Abstract
Transcriptionally active and inactive domains are frequently found adjacent to one another in the eukaryotic nucleus. To better understand the underlying mechanisms by which domains maintain opposing transcription patterns, we performed a systematic genomewide screen for proteins that may block the spread of silencing in yeast. This analysis identified numerous proteins with efficient silencing blocking activities, and some of these have previously been shown to be involved in chromatin dynamics. We isolated subunits of Swi/Snf, mediator, and TFIID, as well as subunits of the Sas-I, SAGA, NuA3, NuA4, Spt10p, Rad6p, and Dot1p complexes, as barrier proteins. We demonstrate that histone acetylation and chromatin remodeling occurred at the barrier and correlated with a block to the spread of silencing. Our data suggest that multiple overlapping mechanisms were involved in delimiting silenced and active domains in vivo.
The eukaryotic nucleus is organized into active and inactive domains, and the modulation of genes within these domains is mediated by different regulatory elements. Enhancers, promoters, and locus control regions activate genes, whereas silencers repress the transcription of genes. Junctions between the active and inactive gene domains commonly occur along chromosomes (reviewed in reference 54), and specific elements and activities that maintain opposing transcription states in adjacent domains have been previously identified.
The budding yeast Saccharomyces cerevisiae also possesses active and inactive chromatin domains, and one of the best-characterized chromatin domains is the silenced HMR locus. The genes present at HMR are flanked by silencer elements, and proteins that bind these elements recruit the Sir proteins that mediate silencing. Following recruitment, the Sir proteins are thought to spread along the DNA to form a specialized chromatin state that is inaccessible to various enzymatic probes. The silent domains extend beyond the silencers and up to barrier elements that block the spread (3, 13, 15, 21, 40, 57).
While the left boundary of the silenced chromatin domain at HMR is not well defined, the right boundary has been studied in great detail. The deletion of this barrier element at the native HMR locus leads to an increased spread of silenced chromatin and to concomitant repression of neighboring euchromatic genes (13, 15), while the ectopic insertion of this barrier between a silencer and a promoter blocks the repressive effects of the silencer. A specific tRNA gene acts as a barrier at HMR, and mutations in the promoter of this gene or in the RNA polymerase III transcription factors weaken the barrier activity mediated by this tRNA gene. In addition, mutations in the acetyltransferases Sas2p and Gcn5p advertently affect barrier activity mediated by the tRNA gene.
The left boundary of the Sir proteins at HML maps to the upstream activation sequence of YCL069w (3, 37), while the right boundary maps to the promoter of the CHA1 gene (37)—though silencing is believed to terminate at HML-I (4). However, the specific proteins that mediate barrier function at these elements have yet to be identified.
The telomeric ends of S. cerevisiae chromosomes also nucleate a silenced domain that spreads along the subtelomeric chromatin, resulting in the formation of an inaccessible chromatin domain. Barrier elements called STARs (signal transduction and activation of RNA), which are found in the subtelomeric sequences, can block this spread. Analysis of these elements revealed that transcription factors Reb1p and Tbf1p, which are bound to specific sites within these elements, are involved in blocking the spread of telomeric silencing (19, 21, 57). Interestingly, while transcription factors are necessary, transcription per se is not required for barrier function (6, 19-21).
While work detailing native yeast barriers suggests that transcription factors and chromatin-modifying activities act as barriers, a recent genetic screen for barrier function at HML revealed that Cse1p, Los1p, Mex67p, Sxm1p, and Gsp2p, which have previously been shown to be involved in nuclear transport (25), can also insulate a reporter gene.
On the basis of these studies, at least two different models for boundary functions have been proposed: the chromatin-modifying model (reviewed in reference 14) and the topological domain model (25). According to the chromatin-modifying model, which is based on studies of native yeast boundaries, barrier proteins bound to DNA create regions of open chromatin that prevent the propagation of silenced chromatin. Boundary proteins are believed to function by recruiting chromatin-modifying enzymes that in turn modify nucleosomes and alter the underlying chromatin substrate to a state that is unfavorable for the binding of the Sir proteins, thereby blocking the propagation of heterochromatin. The topological domain model, on the other hand, maintains that boundary elements tether DNA to a nuclear substructure, which then forms an impediment to the spreading heterochromatin. It further suggests that anchoring DNA to nuclear substructures generates independent and distinct domains, and this may be the mechanism for barrier function. While these two models are mechanistically distinct, the outcome of both is the creation and maintenance of adjacent chromatin domains with opposing transcriptional activities.
To gain further insight into the underlying mechanism of barrier function, we sought to identify proteins that may block the spread of silencing when inserted between a silencer and a promoter. We theorized that an unbiased genomewide screen for barrier function to identify all proteins in yeast with the ability to block heterochromatin will aid in understanding mechanisms for barriers. Screening a genomewide library of yeast fusion proteins allowed us to evaluate the potential of each protein to block the spread of silencing. The screening process led to the isolation of numerous proteins with robust barrier activity. Analysis of some of these proteins suggested that they most likely functioned by remodeling chromatin or modifying histones to form localized regions of open chromatin, and this may be one mechanism of barrier function in yeast.
MATERIALS AND METHODS
Plasmids used in this study.
The base plasmid, pGBK-RC, comprises the following members: pRO586, pGBK-RC plus GBD-Snf6p (open reading frame [ORF]); pRO587, pGBK-RC plus GBD-Taf47p (ORF); pRO588, pGBK-RC plus GBD-Ada1p (ORF); pRO590, pGBK-RC plus GBD-Sas2p (ORF); pRO591, pGBK-RC plus GBD-Sas5p (ORF); pRO594, pGBK-RC plus GBD-Clb1p (ORF); pRO596, pGBK-RC plus GBD-Gds1p (ORF); pRO635, pGBK-RC plus GBD-Nup2p (ORF); pRO637, pGBK-RC plus GBD-Dot1p (ORF); pRO363 (13); pRO486 (15); pRO4 (13); and pRO651, 4xGAL4bs (upstream activation sequence is between HMR-E and I a1).
Silencing blocking screen with Gal4p DNA binding domain-fused S. cerevisiae genes.
ROY1864 was initially transformed with pRO486 (URA3 plasmid) and was then individually cotransformed with GAL4 DNA binding domain (GBD)-fused plasmid DNA. These strains were grown in liquid medium (lacking both Ura and Trp) in 96-well microtiter plates for 2 days at 30°C. Cells were transferred to YMD petri plates containing mating tester lawns (JRY19a) with appropriate selection for diploid colonies. The plates were incubated for 2 to 3 days at 30°C, and the results were catalogued.
Patch mating assays were performed as described previously (13).
Preparation of yeast nuclei and micrococcal nuclease digestion.
