Abstract
Positive responses to combined androgen elimination therapy and radiation therapy have been well documented in the treatment of prostate cancer patients. The detailed mechanisms how androgen-androgen receptor (AR) cross talks to the radiation-related signal pathways, however, remain largely unknown. Here we report the identification of hRad9, a key member of the checkpoint Rad protein family, as a coregulator to suppress androgen-AR transactivation in prostate cancer cells. In vivo and in vitro interaction assays using Saccharomyces cerevisiae two-hybrid, mammalian two-hybrid, glutathione S-transferase pull-down, and coimmunoprecipitation methods prove that AR can interact with the C terminus of hRad9 via its ligand binding domain. The FXXLF motif within the C terminus of hRad9 interrupts the androgen-induced interaction between the N terminus and C terminus of AR. This interaction between AR and hRad9 may result in the suppression of AR transactivation, demonstrated by the repressed AR transactivation in androgen-induced luciferase reporter assay and the reduced endogenous prostate-specific antigen expression in Western blot assay. Addition of small interfering RNA of hRad9 can reverse hRad9 suppression effects, which suggests that hRad9 functions as a repressor of AR transactivation in vivo. Together, our data provide the first linkage between androgen-AR signals and radiation-induced responses. Further studies of the influence of hRad9 on prostate cancer growth may provide potential new therapeutic approaches.
The androgen receptor (AR) is a member of the steroid receptor superfamily and plays a central role in the differentiation, growth, and maintenance of male-specific organs (6, 45, 57, 71). The AR contains three domains: a C-terminal ligand binding domain (LBD), a DNA binding domain (DBD), and an N-terminal transactivation domain (10, 11, 42, 49). In the absence of agonists, the AR protein is found associated with heat shock proteins in the cytosol. Upon binding to ligand, the AR undergoes a conformational change, dissociates from the heat shock proteins, and translocates to the nucleus, where it binds to androgen response elements located in the target genes (9).
Steroid receptors may be modulated by other regulatory proteins in cells by direct or indirect interactions (29, 30, 52, 53). A number of transcriptional coregulators, including coactivators and corepressors, that enhance or suppress the interactions between steroid receptors and the basal transcriptional machinery have been identified (29, 31, 34, 51, 55, 68, 73). The p160/steroid receptor coactivator (SRC) family is the most clearly defined class of coactivators, including SRC-1, SRC-2/TIF2, and SRC-3/AIB1/pCIP/RAC3 (19, 47, 52). Interaction between ligand-activated steroid receptors and the p160 coactivators is mediated by a small α-helical motif containing the LXXLL sequence (where L is leucine and X is any amino acid) (50). Ligand binding leads to realignment of helix 12 in the LBD, revealing a hydrophobic groove where the LXXLL motifs bind (4, 14, 17, 28). In addition to LXXLL motifs, a number of AR coregulators, such as ARA54 and ARA70, interact with AR in an androgen-dependent manner through FXXLF motifs (where F is phenylalanine) (26, 37, 70). Furthermore, the FXXLF motif located in the AR N-terminal region is found to mediate the interaction between the LBD and N terminus of AR (N-terminal-C-terminal [N-C] interaction), which is important for the full AR transactivation capacity (8, 25, 41). The phage display technique demonstrated that the FXXLF motif is a ligand-dependent AR-associated peptide motif (33).
Unrepaired DNA lesions, arising from either intrinsic or exogenous sources, lead to genomic instability and consequently contribute to the development of cancers (24). Cell cycle checkpoints and DNA repair are the primary defenses against genomic instability (21, 24, 56). hRad9, a member of the Rad family of checkpoint proteins, is involved in detection of DNA damage, cell cycle arrest, and DNA repair (3, 20, 44, 67). The N terminus of hRad9 contains a region that is similar to a region in the proliferating cell nuclear antigen (PCNA) and associates with hRad1 and hHus1 in a head-to-tail manner, thus forming a stable heterotrimeric DNA sliding clamp (65, 66, 74). Recent studies suggest that hRad9 may interact with the antiapoptotic Bcl-2 family proteins, Bcl-2 and Bcl-xL, through a BH3 domain at its N terminus (39, 72). Therefore, in addition to its previously reported checkpoint control functions, hRad9 may play a role in regulating apoptosis.
The present studies demonstrate that hRad9 interacts with AR in an androgen-dependent manner. We show that the FXXLF motif at the C terminus of hRad9 mediates the interaction with the AR LBD. The results also show that hRad9 down-regulates AR transcriptional activation through blocking the N-C interaction of AR. These findings may serve as an important model of how checkpoint proteins cross talk with AR signaling in prostate cancers.
MATERIALS AND METHODS
Materials.
MMTV-LUC, pCMV-AR, pCDNA3-Flag, and pCMX-VP16-AR have been described previously (33, 64). pGEX-KG-hRad9 and pCDNA3-AU1-hRad9 were kindly provided by Larry M. Karnitz, Mayo Clinic, Rochester, Minn. Human multiple tissue Northern blot II was purchased from BD Biosciences. Anti-Flag antibody M2 and anti-Rad9 antibody (M-389) were purchased from Sigma and Santa Cruz Biotechnology, Inc., respectively.
Yeast two-hybrid screen.
The DBD and LBD of AR cDNA was amplified and cloned into the NdeI and BamHI sites of pGBKT7 (Clontech). Yeast strain AH109 was transformed with the vector encoding the GAL4DBD-AR-DBD-LBD fusion protein. Transformed AH109 was mated with yeast strain Y187 pretransformed with the human ovary MATCHMAKER cDNA library (Clontech). The yeast clones were selected following the manufacturer's instructions, and positive clones were further confirmed by clone lift assay. Purified plasmids were retransformed into yeast strain AH109 with bait plasmids. The interaction specificity was further confirmed by liquid β-galactosidase assay.
Plasmid constructions.
To clone full-length Flag-tagged hRad9, hRad9 cDNA was amplified and cloned into the BamHI and XbaI sites in pCDNA3-Flag vector. Similarly, the cDNA fragments encoding amino acids (aa) 1 to 270 and aa 269 to 391 of hRad9 were cloned into pCDNA3-Flag to make the vectors expressing the N terminus of hRad9 and the C terminus of hRad9, respectively. To assemble AR fragments into the pGBKT7 vector, fragments covering AR DBD or LBD were inserted at the 5′ end with NdeI and at the 3′ end with BamHI by PCR and cloned into the NdeI and BamHI sites in pGBKT7. The QuikChange site-directed mutagenesis kit (Stratagene) was used to mutate the hRad9 sequence. F361 of hRad9 was converted to Ala residue to yield the AXXLF mutant of hRad9. Similarly, L364 and F365 of hRad9 were converted to Ala residues to yield the FXXAA mutant of hRad9. The mammalian two-hybrid vector of full-length hRad9 was constructed by fusing the hRad9 cDNA in frame to pCMX-GAL4-DBD. The N terminus of hRad9 and C terminus of hRad9 fragments were inserted in frame into the pM vector (Clontech) to generate the GAL4-N-hRad9 and GAL4-C-hRad9 plasmids, respectively. DNA vector-based RNA interference (RNAi) plasmids were used to reduce the endogenous hRad9 expression as previously described (58). RNAi constructs were designed to target the CCCTGTCCCGCATCGGGGACG, GGGGACGAGCTCTACCTGGAA, CCCTTGGAGGACGGGCTCTC, and AAGTCTTTCCTGTCTGTCTT sequences of the hRad9 mRNA and are termed R1, R2, R3, and R4, respectively. The selection of coding sequences was determined empirically and was analyzed by BLAST search to avoid any significant sequence homology with other genes. Vectors that express RNAi under the control of the U6 promoter were constructed by inserting pairs of annealed DNA oligonucleotides into the BS/U6 vector between the ApaI and EcoRI sites. All plasmids were verified by sequencing.
Cell culture and transfections.
PC-3, CWR22R, and LNCaP cell lines were maintained in RPMI 1600 medium supplemented with 10% fetal calf serum (FCS). Transient transfection for luciferase assays was performed in 24-well plates (5 × 104 cells per well) using SuperFect as described previously (46). The DNA mixtures used in transfection assays are indicated in the figures. The total amount of transfected DNA was kept constant (1 μg) by adding the corresponding amount of empty expression plasmids. After transfection, cells were cultured in RPMI 1600 medium supplemented with 5% charcoal-stripped FCS in the presence or absence of 10 nM dihydrotestosterone (DHT) for 24 h. Luciferase assays were performed as previously described (69). In Western blotting assays, CWR22R or LNCaP cells were transfected by electroporation using 5 × 106 cells/0.4 ml of RPMI 1600 medium containing 2% FCS plus 9 μg of the indicated plasmids. One microgram of enhanced green fluorescent protein (EGFP) expression vector was used for transfection efficiency. Electroporation was performed at 250 V and 950 μF using Gene Pulser II (Bio-Rad).
