Abstract
The function of p53 is modulated by several transcriptional coactivators that regulate its tumor suppressor activity. Here we report that human transcriptional coactivator PC4 enhances the DNA binding of p53 to its cognate site in vitro and directly interacts with p53 in vivo. In vitro interaction studies demonstrated that the C-terminal 30 amino acids (364 to 393) of p53 strongly interact with PC4. Surprisingly, PC4 also stimulates the sequence-specific DNA binding of p53 with the C-terminal 30 amino acids deleted (p53Δ30), suggesting that PC4 mediates enhancement of p53 DNA binding by a unique mechanism. We also demonstrated that PC4 can stimulate p53- and p53Δ30-mediated transactivation from a p53-responsive promoter. Furthermore, PC4 enhances p53- and p53Δ30-dependent apoptosis by inducing bax (a p53-targeted proapoptotic gene) gene expression. These results establish the first physiological role of PC4 as a transcriptional coactivator.
p53 exerts its tumor suppressor activity by regulating the transcription of several factors, which in turn modulate cell cycle and apoptotic pathways during genotoxic insult. Several transcription factors and coactivators are involved in the maintenance of stability and the regulation of p53 function. p53 has been shown to interact with a number of proteins implicated in transcriptional control, such as TAF9, TAF6, and hTAF9 (20). Interestingly, the coactivator protein hTAF9 binds to p53 at the amino-terminal region in competition with MDM2 (required for degradation of p53 through the ubiqutin-proteosome pathway) and stabilizes p53 during conditions of stress (4). It is also reported that the interaction of p53 with either recombinant TBP or partially purified TFIID stimulates binding to its cognate site and results in the enhancement of transcription in vivo (7). In another study it has been shown that squelching of in vitro transcription by excess p53 is alleviated by the addition of TFIIB and TFIID (25). This suggests that interaction with these factors plays an important role in p53-regulated transcription.
Apart from interacting with the basal transcription machinery, p53 interacts with several other transcriptional coactivators with functional consequences. These include p300/CBP (2, 10, 24, 32), Zac1 (zinc finger protein which regulates apoptosis and cell cycle) (15), JMY (junction-mediating and regulatory protein) (32), and HMG-B1 (high-mobility-group protein B-1) (16). Zac1 specifically enhances the activity of a p53-responsive promoter in cells expressing wild-type p53. Zac1 was also shown to interact with human papillomavirus protein E6, responsible for p53 degradation in HeLa cells, resulting in stabilization of p53 (15). CBP/p300, which is implicated in cell proliferation and differentiation, has been shown to activate transcription synergistically with p53 (10). JMY forms a complex with p300 during stress conditions and acts as a coactivator for p53 to activate transcription of bax, a p53 target gene that activates apoptosis (32). The highly abundant nonhistone chromatin protein HMGB-1, a transcriptional coactivator, is a unique activator of p53-mediated DNA binding and transcriptional activation (16). Interestingly, the human transcriptional coactivator PC4 (positive coactivator 4) displays significant functional homology to HMGB-1. For example, similar to HMGB-1, PC4 binds to DNA in a sequence-independent manner and is involved in diverse nuclear processes (e.g., polymerase III transcription, elongation, and AP2 self-repression) besides RNA polymerase II-mediated transcription (5, 9, 12, 13, 17, 18, 21, 33, 34).
PC4 is a highly abundant nuclear protein which facilitates activator-dependent transcription by RNA polymerase II through interactions with the basal transcription machinery and transcriptional activators like GAL4-BRCA1, GAL4-VP16, GAL4-CTF1, GAL4-Sp1, and AP2 (9, 14, 18). Presumably by stimulating activator-dependent transcription, PC4 plays a critical role in the maintenance of normal cellular growth. It has been shown that PC4 can inhibit AP2 self-repression in a ras-transformed cell line and thus may act as a putative tumor suppressor (18). Correlating this fact with the observation that PC4 also activates tumor suppressor BRCA1-dependent transcription, we hypothesized that PC4 may be involved in p53-dependent transcriptional regulation to maintain cellular homeostasis.
In this study, we investigated the possible role of PC4 in the modulation of p53 function. We observed that PC4 indeed stimulates the sequence-specific DNA binding ability of p53 in vitro. Immunoprecipitation of p53 from drug (adriamycin)-treated U2OS cells showed that PC4 interacts with p53 in vivo. By employing a glutathione S-transferase (GST) pulldown assay, we determined that the C-terminal 30 amino acids (364 to 393) of p53 are involved in the interaction with PC4. Furthermore, electrophoretic mobility shift assay (EMSA) analysis revealed that PC4 could also stimulate the DNA binding of a consecutively active form of p53, which carries a deletion of the C-terminal 30 amino acids (364 to 393), thus indicating that the PC4-mediated enhancement of p53 DNA binding is unique. Our data also show that PC4 can stimulate both p53- and p53Δ30-mediated reporter gene expression from p53-responsive promoters. Significantly, PC4 activates p53-dependent apoptosis through the induction of bax, a p53-targeted proapoptotic gene. Taken together, the data establish the physiological relevance of stimulation of p53-dependent DNA binding and subsequent transcriptional activation by PC4, a general transcriptional coactivator.
MATERIALS AND METHODS
Plasmid construction.
The mammalian expression clone of full-length PC4 was constructed by PCR-based subcloning, using appropriate primers, into NheI and HindIII sites of the pcDNA 3.1 (+) vector (Invitrogen). The FLAG-tagged bacterial expression clone of p53Δ30 was constructed by PCR-based subcloning, using appropriate primers (containing the nucleotide sequence for the FLAG peptide at the 5′ end), from a full-length p53 clone (11). The same strategy was followed to clone full-length p53 and p53Δ30 into the EcoRI and BamHI sites of the pIRES puro (Clonetech) mammalian expression vector.
Purification of recombinant proteins.
Recombinant PC4 was expressed and purified from Escherichia coli as described elsewhere (9). FLAG-tagged p53 was purified from E. coli as described elsewhere (11) with minor modifications. FLAG-tagged p53Δ30 and Gal4-CTF1 were purified from E. coli by using the same protocol as that described elsewhere (23). The authenticity of both the full-length p53 and p53 with the C terminus deleted (p53Δ30) was confirmed by Western blotting using N-terminus-specific (DO1) and C-terminus-specific (PAb421) p53 monoclonal antibodies.
EMSA.