Yeast cells (4 liters) were grown in YMD medium lacking tryptophan to an absorbance at 600 nm of 1.0 at 30°C. The cells were collected by centrifugation and resuspended in 1 liter of YPD and allowed to grow for 2 h at 30°C. The cells were washed in 100 ml of sorbitol phosphate buffer (1 M sorbitol, 20 mM KPi [pH 7.5]) and resuspended in 80 ml of spheroplasting medium (250 mM sorbitol, 1% glucose, 0.2% yeast nitrogen base, 0.2% Casamino acids, 25 mM HEPES, 50 mM Tris-HCl [pH 7.5], 3 mM dithiothreitol, 2 mM sodium bisulfite). Forty thousand units of lyticase was added, and the cells were incubated at 30°C for 40 min. Following spheroplasting, the cells were washed twice in 200 ml of ice-cold buffer (1 M sorbitol, 50 mM Tris [pH 7.5], 10 mM β-mercaptoethanol, 2 mM sodium bisulfite). The spheroplasts were suspended in 60 ml of F buffer (40 mM KPi, 4 mM EDTA, 1 mM EGTA, 1 mM spermidine, 0.3 mM spermine, 18% Ficoll, 0.2% Triton X-100, 10 mM β-mercaptoethanol, and protease inhibitors [final pH, 6.5 with phosphoric acid]) and homogenized five times with a Dounce homogenizer (tight pestle). The nuclei were washed twice in 20 ml of wash buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA [pH 8.0], 0.5 mM EGTA [pH 8.0], 10 mM β-mercaptoethanol, and protease inhibitors). The pellet was resuspended in nucleus storage buffer (100 mM Tris-acetate [pH 7.9], 50 mM potassium acetate, 20% glycerol, 2 mM EDTA [pH 8.0], 3 mM dithiothreitol, and protease inhibitors). Micrococcal nuclease digestion was performed as described previously (22).
Chromatin immunoprecipitation.
Chromatin immunoprecipitation assays were performed as described previously (11). The primer sequences are available upon request.
RESULTS
Genomewide screen for proteins with barrier function.
We assayed the potential ability of most yeast proteins to block the spread of silencing at the HMR locus. It has previously been established that the HMR-E silencer is essential for repression, with the silenced chromatin initiating at the HMR-E silencer and spreading along the DNA to encompass the MATa1 gene located 1,050 bp from the silencer (41). Though the HMR-I silencer is important for complete silencing, significant silencing is still observed across the HMR locus in its absence (1, 45, 61).
A MATα hmrΔ yeast strain (ROY 1864), carrying a plasmid (pRO486)-borne copy of the HMR locus with a deletion of the HMR-I silencer, was used for the screening procedure. Silencing of the MATa1 gene on the plasmid was mediated solely by the HMR-E silencer (13, 15). Four binding sites for the GBD were inserted ∼600 bp downstream of the silencer between HMR-E and the MATa1 reporter gene (15). We believe that this construct mimicked the native state in yeast in which yeast barriers are located outside the HMR and HML silencers and function by blocking the unidirectional spread of silencing propagating from the silencer (37).
This MATα strain was then individually transformed with a second TRP1-based plasmid. The plasmid contained an ADH1 promoter driving expression of a protein chimera composed of a GBD fused in frame to each yeast ORF (26). A total of 5,551 transformants covering almost the entire yeast genome were obtained. These transformants were grown in liquid culture in microtiter plates, and expression of the MATa1 gene at HMR was monitored by transferring approximately 2 to 3 μl of the cells to plates containing MATa tester lawns. In the absence of a barrier, the silenced domain emanating from HMR-E encompassed MATa1, allowing the MATα strain to mate with the tester lawn and to form diploid colonies on selective plates. A functional barrier blocked the spread of silencing, resulting in the derepression of the reporter MATa1 gene and the concomitant loss of mating ability.
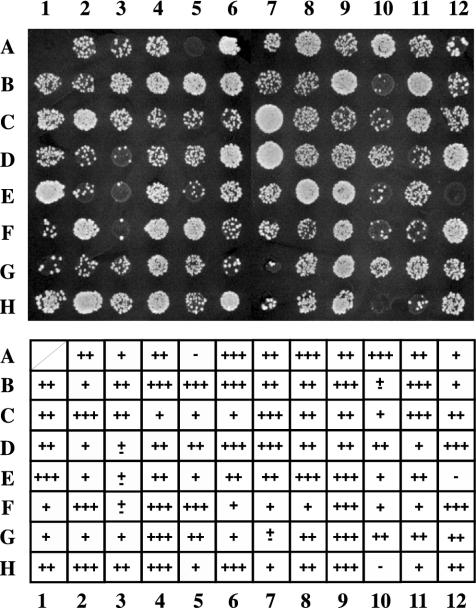
Figure 1 shows a representative result of 95 transformants and their mating abilities. Each transformant was mated in triplicate and analyzed. The results were scored in five categories based on the number of diploid colonies formed. Strains that gave rise to no colonies were classified as bearing protein chimeras with very strong barrier activity, while strains giving rise to one to nine colonies were classified as containing strong barrier proteins. Strains giving rise to 10 to 20 colonies were classified as possessing proteins with moderate barrier activity, and strains with more than 20 colonies had weak barrier proteins. Finally, those that appeared as wild-type strains were considered to have no barrier function. The initial classification resulted in 27 very strong, 102 strong, 182 moderate, and 744 weak barrier proteins, while 4,496 proteins exhibited no barrier function.
FIG. 1.
Systematic screening with GBD-fused library. ROY1864 (MATα hmrΔ::bgl-bcl) containing pRO486 (HMR-EΔI plus GAL4 bs) was individually transformed with a GBD-fused ORF library and grown in selective liquid culture in microtiter plates. Cells were transferred to YMD plates with mating lawns to assay for expression of the MATa1 gene at HMR. The growth of diploid colonies is shown. We classified each clone into five categories. −, 0 diploid colonies (very strong barrier); ±, 1 to 9 colonies (strong barrier); +, 10 to 20 (moderate barrier); ++, >21 diploid colonies (weak barrier); +++, no barrier.
GBD protein-mediated barrier function requires Gal4p binding sites.
During our secondary analysis, we focused on the strong and very strong barrier proteins. Mating assays were repeated by using the patch mating method with four independently isolated transformants, and clones that gave a uniform result were analyzed further.
Plasmids isolated from these yeast strains were retransformed into a different MATα yeast strain (ROY 2042), the HMR reporter cassette of which was integrated in the genome. The transformants were reassayed by patch mating for their ability to block the spread of silencing.
The isolated plasmids were also tested for their ability to disrupt silencing in a yeast strain containing an HMR cassette that had no Gal4p binding sites located between the silencer and the MATa1 reporter gene (ROY 2041). The use of this strain allowed us to distinguish between proteins that specifically blocked the spread of silencing at HMR and those that disrupted silencing (antisilencing) when overexpressed (27, 28, 34, 44, 62).
All positive plasmid clones that blocked the spread of silencing in a Gal4p binding site-dependent manner were then sequenced to determine whether the yeast ORF was fused in frame with the GBD and whether the entire ORF had been cloned (see Fig. S1 in the supplemental material). Following these analyses, 55 clones were isolated, and a majority of these encoded proteins are involved in chromatin dynamics (Table 1).
TABLE 1.
Clones isolated in this study
| Analysis subject | Gene |
|---|---|
| Histone modifications | SPT10, EPL1, YNG1, SAS2, SAS5, ADA5, ADA2, ADA3, HFI1, SGF73, SGF29, DOT1, LGE1 |
| TFIID | TBP1, TAF47, TAF17, TAF90, TAF60, TAF61, TAF30 |
| SWI/SNF | SNF6, SNF5 |
| Mediator | GAL11, MED2, ROX3, MED6, MED8 |
| Transcription factors | ACE1, HSF1, LEU3, RGT1, FIO8, ACA1, SWI5, HMRA1, SPT21, TFA2 |
| Cell cycle | SIW4, CLB1, MND2 |
| Other | LYS5, ICY1, YAP1802, SEC35, SWA2, GDS1, SFP1, GIC1, MRS6 |
| Unknown | YBL081W, YBR271W, YCR076C, YCR082W, YDR031W, YDR223W |
GBD barrier protein chimeras block silencing without eliminating it.