In vitro GST pull-down assays.
The N terminus, DBD, LBD, and DBD-LBD of AR were in vitro translated in the presence of [35S]methionine using T7 polymerase and the coupled transcription-translation kit (Promega). pGEX-KG-hRad9 plasmids expressing glutathione S-transferase (GST)-hRad9 fusion protein were transformed into BL21(DE3) bacterial strain. Isopropyl-β-d-thiogalactopyranoside (0.4 mM) was added to Luria-Bertani (LB) medium containing transformed bacteria when the optical density at 600 nm reached 0.5. Bacteria were further cultured at 30°C for 3 h and lysed by four cycles of freeze-thawing in NETN buffer (20 mM Tris [pH 8.0], 0.5% Nonidet P-40 [NP-40], 100 mM NaCl, 6 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol [DTT], 8% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF]). The GST-hRad9 fusion proteins were purified with glutathione beads at 4°C. Labeled proteins of AR mutants were incubated with equal amounts of GST-hRad9 in binding buffer (50 mM HEPES, 100 mM NaCl, 20 mM Tris-Cl [pH 8.0], 0.1% Tween 20, 10% glycerol, 1 mM DTT, 0.5 mM PMSF, 1 mM NaF, 0.4 mM sodium vanadate) with or without 10 nM DHT at 4°C for 2 h. The beads were then washed with NETN buffer four times and resuspended in sodium dodecyl sulfate (SDS)-polyacrylamide loading buffer, and the proteins were resolved on SDS-10% polyacrylamide gels by electrophoresis followed by autoradiography.
Coimmunoprecipitation assays and Western blotting.
293T cells were transfected in 10-cm-diameter dishes with 2.5 μg of Flag-hRad9 and 7.5 μg of pCMV-AR plasmids in the presence or absence of 10 nM DHT. Total cell extract was prepared in the presence or absence of 10 nM DHT in immunoprecipitation buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 20% glycerol, 0.5% NP-40, 50 mM NaF, 0.4 mM sodium vanadate, 0.5 mM PMSF, 0.5 mM DTT). After centrifugation, supernatants were incubated for 2 h with anti-Flag antibody M2 or normal mouse serum. For CWR22R cells, cell extracts were prepared as described above and supernatants were precipitated by anti-AR antibody (554225; BD Biosciences) or normal mouse serum. Precipitated protein complexes were washed four times in the presence or absence of 10 nM DHT and subsequently analyzed by Western blotting.
Real-time reverse transcription PCR.
Prostate cancer specimens were collected at the time of radical prostatectomy, representing specimens from clinical prostate cancers. All histological diagnoses were confirmed by staining parallel sections with hematoxylin and eosin. Total RNA was isolated using the Trizol (Gibco) reagent, according to the manufacturer's instructions, and 1 μg of RNA was subjected to reverse transcription using Superscript II (Invitrogen, Carlsbad, Calif.). Specific primers for hRAD9, 5′-CGCTGTAAGATCCTGATGAAGTC-3′ (forward) and 5′-TGCCTCCTCCTCGTGGTAG-3′ (reverse), were designed according to Bacon Designer2 software. 18S rRNA primers, 5′-TGCCTTCCTTGGATGTGGTAG-3′ (forward) and 5′-CGTCTGCCCTATCAACTTTCG-3′ (reverse), were used as controls. Real-time PCR was performed with 1 μl of reverse transcription product, 12.5 μl of 2× SYBR green PCR master mix (Bio-Rad), and 0.5 μl of each primer (10 μM), in a total volume of 25 μl. PCR was performed on an iCycler iQ multicolor real-time PCR detection system (Bio-Rad) as follows: (i) 3 min at 94°C and (ii) 40 cycles, with 1 cycle consisting of 15 s at 94°C, 30 s at 60°C, and 30 s at 72°C. Each sample was run in triplicate. Data were analyzed by iCycler iQ software (Bio-Rad).
RESULTS
Ligand-dependent interaction of AR and hRad9 in yeast.
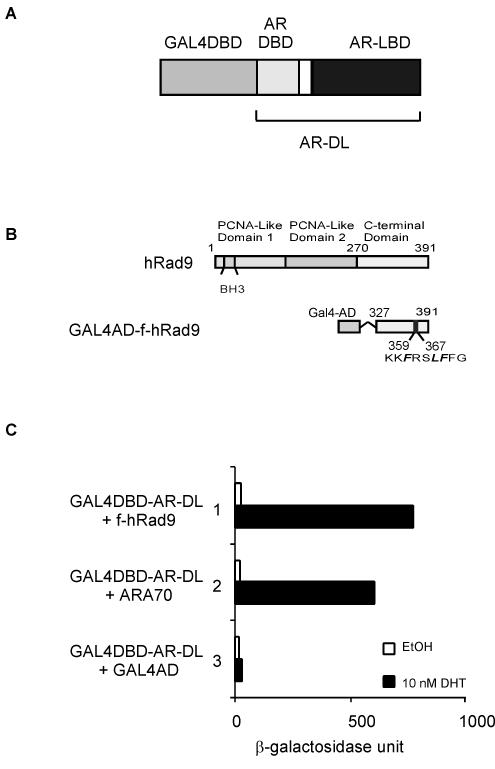
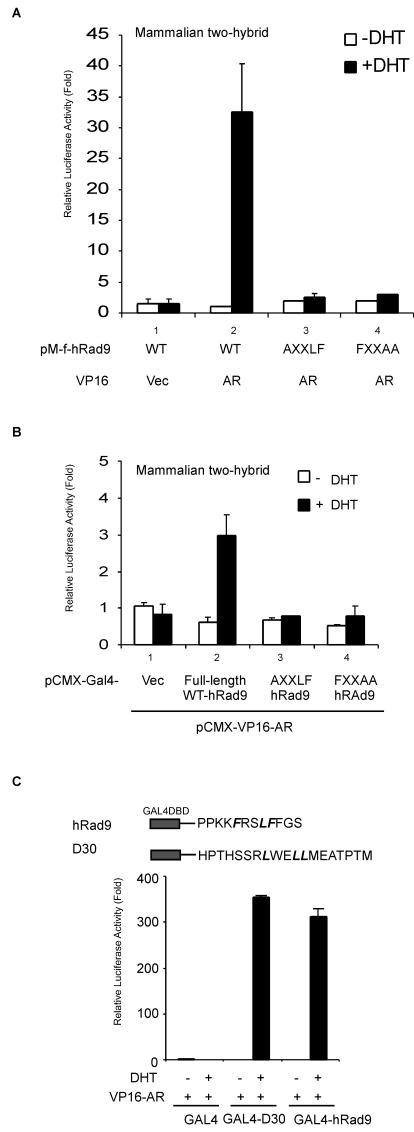
In order to screen proteins with ligand-dependent interaction with AR, the human AR DBD-LBD was fused with the DBD of GAL4 to function as bait in yeast two-hybrid screening (Fig. 1A). A pretransformed normal human ovary cDNA library was screened in the presence of 10 nM DHT. A total of 108 individual yeast clones were first selected by nutrition deprivation and confirmed to activate β-galactosidase by a clone lift assay (data not shown). Sequence analyses showed three clones, each of which encoded aa 327 to 391 of hRad9 in frame with the GAL4 activation domain. This hRad9 fragment from yeast lies in the C terminus of hRad9 and contains an FXXLF motif (aa 361 to 365) that overlaps with the potential nuclear localization sequence motif (aa 356 to 364) (32). This fragment of hRad9 is referred to as f-hRad9 (Fig. 1B). Liquid β-galactosidase assays were performed to quantitatively analyze the interaction between AR and hRad9. Constructs containing either f-hRad9 peptide (aa 327 to 391) or ARA70, an AR coactivator, showed a strong interaction with the AR-DBD-LBD in the presence of DHT (Fig. 1C). As a negative control, the GAL4 activation domain alone was not able to interact with AR. Thus, these results indicate an androgen-dependent interaction between AR and hRad9 in yeast.
FIG. 1.
Isolation of hRad9 as an AR coregulator by the yeast two-hybrid assay. (A) GAL4-DBD-AR-DBD-LBD fusion was used as bait. (B) The structures of the human Rad9 and hRad9 fusion protein isolated from yeast screening. (C) AH109 yeast cells were transformed with GAL4-DBD-AR-DL and GAL4-AD fused with hRad9 (aa 327 to 391). The two-hybrid interaction is determined by β-galactosidase activity expressed in the yeast cells. EtOH, ethanol.
