaged DNA or by telomeric DNA (9). It will be interesting to determine how and when tankyrase 1 is activated.
The present binding studies (Fig. 2 and 3) demonstrated unexpected features of other ARCs. First, ARC I recognized TRF1 at a novel C-terminal locus. This locus does not appear to contain the previously reported ankyrin recognition motif RXXPDG (36). So far, we have failed to identify a minimal sequence for ARC I recognition in TRF1, probably due to improper folding of smaller TRF1 deletion mutants. Thus, it remains to be determined which amino acid residues are crucial for ARC I recognition and whether the motif is conserved in other species. Possible implications for the bipartite tankyrase 1 binding domains on TRF1 include preferential recognition of a specific structure of the telomere-bound TRF1 dimer by tankyrase 1, an allosteric effect on poly(ADP-ribosyl)ation, formation of higher-order complexes of telomeric heterochromatin, and so on. We deleted the whole ARC I domain from FN-tankyrase 1 to assess the importance of the ARC I-TRF1 interaction. Although the resulting mutant lost partial activity when overexpressed in HeLa I.2.11 and HTC75 cells, it retained its TRF1-releasing activity (Fig. 4) and elongated telomeres (+31 bp/PD; data not shown), respectively. Thus, we conclude that ARC I is less important than ARC V for the telomeric function of tankyrase 1. The biological significance of the ARC I-TRF1 interaction still remains to be determined. Meanwhile, ARC III, when exactly trimmed, did not work as a ligand binding site in vitro. One possibility is that proper folding of ARC III depends on the neighboring ANK repeats, as previously suggested for other ANK family proteins (16). In fact, ARC III has fewer ANK repeats (39) and thus may need flanking repeats as scaffolds. Alternatively, ARC III may recognize TRF1 only in certain contexts, since the isolated acidic domain (aa 2 to 69), but not other longer fragments of TRF1, bound ARC III in vitro (Fig. 3C). This implies the presence of a domain within amino acids 69 to 210 of TRF1 that negatively affects ARC III binding. The role of ARC III in TRF1 recognition in intact cells, if one exists, remains elusive.
Tankyrase 1 is present in the cytoplasm and in the nuclear extract at similar levels (39). Anti-TRF1 antibodies coimmunoprecipitate endogenous tankyrase 1, whereas TRF1 is not coimmunoprecipitated by anti-tankyrase 1 antibodies from HeLa cell extracts (9). These observations suggest that the majority of tankyrase 1 resides in nontelomeric locations and is therefore not complexed to TRF1. In fact, tankyrase 1 is present at multiple loci, including centrosomes and nuclear pore and Golgi complexes (7, 44). In addition, overexpression of a catalytically inactive tankyrase 1 mutant does not exhibit a dominant-negative effect on telomere length of HTC75 cells (9). Thus, it is possible that only a small portion of endogenous tankyrase 1 interacts with TRF1 and is sufficient for telomere length regulation.
We have demonstrated herein that tankyrase 1 ARCs work as independent binding sites for TRF1, TAB182, and other ligands. Note that ARC V, the ARC in the most C-terminal position, played a crucial role in the telomeric function of tankyrase 1. This report provides new insight into the molecular mechanisms for macromolecular recognition by ANK repeat proteins and for telomere length regulation by posttranslational modification of telomeric proteins. Since tankyrase 1 ARCs also worked as multiple binding sites for other partners, such as TAB182, it is possible that the functions of those proteins are regulated by tankyrase 1 in a similar fashion.
Acknowledgments
We are grateful to Tomokazu Ohishi for technical assistance.
This work was funded in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and grants of the Uehara Memorial Foundation and the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim.
REFERENCES
- 1.Bianchi, A., S. Smith, L. Chong, P. Elias, and T. de Lange. 1997. TRF1 is a dimer and bends telomeric DNA. EMBO J. 16:1785-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianchi, A., R. M. Stansel, L. Fairall, J. D. Griffith, D. Rhodes, and T. de Lange. 1999. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 18:5735-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 4.Blasco, M. A., H.-W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 5.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C.-P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 6.Chang, W., J. N. Dynek, and S. Smith. 2003. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 17:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi, N.-W., and H. F. Lodish. 2000. Tankyrase is a Golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 275:38437-38444. [DOI] [PubMed] [Google Scholar]
- 8.Chong, L., B. van Steensel, D. Broccoli, H. Erdjument-Bromage, J. Hanish, P. Tempst, and T. de Lange. 1995. A human telomeric protein. Science 270:1663-1667. [DOI] [PubMed] [Google Scholar]
- 9.Cook, B. D., J. N. Dynek, W. Chang, D. Shostak, and S. Smith. 2002. A role for the related poly(ADP-ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22:332-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lange, T. 1992. Human telomeres are attached to the nuclear matrix. EMBO J. 11:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 13.de Lange, T., L. Shiue, R. M. Myers, D. R. Cox, S. L. Naylor, A. M. Killery, and H. E. Varmus. 1990. Structure and variability of human chromosome ends. Mol. Cell. Biol. 10:518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Rycker, M., R. N. Venkatesan, C. Wei, and C. M. Price. 2003. Vertebrate tankyrase domain structure and sterile-alpha motif (SAM) mediated multimerization. Biochem. J. 372:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fairall, L., L. Chapman, H. Moss, T. de Lange, and D. Rhodes. 2001. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8:351-361. [DOI] [PubMed] [Google Scholar]