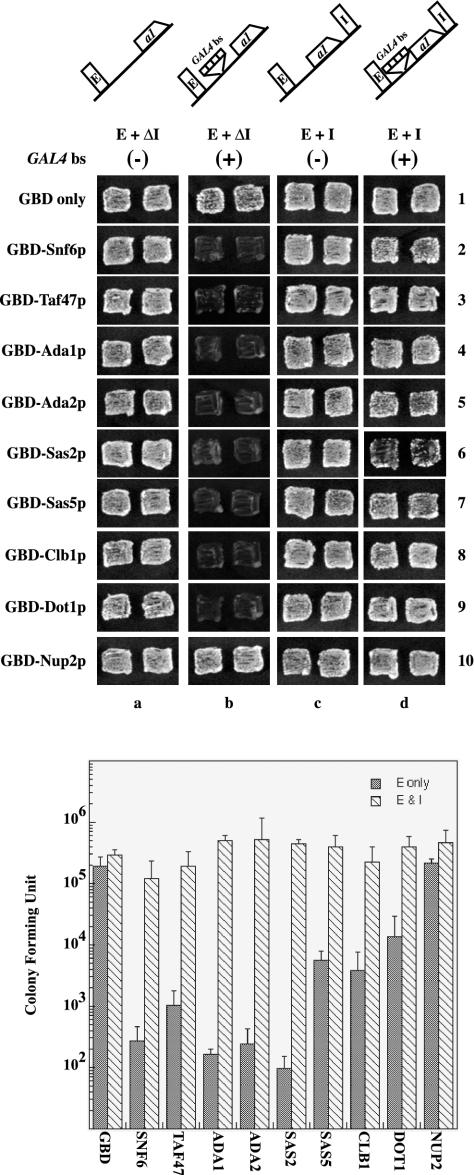
Figure 2 (top) shows the blocking activity of representative clones isolated in our screen. In the absence of any Gal4p binding sites between the HMR-E silencer and MATa1, (column a), the reporter gene is silenced and the strains mate as efficiently as a strain with only the GBD. In the presence of a barrier element (Gal4p binding sites) between HMR-E and the reporter (column b), the GBD chimeras could block the spread of silencing to allow expression of MATa1, resulting in nonmaters (Fig. 2 [top] column b; compare row 1 with rows 2 through 10). Analysis by quantitative mating (Fig. 2 [bottom]) demonstrated that the various barrier proteins efficiently blocked silencing, but to different extents—though none caused the same degree of derepression as that caused by a Sir deletion.
FIG. 2.
(Top) Relative barrier activity of the GBD chimeras. We used patch mating assays to analyze the ability of clones to block silencing but not disrupt silencing. ROY1864 was transformed with any of four different URA3-containing reporter plasmids: pRO363 (HMR-EΔI), pRO486 (HMR-EΔI plus GAL4 bs), pRO4 (HMR-E+I), and pRO651 (HMR-E+I plus GAL4 bs). Each of these strains was cotransformed with a second TRP1-containing plasmid containing variousGBD fusion chimeras as indicated. Cells were grown on selective medium (YMD lacking both Trp and Ura) and were analyzed by patch mating against tester lawns (JRY19a) for silencing of the MATa1 gene. Growth of diploid cells indicates silencing of the reporter gene, while absence of any growth indicates activation of the reporter gene due to blocking of silencing. The chimera plasmids used are GBD alone (pGBK-RC), GBD-Snf6p (pRO586), GBD-Taf47p (pRO587), GBD-Ada1p (pRO588), GBD-Ada2p (pRO592), GBD-Sas2p (pRO590), GBD-Sas5p (pRO591), GBD-Clb1p (pRO594), GBD-Dot1p (pRO637),and GBD-Nup2p (pRO635). (Bottom) Quantitative mating analyses of various GBD chimeras in strain ROY1864 containing either pRO486 (E only) or pRO651 (E & I) were performed as described previously (12, 39). The data are presented as diploid CFU and are mean values from three independent experiments carried out in parallel.
Our results thus far suggested that the proteins identified in our screening process functioned by blocking the spread of silencing that began from the HMR-E silencer. Two scenarios as to how these proteins blocked silencing can be envisioned. In the first scenario, the recruited barrier proteins displaced the Sir proteins from the entire locus to completely abolish silencing. In the second scenario, the GBD chimeras behaved as barriers to block the unidirectional propagation of the Sir proteins from the silencer. We sought to distinguish between the two scenarios by assaying the ability of the barrier GBD chimeras to disrupt silencing from a barrier element flanked by both the HMR-E and HMR-I silencers. We reasoned that if the GBD chimeras displaced the Sir proteins entirely, then recruiting these proteins to Gal4p binding sites placed between the E and I silencers would eliminate the silenced state and lead to expression of the MATa1 reporter gene. None of the 55 positive clones tested was able to disrupt silencing from this locus (Fig. 2 [top and bottom]). This result suggested that the two silencer elements silence the domain and prevent the GBD chimeras from blocking the propagation of the Sir proteins emanating from HMR-E. The observation was not altogether surprising because there are several previous reports showing that the HMR-I element functioned as a silencer in conjunction with HMR-E and is required to stabilize the silenced state (1, 13, 45, 61). It is possible that the two silencers cooperate in spite of the presence of the barrier, thus rendering it inactive.
Nuclear pore proteins are unable to block the spread of silencing at HMR.
An analogous screen recently showed that nuclear transport proteins tethered to barrier elements could insulate a reporter gene present at HML (25). However, we failed to identify any of these proteins in our systematic genomewide screen for tethered barrier protein chimeras at HMR. We retested some of the clones for their blocking activity at HMR by patch and quantitative mating, and the data for one of these, GBD-Nup2p, are shown in Fig. 2 (top), row 10. None of the nuclear pore clones that we retested was able to block the spread of silenced chromatin at HMR.
SAS clones require native SAS-I complex for barrier function.
A significant number of the barrier proteins identified in our screening process have previously been shown to be components of multiprotein complexes. We isolated two subunits of each of the Swi/Snf and SAS-I complexes and eight subunits of the SAGA complex. We were therefore interested in determining whether any of the other subunits of the complex were required for barrier activity mediated by these GBD chimeras or whether they functioned independently of the other subunits of the complex.
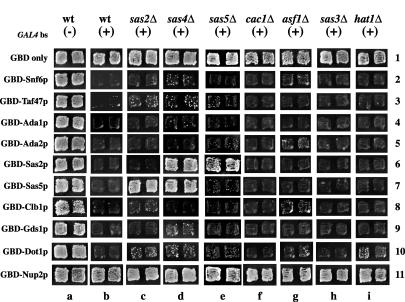
We addressed this question with respect to the SAS-I complex composed of the five proteins Sas2p, Sas4p, Sas5p, Cac1p, and Asf1p. We systematically deleted each of the subunits of the SAS-I complex and were curious as to whether the various barrier GBD chimeras could still block the spread of silencing (Fig. 3). GBD-Sas5p and GBD-Sas2p were unable to block silencing in strains lacking SAS2 and SAS5, respectively (columns c and e). Similarly, neither GBD chimera could function as a barrier in the absence of SAS4 (column d). However, the loss of CAC1 or ASF1 had no effect on the barrier activity of either GBD-Sas2p or GBD-Sas5p (columns f and g).