Analysis of hRad9 expression.
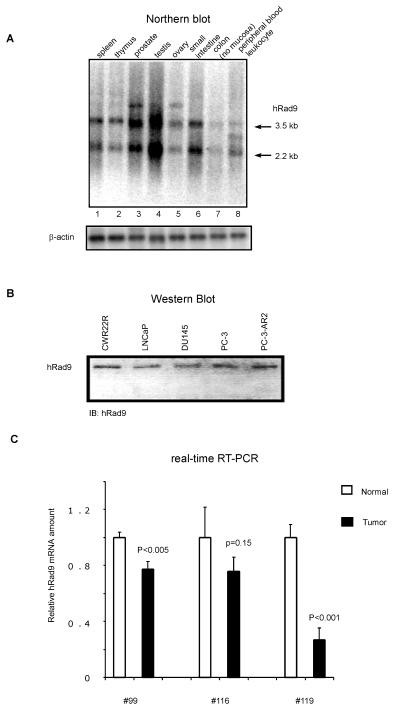
Northern hybridization analyses were performed to determine the expression of hRad9 in various human tissues, especially the reproductive organs. Since the hRad9 N terminus is homologous with PCNA, a specific probe was used covering the last 121 amino acid residues of hRad9 proteins. As shown in Fig. 2A, hRad9 was ubiquitously expressed at variable levels in all eight tissues examined. When normalized to β-actin mRNA levels, the level of hRad9 mRNA was highest in the testis, second highest in the prostate, and lowest in the colon. Interestingly, hHus1 mRNA was found to be most abundant in the testis, where hRad1 also expressed at high levels (22). It is tempting to speculate that hRad9 may contribute to the meiotic checkpoint in the testis where the maintenance of genomic DNA integrity is extremely important. Nonetheless, direct physiological evidence remains to be established.
FIG. 2.
hRad9 expression in human prostate. (A) A human multiple tissue Northern blot (Clontech) containing 2 μg of poly(A+) mRNA from the indicated tissues was hybridized with 32P-labeled probes corresponding to hRad9 and β-actin. (B) Expression of hRad9 proteins in prostate cancer cells. Equal amounts (30 μg) of proteins from the indicated cell lines were analyzed by immunoblotting (IB) with anti-hRad9 antibody. (C) Expression of hRad9 mRNA in tissues from prostate cancer patients. Total RNA was isolated from clinical prostatic carcinoma. Sections of tumors and normal tissues were confirmed by hematoxylin and eosin staining. After cDNA synthesis, real-time reverse transcription (RT)-PCR was performed to analyze the amount of hRad9 in tumor or normal tissues.
The prostate is made up of epithelial glands and a fibromuscular stroma, with prostate cancers arising from the glandular epithelium (16). To determine hRad9 expression in prostate cancers, we performed immunoblot analyses of variable prostate cancer cell lysates, revealing an anti-hRad9-reactive band in all cells examined (Fig. 2B). In agreement with previous reports (20, 32), fluorescence analyses using GFP-hRad9 fusion proteins suggested that hRad9 protein was localized mainly in the nucleus (data not shown). Since AR also translocates into the nucleus upon androgen treatment, hRad9 and AR proteins can be colocalized in the nucleus.
We also analyzed the expression of hRad9 in human prostate samples under normal or pathological situations using quantitative real-time PCR. All three samples were obtained from patients with high-grade prostatic adenocarcinoma. We found that the neoplastic tissues express significantly less hRad9 compared to the adjacent normal area, as revealed by real-time PCR analyses (Fig. 2C), in some patients we examined. Although this result is intriguing, we may need to analyze more samples before we can establish whether hRad9 expression is frequently down-regulated in advanced prostate cancers.
hRad9 associates with AR in vivo.
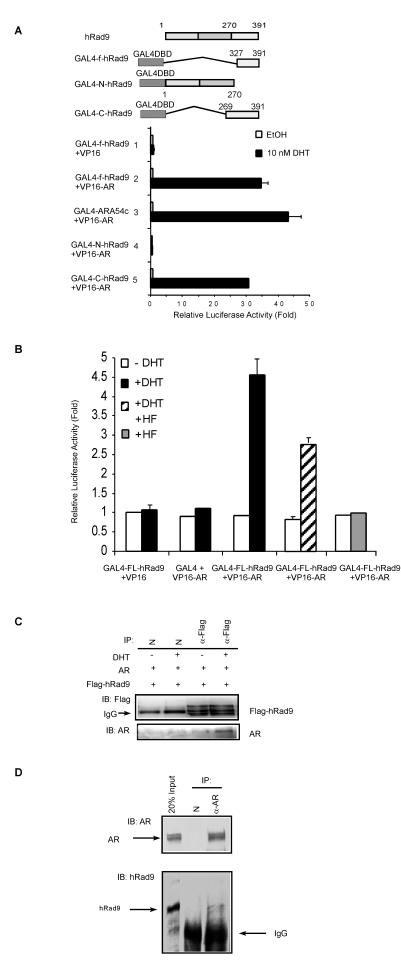
To determine whether hRad9 and AR interact in mammalian cells, the f-hRad9 fragment was subcloned into the mammalian pM expression vector. Mammalian two-hybrid assays were performed in PC-3 cells in the absence and presence of 10 nM DHT. As shown in Fig. 3A, androgen-dependent interactions between GAL4-f-hRad9 and full-length AR were detected (lane 2). The interaction between AR and the C terminus of ARA54 was used as a positive control (Fig. 3A, lane 3). Furthermore, the C terminus of hRad9 (aa 269 to 391) displayed a strong interaction with AR in the presence of androgen (Fig. 3A, lane 5), while the PCNA-like domain of hRad9 (N-hRad9, aa 1 to 270) did not (Fig. 3A, lane 4), suggesting that the C terminus of hRad9 mediates the interaction with AR.
FIG. 3.
hRad9 interacts with AR in mammalian cells. (A) Interaction between AR and the C terminus of hRad9 examined by mammalian two-hybrid assays. PC-3 cells were transiently transfected with 0.4 μg of reporter plasmid pG4-LUC, and 0.3 μg of GAL4-DBD-fused hRad9 mutants with or without 0.3 μg of VP16-fused AR (VP16-AR) as indicated. After 24 h of treatment with 10 nM DHT, the cells were harvested for luciferase (LUC) assay. phRL-tk-LUC expression vector was used as a control for transfection efficiency. Results shown here are the means ± standard deviations for three independent experiments. EtOH, ethanol. (B) The interaction between full-length hRad9 and AR is reduced by HF. PC-3 cells were transfected with a DNA mixture containing pG4-LUC, VP16-AR, and pCMX-GAL4-FL-hRad9, as described for panel A. PC-3 cells were incubated with 10−5 M HF 1 h prior to 10−8 M DHT treatment. Luciferase activities were measured after another 24 h of incubation. Results shown are the means ± standard deviations for three independent experiments. (C) Immunoprecipitation (IP) of AR and hRad9 in 293T cells. 293T cells that overexpressed AR and Flag-hRad9 were treated with (+) or without (−) 10−8 M DHT. Cell extracts were immunoprecipitated with anti-Flag antibody (α-Flag), followed by immunoblotting (IB) with antibody to AR. IgG, immunoglobulin G. (D) Immunoprecipitation of endogenous AR and hRad9. CWR22R cell extracts were prepared in the presence or absence of 10−8 M DHT. Immunoprecipitation (IP) was performed with antibody to AR (NH27) or normal rabbit serum, followed by immunoblotting (IB) with antibodies to hRad9 or AR.
To further investigate the physical association of full-length hRad9 (FL-hRad9) with AR, we performed mammalian two-hybrid assays with FL-hRad9 fused to the DBD of GAL4 and AR fused to VP16. As seen in Fig. 3B, androgen stimulated the interaction between AR and hRad9, while hydroxyflutamide (HF), an antagonist for AR, inhibited the androgen-induced interaction between AR and hRad9. Furthermore, we cotransfected 293T cells with AR and Flag epitope-tagged hRad9 to test whether AR existed in hRad9 immunoprecipitates. An AR band was detected in the Flag-hRad9 immunoprecipitates (Fig. 3C). Finally, coimmunoprecipitation of native proteins from a prostate cancer cell line CWR22R extract confirmed the AR-hRad9 association in vivo (Fig. 3D). Together, the association between AR and hRad9 is unequivocal in mammalian cells.
Domains of AR involved in binding to hRad9.