- 16.Gorina, S., and N. P. Pavletich. 1996. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science 274:1001-1005. [DOI] [PubMed] [Google Scholar]
- 17.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 18.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 19.Hemann, M. T., M. A. Strong, L. Y. Hao, and C. W. Greider. 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67-77. [DOI] [PubMed] [Google Scholar]
- 20.Hussussian, C. J., J. P. Struewing, A. M. Goldstein, P. A. Higgins, D. S. Ally, M. D. Sheahan, W. H. J. Clark, M. A. Tucker, and N. C. Dracopoli. 1994. Germline p16 mutations in familial melanoma. Nat. Genet. 8:15-21. [DOI] [PubMed] [Google Scholar]
- 21.Imai, Y., Y. Matsushima, T. Sugimura, and M. Terada. 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaminker, P. G., S.-H. Kim, R. D. Taylor, Y. Zebarjadian, W. D. Funk, G. B. Morin, P. Yaswen, and J. Campisi. 2001. TANK2, a new TRF1-associated PARP, causes rapid induction of cell death upon overexpression. J. Biol. Chem. 276:35891-35899. [DOI] [PubMed] [Google Scholar]
- 23.Karlseder, J., A. Smogorzewska, and T. de Lange. 2002. Senescence induced by altered telomere state, not telomere loss. Science 295:2446-2449. [DOI] [PubMed] [Google Scholar]
- 24.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. C. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011-2015. [DOI] [PubMed] [Google Scholar]
- 25.Kipling, D., and H. J. Cooke. 1990. Hypervariable ultra-long telomeres in mice. Nature 347:400-402. [DOI] [PubMed] [Google Scholar]
- 26.Lee, H.-W., M. A. Blasco, G. J. Gottlieb, J. W. Horner II, C. W. Greider, and R. A. DePinho. 1998. Essential role of mouse telomerase in highly proliferative organs. Nature 392:569-574. [DOI] [PubMed] [Google Scholar]
- 27.Loayza, D., and T. de Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423:1013-1018. [DOI] [PubMed] [Google Scholar]
- 28.Luh, F. Y., S. J. Archer, P. J. Domaille, B. O. Smith, D. Owen, D. H. Brotherton, A. R. C. Raine, X. Xu, L. Brizuela, S. L. Brenner, and E. D. Laue. 1997. Structure of the cyclin-dependent kinase inhibitor p16Ink4d. Nature 389:999-1003. [DOI] [PubMed] [Google Scholar]
- 29.Lyons, R. J., R. Deane, D. K. Lynch, Z.-S. J. Ye, G. M. Sanderson, H. J. Eyre, G. R. Sutherland, and R. J. Daly. 2001. Identification of a novel tankyrase through its interaction with the adaptor protein Grb14. J. Biol. Chem. 276:17172-17180. [DOI] [PubMed] [Google Scholar]
- 30.Michaely, P., and V. Bennett. 1993. The membrane-binding domain of ankyrin contains four independently folded subdomains, each comprised of six ankyrin repeats. J. Biol. Chem. 268:22703-22709. [PubMed] [Google Scholar]
- 31.Michaely, P., D. R. Tomchick, M. Machius, and G. W. Anderson. 2002. Crystal structure of a 12 ANK repeat stack from human ankyrin R. EMBO J. 21:6387-6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morin, G. B. 1989. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59:521-529. [DOI] [PubMed] [Google Scholar]
- 33.Nugent, C. I., and V. Lundblad. 1998. The telomerase reverse transcriptase: components and regulation. Genes Dev. 12:1073-1085. [DOI] [PubMed] [Google Scholar]
- 34.Pines, J., and T. Hunter. 1991. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 115:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranade, K., C. J. Hussussian, R. S. Sikorski, H. E. Varmus, A. M. Goldstein, M. A. Tucker, M. Serrano, G. J. Hannon, D. Beach, and N. C. Dracopoli. 1995. Mutations associated with familial melanoma impair p16INK4 function. Nat. Genet. 10:114-116. [DOI] [PubMed] [Google Scholar]
- 36.Sbodio, J. I., and N.-W. Chi. 2002. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 277:31887-31892. [DOI] [PubMed] [Google Scholar]
- 37.Sbodio, J. I., H. F. Lodish, and N. W. Chi. 2002. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem. J. 361:451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sedgwick, S. G., and S. J. Smerdon. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24:311-316. [DOI] [PubMed] [Google Scholar]
- 39.Seimiya, H., and S. Smith. 2002. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J. Biol. Chem. 277:14116-14126. [DOI] [PubMed] [Google Scholar]
- 40.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 41.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33:787-791. [DOI] [PubMed] [Google Scholar]
- 42.Sherr, C. J., and R. A. DePinho. 2000. Cellular senescence: mitotic clock or culture shock? Cell 102:407-410. [DOI] [PubMed] [Google Scholar]
- 43.Smith, S. 2001. The world according to PARP. Trends Biochem. Sci. 26:174-179. [DOI] [PubMed] [Google Scholar]
- 44.Smith, S., and T. de Lange. 1999. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112:3649-3656. [DOI] [PubMed] [Google Scholar]
- 45.Smith, S., and T. de Lange. 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10:1299-1302. [DOI] [PubMed] [Google Scholar]
- 46.Smith, S., and T. de Lange. 1997. TRF1, a mammalian telomeric protein. Trends Genet. 13:21-26. [DOI] [PubMed] [Google Scholar]
- 47.Smith, S., I. Giriat, A. Schmitt, and T. de Lange. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484-1487. [DOI] [PubMed] [Google Scholar]
- 48.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 49.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, B., and Z. Peng. 1996. Defective folding of mutant p16INK4 proteins encoded by tumor-derived alleles. J. Biol. Chem. 271:28734-28737. [PubMed] [Google Scholar]