FIG. 3.
Boundary activity in disruption strains. We used patch mating assays to analyze the ability of GBD barrier chimeras to block silencing in strains from which specific genes were deleted. Each strain was transformed with the different plasmids as described in the legend of Fig. 2 and assayed on the same plate. (−) and (+), constructs without and with Aal4p binding sites, respectively; wt, wild type.
The barrier activities of all the other GBD chimeras were unaffected by the loss of SAS-I subunits. Furthermore, since the SAS-I complex possesses histone acetyltransferase activity, we also determined whether loss of any histone acetyltransferase also affected barrier function mediated by these GBD chimeras. Our data revealed that the barrier activity of the clones was not significantly affected in strains lacking Hat1p or Sas3p (Fig. 3, columns h and i).
We did observe that deletions in some of the SAS-I complex reproducibly weakened the barrier activity of GBD-Dot1p. Further experiments will be required in order to determine whether this result was direct or indirect and what its significance might be.
Barrier activity at telomeres.
Silencing mediated by the Sir proteins Sir2p, Sir3p, and Sir4p has been shown to occur at HMR, HML, and telomeres. Since our original screen was performed with HMR, we were interested in determining whether the isolated GBD chimeras would also block the spread of silencing at other loci.
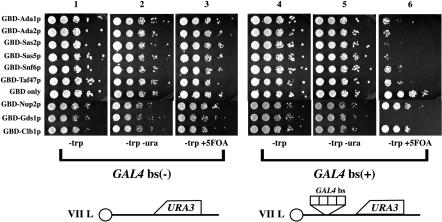
We first tested the ability of the barrier GBD chimeras to block the spread of silencing emanating from telomeres (see Fig. 4 [top]). We used two strains (YDS 631 and YDS 634), the URA3 reporter gene of which was inserted close to the telomere of chromosome VII L (9). Gal4p DNA binding sites were present between the telomere and the URA3 gene in one of the strains. The URA3 gene was stably repressed in approximately 60% of the cells in the population, and these cells could grow in medium containing 5-fluoroorotic acid (5-FOA) but not in medium lacking uracil. In the population's remaining cells, the gene was active; these cells could grow in medium lacking uracil but not in medium containing 5-FOA. The two strains were transformed with plasmids expressing the GBD chimeras, and expression of the URA3 reporter gene was monitored for growth on medium lacking uracil or containing 5-FOA. Most of the GBD chimeras blocked the spread of silencing in a Gal4p binding site-dependent manner to allow URA3 expression in the vast majority of cells (Fig. 4 [top]; compare columns 3 and 6).
FIG. 4.
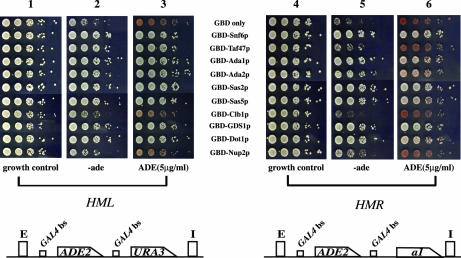
(Top) Barrier activity at telomere VIIL. Strains YDS631 (adh4::URA3-telVII-L) and YDS634 (adh4::URA3-4xGAL4 bs-telVII-L) (9) were transformed with GBD fusion chimera plasmids as described in the legend of Fig. 2. Cells were grown on selective liquid media (YMD lacking Trp) and spotted as 10-fold serial dilutions on YMD plates lacking uracil and tryptophan or containing 5-FOA as described previously (28). (Bottom) Strain KIY54 (25) and ROY 2770 were transformed with the GBD fusion chimera plasmids described in the legend of Fig. 2A. The strains were grown in liquid YMD-HC medium with selection (−trp) overnight, and the cells were spotted as 10-fold serial dilutions on YMD-HC plates lacking adenine and tryptophan or containing 5 μg of adenine per ml but lacking tryptophan as described previously (28). The plates were photographed after 2 days.
Similar to the results observed at HMR (Fig. 2 [top], column b), GBD-Nup2p was unable to block the spread of silencing at the telomeres. Furthermore, GBD-Clb1p, which was able to block the unidirectional spread of silencing at HMR, was unable to block telomeric silencing (Fig. 4 [top]).
Insulation of genes from flanking silencers at HML and HMR.
Since we observed some differences in the ability of the GBD chimeras to block the spread of silencing at HMR and telomeres, we also investigated the ability of these proteins to function as barriers at HML by using a strain (KIY54) that had previously been used to study insulator function at this locus (25). This strain has a modified HML locus, with the ADE2 gene replacing the divergently transcribed MATα1 and MATα2 genes, including the protosilencers. Gal4p binding sites flank the ADE2 gene. Transcription of ADE2 occurs if the gene is insulated from silencing emanating from the HML-E and HML-I silencers.
Expression was assayed in the presence of various GBD chimeras in order to analyze their ability to insulate this gene at this locus (Fig. 4 [bottom], columns 1 to 3). All the chimeras isolated in our HMR barrier screen effectively blocked silencing to allow expression of ADE2. Interestingly, while GBD-Nup2p and GBD-Clb1p were able to insulate the ADE2 gene at this locus, the insulation of the gene by these proteins was not very robust.
The assay for insulator function at HML measured the ability of GBD chimeras located on either side of a reporter gene to insulate from silencing a gene that was propagating from two flanking silencers (as shown in Fig. 4 [bottom]). This arrangement was different from that used for the assays that we used to analyze the ability of proteins to block the unidirectional spread of silencing from a single silencer at HMR and the telomeres (Fig. 2 [top] and Fig. 4[top]). Thus, we could not directly compare the results observed at HML with our data from HMR and telomeres.
To directly determine whether the differences we observed were genuine or were due to the differences in the assays used, we generated a strain where the ADE2 locus with the flanking Gal4p binding sites was inserted into the HMR locus such that it was flanked by the HMR-E and HMR-I silencers. The flanking Gal4p binding sites were in exactly the same context with respect to ADE2 as those that were used to study insulator function at HML, since the construct was PCR amplified from the HML reporter strain (KIY54 [25]) and integrated at HMR. The HMR::Gbs-ADE2-Gbs strain (ROY2770) was transformed with plasmids expressing the various GBD chimera proteins, and expression of the ADE2 gene was monitored (Fig. 4 [bottom], columns 4 to 6). Once again, we observed that some of the chimeras insulated the reporter gene from silencing better than did others, such as GBD-Clb1p, GBD-Nup2p, and GBD-Taf47p, which were not able to block silencing efficiently (as determined by assays for growth in medium lacking adenine).
Our results with HMR confirm previous reports, which demonstrated that HMR-I functions as a silencer in conjunction with HMR-E (1, 13, 45, 61). The results depicted in Fig. 2 (top) and 4 (bottom) show that in the presence of HMR-I, insulation of genes required barriers on both sides of the gene.
Analysis of chromatin structure around a barrier.
Identification of subunits of the Swi/Snf complex as barrier proteins led to the intriguing possibility that this complex may be mediating barrier function by remodeling the chromatin around the barrier element. We were therefore interested in determining whether this complex remodeled chromatin, and if so, whether the remodeling was localized to the barrier element or affected the entire chromatin domain.