A 40-mer oligonucleotide containing p53 binding sites (5′-AATTCTCGAGCAGAACATGTCTAAGCATGCTGGGCTCGAG-3′) derived from the GADD45 promoter was labeled by using T4 polynucleotide kinase (New England Biolabs). The radiolabeled strand was annealed with the complementary strand, and an EMSA was performed as described previously (16). For the cold oligonucleotide challenge experiment, the double-stranded unlabeled 40-mer oligonucleotide containing wild-type or mutant p53 binding sites (5′-AATTCTCGAGCAGAAAATTTCTAAGAATTCTGGGCTCGAG-3′) was used in excess (100- and 200-fold) over radiolabeled oligonucleotide containing wild-type p53 binding sites.
For supershift assays, the indicated amounts of antibodies were added into the reaction mix, and an EMSA was performed. In order to further elucidate the role of PC4 in p53 DNA binding, the indicated amounts of p53 and its C-terminus-specific antibody, PAb421, were incubated for 30 min prior to EMSA. Reaction mixtures (40 μl) contained 8 μl of 5× EMSA buffer (100 mM HEPES at pH 7.9, 125 mM KCl, 0.5 mM EDTA, 50% glycerol, 10 mM MgCl2), 2 μl of 60-μg/ml double-stranded poly(dI-dC), 4 μl of bovine serum albumin (BSA) (1 mg/ml), 32 P-labeled probe DNA (3 ng), proteins as indicated, and water to make up the volume of 40 μl. The reaction mixtures were incubated at 30°C for 30 min, and the total volume of each reaction mixture was then loaded on a native 4% polyacrylamide gel containing 0.5× Tris-borate-EDTA buffer and 0.05% NP-40 and electrophoresed at 4°C for 2 h (250 to 260 V). Similarly, for the DNA binding experiment with Gal4-CTF1, the oligonucleotide containing the Gal4 binding site (5′-GTCTCTAGGTCGGAGTACTGTCCTCCGCGGAGACTCTAGA- 3′) was radiolabeled and annealed with a complementary oligonucleotide. The EMSA was performed as described previously (6). Briefly, the reaction mixture (40 μl), containing 20 mM HEPES (pH 7.5), 50 mM KCl, 5 mM MgCl2, 10 μM ZnCl2, 6% glycerol, 200 μg of BSA/ml, 50 μg of poly(dI-dC)/ml, and 3 ng of 32P-labeled DNA fragment, was incubated with proteins as indicated at 30°C for 30 min, and the total reaction mixture was then loaded on a native 6% polyacrylamide gel containing 0.5× Tris-borate-EDTA buffer. The gels were then dried and exposed for autoradiography. DNA-protein complexes were quantified with a PhosphorImage Analyzer (Fuji) using Image Guage software. Quantitation of DNA binding data represents an average of three independent experiments.
Immunoprecipitation and immunoblotting.
Immunoprecipitation was performed as described previously (32). Briefly, U2OS cells (p53+/+) were treated with 2 μg of adriamycin/ml for 24 h. After adriamycin treatment, the cells were lysed in a buffer containing 10 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 150 mM NaCl, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 0.2 mM sodium orthovanadate, 0.5 μg of leupeptin/ml, 1.0 μg of trypsin inhibitor/ml, 0.5 μg of aprotinin/ml, and 40 μg of pepstatin A/ml and incubated on ice for 1 h. The cell debris was removed by centrifugation. The protein content in the lysates was estimated by Bradford assay using a Bio-Rad protein assay reagent (catalog no. 500-0006). An equal amount of protein from each sample was subjected to immunoprecipitation by protein A-agarose beads (GIBCO BRL) conjugated with either anti-p53 monoclonal antibody DO1 (Oncogene) or goat anti-PC4 polyclonal antibody N17 (Santa Cruz). Mouse preimmune serum and goat preimmune serum were used as controls. The immunoprecipitated proteins were analyzed by Western blotting with the above-mentioned antibodies as indicated in the figure legends.
GST pulldown assay.
The GST pulldown assay was performed as described previously (22). GST, GST-p53 (full-length), GST-p53(1-73) (p53 with amino acids 1 to 73), GST-p53(120-290), GST-p53(284-330), GST-p53(328-368), and GST-p53(364-393) were expressed in E. coli and conjugated to glutathione-agarose beads as described elsewhere (11). The amounts of GST and GST fusion proteins on the beads were normalized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Coomassie brilliant blue R-250 with BSA as a standard. Typically, 1 μg of GST and GST-fusion proteins was incubated with the E. coli lysate containing 200 ng of recombinant PC4 in a final volume of 200 μl of BC buffer (20 mM Tris HCl [pH 7.4], 20% glycerol, 0.2 mM EDTA, 0.1% NP-40) containing 150 mM KCl at 4°C for 2 h. The beads were washed five times (1 ml each) with the incubation buffer and subjected to SDS-PAGE analysis followed by immunoblotting with anti-PC4 polyclonal antibody N17 (Santa Cruz).
To analyze the nature of the PC4-induced p53-DNA complex, GST pulldown assays were also performed with purified GST-p53 (500 ng) and PC4 (2 μg) either in the presence or absence of oligonucleotides containing p53 binding sites (30 ng) in a total reaction volume of 400 μl of BC buffer containing 60 mM KCl at 4°C for 2 h. The presence of PC4 was detected by immunoblotting using affinity-purified anti-PC4 polyclonal antibody.
Transfection assay.
H1299 (p53 null) cells were maintained at 37°C in Dulbecco's modified Eagle medium with 10% fetal bovine serum. The mammalian expression constructs of p53, p53Δ30, Gal4-VP16, and PC4 used in this study were placed under a cytomegalovirus (CMV) promoter. The p53-responsive promoter PG13Luc contains 13 consensus p53-binding sites in tandem followed by a polyomavirus promoter, which drives the luciferase gene (19). MDM2Luc contains an MDM2 promoter-driven luciferase gene (8). These two constructs were used to monitor the effect of PC4 on p53-mediated gene expression in vivo. The CMV-driven β-galactosidase (pCMV-βgal) construct was used as an internal control. Prior to transfection, cells were seeded at 0.6 × 106 cells per 60-mm-diameter dishes and transfected with different plasmid constructs, using Lipofectamine 2000 Plus (Invitrogen) according to the manufacturer's protocol. An empty vector was used to keep the total amount of DNA constant in each transfection. Luciferase and β-galactosidase activities were measured 24 h after the transfection with luciferase assay and β-galactosidase assay systems according to the procedure provided by manufacturer (Promega). The transient transfection assay was performed three times independently to monitor average relative luciferase activity.
Apoptosis assay.