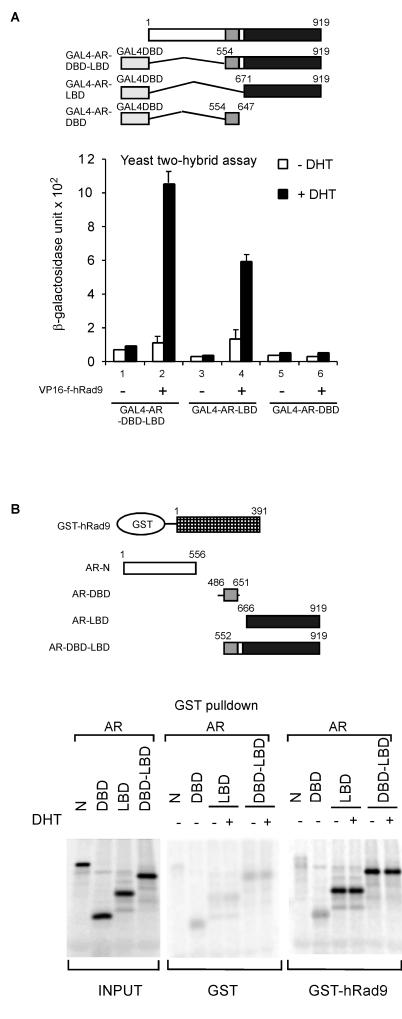
While the C terminus of hRad9 associates with AR, we were interested in determining which domain(s) of AR is responsible for the interaction. Yeast two-hybrid assays were performed first. In AH109 yeast cells, different regions of AR fused with GAL4-DBD were cotransformed with the plasmid containing VP16 activation domain (VP16-AD) or VP16-AD fused with amino acids 327 to 391 of hRad9 (VP16-f-hRad9) in the presence or absence of 10 nM DHT. In the absence of androgen, there was little interaction between VP16-hRad9 and various GAL4-AR fusion proteins (Fig. 4A). However, with 10 nM DHT treatment (Fig. 4A), coexpression of VP16-f-hRad9 and GAL4-AR-DBD-LBD yielded an increased reporter activity by ∼10-fold over that with GAL4-AR-DBD-LBD and VP16 AD (Fig. 4A, lane 2 versus lane 1). As expected, VP16-f-hRad9 also interacted with AR LBD in the presence of androgen (Fig. 4A, lane 4). Though GAL4-AR-DBD did not interact with hRad9 (Fig. 4A, lane 6), the interaction between hRad9 and AR LBD was weaker than the association between AR DBD-LBD and hRad9, suggesting that AR DBD might also contribute to the proper folding of AR-DBD-LBD in yeast.
FIG. 4.
Mapping the domains of AR that are responsible for hRad9 interaction. (A) AH109 yeast cells were transformed with GAL4-DBD fused with various AR domains and GAL4-AD fused with hRad9 (aa 327 to 391). Liquid β-galactosidase assays were performed as described in the legend to Fig. 1A. (B) A series of 35S-labeled mutant ARs were incubated with purified GST-hRad9 or GST alone in the presence (+) or absence (−) of 10−8 M DHT. The results indicated that AR LBD mediates the interaction with hRad9.
Since two-hybrid assays provide an indirect measurement of protein interactions, to investigate whether hRad9 interacts directly with AR LBD, GST pull-down assays were performed using GST protein alone or GST-hRad9 fusion protein. The various domains of AR were labeled with [35S]methionine by in vitro translation and incubated with GST-hRad9-bound beads. As shown in Fig. 4B, the AR LBD and AR-DBD-LBD interacted specifically with hRad9. Unlike the interaction observed in the yeast two-hybrid system or the mammalian two-hybrid system, the presence or absence of androgen did not robustly influence the interaction between AR and hRad9. Consistent with previous studies, this discrepancy may be because the high concentration of proteins in GST pull-down assays may change the binding sensitivity between AR and its coregulators (36, 54). Another possible mechanism is that AR might be associated with many other proteins that interrupt the AR-hRad9 association in the absence of ligand. Neither the N terminus of AR (aa 1 to 556) nor the DBD alone adhered to GST-hRad9. Therefore, these results are consistent with the yeast two-hybrid experiments and suggest that the AR LBD is required for interaction with hRad9.
The FXXLF motif mediates AR-hRad9 interaction.
The LXXLL motif was first identified in some SRCs (28). However, among steroid receptors, AR appears to be relatively unique, as it interacts with only a very limited subset of LXXLL sequences (12). Previous studies showed that the FXXLF motif plays important roles in mediating the interaction of the AR LBD with several FXXLF-containing AR coregulators (25, 26). Interestingly, one FXXLF motif is located at the carboxyl terminus of hRad9 (aa 361 to 365). To investigate whether this FXXLF motif contributes to the association between AR and hRad9, hRad9 mutants with mutations in the FXXLF motif were tested in mammalian two-hybrid assays. These mutations dramatically decreased the interaction between AR and the fragment of hRad9 (aa 327 to 391), as shown by either the AXXLF or FXXAA mutant (Fig. 5A, lanes 3 and 4 versus lane 2, black bars). Similarly, the AXXLF or FXXAA mutant reduced the interaction between AR and full-length hRad9 (Fig. 5B, lanes 3 and 4 versus lane 2, black bars), which suggests that this FXXLF motif is critical for hRad9 interaction with AR.
FIG.5.
FXXLF motif in hRad9 mediates the AR-hRad9 interaction. (A) Mutants of hRad9 were constructed using the QuikChange kit. Mammalian two-hybrid assays were performed in PC-3 cells using 0.3 μg of GAL4-f-hRad9 coding for the GAL4 DBD fused to the fragment of hRad9 isolated from yeast containing residues 327 to 391 with the wild-type (WT) or indicated mutant sequences. GAL4-f-hRad9 was cotransfected with the 0.4 μg of pG4LUC reporter vector and 0.3 μg of VP16-AR. phRL-tk-LUC expression vector was used as a control for transfection efficiency. Assays were performed with PC-3 cells in the presence (+DHT) or absence (−DHT) of 10 nM DHT. Vec, vector. (B) Full-length wild-type or mutant hRad9 proteins were fused with GAL4-DBD and tested in mammalian two-hybrid assays as described for panel A. (C) Mammalian two-hybrid assays were performed with PC-3 cells by coexpressing GAL4-hRad9 peptides, which contained the GAL4 DBD and the hRad9 FXXLF motif, with VP16-AR.
However, the interaction of the LXXLL or FXXLF motif with AR cannot be predicted precisely from their sequences alone. For example, FXXLF motif peptides derived from the CBP (FGSLF) and p300 (FGSLF) fail to interact with AR (26). Moreover, the mutant proteins with mutations in the FXXLF motif in hRad9 might eliminate the AR-hRad9 interaction because of the whole conformation change of hRad9, not limited to the FXXLF α-helix. Thus, we were interested in determining whether the FXXLF motif in hRad9 can directly interact with AR. Therefore, a small peptide containing the FXXLF motif of hRad9 was fused with GAL4-DBD (Fig. 5C), cotransfected with VP16-AR, and tested in the absence and presence of 10 nM DHT in two-hybrid peptide assays. Androgen-dependent interactions were demonstrated between VP16-AR and the GAL4-FXXLF (hRad9) fusion peptides (Fig. 5C). For a positive control, we observed the DHT-dependent interaction of AR with a GAL4-D30 peptide, which contains a LXXLL motif that interacts with AR as described previously (12). Together, our data demonstrate that the FXXLF motif in C terminus of hRad9 mediates the interaction with the AR.
hRad9 selectively represses AR-mediated transactivation.