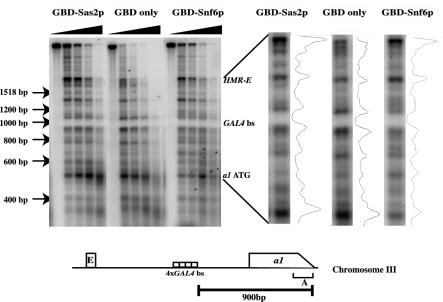
To study the changes in chromatin structure at the barrier, we performed a micrococcal nuclease analysis followed by indirect end labeling (70) that was performed by using nuclei isolated from three yeast strains for which barrier function was mediated by either the GBD alone, GBD-Snf6p, or GBD-Sas2p. The nuclei were digested for various times with micrococcal nuclease, and the purified DNA was subsequently digested with BglII, resolved on agarose gels, and analyzed by DNA blotting with probes specific to either HMR (Fig. 5) or the ACT1 gene (data not shown). Using markers, we were able to locate the start site of the MATa1 reporter gene, the four Gal4p binding site barriers, and the HMR-E silencer. Analysis of the indirect end-labeling profile indicated that the four Gal4p binding sites were inaccessible to the enzyme in all three strains, suggesting either that the sites were bound by GBD chimeras in all three strains or, alternatively, were packaged in a positioned nucleosome. The HMR-E silencer and the start of the MATa1 gene were in a region that was highly accessible to the nuclease, consistent with previously published data (50, 58). Interestingly, when GBD-Snf6p was present at the barrier element, there was increased digestion by micrococcal nuclease immediately around the barrier, extending a few hundred base pairs to either side of the barrier. While this increased cutting by the enzyme was not very dramatic, it was reproducible and manifested itself only in regions flanking the barrier. This increased digestion was not observed in strains in which only GBD or the GBD-Sas2p fusion proteins were present. These experiments were performed multiple times with multiple independently isolated nucleus preparations, and essentially the same result was obtained in each case. When the analysis was performed on the same digested samples but with the ACT1 probe (data not shown), no difference was observed around the promoter of this gene in all three strains, suggesting that these differences were specific to the HMR locus and specific to GBD-Snf6p.
FIG. 5.
Analysis of chromatin structure around the barrier. Strain ROY2042 containing HMRΔI with four Gal4p binding sites located between HMR-E and the MATa1 gene was transformed with pGBK-RC (GBD alone), pRO586 (GBD-Snf6p), or pRO590 (GBD-Sas2p). Nuclei were prepared from these strains and digested for various lengths of time with micrococcal nuclease. The deproteinized DNA was then digested with BglII, and the DNA was resolved on an agarose gel, blotted onto membranes, and probed with specific probes. This figure shows five time points of digestion by micrococcal nuclease for each strain. The region between HMR-E and the start of the MATa1 gene is shown on the right with a densitometric scan of the digestion profile.
Mapping histone acetylation at the barrier.
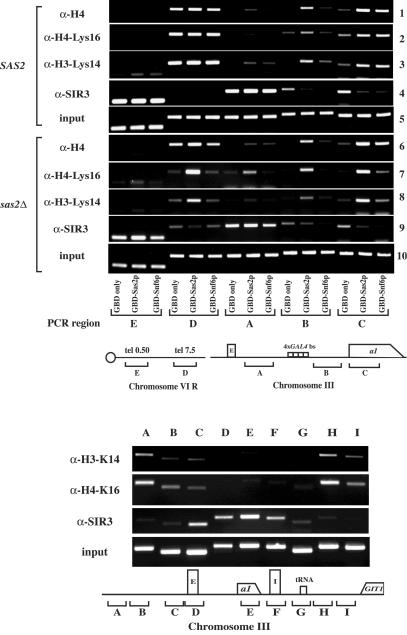
The identification of subunits of several acetyltransferase complexes (we identified subunits of TFIID, SAS-I, SAGA, NuA3, NuA4, and Spt10p) begged the question of whether acetylation of histones accompanied barrier activity mediated by these proteins. We therefore performed chromatin immunoprecipitation analysis on three strains for which barrier function was mediated by GBD alone, GBD-Snf6p, or GBD-Sas2p. We obtained logarithmically growing cells from each of these cells and cross-linked the proteins to the DNA by using formaldehyde; after shearing, the cross-linked protein-DNA complexes were immunoprecipitated with various antibodies (Fig. 6 [top]). We used antibodies that recognized Sir3p, acetylated histone H4, acetylated H4 K16, and acetylated H3 K14. We chose to analyze these two specific modifications since Sas2p has been shown to modify these residues in vitro (31, 64, 67). Following the immunoprecipitation, we analyzed the coimmunoprecipitated DNA by PCR with probes specific to various regions of the yeast genome, including sites flanking the HMR barrier. The results of this analysis are presented in Fig. 6 (top).
FIG. 6.
(Top) Histone H4 acetylation analysis around the Gal4p binding site barrier. Chromatin immunoprecipitation analysis was performed with antibodies against Sir3p, acetyl-histone H4, acetyl-H4K16, or acetyl-H3 K14. Strains ROY2042 (SAS2) and ROY2243 (sas2Δ) containing HMRΔI with four Gal4p binding sites located between HMR-E and the MATa1 gene were transformed with pGBK-RC (GBD only), pRO586 (GBD-Snf6p), or pRO590 (GBD-Sas2p). All six strains were grown in liquid culture prior to cross-linking and immunoprecipitations. PCR probe A analyzed a region between HMR-E and the barrier, probe B was located between the barrier and the MATa1 gene, and probe C was located in the coding region of the MATa1 gene. Probe D analyzed a region 7.5 kb from telomere 6R, and probe E analyzed a region 500 bp from telomere 6R. (Bottom) Strain JRY4013 was grown in liquid culture prior to cross-linking. Immunoprecipitations were performed with antibodies against Sir3p, acetyl-H4K16, and acetyl-H3K14. The probes used were as described previously (73).
We mapped the modifications of the histones at the native yeast telomeres as a control. As previously reported (31, 32, 64), acetylated histones were present 7.5 kb (Fig. 6 [top], probe D) from telomere 6R but were not present 500 bp from this telomere (Fig. 6 [top], probe E), while Sir3p had a reciprocal distribution, being present adjacent to the telomere but not distant from it.
When the same immunoprecipitated samples were analyzed for acetylation around HMR, we observed significant differences in the different strains. In strains in which the GBD was bound to the barrier, we observed almost no acetylation on either side of the barrier (rows 1 to 3, probes A and B). In strains in which barrier function was mediated by GBD-Snf6p, we observed significant levels of acetylation in the transcribed region of the reporter gene (rows 1 to 3, probe C) but very little acetylation with probes immediately flanking the barrier sites (probes A and B). When GBD-Sas2p was present at the barrier, we observed significant levels of acetylation in the coding region of the reporter gene (probe C) and in the region between the barrier and the reporter gene (probe B); we also observed reduced amounts of acetylation between the silencer and the barrier (probe A).
We also mapped the distribution of Sir3p by using antibodies specific to this protein. At HMR, Sir3p was present in the region between the silencer and the barrier (row 4, probe A) in all three strains. However, Sir3p was absent beyond the barrier when GBD-Sas2p and GBD-Snf6p were present at the barrier (row 4, probes B and C) but was present beyond the barrier in strains where only GBD was bound to the barrier. The results with Sir3p directly demonstrated that the protein chimeras (GBD-Sas2p and GBD-Snf6p) functioned by blocking the unidirectional spreading of Sir proteins and not by disrupting silencing over the entire HMR locus.