H1299 cells were seeded at 0.6 × 106 cells per 60-mm-diameter dish 14 h prior to transfection. The cells were then transfected with the indicated amounts (see figure legends) of mammalian expression constructs of p53, p53Δ30, and PC4 (either alone or in combination), using Lipofectamine 2000 Plus (Invitrogen) according to the manufacturer's protocol. After 24 h of transfection, the cells were fixed with 2.5% paraformaldehyde, stained with Hoechst 33258, and observed under a fluorescence microscope by a blind approach. The quantitation of the number of apoptotic nuclei was performed by counting 300 nuclei each time for three independent experiments.
Endogenous gene expression assay.
H1299 cells were seeded at 1.5 × 106 cells per 100-mm-diameter dish 14 h prior to transfection. Cells were transfected with the indicated amounts of mammalian expression vectors containing full-length p53, p53Δ30, and PC4 either alone or in combination as described above. After 48 h, cells were lysed in a buffer containing 10 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 250 mM NaCl, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 0.2 mM sodium orthovanadate, 0.5 μg of leupeptin/ml, 1.0 μg of trypsin inhibitor/ml, 0.5 μg of aprotinin/ml, and 40 μg of pepstatin A/ml and incubated on ice for 1 h. The cell debris was removed by centrifugation. The protein content of the lysate was estimated by Bradford assay as described above. The cell lysates (containing 600 μg of total cellular proteins) were analyzed by SDS-PAGE (12%) followed by Western blotting using Bax polyclonal antibody Ab4 (Oncogene), p53 monoclonal antibody DO1, and affinity-purified PC4 polyclonal antibody.
RESULTS
Effect of PC4 on sequence-specific DNA binding of p53.
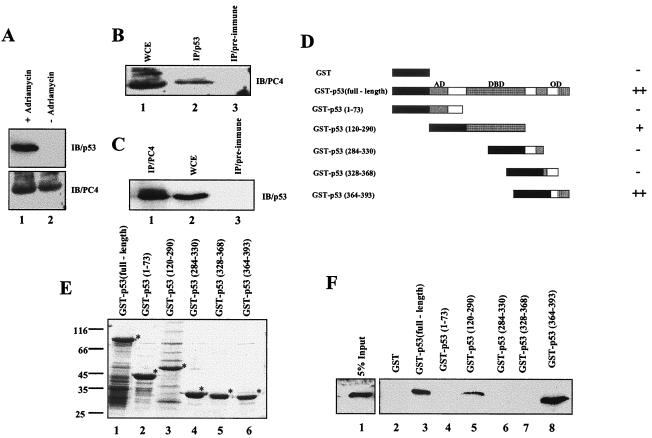
Most of the proteins that activate p53 function stimulate its DNA binding to the cognate site. In our attempt to determinewhether PC4 influences the sequence-specific DNA binding of p53, we purified recombinant PC4 and FLAG-tagged p53 from E. coli (Fig. 1A, lane 1, and 1B, lane 1). The authenticity of these proteins was verified by Western blot analysis using the respective antibodies (Fig. 1A, lane 2, and 1B, lane 2). We tested the effect of PC4 on the DNA binding of p53 by EMSA using a 40-mer double-stranded oligonucleotide containing the p53 binding sites from the GADD45 promoter (16). The EMSA analysis showed that 200 or 300 ng of PC4 in combination with 50 ng of p53 (the molar ratios for tertrameric p53 and dimeric PC4 were 1:7.7 and 1:11.5, respectively) efficiently enhances (about two- and eightfold, respectively) p53 DNA binding to its cognate site as compared to p53 alone (Fig. 1C, lane 2 versus lanes 5 and 6). Apart from the predominant p53 complex, we also observed another complex when higher concentrations of PC4 (200 and 300 ng) were incubated with p53 (Fig. 1C, lanes 5 and 6). This could be attributed to a minor degradation product of p53 whose DNA binding may be activated at the higher concentrations of PC4. However, we did not observe an enhancement of p53 DNA binding when 50 and 100 ng of PC4 were added in combination with 50 ng of p53 (molar ratios for tertrameric p53 and dimeric PC4 were 1:1.9 and 1:3.8, respectively) (Fig. 1C, lane 2 versus lanes 3 and 4). Since PC4 is a non-sequence-specific DNA binding protein, low amounts are engaged in forming the probe-PC4 complex (Fig. 1C and D, arrows; see also Discussion).
FIG. 1.
PC4 activates sequence-specific DNA binding of p53. (A and B) Recombinant proteins were used in different experiments. (A) Lane 1, 300 ng of recombinant PC4 was analyzed by SDS-15% PAGE. Lane 2, authenticity of the PC4 was analyzed by Western blotting using anti-PC4 polyclonal antibody N17. (B) Lane 1, 100 ng of recombinant FLAG-tagged p53 was analyzed by SDS-10% PAGE. Lane 2, authenticity of the p53 was analyzed by Western blotting using anti-p53 monoclonal antibody PAb421. (C and D) Effect of PC4 in p53-mediated DNA binding.. (C) Three nanograms of a 32P-labeled oligonucleotide containing a p53 binding site was incubated with 50 ng of p53 either in the absence of PC4 (lane 2) or in the presence of 50 ng (lane 3), 100 ng (lane 4), 200 ng (lane 5), or 300 ng (lane 6) of PC4. The 32P-labeled oligonucleotide was also incubated with 50 ng (lane 7), 100 ng (lane 8), 200 ng (lane 9), and 300 ng (lane 10) of PC4 alone. Lane 1 contains the 32P-labeled oligonucleotide alone. (D) EMSA was done as described in the text in the presence of p53 (50 ng) either alone (lane 2) or in combination with PC4 (300 ng) (lane 3) and competitor DNA (100× [lanes 4 and 6] and 200× [lanes 5 and 7]) containing wild-type (WT) (lanes 4 and 5) and mutant (mt) (lanes 6 and 7) p53 binding sites. Lane 1 contains 32P-labeled oligonucleotide alone.
Under certain circumstances, p53 may also bind to the DNA without any sequence specificity; hence, it is essential to investigate the specificity of p53 DNA binding to its cognate site. In order to demonstrate that the activated complex is due to sequence-specific binding of p53, we performed an EMSA with an excess of cold competitor oligonucleotide containing wild-type or mutant p53 binding sites (see Materials and Methods). We observed that in the presence of cold oligonucleotide (100-fold excess over radiolabeled oligonucleotide) containing wild-type p53 binding sites, the formation of activated p53-DNA complex was dramatically reduced (Fig. 1D, compare lanes 3 and 4). Further, increasing the wild-type cold oligonucleotide (200-fold excess) almost completely abolished the DNA binding of p53 to its radiolabeled cognate sites (Fig. 1D, compare lanes 3 and 5). On the other hand, even a 200-fold excess of cold oligonucleotide containing mutant binding sites did not affect the DNA binding of p53 to the radiolabeled probe (Fig. 1D, lane 3 versus lanes 6 and 7). These observations suggest that the PC4-mediated enhancement of p53 DNA binding to its cognate sites is sequence specific.