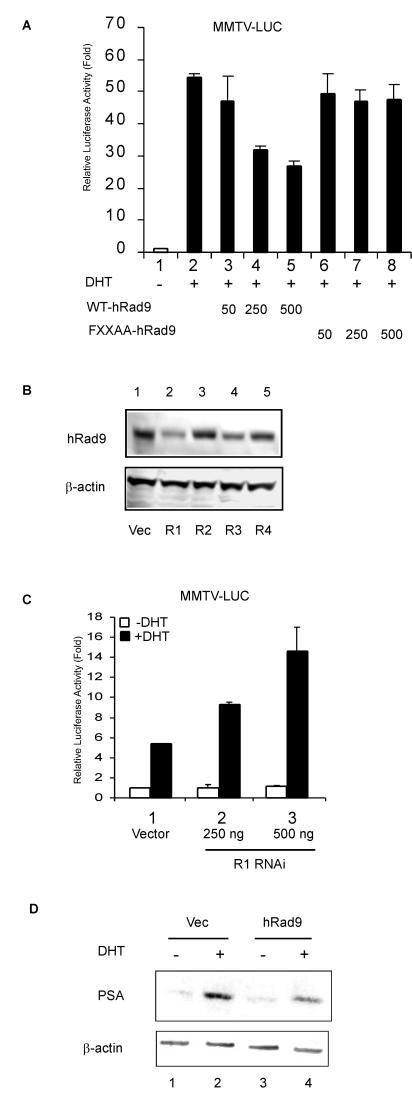
To understand the consequence of hRad9 binding to the AR, AR transactivation was studied with the MMTV-LUC reporter in PC-3 cells. The promoter of MMTV-LUC is a naturally occurring mouse mammary tumor virus (MMTV) long terminal repeat which contains androgen-responsive elements. Cotransfection of wild-type hRad9 with AR decreased the transcriptional activity of AR in a dose-dependent manner (Fig. 6A, lanes 3 to 5), whereas FXXAA mutants had only marginal effects on AR transactivation (Fig. 6A, lanes 6 to 8). Wild-type hRad9 and FXXAA mutant hRad9 did not have an effect on the transcriptional activity in the absence of 10 nM DHT (data not shown), suggesting that they do not affect the basal transcriptional activity. Similar results were observed when we replaced PC-3 cells with LNCaP cells (data not shown).
FIG. 6.
hRad9 suppresses AR transcriptional activity. (A) hRad9 suppresses AR transactivation of MMTV-LUC reporter. PC-3 cells were cotransfected with 100 ng of pCMV-AR and the indicated amount (in nanograms) of pCDNA3-Flag vectors expressing wild-type (WT) hRad9 or hRad9 mutant (FXXAA-hRad9) and MMTV-LUC reporter vector using SuperFect. phRL-tk-LUC expression vector was used as a control for transfection efficiency. Cells were treated with ethanol or DHT and then lysed for luciferase (LUC) activities. The MMTV-LUC reporter activity was normalized to control LUC activity. The LUC activity relative to lane 1 was calculated, and results shown are the means ± standard deviations for three independent experiments. (B) RNAi constructs of hRad9 block hRad9 expression. CWR22R cells were transfected with the indicated RNAi plasmids targeting hRad9 by electroporation. Cell lysates were collected and tested by immunoblotting with antibodies to hRad9 or β-actin. Vec, vector. (C) CWR22R cells were transfected as described for panel A to determine the effect of blocking endogenous hRad9 on AR transcriptional activity. (D) LNCaP cells were transfected with pCDNA vector or pCDNA-hRad9 by electroporation. After 24 h, cells were treated with ethanol or 10 nM DHT for another 48 h, and 50-μg samples of cell extracts from LNCaP cells were loaded on SDS-10% polyacrylamide gel and analyzed by Western blotting using PSA antibodies.
To determine the effect of endogenous hRad9 on AR, CWR22R cells were transfected with several small interfering RNA (siRNA) constructs targeting hRad9 (R1, R2, R3, and R4) or mock transfected for controls. After 1 days of transfection, the protein levels of hRad9 were evaluated by immunoblot analyses with anti-hRad9 antibodies. Whereas R2 andR4 siRNA constructs only marginally reduced endogenous hRad9 expression and R3 moderately decreased hRad9 expression (Fig. 6B, lanes 3, 5, and 4), R1 siRNA dramatically reduced the hRad9 protein in CWR22R cells (Fig. 6B, lane 2). Therefore, we tested the influence of siRNA R1 on AR transcriptional activity in CWR22R cells. R1 siRNA increased the DHT-induced activation of the MMTV-LUC reporter in a dose-dependent manner (Fig. 6C), suggesting the repressive effect of endogenous hRad9 on AR. Similar results were observed when we replaced CWR22R cells with PC-3 cells (data not shown).
Prostate-specific antigen (PSA) is a clinically significant androgen-stimulated gene that is used to monitor response to treatment and progression of prostate cancer (15). Endogenous PSA protein expression was induced by the DHT treatment in LNCaP cells (Fig. 6D, lane 2). Addition of hRad9 potently inhibited the DHT-mediated induction of PSA (Fig. 6D, lane 4). Taken together, these data showed, for the first time, an involvement of hRad9 in AR transcriptional activation.
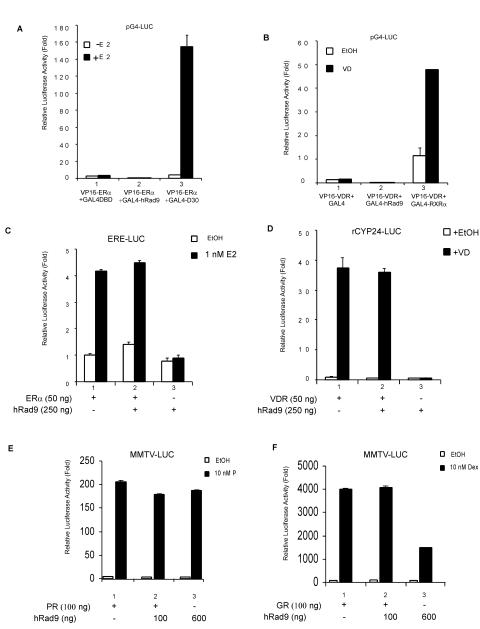
To determine whether hRad9 can interact with other steroid receptors and further affect their transactivation, we examined the possible association of hRad9 with the estrogen receptor α (ERα) or the vitamin D receptor (VDR) in a mammalian two-hybrid system. In the presence of estrogen, ERα showed strong interaction with GAL4-D30 (Fig. 7A, lane 3), whereas there was no interaction with hRad9 (Fig. 7A, lane 2). Similarly, VDR associated with GAL4-RXRα (Fig. 7B, lane 3); however, hRad9 did not interact with VDR (Fig. 7B, lane 2). As previous studies reported that FXXLF is a motif specific for AR coregulators (26), it is not surprising that hRad9 showed more specificity for AR than for other steroid receptors, since our studies showed that the FXXLF motif in hRad9 mediates its interaction with AR. ERE-LUC and rCyp24-LUC reporter plasmids were used to determine the transcriptional activity of ERα and VDR, respectively. As shown in Fig. 7C and D, whereas the ER and VDR could induce luciferase activity in the presence of their cognate ligands in PC-3 cells, cotransfection of hRad9 had little inhibitory effect on their transcriptional activity. Furthermore, hRad9 showed marginal effect on the progesterone receptor activity but could inhibit the glucocorticoid receptor transactivation (Fig. 7E and F).
FIG. 7.
Effects of hRad9 on other nuclear receptors. (A) No interaction between hRad9 and ERα. PC-3 cells were transfected with DNA mixtures of pG4-LUC, pM-f-hRad9, and VP16-ERα as indicated. GAL4-D30 was used as a positive control for VP16-ERα. E 2, 17β-estradiol. (B) No interaction between hRad9 and VDR. PC-3 cells were transfected as described for panel A, using VP16-VDR instead of VP16-ERα. GAL4-RXRα was used as a positive control for VP16-VDR. EtOH, ethanol; VD, 1α,25-dihydroxy-vitamin D3. (C) hRad9 has little effect on ER transactivation. PC-3 cells were transfected with a DNA mixture containing pSG5-ERα, ERE-Luc reporter, and pCDNA3-Flag-hRad9. Luciferase activity was measured after 24 h of treatment with E2. Results are the means ± standard deviations for three independent experiments. (D) hRad9 has little effect on VDR transactivation. pSG5-VDR, pCDNA3-Flag-hRad9, and rCYP24-LUC reporter plasmids were used in luciferase assays as indicated. (E) hRad9 has marginal effect on progesterone receptor (PR) transactivation. PSG5-PR, pCDNA3-Flag-hRad9, and MMTV-LUC reporter plasmids were transfected into PC-3 cells as indicated. P, progesterone. (F) hRad9 could suppress glucocorticoid receptor (GR) activity. PSG5-GR, pCDNA3-Flag-hRad9, and MMTV-LUC reporter plasmids were used in luciferase assays as indicated. Dex, dexamethasome.
hRad9 suppressed the AR N-C interaction.