Our functional analysis had indicated that GBD-Snf6p could block silencing even in the absence of Sas2p (Fig. 3). We therefore investigated the distribution of acetylated histones as well as Sir3p in a strain lacking Sas2p. At native telomeres, we observed that while histone H4 was still acetylated, acetylation of H4 K16 was abolished and acetylation of H3 K14 was reduced in the region 7.5 kb from the telomere. The presence of acetylated H4 and histone H3 K14 (albeit at a reduced level) in a sas2Δ strain was presumably due to Esa1p and Gcn5p, respectively. At the HMR locus, we similarly observed a loss of acetylated H4 K16 and a reduction of acetylated H3 K14 in strains in which GBD-Snf6p was bound to the barrier (probe C). As expected, strains expressing GBD-Sas2p did not show this change in acetylation because this chimera complemented the sas2 deletion. While Sas2p has previously been shown to acetylate H4 K16 in vitro and in vivo, Sas2p has been shown to acetylate H3 K14 only in vitro. Our results demonstrated that Sas2p was also required (either directly or indirectly) to modify H3 K14 in vivo.
Mapping the distribution of Sir3p in a sas2Δ strain indicated that GBD-Snf6p was indeed still able to block the spread of this protein beyond the barrier, indicating that it functioned via a mechanism distinct from GBD-Sas2p.
Mapping histone modifications at HMR.
The studies described above were all performed at the synthetic locus, i.e., where a barrier element comprising four Gal4p binding sites was inserted between the HMR-E silencer and the reporter gene. Consistent with previously published data, our chromatin immunoprecipitation data indicated that the distribution of Sir3p and acetylated histones was mutually exclusive. We decided to analyze the distribution of these proteins at the native HMR locus, where a tRNA gene has previously been shown to function as a barrier (13) in wild-type W-303 strains (Fig. 6 [bottom]). This analysis demonstrated that Sir3p was localized to the previously mapped silenced domain (probes C, D, E, and F) and extended up to the native barriers (probes C and G) but not beyond (probes A, B, H, and I). Histone H3 acetylated at K14 and histone H4 acetylated at K16 were present in regions flanking the silenced domain that was not occupied by Sir3p. These mapping analyses of the native HMR locus mirrored the results obtained at the synthetic locus with GBD-Sas2p.
DISCUSSION
Silencers and telomeres mediate silencing by recruiting the Sir2p-, Sir3p-, and Sir4p-containing complexes. The silencing that emanates from silencers at HML and HMR is not restricted to the region between the silencers but spreads beyond the silencers (3, 13, 37, 40, 47, 73). The extent of the silenced chromatin domains has been mapped, and elements that block the unidirectional spread of silencing have been identified.
While work involving native yeast barriers suggests that promoters of certain genes act as barriers, a recent genetic screen for barrier function has identified proteins involved in nuclear transport (Cse1p, Los1p, Mex67p, Sxm1p, and Gsp2p) as having the ability to insulate a reporter gene from silencing (25).
Analysis of proteins with barrier function.
To better understand the underlying mechanisms by which domains maintained distinct transcription patterns, we performed a systematic genomewide screen for proteins that might block the spread of silencing. This analysis involved recruiting GBD chimeras to a synthetic barrier element (four Gal4p binding sites) located between a silencer and a reporter gene and assaying the ability of various chimeric proteins to block the unidirectional spread of silencing. We believe that this construct mimics the native state in yeast wherein yeast barriers are located outside of the HMR and HML silencers and function by blocking the unidirectional spread of silencing propagating from the silencer (37). This systematic screen of more than 5,500 yeast ORFs identified numerous proteins with efficient silencing blocking activities, a significant majority of which had previously been shown to be involved in chromatin dynamics. Some of the proteins isolated in this screen (members of SAGA and SAS-I) had previously been shown to be required for the function of native heterochromatic barriers (15). These results suggest that the proteins identified by this screen-mediated barrier function by using mechanisms similar to those observed in vivo, thus validating our screening method.
While we did identify subunits of large protein complexes that might function by merely generating a large impediment to the spread of the Sir proteins, we believe that this is unlikely since only some large complexes could block silencing, while others were unable to efficiently do so.
Since our screen did not identify all of the known chromatin-modifying enzymes, the question of specificity is raised. While we isolated subunits of only some complexes, this question's significance or lack thereof is unclear. The inability of a protein to block silencing may be due to trivial reasons such as loss of activity during the formation of the GBD chimera, inability of the fusion to form a functional complex, or the instability of the fusion protein itself. Additional complementation experiments with various chimeric proteins will be necessary in order to resolve this issue.
Some of the barrier proteins identified by us are present in multiple complexes. For example, we isolated GBD-Taf90p, and Taf90p has been shown to be present in both the SAGA and TFIID complexes (23). At present, we are unable to unambiguously state whether these proteins recruit either or both complexes to the barrier and whether both are necessary for barrier function.
While our studies demonstrate the potential of proteins to block the spread of silencing, it is not clear whether all of these proteins actually function at any or all native barriers. Some of the proteins we have identified have indeed been shown to restrict the spread of silencing at native loci. Genetic data demonstrate that Sas2p and Gcn5p influence barrier function at the native HMR locus (15), and molecular mapping data indicate that these enzymes and other histone-modifying enzymes (Dot1p) play a role in restricting the spread of the silenced domains in vivo (31, 32, 51, 64, 68).
The entire Sas-I enzymatic complex was necessary for barrier function.
A large number of the barrier protein chimeras that we isolated in our screen exist in multiprotein complexes in vivo. We were therefore interested in determining whether the GBD chimeras required the entire protein complex for their barrier function or whether they acted on their own. We addressed this question by performing a mutational analysis on one complex—the SAS-I complex. Sas2p is a histone acetyltransferase (17, 59) that interacts with Sas4p and Sas5p (46, 55, 71, 72). Two additional proteins that coimmunoprecipitate with the Sas proteins are the regulatory proteins Asf1p and Cac1p. We found that the barrier activity of GBD-Sas2p and GBD-Sas5p was dependent on the integrity of the SAS-I complex since loss of SAS2, SAS4, or SAS5 from cells severely compromised the barrier function of these chimeras. However, neither Asf1p nor Cac1p was required for the barrier activity of GBD-Sas2p or GBD-Sas5p. Interestingly, Sas2p requires Sas4p and Sas5p for its acetyltransferase activity in vitro, whereas Asf1p and Cac1p regulate the activity of the complex (67). These data suggest that at least for the SAS-I complex, the entire enzymatic complex was being recruited to the barrier element via interactions with GBD-Sas2p or GBD-Sas5p.
We also demonstrated that subunits of the SAS-I complex were not required for the barrier activity of other chimeras such as GBD-Snf6p that was presumably functioning as part of the Swi/Snf complex. This observation was further supported by our data regarding histone modification mapping, wherein we found that in strains lacking Sas2p, GBD-Snf6p still blocked the spread of Sir3p. Taken together, our results raise the possibility that there are overlapping but distinct mechanisms that restrict the spread of silencing.
Nuclear transport proteins as barriers.
We isolated no proteins involved in nuclear transport as strong blockers of silencing at HMR. Upon close examination of their barrier activity, we found that these proteins were unable to block the unidirectional spread of silencing emanating from HMR-E or the telomeres and that the proteins weakly insulated genes from silenced chromatin at HML and HMR.