In vivo and in vitro interaction of PC4 and p53.
PC4 interacts with several activators, namely VP16, CTF1, Sp1, and BRCA1, resulting in stimulation of activator-dependent transcription (9, 14, 18). On the basis of these observations, we hypothesized that PC4 may interact with p53 to facilitate recruitment to its cognate DNA binding site. In order to investigate the possible interaction between the two proteins, p53 expression was induced in U2OS cells by adriamycin treatment (2 μg/ml for 24 h) (Fig. 2A, upper panel, compare lanes 1 and 2). It was observed that the amount of PC4 remained almost the same before and after adriamycin treatment (Fig. 2A, lower panel, lanes 1 and 2). Immunoprecipitation was performed by incubating the drug-treated U2OS cell lysate with protein A-agarose beads conjugated to either anti-p53 mouse monoclonal antibody DO1 (Oncogene) or anti-PC4 goat polyclonal antibody N17 (Santa Cruz). Immunoprecipitation with anti-p53 or anti-PC4 antibodies pulled down PC4 and p53, respectively (Fig. 2B, lane 2, and 2C, lane 1). In contrast, immunoprecipitation with the respective mouse and goat preimmune sera did not pull down PC4 or p53 nonspecifically (Fig. 2B, lane 3, and 2C, lane 3). These data suggest that PC4 interacts with p53 in vivo.
FIG. 2.
PC4 directly interacts with p53 in vitro and in vivo. (A) Induction of p53 expression in the U2OS cell line. The levels of p53 and PC4 present in adriamycin (2 μg/ml)-treated U2OS cells were assessed by Western blotting using anti-p53 (upper panel) and anti-PC4 (lower panel) antibodies. (B and C) In vivo interaction of PC4 with p53. (B) Lane 1, an adriamycin (2 μg/ml)-induced U2OS cell extract was immunoblotted with polyclonal PC4 antibody N17. Lane 2, immunoprecipitation of endogenous PC4 from an induced U2OS cell lysate was performed using anti-p53 monoclonal antibody DO1 followed by immunoblotting with anti-PC4 polyclonal antibody. Lane 3, immunoprecipitation using mouse preimmune serum used as a control. (C) Lane 1, immunoprecipitation of endogenous p53 from an induced U2OS cell extract, using anti-PC4 polyclonal antibody N17 followed by immunoblotting with anti-p53 monoclonal antibody DO1. Lane 2, adriamycin-induced U2OS cell extract immunoblotted with monoclonal p53 antibody DO1. Lane 3, immunoprecipitation reaction with goat preimmune serum used as a control. (D, E, and F) Interaction of PC4 with p53 in an in vitro GST pulldown assay. (D) Schematic representation of GST and GST-p53 fusion proteins. ++, strong interaction of PC4 with respective GST-p53 fusion protein; +, weaker interaction; −, no interaction. (E) SDS-PAGE (10%) and Coomassie blue R250 staining of immobilized GST-p53 fusion proteins. Lane 1, GST-p53 (full length); lane 2, GST-p53(1-73), lane 3, GST-p53(120-290); lane 4, GST-p53(284-330), lane 5, GST-p53(328-368); lane 6, GST-p53(364-393). GST fusion proteins predominantly contain intact proteins (indicated with asterisks) with minimum low-molecular-weight breakdown products. (F) One microgram of GST (lane 2) or GST-p53 fusion proteins (lanes 3 to 8) was incubated with bacterial extract containing 200 ng of PC4 and analyzed by immunoblotting with anti-PC4 N17 antibody. Lane 1, 5% input of bacterial cell lysate. IP, immunoprecipitation; IB, immunoblot; WCE, whole-cell extract; AD, activation domain; DBD, DNA binding domain; OD, oligomerization domain.
As p53 is a protein with distinct functional domains, we investigated the domain of p53 involved in the interaction with PC4. For this purpose different GST-fused constructs of p53 (Fig. 2D) were expressed in bacteria and immobilized on glutathione-agarose beads (Fig. 2E). These proteins were subjected to GST pulldown assays (see Materials and Methods) using a bacterial lysate containing full-length recombinant PC4. It was observed that PC4 indeed interacts with p53, as observed in our previous immunoprecipitation experiments. This further confirmed that the interaction is direct (Fig. 2F, lane 3). We also found that residues 364 to 393 of the C-terminal oligomerization domain of p53 strongly interacts with PC4 (Fig. 2F, lane 8) while residues 120 to 290 of p53, spanning the sequence-specific DNA binding domain, weakly interact with PC4 (Fig. 2F, lane 5). Thus, PC4 directly interacts with p53, consistent with similar observations for a number of other transcriptional activators (as discussed above), for which it acts as a general coactivator.
Involvement of PC4 in the activated complex.