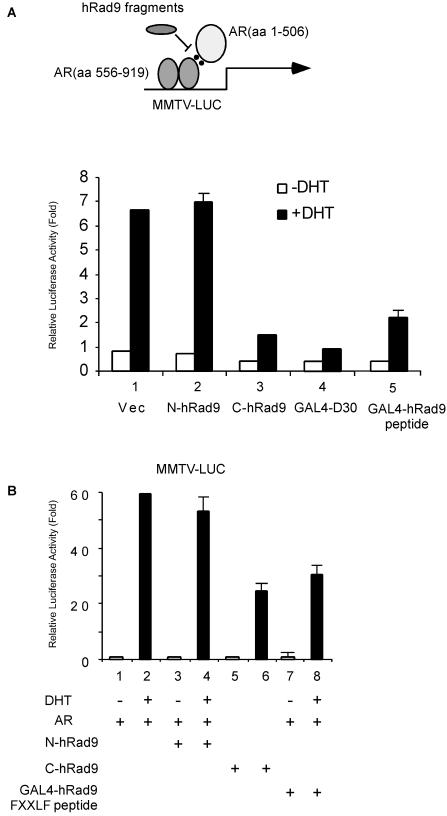
Early reports suggested that the FXXLF motif in the N terminus of AR is important for interacting with the C terminus of AR and that this N-C interaction is required for full capacity of AR transactivation (33). While the C-terminal hRad9 contains the FXXLF motif and interacts with the AR LBD, it is possible that hRad9 may influence the AR N-C interaction. As previously described (12), we used a reconstituted AR transcription assay to address this possibility (Fig. 8A, upper panel). In PC-3 cells, the AR DBD-LBD (aa 556 to 919) displayed minimal transactivation even in the presence of DHT (data not shown), consistent with previous studies showing that AR LBD has only minimal transcriptional activity. However, coexpression of the N terminus of AR (aa 1 to 506) with AR DBD-LBD restores agonist-induced transactivation (Fig. 8A, lower panel, lane 1). The GAL4-D30 protein was used as a positive control in this experiment to show the blockage of the N-C interaction in AR (Fig. 8A, lane 4). The C terminus of hRad9 can potently inhibit the interaction between the N and C termini of AR in the presence of androgen (Fig. 8A, lane 3), whereas the N terminus of hRad9, which cannot interact with AR, has no inhibitory effect on AR N-C interaction (Fig. 8A, lane 2). Furthermore, we applied the full-length AR to test whether the C terminus of hRad9 can block intact AR transactivation. Our data demonstrated that only the C terminus of hRad9, not the N terminus of hRad9, suppressed AR-mediated transactivation (Fig. 8B). Interestingly, the small peptide containing the hRad9 FXXLF motif alone can also suppress the AR N-C interaction and AR transactivation (Fig. 8A, lane 5 and B, lane 8). Together, these results suggest one mechanism by which disruptions of AR N-C interaction by the hRad9 FXXLF motif might contribute to the inhibitory role of hRad9 on AR. Consequently, the binding between other coactivators and AR may be blocked due to the lack of a stabilized N-C interaction which is necessary for AR activation.
FIG. 8.
The C terminus of hRad9 interrupts AR N-C interaction. (A) The FXXLF-containing fragment of hRad9 efficiently blocked the interaction between the N terminus of AR and the AR-LBD. The reconstituted AR transcription assay to determine the AR N-C interaction is shown at the top. PC-3 cells were transfected with MMTV-LUC, pRL-tk-LUC, AR mutants, and hRad9 as indicated in the bar graph. After transfection, cells were treated with 10 nM DHT (+DHT) for 24 h before harvesting. The luciferase (LUC) activity relative to lane 1 was calculated, and results are the means ± standard deviations for three independent experiments. Vec, vector. (B) The C terminus or FXXLF-containing peptide, not the N terminus, of hRad9 inhibits AR transactivation. PC-3 cells were transfected as described for panel A, except pCMV-AR that expresses intact AR was used.
DISCUSSION
Studies in Schizosaccharomyces pombe and human cells have demonstrated a conserved checkpoint pathway, including hRad9, hHus1, and hRad1, capable of causing cell cycle arrest in response to incomplete DNA replication or DNA damage (1, 18, 38, 40). Later studies demonstrated that hRad9, hHus1, and hRad1 form a stable heterotrimeric complex, called the 9-1-1 complex, with a clamp structure similar to that of PCNA (63). Biochemical, biophysical, and molecular modeling studies suggest that Rad17 may help load the 9-1-1 complex onto sites of DNA damage in the checkpoint signaling pathway (59). Since DNA damage induces hRad17-dependent association of 9-1-1 with chromatin, it is believed that the 9-1-1 complex is involved in the direct recognition of DNA lesions and initiates the checkpoint responses (74). Nonetheless, the hRad9 C-terminal region is not involved in the interaction with hRad1 or hHus1 and is exposed outside the 9-1-1 clamp structure (60). This flexible structure of the C terminus of hRad9 leads to the possibility that it may play important roles in interacting with other proteins and subsequently regulate other signal transduction pathways. Indeed, the C-terminal region of hRad9 (aa 270 to 391) contains a predicted nuclear localization sequence (aa 356 to 364) that may act to guide the 9-1-1 complex into the nucleus (32). The SH3 domain of c-Abl also interacts directly with the C-terminal region of hRad9 (72). Recently, hRad9 was found to interact with replication and checkpoint protein topoisomerase II beta binding protein 1 through the C-terminal 17 amino acids of hRad9 (20). Furthermore, several phosphorylation sites that may play critical roles in the transduction of downstream checkpoint signals in the hRad9 C-terminal region were identified (60, 62). The results of the present studies offer support for a new role of the hRad9 C terminus in the modulation of AR transcriptional activity through its interaction with the AR LBD via its FXXLF motif (Fig. 5), which directly links a key player in DNA damage detection and repair with AR-mediated transcription in prostate cancer.
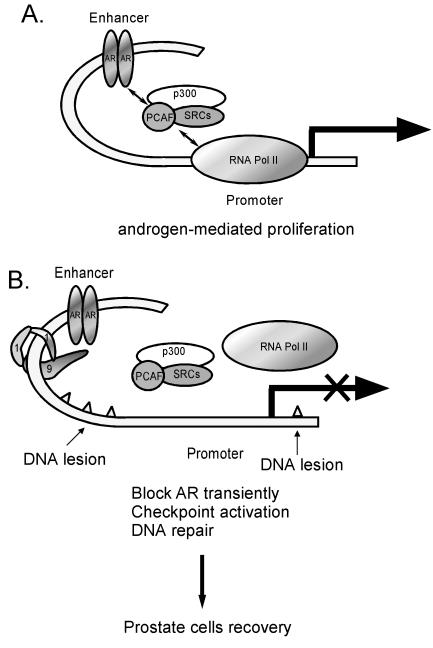
Clinical studies have shown that eliminating androgen improved the survival of patients with locally advanced prostate cancer when combined with radiation therapy (5). Furthermore, the use of animal models has suggested that androgen may protect prostate cancer from apoptosis induced by radiotherapy (35). Studies using prostate cancer cell lines also demonstrate that androgen plays protective roles in LNCaP cells exposed to radiation or chemotherapeutic agents (2, 13). Intriguingly, irradiation can selectively inhibit transcription from the androgen-dependent Pem homeobox gene promoter in AR-positive Sertoli cells, without changing most other genes studied, including FSHR, SGP1, AR, and CREB (48). However, the mechanism underlying the selective inhibition of AR activity and the protective effect of androgen remains largely unknown. Our findings that hRad9 functions as a corepressor for AR may open up several avenues of investigation. Though prostate cancer has a low proliferative index, it is noteworthy that prostate cancer cells show high rates of mutation, which suggests that DNA lesions occurs frequently in prostate cancer cells (23). With evidence showing that hRad9 functions as a negative regulator of the AR-mediated transcription (Fig. 6), it is possible that prostate cells may utilize hRad9 to reduce AR-mediated cell proliferation at the moment when cells are repairing the DNA lesions. Loss of hRad9 in cells may decrease the checkpoint activation, reduce DNA repair, and increase cell proliferation mediated by androgen or AR (Fig. 9). Interestingly, our preliminary analyses using a few prostate cancer samples show that the expression of hRad9 is reduced in prostate tumors compared to normal prostatic tissues (Fig. 2C). This fits our hypothesis and suggests that dysregulated expression of hRad9 may be involved in the progression of prostate cancer. Early studies also showed that hRad9 may play roles in the modulation of cell cycle progression (61). Blocking of hRad9 expression showed reduced ionizing radiation-induced accumulation of G2-M cells (32). Furthermore, previous reports demonstrate that hRad9 and hHus1 might act as tumor suppressors through their functions of maintaining chromosome integrity (7). Therefore, these two functions of hRad9, repressing AR activity and DNA damage checkpoint, could interdependently prevent cell transformation in prostate cancer development.
FIG. 9.
Model for the role of hRad9 in AR signaling. See text for discussion. RNA Pol II, RNA polymerase II.