The differences in the abilities of these proteins to block silencing at HML and HMR are interesting because the repressed state at these loci utilizes the same factors (10), although the chromatin topology at HMR and HML has been shown to be distinct (5, 8). These differences may also be manifested in the various abilities of GBD-Clb1p and GBD-Nup2 to act as barriers at HMR and HML, respectively, versus telomeres and are consistent with previous genetic experiments, the results of which suggested differences in these loci (16, 17, 30, 48, 59).
It has been proposed that the barrier function of the nuclear transport proteins may result from their ability to form loops and tether chromatin to nuclear substructures, resulting in the formation of topologically distinct active and inactive domains (25). While loop formation by insulators has also been observed in other eukaryotes (7) and may be important in terms of barrier function in yeast, we are unable to support or refute this hypothesis with our present data. It should, however, be noted that nuclear pore proteins can regulate a reporter gene in a one-hybrid assay (29). A clear understanding of barrier function by these proteins at HML awaits further experiments coupled with mutational studies defining the role of these proteins at native yeast barrier elements.
Chromatin structure changes at barriers.
Our studies involving changes in chromatin structure of the HMR domain suggest that GBD-Snf6p remodeled nucleosomes immediately around the barrier. The changes in digestion patterns induced by GBD-Snf6p, while subtle, were not observed when only GBD or GBD-Sas2p was present at the barrier. The observed difference in chromatin structure between GBD-Snf6p and GBD-Sas2p suggests that one mechanism by which the spread of silencing may be disrupted is by altering the conformation of nucleosomes around the barrier or altering the even spacing of nucleosomes (58, 65, 69). Changes in nucleosomal configurations may weaken the binding of the Sir proteins to nucleosomes, thus blocking the spread of silencing. Barrier studies with Rap1p (6) have also suggested a similar model. Obviously, alternative possibilities such as recruitment of specific histone-modifying activities by the Swi/Snf complex to aid in barrier function also exist, and these possibilities may or may not be mutually exclusive.
The chromatin immunoprecipitation experiments with antibodies against acetylated histones are revealing. Acetylation in the coding region of the reporter gene occurred in all instances in which a barrier was functional (GBD-Sas2p, GBD-Snf6p, GBD-Clb1p, and GBD-Dot1p [data not shown]), but in regions immediately flanking the barrier, acetylation of H4 K16 and H3 K14 was observed only when GBD-Sas2p acetyltransferase was recruited. We did not observe significant acetylation of these residues when GBD-Snf6p was functioning at the barrier to block the spread of Sir proteins. These results, coupled with those of the quantitative mating analysis (Fig. 2 [bottom]) and the deletion analysis of various SAS-I subunits (Fig. 3), suggest that the SAS-I complex is able to block silencing by acetylating the residues of histones H3 K14 and H4 K16.
We do not, however, believe that acetylation of these lysines is necessary for all barriers to function. This conclusion is based on the observations that histone H4 K16 is not significantly acetylated when GBD-Snf6p is present at the barrier, and while Sas2p was required for the acetylation of histone H4 on K16, loss of Sas2p did not affect GBD-Snf6p-mediated barrier function. At the least, these observations suggest that these two protein complexes (GBD-Snf6p and GBD-Sas2p) function by two distinct molecular mechanisms, one of which (GBD-Sas2p) utilized acetylation of these residues to block the spread of silencing.
Competition delimits chromatin domains.
The interface between active and silenced chromatin can be visualized as a junction of opposing activities (reviewed in reference 14), with competition between activities that aid in the spread of silencing and activities that prevent the spread of silenced chromatin.
Specific DNA elements help block the spread of silencing by recruiting and stably binding transcription complexes. These transcription complexes block the spread of silencing (6, 15, 19, 20) either by creating gaps in the regular nucleosomal arrays or by recruiting histone-modifying activities that disrupt silencing.
At the native HMR barrier, the presence of a stably bound RNA polymerase complex is necessary to block the spread of silencing (13, 15). Robust barrier activity also requires histone-modifying activities (15), and the demonstration that active domains are enriched in Sas2p (31, 64)- and Dot1p (51)-catalyzed modifications suggests that these enzymes may aid in the blocking of silencing.
Activities such as Sir2p-mediated histone deacetylation (24) and chromatin remodeling mediated by Esc8p (10) may aid in the spread of silenced chromatin, while activities such as acetylation (31, 33, 36, 38, 49, 64, 66), along with methylation (18, 51-53, 56, 68) and other modifications, may aid in the formation of active chromatin and act as a block to silenced chromatin.
This model is also consistent with data demonstrating that increasing the levels of repressor proteins increased the spread of silenced domains (42, 43, 60, 63), while increasing the levels of activator proteins abolished silencing (2, 31, 35, 51, 64).
Within this competition model, different chromatin factors presumably block the spread of silencing to different extents by different molecular mechanisms. It is likely that chromatin-remodeling complexes such as Swi/Snf block the propagation of silencing by affecting either the position or conformation of nucleosomes, while changes in histone modification—such as ubiquitination; acetylation; and methylation by Rad6p, Dot1p, Sas2p, or Gcn5p—block the spread of silencing by creating a nucleosomal substrate with a reduced affinity for Sir proteins. At native barriers, it is likely that these and other factors cooperate to efficiently block the spread of silencing.
Our results set the stage for a thorough analysis to discern the role and extent of each of the proteins we have isolated in blocking the spread of silenced chromatin.
Supplementary Material
Acknowledgments
We thank U. Laemmli, D. Shore, and J. Workman for providing yeast strains. We also thank Namrita Dhillon for her editing help, advice, comments, criticisms, and encouragement; finally, we thank Genevieve Fourel and Brad Cairns for comments on the manuscript.