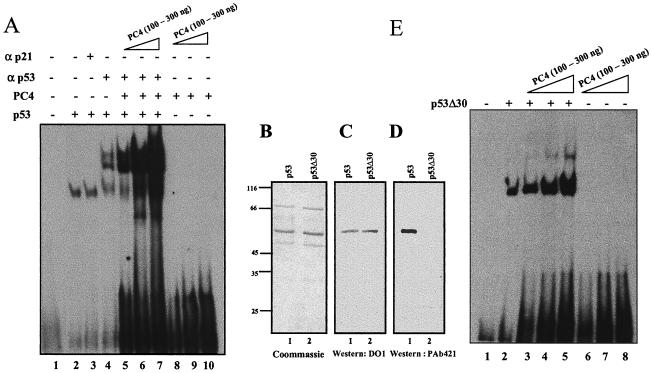
After establishing the fact that the direct interaction between PC4 and p53 stimulates the sequence-specific DNA binding of the latter, we were interested in investigating whether PC4 is a part of the activated complex. In order to address this possibility, the components of the enhanced DNA-protein complex were analyzed by supershift assay using different antibodies against p53 and PC4. The EMSA results showed that monoclonal p53 antibody PAb421 shifted the complex (Fig. 3A, lanes 4 and 7) whereas antibodies specific to PC4 (N-terminus-specific antibody N17, C-terminus-specific antibody D18, and antibody raised against full-length PC4) had a very minimal effect on complex mobility. Among the three different antibodies against PC4, D18 and polyclonal antibodies could shift the complex distinctly, but the relative amount of the shifted complex was found to be less than 10% compared to the total activated complex (Fig. 3A, lanes 9 and 10). These results suggest that PC4 may be a part of the DNA-protein complex, which is either very poorly accessed by the antibodies or the protein gets dissociated during electrophoresis. Yet another possibility could be that PC4 forms a transient ternary complex. In order to clarify this ambiguity, we performed a GST pulldown assay in the presence or absence of oligonucleotide containing p53-binding sites. If PC4 forms a transient ternary complex and falls apart after recruitment of p53 to the cognate site, a GST pulldown assay with GST-p53 in the presence of oligonucleotide should not be able to detect PC4. However, the results showed that GST-p53 efficiently pulls down PC4 even in the presence of the oligonucleotide, which argues in favor of the formation of a p53-DNA-PC4 ternary complex (Fig. 3B, compare lanes 1 and 2). Furthermore, a nonspecific antibody against p21 could not shift the complex (Fig. 3A, lanes 3 and 6), which confirmed the specificity of the supershift brought about by the p53 antibody. Taken together, the data suggest that PC4 stimulates the DNA binding of p53 to its cognate site through the formation of a stable ternary complex with p53 and DNA.
FIG. 3.
PC4 forms a ternary complex with p53 and DNA. (A) EMSA was done in presence of p53 (50 ng) and 32P-labeled oligonucleotide (3 ng) containing a p53 binding site without antibody (lane 2), with p21 antibody (500 ng) (lane 3), with p53 antibody (500 ng) (lane 4), with 250 ng of PC4 in the absence of any antibody (lane 5), or with 250 ng of PC4 with p21 antibody (500 ng) (lane 6), p53 antibody (500 ng) (lane 7), N-terminus-specific PC4 antibody N17 (2.5 μg) (lane 8), C-terminus-specific PC4 antibody D18 (2.5 μg) (lane 9), or affinity-purified anti-PC4 polyclonal antibody raised against full-length PC4 (2.5 μg) (lane 10). Lane 1 contains 32P-labeled oligonucleotide alone. (B) A GST pulldown assay was performed (as described in the legend to Fig. 2) with p53 (500 ng) and PC4 (2.5 μg) either in the absence (lane 1) or presence (lane 2) of cold oligonucleotide containing a wild-type p53 binding site from the GADD45 promoter. PC4 was detected by Western blot analysis using affinity-purified anti-PC4 polyclonal antibody.
Effect of PC4 on sequence-specific DNA binding of p53 with the C-terminal domain deleted.
The C-terminal 30 amino acid residues of p53, which strongly interact with PC4, negatively regulate p53-mediated DNA binding (1, 16). Most of the p53 activators relieve the repression of this domain to activate p53 function (16) and thereby cannot activate p53 lacking the C-terminal 30 amino acids. We investigated the role of this domain in PC4-mediated enhancement of p53 DNA binding. The 30 amino acid residues of p53 were masked by PAb421 (Oncogene), and p53 was then subjected to EMSA using increasing concentrations of PC4. Surprisingly, PC4 could enhance the DNA binding of the antibody-masked p53 significantly (up to about eightfold) (Fig. 4A, lane 4 versus lanes 5, 6, and 7). This could indicate that PC4 stimulates DNA binding of p53 independently of its interaction with the C-terminal domain. In order to confirm this observation, we cloned, expressed, and purified FLAG-tagged p53Δ30 (Fig. 4B, lane 2). The authenticity of p53Δ30 was confirmed by Western blot analysis using an N-terminus-specific antibody, DO1 (Fig. 4C, lanes 1 and 2), and C-terminus-specific antibody, PAb421 (Fig. 4D, lanes 1 and 2). While the full-length p53 interacted with both antibodies (Fig. 4C, lane 1, and 4D, lane 1), p53 with the C terminus deleted was Western blotting positive only with DO1 (Fig. 4D, compare lanes 1 and 2). Our EMSA analysis using 25 ng of p53Δ30 either alone or in combination with indicated amounts of PC4 showed that the sequence-specific DNA binding of recombinant p53Δ30 was significantly enhanced (up to about sixfold) (Fig. 4E, lane 2 versus lanes 3, 4, and 5). Though PC4-mediated stimulation of p53 DNA binding does not seem to require direct interaction between the C-terminal domain of p53 and PC4, the role of the weaker interaction with the DNA binding domain cannot be ruled out. Presumably, this interaction plays an important role in the activation process during the absence of the C-terminal domain of p53. This observation suggests that similar to HMGB-1 (16), PC4 is also a unique activator of p53-mediated DNA binding and activates p53 through a novel mechanism.
FIG. 4.
Effect of PC4 on constitutively active p53. (A) PC4 activates p53 with the C-terminal domain masked. p53 (25 ng) and 32P-labeled oligonucleotide containing a p53 binding site (3 ng) were incubated without any antibody (lane 2) or with nonspecific p21 antibody (250 ng) (lane 3) in an EMSA reaction (as described in the legend to Fig. 1). The C terminus of p53 (25 ng) was masked with p53 antibody PAb421 (250 ng) and further incubated in an EMSA reaction (as described in the legend to Fig. 1) either alone (lane 4) or in combination with 100, 200, and 300 ng of PC4 (lanes 5, 6, and 7, respectively). One hundred, 200, and 300 ng of PC4 alone (lanes 8, 9, and 10, respectively) were also incubated with 32P-labeled probe. Lane 1 contains only probe. (B) Purification of p53Δ30. FLAG-tagged p53 (lane 1) and p53Δ30 (lane 2) were purified from E. coli BL21/pLys and stained with Coomassie brilliant blue R-250. (C and D) Western blot analyses were performed with N-terminus-specific antibody DO1 and C-terminus-specific antibody PAb421 to check the authenticity of p53 (lane 1) and p53Δ30 (lane 2). (E) Effect of PC4 on p53Δ30 DNA binding. p53Δ30 (10 ng) was incubated either alone (lane 2) or in the presence of 100, 200, and 300 ng of PC4 (lanes 3, 4, and 5, respectively) along with 32P-labeled oligonucleotide containing a p53 binding site. One hundred, 200, and 300 ng of PC4 were incubated with the 32P-labeled oligonucleotide (lanes 6, 7, and 8, respectively). Lane 1 contains 32P-labeled oligonucleotide probe alone.