Finally, our data (Fig. 8) demonstrated that hRad9 may suppress AR transcriptional activity via interrupting the AR N-C interaction. Previous studies suggested that AR N-C interaction might play essential roles for AR transcriptional activity. Several AR coactivators, such as SRC-1 and CBP, were able to promote AR N-C interaction (51). Conversely, SMRT and filamin A, two AR corepressors, were shown to inhibit AR activity through disruption of the AR N-C interaction and/or competition with the p160 coactivators (43, 54). However, whether these coregulators utilize their LXXLL or FXXLF motif to affect AR N-C interaction is not clear. The potential reasons why the FXXLF motif in hRad9 strongly interacts with the AR LBD follow: (i) two positive amino acid residues (K359 and K360) lie at the N terminus of FXXLF; (ii) no positively charged amino acid residues are located near the C terminus of FXXLF; (iii) there are no amino acid residues, such as glycine and proline, which can interrupt the FXXLF α-helix structure in FXXLF; and (iv) F366 is located in the C-terminal flanking area of the FXXLF motif. Thus, hRad9 fits quite well in the model recently proposed for FXXLF motif binding to AR LBD (27).
In summary, we have identified hRad9 as a novel corepressor of AR. hRad9 interacts with AR LBD through its C terminus and reduces AR transcriptional activity by interrupting the AR N-C interaction. Further studies may help us to better understand the connection between hRad9 and AR in prostate cancers.
Acknowledgments
We thank Larry M. Karnitz for kindly providing plasmids. We thank K. Wolf for help in manuscript preparation.
REFERENCES
- 1.al-Khodairy, F., E. Fotou, K. S. Sheldrick, D. J. Griffiths, A. R. Lehmann, and A. M. Carr. 1994. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell 5:147-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berchem, G. J., M. Bosseler, L. Y. Sugars, H. J. Voeller, S. Zeitlin, and E. P. Gelmann. 1995. Androgens induce resistance to bcl-2-mediated apoptosis in LNCaP prostate cancer cells. Cancer Res. 55:735-738. [PubMed] [Google Scholar]
- 3.Bessho, T., and A. Sancar. 2000. Human DNA damage checkpoint protein hRAD9 is a 3′ to 5′ exonuclease. J. Biol. Chem. 275:7451-7454. [DOI] [PubMed] [Google Scholar]
- 4.Bledsoe, R. K., V. G. Montana, T. B. Stanley, C. J. Delves, C. J. Apolito, D. D. McKee, T. G. Consler, D. J. Parks, E. L. Stewart, T. M. Willson, M. H. Lambert, J. T. Moore, K. H. Pearce, and H. E. Xu. 2002. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93-105. [DOI] [PubMed] [Google Scholar]
- 5.Bolla, M., D. Gonzalez, P. Warde, J. B. Dubois, R. O. Mirimanoff, G. Storme, J. Bernier, A. Kuten, C. Sternberg, T. Gil, L. Collette, and M. Pierart. 1997. Improved survival in patients with locally advanced prostate cancer treated with radiotherapy and goserelin. N. Engl. J. Med. 337:295-300. [DOI] [PubMed] [Google Scholar]
- 6.Brown, T. R. 1995. Human androgen insensitivity syndrome. J. Androl. 16:299-303. [PubMed] [Google Scholar]
- 7.Cai, R. L., Y. Yan-Neale, M. A. Cueto, H. Xu, and D. Cohen. 2000. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem. 275:27909-27916. [DOI] [PubMed] [Google Scholar]
- 8.Chang, C., J. D. Norris, H. Gron, L. A. Paige, P. T. Hamilton, D. J. Kenan, D. Fowlkes, and D. P. McDonnell. 1999. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta. Mol. Cell. Biol. 19:8226-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, C., A. Saltzman, S. Yeh, W. Young, E. Keller, H. J. Lee, C. Wang, and A. Mizokami. 1995. Androgen receptor: an overview. Crit. Rev. Eukaryot. Gene Expr. 5:97-125. [DOI] [PubMed] [Google Scholar]
- 10.Chang, C. S., J. Kokontis, and S. T. Liao. 1988. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science 240:324-326. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C. S., J. Kokontis, and S. T. Liao. 1988. Structural analysis of complementary DNA and amino acid sequences of human and rat androgen receptors. Proc. Natl. Acad. Sci. USA 85:7211-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, C. Y., and D. P. McDonnell. 2002. Evaluation of ligand-dependent changes in AR structure using peptide probes. Mol. Endocrinol. 16:647-660. [DOI] [PubMed] [Google Scholar]
- 13.Coffey, R. N., R. W. Watson, A. J. O'Neill, K. McEleny, and J. M. Fitzpatrick. 2002. Androgen-mediated resistance to apoptosis. Prostate 53:300-309. [DOI] [PubMed] [Google Scholar]
- 14.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debes, J. D., and D. J. Tindall. 2002. The role of androgens and the androgen receptor in prostate cancer. Cancer Lett. 187:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Feldman, B. J., and D. Feldman. 2001. The development of androgen-independent prostate cancer. Nat. Rev. Cancer 1:34-45. [DOI] [PubMed] [Google Scholar]
- 17.Feng, W., R. C. Ribeiro, R. L. Wagner, H. Nguyen, J. W. Apriletti, R. J. Fletterick, J. D. Baxter, P. J. Kushner, and B. L. West. 1998. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science 280:1747-1749. [DOI] [PubMed] [Google Scholar]
- 18.Freire, R., J. R. Murguia, M. Tarsounas, N. F. Lowndes, P. B. Moens, and S. P. Jackson. 1998. Human and mouse homologs of Schizosaccharomyces pombe rad1+ and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev. 12:2560-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 20.Greer, D. A., B. D. Besley, K. B. Kennedy, and S. Davey. 2003. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 63:4829-4835. [PubMed] [Google Scholar]
- 21.Hagmann, M. 1999. Checkpoint gene linked to human cancer. Science 286:2433-2434. [DOI] [PubMed] [Google Scholar]
- 22.Hang, H., and H. B. Lieberman. 2000. Physical interactions among human checkpoint control proteins HUS1p, RAD1p, and RAD9p, and implications for the regulation of cell cycle progression. Genomics 65:24-33. [DOI] [PubMed] [Google Scholar]
- 23.Hara, T., J. Miyazaki, H. Araki, M. Yamaoka, N. Kanzaki, M. Kusaka, and M. Miyamoto. 2003. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer Res. 63:149-153. [PubMed] [Google Scholar]
- 24.Hartwell, L. H., and M. B. Kastan. 1994. Cell cycle control and cancer. Science 266:1821-1828. [DOI] [PubMed] [Google Scholar]
- 25.He, B., J. A. Kemppainen, and E. M. Wilson. 2000. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 275:22986-22994. [DOI] [PubMed] [Google Scholar]
- 26.He, B., J. T. Minges, L. W. Lee, and E. M. Wilson. 2002. The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J. Biol. Chem. 277:10226-10235. [DOI] [PubMed] [Google Scholar]
- 27.He, B., and E. M. Wilson. 2003. Electrostatic modulation in steroid receptor recruitment of LXXLL and FXXLF motifs. Mol. Cell. Biol. 23:2135-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 29.Heinlein, C. A., and C. Chang. 2002. Androgen receptor (AR) coregulators: an overview. Endocrinol. Rev. 23:175-200. [DOI] [PubMed] [Google Scholar]
- 30.Heinlein, C. A., and C. Chang. 2002. The roles of androgen receptors and androgen-binding proteins in nongenomic androgen actions. Mol. Endocrinol. 16:2181-2187. [DOI] [PubMed] [Google Scholar]
- 31.Hermanson, O., C. K. Glass, and M. G. Rosenfeld. 2002. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 13:55-60. [DOI] [PubMed] [Google Scholar]
- 32.Hirai, I., and H. G. Wang. 2002. A role of the C-terminal region of human Rad9 (hRad9) in nuclear transport of the hRad9 checkpoint complex. J. Biol. Chem. 277:25722-25727. [DOI] [PubMed] [Google Scholar]
- 33.Hsu, C. L., Y. L. Chen, S. Yeh, H. J. Ting, Y. C. Hu, H. Lin, X. Wang, and C. Chang. 2003. The use of phage display technique for the isolation of androgen receptor interacting peptides with (F/W)XXL(F/W) and FXXLY new signature motifs. J. Biol. Chem. 278:23691-23698. [DOI] [PubMed] [Google Scholar]
- 34.Jepsen, K., O. Hermanson, T. M. Onami, A. S. Gleiberman, V. Lunyak, R. J. McEvilly, R. Kurokawa, V. Kumar, F. Liu, E. Seto, S. M. Hedrick, G. Mandel, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102:753-763. [DOI] [PubMed] [Google Scholar]