This work was supported by an intramural NIH grant to R.T.K. (grant number ZO1 HD01904). M.O. was supported by a JSPS research fellowship for Japanese biomedical and behavioral researchers.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Abraham, J., K. A. Nasmyth, J. N. Strathern, A. J. Klar, and J. B. Hicks. 1984. Regulation of mating-type information in yeast. Negative control requiring sequences both 5′ and 3′ to the regulated region. J. Mol. Biol. 176:307-331. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio, O. M., and D. E. Gottschling. 1994. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 8:1133-1146. [DOI] [PubMed] [Google Scholar]
- 3.Bi, X. 2002. Domains of gene silencing near the left end of chromosome III in Saccharomyces cerevisiae. Genetics 160:1401-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi, X., M. Braunstein, G. J. Shei, and J. R. Broach. 1999. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc. Natl. Acad. Sci. USA 96:11934-11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, X., and J. R. Broach. 1997. DNA in transcriptionally silent chromatin assumes a distinct topology that is sensitive to cell cycle progression. Mol. Cell. Biol. 17:7077-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi, X., and J. R. Broach. 1999. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 13:1089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanton, J., M. Gaszner, and P. Schedl. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17:664-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, T. H., and M. R. Gartenberg. 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14:452-463. [PMC free article] [PubMed] [Google Scholar]
- 9.Chien, C. T., S. Buck, R. Sternglanz, and D. Shore. 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75:531-541. [DOI] [PubMed] [Google Scholar]
- 10.Cuperus, G., and D. Shore. 2002. Restoration of silencing in Saccharomyces cerevisiae by tethering of a novel Sir2-interacting protein, Esc8. Genetics 162:633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhillon, N., and R. T. Kamakaka. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6:769-780. [DOI] [PubMed] [Google Scholar]
- 12.Dillin, A., and J. Rine. 1997. Separable functions of ORC5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics 147:1053-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donze, D., C. R. Adams, J. Rine, and R. T. Kamakaka. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donze, D., and R. T. Kamakaka. 2002. Braking the silence: how heterochromatic gene repression is stopped in its tracks. Bioessays 24:344-349. [DOI] [PubMed] [Google Scholar]
- 15.Donze, D., and R. T. Kamakaka. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20:520-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrenhofer-Murray, A. E., R. T. Kamakaka, and J. Rine. 1999. A role for the replication proteins PCNA, RF-C, polymerase epsilon and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics 153:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrenhofer-Murray, A. E., D. H. Rivier, and J. Rine. 1997. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics 145:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst, K. Struhl, and Y. Zhang. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12:1052-1058. [DOI] [PubMed] [Google Scholar]
- 19.Fourel, G., C. Boscheron, E. Revardel, E. Lebrun, Y. F. Hu, K. C. Simmen, K. Muller, R. Li, N. Mermod, and E. Gilson. 2001. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2:124-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fourel, G., T. Miyake, P. A. Defossez, R. Li, and E. Gilson. 2002. General regulatory factors (GRFs) as genome partitioners. J. Biol. Chem. 277:41736-41743. [DOI] [PubMed] [Google Scholar]
- 21.Fourel, G., E. Revardel, C. E. Koering, and E. Gilson. 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18:2522-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghidelli, S., D. Donze, N. Dhillon, and R. T. Kamakaka. 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20:4522-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 24.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 25.Ishii, K., G. Arib, C. Lin, G. Van Houwe, and U. K. Laemmli. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109:551-562. [DOI] [PubMed] [Google Scholar]
- 26.Ito, T., K. Tashiro, S. Muta, R. Ozawa, T. Chiba, M. Nishizawa, K. Yamamoto, S. Kuhara, and Y. Sakaki. 2000. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 97:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivy, J. M., A. J. Klar, and J. B. Hicks. 1986. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 6:688-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamakaka, R. T., and J. Rine. 1998. Sir- and silencer-independent disruption of silencing in Saccharomyces by Sas10p. Genetics 149:903-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasper, L. H., P. K. Brindle, C. A. Schnabel, C. E. Pritchard, M. L. Cleary, and J. M. van Deursen. 1999. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 19:764-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kayne, P. S., U. J. Kim, M. Han, J. R. Mullen, F. Yoshizaki, and M. Grunstein. 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55:27-39. [DOI] [PubMed] [Google Scholar]
- 31.Kimura, A., T. Umehara, and M. Horikoshi. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32:370-377. [DOI] [PubMed] [Google Scholar]
- 32.Kristjuhan, A., B. O. Wittschieben, J. Walker, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2003. Spreading of Sir3 protein in cells with severe histone H3 hypoacetylation. Proc. Natl. Acad. Sci. USA 100:7551-7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladurner, A. G., C. Inouye, R. Jain, and R. Tjian. 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11:365-376. [DOI] [PubMed] [Google Scholar]
- 34.Le, S., C. Davis, J. B. Konopka, and R. Sternglanz. 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13:1029-1042. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S., and D. S. Gross. 1993. Conditional silencing: the HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol. Cell. Biol. 13:727-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 37.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 38.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loo, S., P. Laurenson, M. Foss, A. Dillin, and J. Rine. 1995. Roles of ABF1, NPL3, and YCL54 in silencing in Saccharomyces cerevisiae. Genetics 141:889-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loo, S., and J. Rine. 1994. Silencers and domains of generalized repression. Science 264:1768-1771. [DOI] [PubMed] [Google Scholar]
- 41.Loo, S., and J. Rine. 1995. Silencing and heritable domains of gene expression. Annu. Rev. Cell Dev. Biol. 11:519-548. [DOI] [PubMed] [Google Scholar]
- 42.Lustig, A. J., C. Liu, C. Zhang, and J. P. Hanish. 1996. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2483-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcand, S., S. W. Buck, P. Moretti, E. Gilson, and D. Shore. 1996. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev. 10:1297-1309. [DOI] [PubMed] [Google Scholar]
- 44.Marshall, M., D. Mahoney, A. Rose, J. B. Hicks, and J. R. Broach. 1987. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:4441-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNally, F. J., and J. Rine. 1991. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5648-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meijsing, S. H., and A. E. Ehrenhofer-Murray. 2001. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 15:3169-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreira, J. M., and S. Holmberg. 1998. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 17:6028-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullen, J. R., P. S. Kayne, R. P. Moerschell, S. Tsunasawa, M. Gribskov, M. Colavito-Shepanski, M. Grunstein, F. Sherman, and R. Sternglanz. 1989. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mutskov, V. J., C. M. Farrell, P. A. Wade, A. P. Wolffe, and G. Felsenfeld. 2002. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 16:1540-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nasmyth, K. A. 1982. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell 30:567-578. [DOI] [PubMed] [Google Scholar]
- 51.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 54.Oki, M., and R. T. Kamakaka. 2002. Blockers and barriers to transcription: competing activities. Curr. Opin. Cell Biol. 14:299-304. [DOI] [PubMed] [Google Scholar]
- 55.Osada, S., A. Sutton, N. Muster, C. E. Brown, J. R. Yates III, R. Sternglanz, and J. L. Workman. 2001. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev. 15:3155-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park, J. H., M. S. Cosgrove, E. Youngman, C. Wolberger, and J. D. Boeke. 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 32:273-279. [DOI] [PubMed] [Google Scholar]
- 57.Pryde, F. E., and E. J. Louis. 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18:2538-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ravindra, A., K. Weiss, and R. T. Simpson. 1999. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol. Cell. Biol. 19:7944-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reifsnyder, C., J. Lowell, A. Clarke, and L. Pillus. 1996. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat. Genet. 14:42-49. (Erratum, 16:109, 1997.) [DOI] [PubMed] [Google Scholar]
- 60.Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani, and D. E. Gottschling. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7:1133-1145. [DOI] [PubMed] [Google Scholar]
- 61.Rivier, D. H., J. L. Ekena, and J. Rine. 1999. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 151:521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 64.Suka, N., K. Luo, and M. Grunstein. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32:378-383. [DOI] [PubMed] [Google Scholar]
- 65.Sun, F. L., M. H. Cuaycong, and S. C. Elgin. 2001. Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol. Cell. Biol. 21:2867-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun, Z. W., and M. Hampsey. 1999. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics 152:921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sutton, A., W. J. Shia, D. Band, P. D. Kaufman, S. Osada, J. L. Workman, and R. Sternglanz. 2003. Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J. Biol. Chem. 278:16887-16892. [DOI] [PubMed] [Google Scholar]
- 68.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 69.Weiss, K., and R. T. Simpson. 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol. Cell. Biol. 18:5392-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, C. 1980. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286:854-860. [DOI] [PubMed] [Google Scholar]
- 71.Xu, E. Y., S. Kim, K. Replogle, J. Rine, and D. H. Rivier. 1999. Identification of SAS4 and SAS5, two genes that regulate silencing in Saccharomyces cerevisiae. Genetics 153:13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu, E. Y., S. Kim, and D. H. Rivier. 1999. SAS4 and SAS5 are locus-specific regulators of silencing in Saccharomyces cerevisiae. Genetics 153:25-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, Z., M. K. Hayashi, O. Merkel, B. Stillman, and R. M. Xu. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21:4600-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.