Role of PC4 in p53- and p53Δ30-mediated gene expression in vivo.
It is a well-established fact that DNA binding by a sequence-specific transcription factor results in the recruitment of RNA polymerase II machinery, leading to transcriptional activation. It is therefore important to functionally correlate the PC4-mediated stimulation of p53 DNA binding in the context of transcriptional activation. In order to address this, we performed transient transfection assays using the H1299 (p53 null) cell line with two different p53-responsive promoters, PG13Luc and MDM2Luc. Consistent with the results obtained in DNA binding experiments, PC4 in the presence of p53 could stimulate transactivation from both promoters significantly (about sixfold for PG13Luc and about eightfold for MDM2Luc) (Fig. 5A and B). However, the vector control and PC4 alone showed negligible luciferase activity (Fig. 5A and B). Since, PC4 also enhanced the DNA binding of constitutively active p53Δ30, we tested the ability of PC4 to stimulate the p53Δ30-mediated transactivation of PG13Luc and MDM2Luc. Significantly, cotransfection of PC4 with p53Δ30 was found to activate transcription from both templates (about fivefold for PG13Luc and about eightfold for MDM2Luc) as compared to transfection of p53Δ30 alone (Fig. 5C and D). This observation further supports the DNA binding data and suggests that PC4-mediated activation of p53 DNA binding is unique and distinct from that of other p53 modulators.
FIG. 5.
PC4 stimulates p53- and p53Δ30-mediated reporter gene expression in vivo. (A) H1299 (p53 null) cells were transiently transfected with the mammalian expression constructs of p53 (250 ng) or PC4 (2.5 μg) either alone or in combination. The p53-responsive promoter construct pG13Luc (1 μg) was used to measure the luciferase activity driven by p53. (B) H1299 cells were cotransfected with p53 (250 ng) or PC4 (4.5 μg) either alone or in combination. The reporter construct used was MDM2Luc (1 μg). (C) p53Δ30 (100 ng) or PC4 (2.5 μg) was cotransfected either alone or in combination with PG13Luc. (D) H1299 cells were cotransfected with p53Δ30 (100 ng) or PC4 (4.5 μg) either alone or in combination. MDM2Luc (1 μg) was used as the reporter construct. Equivalent amounts of control empty vectors were used to normalize the amount of transfected DNA. pCMV-βgal (1 μg) was used as an internal control for all the transfection experiments to normalize luciferase activity. After normalization, relative luciferase activity was plotted (y axis). The relative luciferase activity is represented as an average of triplicate samples.
Presumably, in spite of being a unique activator of p53, the mechanism of PC4-mediated transcriptional activation is global. Consistent with this view, we found that PC4 could indeed induce sequence-specific DNA binding of Gal4-CTF1 (up to about threefold) (Fig. 6A, lane 2 versus lanes 3 and 4) in our system, as reported previously (9). Furthermore, PC4 could also stimulate Gal4-VP16-mediated reporter gene expression (up to about fourfold) from a Gal4-responsive promoter, G10Luc (Fig. 6B).
FIG. 6.
PC4 is a global transcriptional coactivator. (A) PC4 stimulates sequence-specific DNA binding of Gal4-CTF1. 32P-labeled oligonucleotide (3 ng) containing a Gal4 binding site was incubated with 20 ng of Gal4- CTF1 either in the absence (lane 2) or presence of 100 ng (lane 3) or 200 ng (lane 4) of PC4. The 32P-labeled oligonucleotide was also incubated with 100 ng (lane 5) or 200 ng (lane 6) of PC4 alone. Lane 1 contains 32P-labeled oligonucleotide alone. (B) PC4 activates Gal4-VP16-mediated reporter gene expression. H1299 cells were transiently transfected with Gal4-VP16 (100 ng) or PC4 (4.5 μg) either alone or in combination. The Gal4-responsive promoter construct G10Luc (1 μg) was used to measure the luciferase activity driven by Gal4-VP16. An equivalent amount of control parental vectors was used to normalize the amount of transfected DNA. pCMV-βgal (1 μg) was used as an internal control for all the transfection experiments to normalize luciferase activity. After normalization, relative luciferase activity was plotted (y axis). The relative luciferase activity is represented as an average of three individual experiments.
PC4, being a global transcriptional coactivator, interacts with a diverse and a constantly expanding array of transcriptional activators and enhances activator-dependent transcription. In the present study, we identified p53 as another activator whose function is regulated by PC4 through a unique mechanism.
Physiological significance of PC4-mediated stimulation of p53 function.
p53 acts as a transcription factor by activating several genes which facilitate the maintenance of normal cellular growth and function (28, 29, 30). Thus, the enhancement of p53-mediated transcriptional activity should lead to induction of either apoptosis or cell stasis as downstream physiological effects. In order to validate the effects of PC4 on p53- and p53Δ30-mediated transcriptional activation of p53-responsive promoters in a physiological environment, we wished to investigate the role of PC4 in induction of p53- and p53Δ30-dependent apoptosis. For this purpose, the H1299 (p53 null) cell line was cotransfected with the indicated amounts of PC4 and p53 mammalian expression clones. After 24 h of transfection, cells were stained with Hoechst 33258 and observed under a fluorescence microscope to score the extent of nuclear fragmentation (Fig. 7A to F). The results showed that p53-dependent apoptosis (as measured by the number of fragmented nuclei) was enhanced by about twofold upon cotransfection of PC4 and p53 as compared to transfection of p53 alone (Fig. 7G). Interestingly, we also observed that PC4 alone could minimally stimulate apoptosis (Fig. 7G). In order to further confirm the effect of PC4 on p53- or p53Δ30-mediated apoptosis, we analyzed the expression of Bax (a proapoptotic target protein of p53). Significantly, we observed that PC4 could induce p53- and p53Δ30-dependent Bax expression (up to about three- and twofold, respectively) (Fig. 8, lane 5 versus lane 6 and lane 3 versus lane 4). Although our nuclear fragmentation assay data showed that PC4 induces apoptosis minimally, we did not observe PC4-induced Bax expression. It is possible that higher concentrations of PC4 may stimulate apoptosis through an alternative pathway which does not involve Bax. These data strongly correlate PC4-mediated activation of p53 DNA binding and reporter gene expression with their functional consequence. This correlation leads to a very significant clue that transcriptional coactivator PC4 may be involved in regulation of p53-mediated apoptosis during DNA damage or cellular stress.
FIG.7.