- 35.Joon, D. L., M. Hasegawa, C. Sikes, V. S. Khoo, N. H. Terry, G. K. Zagars, M. L. Meistrich, and A. Pollack. 1997. Supraadditive apoptotic response of R3327-G rat prostate tumors to androgen ablation and radiation. Int. J. Radiat. Oncol. Biol. Phys. 38:1071-1077. [DOI] [PubMed] [Google Scholar]
- 36.Kang, H. Y., H. K. Lin, Y. C. Hu, S. Yeh, K. E. Huang, and C. Chang. 2001. From transforming growth factor-beta signaling to androgen action: identification of Smad3 as an androgen receptor coregulator in prostate cancer cells. Proc. Natl. Acad. Sci. USA 98:3018-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang, H. Y., S. Yeh, N. Fujimoto, and C. Chang. 1999. Cloning and characterization of human prostate coactivator ARA54, a novel protein that associates with the androgen receptor. J. Biol. Chem. 274:8570-8576. [DOI] [PubMed] [Google Scholar]
- 38.Kaur, R., C. F. Kostrub, and T. Enoch. 2001. Structure-function analysis of fission yeast Hus1-Rad1-Rad9 checkpoint complex. Mol. Biol. Cell 12:3744-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komatsu, K., T. Miyashita, H. Hang, K. M. Hopkins, W. Zheng, S. Cuddeback, M. Yamada, H. B. Lieberman, and H. G. Wang. 2000. Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat. Cell Biol. 2:1-6. [DOI] [PubMed] [Google Scholar]
- 40.Kostrub, C. F., K. Knudsen, S. Subramani, and T. Enoch. 1998. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 17:2055-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langley, E., Z. X. Zhou, and E. M. Wilson. 1995. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J. Biol. Chem. 270:29983-29990. [DOI] [PubMed] [Google Scholar]
- 42.Lee, D. K., and C. Chang. 2003. Molecular communication between androgen receptor and general transcription machinery. J. Steroid Biochem. Mol. Biol. 84:41-49. [DOI] [PubMed] [Google Scholar]
- 43.Liao, G., L. Y. Chen, A. Zhang, A. Godavarthy, F. Xia, J. C. Ghosh, H. Li, and J. D. Chen. 2003. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J. Biol. Chem. 278:5052-5061. [DOI] [PubMed] [Google Scholar]
- 44.Lieberman, H. B., K. M. Hopkins, M. Nass, D. Demetrick, and S. Davey. 1996. A human homolog of the Schizosaccharomyces pombe rad9+ checkpoint control gene. Proc. Natl. Acad. Sci. USA 93:13890-13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, H.-K., Y.-C. Hu, L. Yang, S. Altuwaijri, Y.-T. Chen, H.-Y. Kang, and C. Chang. 2003. Suppression versus induction of androgen receptor functions by the phosphatidylinositol 3-kinase/Akt pathway in prostate cancer LNCaP cells with different passage numbers. J. Biol. Chem. 278:50902-50907. [DOI] [PubMed] [Google Scholar]
- 46.Lin, H. K., L. Wang, Y. C. Hu, S. Altuwaijri, and C. Chang. 2002. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 21:4037-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Llopis, J., S. Westin, M. Ricote, Z. Wang, C. Y. Cho, R. Kurokawa, T. M. Mullen, D. W. Rose, M. G. Rosenfeld, R. Y. Tsien, C. K. Glass, and J. Wang. 2000. Ligand-dependent interactions of coactivators steroid receptor coactivator-1 and peroxisome proliferator-activated receptor binding protein with nuclear hormone receptors can be imaged in live cells and are required for transcription. Proc. Natl. Acad. Sci. USA 97:4363-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maiti, S., M. L. Meistrich, G. Wilson, G. Shetty, M. Marcelli, M. J. McPhaul, P. L. Morris, and M. F. Wilkinson. 2001. Irradiation selectively inhibits expression from the androgen-dependent Pem homeobox gene promoter in Sertoli cells. Endocrinology 142:1567-1577. [DOI] [PubMed] [Google Scholar]
- 49.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McInerney, E. M., M. J. Tsai, B. W. O'Malley, and B. S. Katzenellenbogen. 1996. Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone receptor coactivator. Proc. Natl. Acad. Sci. USA 93:10069-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 53.McKenna, N. J., and B. W. O'Malley. 2002. Nuclear receptor coactivators—an update. Endocrinology 143:2461-2465. [DOI] [PubMed] [Google Scholar]
- 54.Ngan, E. S., Y. Hashimoto, Z. Q. Ma, M. J. Tsai, and S. Y. Tsai. 2003. Overexpression of Cdc25B, an androgen receptor coactivator, in prostate cancer. Oncogene 22:734-739. [DOI] [PubMed] [Google Scholar]
- 55.Nishimura, K., H. J. Ting, Y. Harada, T. Tokizane, N. Nonomura, H. Y. Kang, H. C. Chang, S. Yeh, H. Miyamoto, M. Shin, K. Aozasa, A. Okuyama, and C. Chang. 2003. Modulation of androgen receptor transactivation by gelsolin: a newly identified androgen receptor coregulator. Cancer Res. 63:4888-4894. [PubMed] [Google Scholar]
- 56.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 57.Quigley, C. A., A. De Bellis, K. B. Marschke, M. K. el-Awady, E. M. Wilson, and F. S. French. 1995. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocrinol. Rev. 16:271-321. [DOI] [PubMed] [Google Scholar]
- 58.Rahman, M. M., H. Miyamoto, H. Lardy, and C. Chang. 2003. Inactivation of androgen receptor coregulator ARA55 inhibits androgen receptor activity and agonist effect of antiandrogens in prostate cancer cells. Proc. Natl. Acad. Sci. USA 100:5124-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rauen, M., M. A. Burtelow, V. M. Dufault, and L. M. Karnitz. 2000. The human checkpoint protein hRad17 interacts with the PCNA-like proteins hRad1, hHus1, and hRad9. J. Biol. Chem. 275:29767-29771. [DOI] [PubMed] [Google Scholar]
- 60.Roos-Mattjus, P., K. M. Hopkins, A. J. Oestreich, B. T. Vroman, K. L. Johnson, S. Naylor, H. B. Lieberman, and L. M. Karnitz. 2003. Phosphorylation of human Rad9 is required for genotoxin-activated checkpoint signaling. J. Biol. Chem. 278:24428-24437. [DOI] [PubMed] [Google Scholar]
- 61.Siede, W., A. S. Friedberg, and E. C. Friedberg. 1993. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:7985-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.St. Onge, R. P., B. D. Besley, J. L. Pelley, and S. Davey. 2003. A role for the phosphorylation of hRad9 in checkpoint signaling. J. Biol. Chem. 278:26620-26628. [DOI] [PubMed] [Google Scholar]
- 63.Thelen, M. P., C. Venclovas, and K. Fidelis. 1999. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell 96:769-770. [DOI] [PubMed] [Google Scholar]
- 64.Thin, T. H., L. Wang, E. Kim, L. L. Collins, R. Basavappa, and C. Chang. 2003. Isolation and characterization of androgen receptor mutant, AR(M749L), with hypersensitivity to 17-beta estradiol treatment. J. Biol. Chem. 278:7699-7708. [DOI] [PubMed] [Google Scholar]
- 65.Venclovas, C., and M. P. Thelen. 2000. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 28:2481-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Volkmer, E., and L. M. Karnitz. 1999. Human homologs of Schizosaccharomyces pombe rad1, hus1, and rad9 form a DNA damage-responsive protein complex. J. Biol. Chem. 274:567-570. [DOI] [PubMed] [Google Scholar]
- 67.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 68.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 69.Yang, L., H. K. Lin, S. Altuwaijri, S. Xie, L. Wang, and C. Chang. 2003. APPL suppresses androgen receptor transactivation via potentiating Akt activity. J. Biol. Chem. 278:16820-16827. [DOI] [PubMed] [Google Scholar]
- 70.Yeh, S., and C. Chang. 1996. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl. Acad. Sci. USA 93:5517-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeh, S., Y.-C. Hu, P.-H. Wang, C. Xie, Q. Xu, M.-Y. Tsai, Z. Dong, R.-S. Wang, T.-H. Lee, and C. Chang. 2003. Abnormal mammary gland development and growth retardation in female mice and MCF7 breast cancer cells lacking androgen receptor. J. Exp. Med. 278:1899-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshida, K., K. Komatsu, H. G. Wang, and D. Kufe. 2002. c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol. Cell. Biol. 22:3292-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, Y., Y. Yang, S. Yeh, and C. Chang. 2003. ARA67 functions as a repressor to suppress androgen receptor transactivation. Mol. Cell. Biol. 24:1044-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]