Effect of PC4 on p53-dependent apoptosis. (A to F) H1299 cells were transfected with p53 (2.5 μg) and p53Δ30 (1.25 μg) either alone or in combination with PC4 (5 μg). After 24 h, cells were stained with Hoechst 33258 and observed under a fluorescence microscope. Apoptotic nuclei are indicated with arrowheads. (G) Percentage of apoptotic nuclei represents the number of apoptotic nuclei present in 300 nuclei counted each time for three independent experiments. The statistical value is represented as an average percentage of apoptotic nuclei.
FIG. 8.
PC4 stimulates p53-dependent Bax expression. H1299 cells were transfected with p53 (5 μg) and p53Δ30 (2.5 μg) either alone or in combination with PC4 (20 μg). After 48 h cells were lysed and total cellular proteins were subjected to SDS-PAGE followed by Western blotting with antibodies against Bax, p53, p53Δ30, PC4, and actin.
DISCUSSION
p53 functions as a transcription factor to activate expression of its target genes, which either induce cell cycle arrest or apoptosis. To carry out its function, p53 interacts with several coactivators, namely p300/CBP, PCAF, JMY, Zac1, HMGB-1, and hTAF9, with distinct functional consequences (10, 11, 15, 31). In this study, we report that (i) PC4 (a transcriptional coactivator in RNA polymerase II-mediated transcription) activates sequence-specific DNA binding of p53 and interacts with p53 in vitro and in vivo; (ii) the activated complex is ternary in nature, containing PC4, p53, and DNA; (iii) it can induce sequence-specific DNA binding of p53Δ30; (iv) gene expression regulated by p53 and p53Δ30 is stimulated by PC4 in transient transfection assays from both synthetic and natural p53-responsive promoters, which is reflected in the fact that PC4 could induce p53-dependent apoptosis by stimulating bax. These results have been discussed in relation to the fact that this is the first report demonstrating a functional interaction of PC4 with p53 and its physiological consequence.
The efficiency of p53 in functioning as a transcription factor is modulated by its ability to bind its cognate DNA site, which can be regulated by several interacting factors. PC4 interacts with a diverse group of transcriptional activators and not only enhances their DNA binding ability but also their transcriptional activation (9). Surprisingly, p53, being one of the most significant transcription factors for the maintenance of cellular homeostasis, was not studied in conjunction with PC4. We were interested in investigating the possible functional interaction of PC4 with p53 and found that PC4 could stimulate sequence-specific DNA binding of p53 by about eightfold. However, we found that a molar excess of PC4 was required for the stimulation of p53 DNA binding. This requirement of high amounts of PC4 can be easily explained by a careful observation of the results of EMSA (Fig. 1C). PC4 is known to have a non-sequence-specific DNA binding ability (9), which is ratified by our data (Fig. 1C, lanes 7 to 10). The unbound DNA probe in our experiments titrates out the PC4 that is required to show an enhancement of p53 DNA binding. Therefore, the excess of PC4 that remains after saturation of the free probe is what causes the dramatic increase in p53 DNA binding.
PC4 also directly interacts with p53 to form a ternary complex with p53 and DNA. Both the DNA binding (amino acid residues 120 to 290) and the C-terminal (residues 364 to 393) domains of p53 interact with PC4. The PC4-mediated stimulation of p53 DNA binding is quite similar to that of several other coactivators, as reported previously (9). Our data also showed that PC4 could stimulate proline-rich activator Gal4-CTF1- and the acidic activator Gal4-VP16-mediated DNA binding and transactivation, respectively. These results suggest that PC4-mediated activation of various transcription factors is a global phenomenon. However, most interestingly, PC4 could stimulate the sequence-specific DNA binding of p53 with the C terminus deleted. Although the activation of p53Δ30 DNA binding by PC4 suggests that the stronger interaction between residues 364 to 393 of p53 and PC4 is not essential for the induction of p53-mediated DNA binding, the weaker interaction between amino acid residues 120 to 290 of p53 and PC4 may play a significant role in this phenomenon. The weaker interaction through the DNA binding domain of p53 with PC4 may also help in the recruitment of p53, especially in the absence of the C-terminal 30 residues of p53. Presumably, PC4 changes the DNA structure to a favorable conformation for p53 binding and recruits it through direct interaction. Mechanistically, it might increase the specificity of p53 binding or help it to monitor the DNA sequence for cognate sites.
In correlation with our DNA binding observations, we found that PC4 could stimulate p53-dependent reporter gene expression from the synthetic (PG13Luc) as well as natural (MDM2Luc) p53-responsive promoters. Interestingly, PC4 could also stimulate p53Δ30-mediated transactivation from both the p53-responsive promoters. Similar to findings for HMGB-1 (16), these observations suggest that the PC4-mediated activation of p53 function is not an ordinary activation process wherein p53 activators negate the autoinhibitory function of the C-terminal domain and thus cannot stimulate DNA binding of p53Δ30. It would be interesting to elucidate further the mechanism of PC4-mediated activation of p53.
HMGB-1 is a nonhistone chromatin protein that belongs to the HMG superfamily. HMGB-1 is composed of two homologous HMG boxes (A and B) which are connected to an extremely acidic C-terminal domain. Recently, we have shown that both the A-box and the C-terminal domain are essential to stimulate p53-mediated DNA binding, transactivation, and apoptosis (3). HMGB-1 has an intrinsic DNA-bending property. It provides prebent DNA to p53 and thereby enhances the efficiency of p53 DNA binding to its cognate site (27). Like HMGB-1, PC4 is also a nonspecific DNA binding protein which can bind to both single-stranded and double-stranded DNA. The double-stranded DNA binding domain of PC4 is essential for transcriptional coactivation (9). It would be interesting to investigate the requirement of different domains of PC4 for stimulation of p53 DNA binding and whether the DNA bending is responsible for this activation process.
Though PC4 functions as a general transcriptional coactivator for activator-dependent transcription, its role in a physiological context has not yet been demonstrated. In the present report, we have addressed the biological relevance of PC4-mediated stimulation of p53 DNA binding and transactivation. Most of the p53 modulators induce p53-dependent apoptosis by regulating its target apoptotic genes (28, 29, 30). We observed that transiently expressed p53 and p53Δ30, in combination with PC4, markedly enhanced p53- and p53Δ30-dependent apoptosis. Furthermore, it was also observed that transient expression of p53 and p53Δ30 in combination with PC4 induces the expression of the p53-responsive proapoptotic gene bax. These data are highly suggestive of a link between PC4-mediated transcriptional activation and its physiological effect in the cellular environment. Apart from the inhibition of self-repression of AP2 (in a ras-transformed cell line) (18), our study shows that PC4 might also play a key role in tumor suppression by stimulating p53 function.
PC4 and p53 undergo posttranslational modification (phosphorylation and acetylation) with several functional consequences (22, 26, 31). Phosphorylation of PC4 negatively regulates its acetylation and coactivator function (22, 26), whereas both acetylation and phosphorylation synergistically activate p53 function (31). The posttranslational modifications of PC4 and p53 could be the key regulatory switch for PC4-mediated p53 activation during genotoxic insult. The present study has revealed a new role of PC4 in the regulation of tumor suppression. Further understanding of this novel function of PC4 will establish a link between direct transcriptional activation and its physiological consequences in greater detail.
Acknowledgments
We thank Robert G. Roeder, Elliot Androphy, Bert Vogelstein, K. Vousden, and Chandrima Das for providing valuable reagents used in this study and K. Somasundaram for helpful discussion.
This work is supported by the Department of Science & Technology, Government of India.
REFERENCES
- 1.Ahu, J., and C. Prives. 2001. The C-terminal of p53: the more you learn the less you know. Nat. Struct. Biol. 8:730-731. [DOI] [PubMed] [Google Scholar]
- 2.Avntaggiati, M. L., V. Ogryzko, K. Gradner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, S., and T. K. Kundu. 2003. The acidic C-terminal domain and A-box of HMGB-1 regulates p53-mediated transcription. Nucleic Acids Res. 31:3236-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buschmann, T., Y. Lin, N. Aithimitti, S. Y. Fuchs, H. Lu, L. Resnick-Silverman, J. J. Manfredi, Z. Ronai, and X. Wu. 2001. Stabilization and activation of p53 by the coactivator protein TAFII31. J. Biol. Chem. 276:13852-13857. [DOI] [PubMed] [Google Scholar]
- 5.Calvo, O., and J. L. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7:1013-1023. [DOI] [PubMed] [Google Scholar]
- 6.Carey, M., H. Kakidani, J. Leatherwood, F. Mostashari, and M. Ptashne. 1989. An amino-terminal fragment of GAL4 binds DNA as a dimer. 209:423-432. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., G. Farmer, H. Zhu, R. Prywes, and C. Prives. 1993. Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 7:1837-1849. [DOI] [PubMed] [Google Scholar]
- 8.Friedlander, P., Y. Haupt, C. Prives, and M. Oren. 1996. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Mol. Cell. Biol. 16:4961-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge, H., and R. G. Roeder. 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78:513-523. [DOI] [PubMed] [Google Scholar]
- 10.Gu, W., X. L. Shi, and R. G. Roeder. 1997. Synergistic activation of transcription by CBP and p53. Nature 387:819-823. [DOI] [PubMed] [Google Scholar]
- 11.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 12.Gu, W., S. Malik, M. Ito, C. X. Yuan, J. D. Fondell, X. Zhang, E. Martinez, J. Qin, and R. G. Roeder. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 3:97-108. [DOI] [PubMed] [Google Scholar]
- 13.Guermah, M., S. Malik, and R. G. Roeder. 1998. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-κB and Sp1. Mol. Cell. Biol. 18:3234-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haile, D. T., and J. D. Parvin. 1999. Activation of transcription in vitro by the BRCA1 carboxyl-terminal domain. J. Biol. Chem. 274:2113-2117. [DOI] [PubMed] [Google Scholar]
- 15.Huang, S. M., A. H. Schonthal, and M. R. Stallcup. 2001. Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1. Oncogene 20:2134-2143. [DOI] [PubMed] [Google Scholar]
- 16.Jayaraman, L., N. C. Moorthy, K. G. Murthy, J. L. Manley, M. Bustin, and C. Prives. 1998. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 12:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser, K., and M. Meisternst. 1996. The human general co-factors. Trends Biochem. Sci. 21:342-345. [PubMed] [Google Scholar]
- 18.Kannan, P., and M. A. Tainsky. 1999. Coactivator PC4 mediates AP-2 transcriptional activity and suppresses ras-induced transformation dependent on AP-2 transcriptional interference. Mol. Cell. Biol. 19:899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kern, S. E., J. A. Pietenpol, S. Thiagalingam, A. Seymour, K. W. Kinzler, and B.Vogelstein. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256:827-830. [DOI] [PubMed] [Google Scholar]
- 20.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 21.Kretzschman, M., K. Kaiser, F. Lottspeich, and M. Meisterernst. 1994. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell 78:525-534. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, P. B. R., V. Swaminathan, S. Banerjee, and T. K. Kundu. 2001. p300-mediated acetylation of human transcriptional coactivator PC4 is inhibited by phosphorylation. J. Biol. Chem. 276:16804-16809. [DOI] [PubMed] [Google Scholar]
- 23.Kundu, T. K., V. B. Palhan, Z. Wang, W. Au, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 24.Lill, N. L., S. R. Grossman, D. Ginsberg, J. De Caprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 25.Liu, X., and A. J. Berk. 1995. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol. Cell. Biol. 15:6474-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik, S., M. Guermah, and R. G. Roeder. 1998. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc. Natl. Acad. Sci. USA 95:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinney, K., and C. Prives. 2002. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol. Cell. Biol. 22:6797-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 27:293-299. [DOI] [PubMed] [Google Scholar]
- 29.Nakano, K., and K. H. Vousden. 2001. PUMA, a novel proapoptotic gene is induced by p53. Mol. Cell 7:683-694. [DOI] [PubMed] [Google Scholar]
- 30.Oda, E., R. Okhi, H. Murasawa, J. Nemoto, T. Shibue, T. Yamashita, T. Tokino, T. Taniguchi, and N. Tanaka. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288:1053-1058. [DOI] [PubMed] [Google Scholar]
- 31.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shikama, N., C. W. Lee, S. France, L. Delavaine, J. Lyon, K. M. Demonacus, and N. B. La Thangue. 1999. A novel cofactor for p300 that regulates the p53 response. Mol. Cell 4:365-376. [DOI] [PubMed] [Google Scholar]
- 33.Stelzer, G., A. Goppelt, F. Lottspeich, and M. Meisterernst. 1994. Repression of basal transcription by HMG2 is counteracted by TFIIH-associated factors in an ATP-dependent process. Mol. Cell. Biol. 14:4712-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, S. Y., and C. M. Chiang. 1998. Properties of PC4 and an RNA polymerase II complex in directing activated and basal transcription in vitro J. Biol. Chem. 273:12492-12498. [DOI] [PubMed] [Google Scholar]